Abstract
FAD Synthetase (FADS) [EC 2.7.7.2], the second enzyme in flavin cofactor biosynthetic pathway converts FMN to FAD, plays an important role in many redox reactions. Neurospora crassa FADS (NcFADS) was cloned and overexpressed in E. coli cells. Recombinant NcFADS was purified in high yields of ∼8 mg per liter of bacterial culture using a single step glutathione sepharose affinity chromatography. SDS-PAGE and MALDI-MS revealed that NcFADS has a molecular mass of ∼31 kDa. Enzyme kinetic analysis monitored by reverse phase HPLC demonstrate a specific activity and kcat of 1356 nmol/min/mg and 0.69sec−1 respectively. Steady state kinetic analysis of NcFADS exhibited a Km of NcFADS for FMN is 2.7 μM and for MgATP−2 is 88.7 μM. Isothermal titration calorimetry experiments showed that the recombinant protein binds to the substrates with apparent Kd of 20.8 μM for FMN and 16.6 μM for MgATP−2. Biophysical characterization using intrinsic fluorescence suggests that the enzyme is in folded conformation. Far-UV CD data suggest that the backbone of the enzyme is predominantly in a helical conformation. Differential scanning calorimetry data shows that the Tm is 53 °C ± 1. This is the first report on cloning, purification and characterization of FADS from N. crassa. The specific activity of NcFADS is the highest than any of the reported FADS from any other source. The results obtained in this study is expected to pave way for intensive research aimed to understand the molecular basis for the extraordinarily high turnover rate of NcFADS.
Keywords: Enzyme kinetics, Flavo proteins, Intrinsic fluorescence
Highlights
-
•
rNcFADS active at pH7.5.
-
•
High turnover, more thermal stability.
-
•
2ostructure-helical.
1. Introduction
Flavoproteins play a crucial role in many biological processes [1]. Flavin mononucleotide (FMN) and Flavin adenine dinucleotide (FAD) derived from the precursor riboflavin, act as coenzymes for various flavoproteins. Therefore, flavin metabolism must be tightly regulated in the cell [2]. Most of the higher eukaryotes cannot synthesize riboflavin de novo so they depend on external dietary sources. Cells need to efficiently acquire them from absorbed food with specific transporter(s) of plasma membrane [3]. Riboflavin kinase (RFK) [ATP: riboflavin 5'phosphoryl transferase (EC2.7.1.26)] catalyzes the first step in biosynthesis of flavin cofactors by ATP-dependent phosphorylation of riboflavin to form FMN. Subsequent catalysis of ATP dependent adenylation of FMN to form FAD is carried out by FAD synthetase (FADS) or FMN adenylyl transferase (FMNAT) [ATP: FMN adenylyl transferase (EC2.7.7.2)] [4,5]. These two reactions follow a sequential bi-bi ordered mechanism [6,7].
In prokaryotes, RFK and FADS are encoded by C-terminal domain in the same gene (ribF or ribC) [8]. The gene product is a single polypeptide in the form of a bifunctional enzyme (RFK/FADS), with C-terminal showing riboflavin kinase activity and N-terminus showing adenylation activity. RFK/FADS bifunctional enzyme was purified and characterized from certain bacteria [[9], [10], [11]]. Apart from the mentioned bifunctional enzyme prokaryotic monofunctional RFK's were also reported [12,13]. In eukaryotes, both RFK and FADS are encoded by two separate genes resulting in two independent monofunctional enzymes [7,14]. Monofunctional enzymes, RFK and FADS were purified and characterized from yeast and rat tissues [4,5,[15], [16], [17]]. Eukaryotic RFK shares sequence similarities with the C-terminus of RFK/FADS bifunctional enzyme of prokaryotes. N-terminus of prokaryotic bifunctional RFK/FADS showed sequence similarity to the proteins of “nucleotidylyl transferase family” unlike the eukaryotic FADS that showed similarity to proteins of “PAPS reductase like family” [8,18].
Eukaryotic FADS were cloned and over-expressed from yeasts [19], mammals [5,16,20] and plants [21]. Saccharomyces cerevisiae monofunctional FADS was designated as FAD1. The knockout of FAD1 gene was lethal to S. cerevisiae indicating the importance of FADS to various flavoproteins involved in key metabolic reactions of the cell [19]. Eukaryotic FADS is present in the cytosol or in the mitochondria. In humans and plants two isoforms of FADS were reported. Human isoforms are the products of a single gene FLAD1. The two isoforms are the alternately spliced transcripts of the gene. Isoform-1 has a signal peptide to mitochondria and the Isoform-2 do not have an any signal peptide (iPSORT, TarfetP and MITO-PROT) [20]. A genome of Arabidopsis thaliana codes for two proteins AtRibF1 and AtRibF2, with the organelle targeting peptide [21]. These isoforms were differentiated based on the N-terminus organelle targeted signal peptide [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28]]. Studies on the human FADS emphasized its importance in the catalysis of FAD synthesis and delivery of FAD to apoproteins [29]. To date the crystal structures of eukaryotic FADS were reported from Candida glabrata [8] and S. cerevisiae [30].
Genes for more than 100 different flavoproteins were reported in the filamentous fungi N. crassa. Most of these flavoproteins are oxidases and dehydrogenases that help in accessing the organic matter from the environment to support its saprophytic life style [31]. Also, earlier reports of RFK from N. crassa showed a high turnover number and a very low Km for its substrates [32] that might be supporting the life style of the fungi. Therefore, we hypothesize that the sequential enzyme FADS, in flavin biosynthesis pathway, may also be showing these exceptional properties to maintain the flavin content of the cell. Structural studies are the lacuna because of the non-availability of the FADS clone from N. crassa. The over expression of the enzyme would pave way for detailed analysis to understand the molecular intricacies responsible for its high turnover number and high specific activity compared to FADS from any other sources. In the present research, we successfully cloned, over expressed and purified the enzyme in high yields that helped in studying the kinetic and preliminary biophysical aspects of the enzyme.
2. Materials and methods
Materials: Taq polymerase, glutathione sepharose beads, thrombin, were purchased from Sigma-Aldrich, Saint Louis, USA. Restriction enzymes from New England Biolabs, Inc, USA. pGEX-KG vector was a gift from Dr. Junnutula's lab Genentech (Amersham Biosciences, United Kingdom). Qiaquick Gel extraction kit (#28704) was obtained from Qiagen, Hilden. Protease inhibitor cocktail tablets, DNA rapid ligation kit (#K1422) and first stand cDNA synthesis kit (#K1612) were from MBI-Fermentas, USA. SeeBlue-Plus2 pre-stained protein molecular weight standards were purchased from Invitrogen Life Technologies, USA. Other fine chemicals were obtained from Amersham Pharmacia Biotech, USA; Sigma-Aldrich, USA and GIBCO-BRL Chemicals, USA. Components of Luria-Bertani media were purchased from Himedia, Mumbai. Inorganic salts and solvents used were purchased from Qualigens and Merck, India. All other chemicals used in this study were analytical grade. Glassware was purchased from Borosil, India.
Cloning of FADS from N. crassa (FGSC #2489): A coding sequence of FAD synthetase gene from Neurospora data base (BROAD MIT, N. crassa OR74A (NC10): NCU09233 - FAD synthetase) was identified. N. crassa (FGSC #2489) was grown in 1X Vogel's supplemented medium at 28 °C for 48 h with shaking (100 rpm) in an environmental incubator. The vegetative mycelium obtained was used to isolate total RNA using TRI reagent (#T9424; Sigma, Saint Louis, USA). cDNA was synthesized from the isolated RNA using First Strand synthesis kit (#K1612, MBI-Fermentas, USA). cDNA coding for FADS of N. crassa was amplified using gene-specific primers; ncFADS-Forward primer 5′ACGTGGATCCGCTACCTCATCGGTCTC3’ (BamHI) and ncFADS-Reverse primer 5′ACGTCTCGAGCTATCGATCGCGACCTAGC3’ (XhoI). Amplified product was cloned into pGEX-KG vector which was already pre-digested using the same restriction enzymes. Authenticity of the clone was confirmed by nucleotide sequencing.
Expression of GST-NcFADS fusion protein in E. coli:E. coli BL21 (DE3) expression host cells were transformed with pGEX-KG-ncFADS. Selection of transformed colonies was performed on Luria broth (LB) agar plates containing ampicillin (100 μg/ml). Transformed bacterial cells with the recombinant pGEX-KG-ncFADS plasmid was inoculated in 25 ml LB containing ampicillin (100 μg/ml) and grown for overnight at 37 °C with rotary shaking (250 rpm). 0.5L of LB was inoculated with overnight culture (2.5%) in the presence of ampicillin (50 μg/ml). Protein over-expression was achieved by induction with 0.6 mM IPTG when the optical density of the culture at 600 nm (A600) reached 0.6. The culture was further incubated at 37 °C for 3 h and the cells were harvested by centrifugation at 6000 rpm (Kubota 7820 RA-6 rotor). Harvested cells were subjected to sonication (3 × 10 s with 10 s pauses; amplitude 50% on Vibra-Cell VCX 750, TX, USA). Expression of NcFADS was checked by SDS-PAGE [33].
Purification of recombinant NcFADS: Glutathione-agarose affinity chromatography was employed for the purification of GST tagged NcFADS. Cell pellet was resuspended in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4), pH 7.5 and subjected to sonication. The supernatant obtained after centrifugation was loaded on glutathione-agarose column (Sigma, USA) that was pre-equilibrated with 1xPBS pH 7.5 and further washed with 5 column volumes of 1xPBS pH 7.5. The bound GST-NcFADS was subjected to on-column cleavage with 1Unit of thrombin for every 200 μg of the fusion protein and incubated at 22 °C for 8 h. The eluates were collected using 1xPBS pH 7.5 followed by concentration of NcFADS using 10/30 kDa cut-off Amicon Ultra protein concentrating filters (Millipore, Bedford, MA, USA) by centrifugation at 3000 rpm (1025.4×g). Homogeneity of the protein was assessed using SDS-PAGE (Table 2). NcFADS was obtained in homogeneity with a yield of 8 mg/L of culture. The authenticity of the protein sample was further verified by MALDI-TOF mass analysis. Concentration of the protein was estimated by dye-binding method according to Bradford [34] using BSA as standard.
Table 2.
Purification of NcFADS from overexpressing E. coli.
| Purification step | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | % recovery | Fold enrichment (Sp. Act) |
|---|---|---|---|---|---|
| Crude cell lysate | 862 ± 5.36 | 10.76 ± 0.17 | 80 ± 1.45 | 100 | 1 |
| Clarified supernatant | 710 ± 6.35 | 6.46 ± 0.12 | 110 ± 2.51 | 81 ± 0.73 | 1.4 |
| Thrombin cleaved NcFADS | 502 ± 9.07 | 0.37 ± 0.01 | 1356 ± 5.48 | 57 ± 1.05 | 17 |
Note: The starting volume of culture for each experiment was 50 mL. Values for total activity, specific activity and % recovery have been rounded off to the nearest integer. Data are the Mean ± SE of three triplicate experiments.
Measurement of NcFADS activity: Enzyme aliquots were incubated with assay mixture (0.1 M Tris.HCl buffer pH 7.5, in the presence of 10 μM FMN, 1 mM ATP with 1 mM MgCl2) at 37 °C for 15 min, in a final volume of 0.1 mL. Reactions were terminated by the addition of formic acid (2 μL of 7 N stock). FMN and FAD were separated on Nucleosil C18 column and samples were analyzed by RP-HPLC (Gilson HPLC model 321 Pump, with online UV/VIS-156 uv–vis spectrophotometric detector and SOMA S-3370 fluorescence detector). Under test conditions 1 pmol of FAD resulted in a peak area of 20.028 units. One unit of enzyme activity was defined as that producing 1 nmol of FAD per min at 37 °C (one milliunit (mU) produces 1 pmol of FAD per min. at 37 °C). The kinetic parameters were calculated from the saturation experiments carried out at pH7.5. For FMN saturation, initial velocity (amount of FAD formed) of the catalytic reaction was determined by incubating fixed amount of NcFADS with saturating levels of MgATP2−(1 mM) and varying the concentration of FMN (0.1–10 μM). For MgATP2− saturation, initial velocity of the reaction was determined by incubating the fixed amount of NcFADS with varying level of MgATP2− (10–1000 μM) in the presence of saturating concentrations of FMN (10 μM).
Circular Dichorism Spectroscopy: Spectra were acquired using JASCO-720 spectropolarimeter using 0.1 mm path length quartz cell at room temperature. Far-UV spectra was measured from 200 to 250 nm averaged over 10 scans at a speed of 50 nm/min. NcFADS was diluted to final concentration of 33.3 μM in 1xPBS (pH 7.5). Necessary background corrections were made.
Steady state fluorescence: All fluorescence measurements were performed using Hitachi F-250 fluorescence spectrophotometer at 25 °C. The excitation wavelength was fixed at 280 nm and emission spectra were recorded from 300 to 450 nm wavelength, and the bandwidth for excitation and emission were set to 2.5 nm and 10 nm respectively. Spectrum was measured at protein concentration of 3.3 μM in 1xPBS (pH 7.5). Necessary background corrections were made.
Equilibrium unfolding: Guanidinium hydrochloride (GdnCl) induced equilibrium unfolding of NcFADS was performed at a protein concentration of 3.3 μM in 1xPBS (pH7.5). The excitation wavelength was fixed at 280 nm and emission maxima was recorded based on the intrinsic tryptophan fluorescence changes with increasing GdnCl concentration, and the bandwidth for excitation and emission were set to 2.5 nm and 10 nm respectively. Necessary background corrections were made.
Differential Scanning Calorimetry: Calorimetric scans were performed using CSC 6300 Nano DSC III differential scanning calorimeter (Calorimetry Sciences Corporation). NcFADS with a concentration of 33.3 μM in 1xPBS (pH 7.5) was degassed under vacuum for 10min.Calorimetric scans were conducted at 1 °C/min under a pressure of 3atm.The CSC DSC software was used for the baseline subtraction and determination of the transition temperature of NcFADS.
Isothermal Calorimetry: Binding parameters of NcFADS to its substrates FMN and ATP was assessed by measuring the heat changes during the titration using MicroCalTMiTC200 (GE Healthcare). Protein and the ligand solutions were degassed under vacuum and equilibrated at 25 °C prior to titration. The sample cell (∼220 μL) contained 40–50 μM NcFADS in 1xPBS (pH 7.5). The reference cell contained MilliQ water and the substrates were in the same buffer conditions. Ligands FMN (1.5 mM), ATP (1 mM) with MgCl2 (5 mM) were titrated with a stirring speed of 1000 rpm. A typical experiment had 30 injections with 1.2 μL each with 120sec equilibration time involved. The resulting titration curves were corrected using protein free buffer controls. Ka, ΔH and N were obtained from Origin ITC software supplied by MicroCalTMiTC200 (GE Healthcare).
3. Results and discussion
Identification and Cloning of theFADSGene from N. crassa FGSC #2489: A search of whole genome database of N. crassa (FGSC #2489, BROAD Institute, MIT) yielded a putative “FAD Synthetase” gene at locus NCU09233.1 (presently denoted as NCU09233.5) on chromosome I, coding for a hypothetical protein of 272aa (Table 1). The locus has a gene of 952 nt with three exons forming a coding region of 816 nt. This putative NcFADS is a purified from the soluble fraction to homogeneity using a single step purification. NGGKDC is the conserved motif at position 71–76 for FADS but not the signal peptide and it shows single form (Fig. 1 A & B), this was a characteristic signature motif observed in all the eukaryotic FADS reported [30]. Expression of this construct resulted in the production of enzymatically active NcFADS protein in transformed E. coli expression cells. Total RNA (∼2 μg) was isolated from N. crassa mycelia. The PCR amplified fragment cloned into pGEX-KG expression vector generated a recombinant plasmid pGEX-KG-ncFADS. The presence of insert was confirmed by double digestion with BamHI and XhoI further ascertained by DNA sequencing. This fragment cloned in-frame with the GST coding sequence of the expression vector allowed the expression of a GST-NcFADS fusion protein from the pTac promoter (T7 and Lac promoter) of the vector. pGEX-KG-ncFADS plasmid was used to transform E. coli BL-21(DE3) cells.
Table 1.
Amino acid sequence of putative FADS from N. crassa.
| >gi|85111171|ref|XP_963810.1| hypothetical protein [N. crassa OR74A]-272aa MATSSVSKIVSQPRSLAEVCAALRAKLLAFLALHSNDETVQGTQRRARDAMEVIEEALRRYRPEELSLSYNGGKDCLVLLILILACWPASVQPPSSSSSSSSSSWASNSSKQTLPRLQCIYIAPPD PFQEVEDFVATTTEEYHLDLARYALPMRQALDSYLDEKPHVKAVFMGTRRTDPHSEFLNNFTPTDKGWPQFMRINPVLDWHYVEIWTFIRQLDIPFCSLYSQGFSSLGGTKDTRPNPALAL NAEGNKFRPAYELTRDDEERLGRDR |
Fig. 1.
(A) Cell lysates obtained before and after induction with IPTG was resolved on 12% SDS PAGE and was stained with Coomassie Blue. 1: lysate of uninduced culture; 2: lysate of induced culture showing an over-expressed band at ∼56 kDa; 3: NcFADS recovered after treatment with thrombin. (B) MALDI MS Analysis of NcFADS showed a peak that corresponds to a monoisotopic mass of 30662. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Expression of NcFADS in E. coli: The recombinant plasmid pGEX-KG-ncFADS encoded the protein NcFADS with the GST tag at the N-terminus. SDS-PAGE of the plasmid containing E. coli BL-21 (DE3) cell cultures induced by IPTG showed an intense band at ∼56 kDa corresponding to GST-NcFADS (Fig. 1A). GST tag at the N-terminus facilitated the purification of recombinant NcFADS using the glutathione agarose affinity chromatography. This protein was purified to homogeneity by a single-step purification procedure. The purified NcFADS migrated as a single intense band of ∼31 kDa on SDS-PAGE under reduced conditions (Fig. 1A). MALDI-MS analysis of the purified NcFADS showed a molecular mass of 30.6 kDa (Fig. 1B) that was also in agreement with the theoretical mass of 30.7 kDa. The yield of the protein was ∼8 mg per liter of the culture. Such high yields of over-expressed protein would enable us to carry out the structural studies of enzyme.
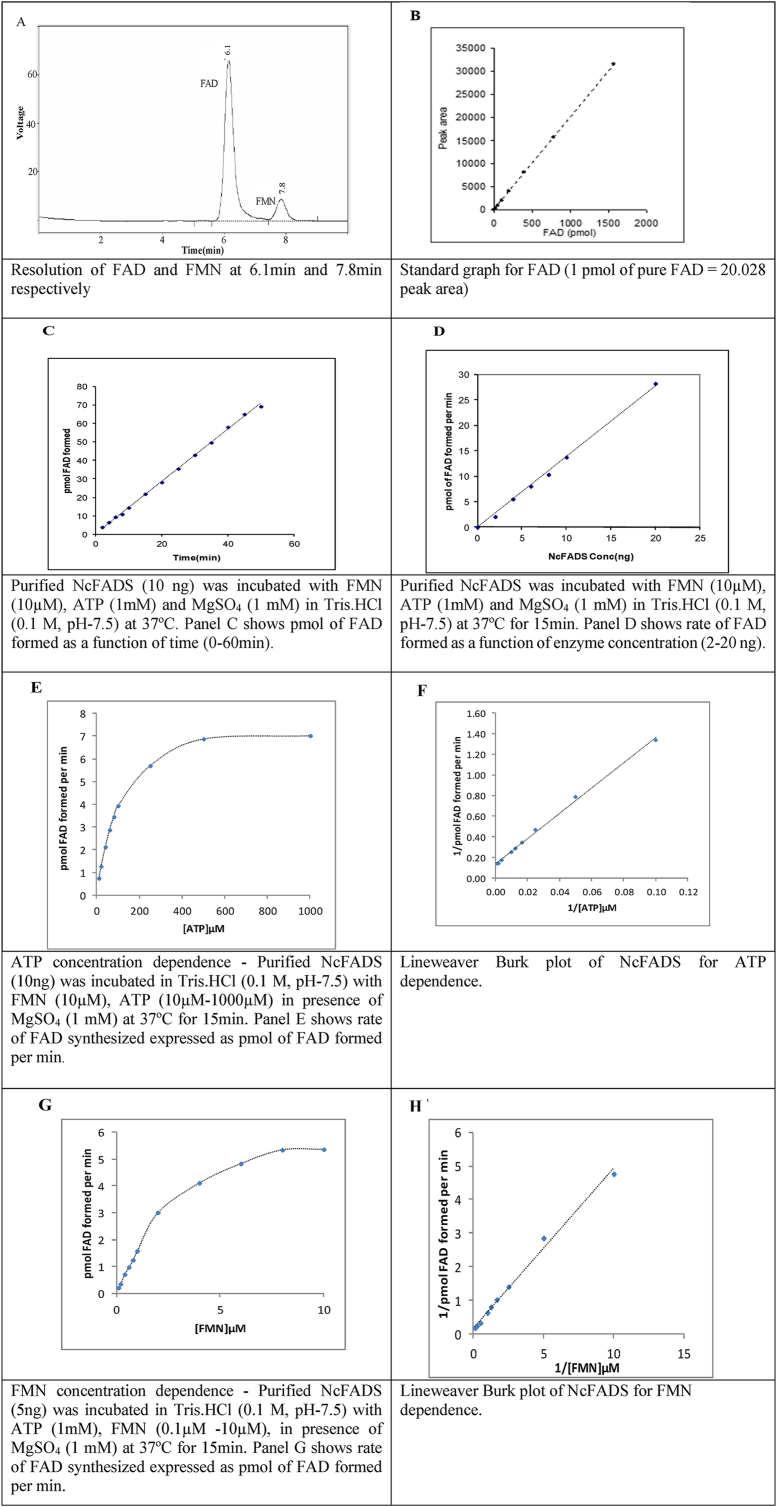
Functional characterization of NcFADS: NcFADS activity was tested using a standard RP-HPLC based enzyme assay. Under standard assay conditions, the rate of production of FAD increased linearly with time up to 60 min (Fig. 2C) and protein concentration (Fig. 2D) up to 20 ng. The purified NcFADS showed a specific activity of 1356 U/mg with 17-fold purification (Table: 2). The steady state kinetic parameters of NcFADS were determined using Lineweaver Burk plot of 1/v (initial velocity) versus 1/[FMN] or 1/[ATP]. FMN adenylation reaction was shown to follow a sequential ordered bi-bi mechanism in eukaryotes [6,7]. NcFADS showed a typical hyperbolic saturation curve for both the substrates (Fig. 2 E, G). The Km for FMN, calculated from Lineweaver Burk plots (Fig. 2H) was 2.23 ± 0.2 μM and for MgATP2 was found to be 84.8 ± 2 μM (Fig. 2 F). NcFADS showed a kcat value of 0.69 sec−1. The catalytic efficiency for FMN and MgATP2− obtained for NcFADS were 0.256 μM s−1 and 0.0077 μM s−1 respectively. As shown in the Table 3, FADS from other eukaryotic sources like yeast showed the Km values for FMN and ATP as 0.76 μM and 10.7 μM respectively and kcat value as 0.087 sec−1 [8]. In humans Km value for FMN reported was 0.36 μM and the kcat value was 0.0036 sec−1 [28]. Km values for NcFADS were in comparison with other FADS reported as they were all in μM range but the kcat of NcFADS was ∼ 8-fold high compared to yeast and ∼200 times higher than human enzymes. Catalytic efficiency of NcFADS for FMN is 25-fold higher to the human and 2-fold higher to C. glabrata FADS. Earlier reports also indicate the high turnover number of RFK from N. crassa [32]. In our present study NcFADS also showed a very high turnover number compared to other eukaryotic FADS (Table 3). The high turn-over numbers and the catalytic efficiencies of both the enzymes RFK and FADS from N. crassa indicate the requisite of the flavins in this organism that also supports its lifestyle.
Fig. 2.
A) Resolution of FAD and FMN on Nucleosil C18 Reverse phase column using at 6.1min and 7.8min respectively; B) Standard graph for FAD (1 pmol of pure FAD = 20.028 peak area). HPLC buffer used for resolving the peaks contain 0.1 M ammonium formate, 0.1 M Formic acid and 30%methanol.
Table 3.
Comparison of the kinetic parameters.
| Source organism | Km(μM) |
Catalytic efficiency (μMsec−1) |
kcat (sec−1) | ||
|---|---|---|---|---|---|
| FMN | MgATP | FMN | MgATP | ||
| Nc FADS | 2.23 | 84.8 | 0.256 | 0.0077 | 0.69 |
| Yeast | 0.76 | 10.7 | NA | NA | 0.087 |
| Human | 0.36 | NA | NA | NA | 0.0036 |
Biophysical characterization of NcFADS: Structural analysis of recombinant NcFADS was determined using various biophysical techniques such as Far-UV CD, fluorescence spectroscopy, differential scanning calorimetry. Far-UV CD provides valuable information about the gross secondary structure of proteins. Spectrum of NcFADS showed a double minimum at 208 nm and 222 nm (Fig. 3A) indicates that majority of the secondary structure in NcFADS is α-helix. According to SCOP classification eukaryotic FADS belongs to α/β family of proteins. Further confirmation of the secondary structure was done by submitting the NcFADS absorbance values to the K2D3 software [35]. The resulting percentage of α-helix and β-sheet is ∼65 and 2 respectively. The secondary structural information from both the reported crystal structures of FADS from C. glabrata (3FWK) and S. cerevisiae (2WSI) [8,30] showed 40% of the helical content and <9% of total β-content. NcFADS shows ∼70% sequence similarity with the reported PDB structures, thus the NcFADS experimental data is also in agreement with the SCOP classification.
Fig. 3.
Structural analysis of recombinant NcFADS was determined using various biophysical techniques such as Far-UV CD, fluorescence spectroscopy, differential scanning calorimetry.
Intrinsic tryptophan fluorescence would be an excellent probe to measure the tertiary structure of proteins. Changes in the fluorescence emission of tryptophans in proteins have been widely reported as a consequence of conformational transitions, subunit association, ligand interactions or denaturation. NcFADS has five tryptophan residues at position 87,105,194,206 and 212 of the primary amino-acid sequence (Table 1). NcFADS in its native state showed an emission maxima around 338 nm indicate that the tryptophan residues are buried in the hydrophobic environment of the protein. Guanidinium chloride induced equilibrium unfolding of NcFADS is monitored by the changes in the tryptophan fluorescence, showed that NcFADS completely unfolds at 6MGdnCl. Concentration of the denaturant at which 50% of the molecules remain in the denatured state is estimated to be 3.2 M. The free energy of unfolding of NcFADS is estimated to be 748.8calmol-1. These studies indicate that NcFADS is in stable native conformation.
This denaturation was not successful with GdnCl so we later used Guanidinium thio-cynate that resulted in complete unfolding. Guanidium thiocyanate induced equilibrium unfolding of NcFADS is monitored by the changes in the tryptophan fluorescence shows that NcFADS completely unfolds at 6 M GdnSCN. Concentration of the denaturant at which 50% of the molecules remain in the denatured state is estimated to be 1.59 M. The free energy of unfolding of NcFADS is estimated to be 1.4kCalmol−1(Fig. 3E).
Thermodynamic stability of NcFADS was measured by subjecting the enzyme to denaturation with gradual increase in temperature. Differential scanning calorimetry (DSC), directly measures the enthalpic changes, accompanied by the folding and unfolding of proteins which are most commonly exothermic in nature. It directly measures the Tm of a protein indicating the temperature at which half the protein population was in equilibrium with the native and denatured state(s). Tm of NcFADS was found to be ∼53.3 °C. The heat capacity (Cp) observed at this temperature was 4.99 kcal/mol K. The ΔH and ΔS obtained were −222.43 kcal/mol and 0.68 kcal/K mol respectively, showing that NcFADS is a fairly stable enzyme.
ITC Analysis for the Interaction of NcFADS with FMN and ATP: Isothermal titration calorimetry is the direct measure of the binding affinity of a protein with its ligand and also a tool for studying the thermodynamic properties of the biological molecules. In the present study ITC was used to measure the binding affinities of both the substrates FMN and MgATP2− to NcFADS. Isothermograms obtained were hyperbolic. Isothermograms represent the binding of ligands as well the heat changes occurring during binding. A single binding site was observed for both the substrates. kd values for FMN was 20.8 μM and for MgATP-216.6 μM respectively (Fig. 4). The lower kd value of MgATP−2 compared to FMN is consistent with the MgATP−2 being the first substrate as FADS follows an ordered bi-bi mechanism [18], it's possible that a better site of binding for FMN could be created after the binding of the nucleotide moiety.
Fig. 4.
Isothermal colorimetric profile of NcFADS: Upper panel represents the raw thermograms and the lower panel represents the binding isotherms with integrated heats for NcFADS titrations. Both the titrations were performed in 1xPBS (pH7.5). A. NcFADS (50 μM) in the cell binds to 1 mM ATP with 5 mM MgCl2 present in the syringe. B. NcFADS (40 μM) in the cell interacts with 1.5 mM FMN present in the syringe.
This is the first report of cloning, over-expression and purification of NcFADS in high yields due to the single step affinity purification method. Such high yields of the protein can be used for the structural studies to understand the molecular details of the enzyme that could reveal the reason for high turnover number and greater catalytic efficiency compared to other eukaryotic sources. N. crassa being filamentous-fungi with a saprophytic life style requires high amount of flavoproteins to metabolize the contents present in the organic environment and this organism is also known for the highest number of flavo-proteins. Most of the flavoenzymes in N. crassa plausibly contribute to multitude of biosynthetic pathways resulting in the secondary metabolites. The saprotrophic nature of the filamentous fungi requires the synthesis of oxidases and dehydrogenases to access organic matter in the environment. Members of this family are typically oxidases that are capable of performing a wide range of substrate (e.g., sugars and alcohols) oxidation reactions [31]. RFK isolated from N. crassa also showed high turnover number [32], both the enzymes (RFK and FADS) involved in the flavin biosynthesis pathway show an exceptional kinetic property that directly implies the necessity of the flavins in the cell. The high yield and turn over number of the FADS could be a therapeutic alternate for certain diseased conditions like the Fraxetin deficiency that could be resolved by improving the mitochondrial respiration supplementing with the FAD [36]. Based on the recent analysis of the FADS isoforms isolated from humans resulting as an emergency protein used for the patients suffering from the deficiencies caused by FADS [37]. Virtual screening protocols with respect to certain ligands interacting with FADS from prokaryotes (Corynebacterium ammoniagenes) resulted in therapeutic strategy, screening for antimicrobial resistant pathogens causing tuberculosis and pneumonia indicate the potentiality of FADS as drug target [38,39]. Therefore the recombinant NcFADS could provide an opportunity to go for the detailed structural analysis using X-ray crystallography or multi-dimensional NMR, also synthesis of the side-directed mutants at the active site can further help in understanding the kinetic and structural aspects of the enzyme.
References
- 1.Fraaije M.W., Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 2.Maruta T., Yoshimoto T., Ito D., Ogawa T., Tamoi M., Yoshimura K., Shigeoka S. An Arabidopsis FAD pyrophosphohydrolase, AtNUDX23, is involved in flavin homeostasis. Plant Cell Physiol. 2012;53:1106–1116. doi: 10.1093/pcp/pcs054. [DOI] [PubMed] [Google Scholar]
- 3.Foraker A.B., Khantwal C.M., Swaan P.W. Current perspectives on the cellular uptake and trafficking of riboflavin. Adv. Drug Deliv. Rev. 2003;55:1467–1483. doi: 10.1016/j.addr.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Bowers-Komro D.M., Yamada Y., McCormick D.B. Substrate specificity and variables affecting efficiency of mammalian flavin adenine dinucleotide synthetase. Biochemistry. 1989;28:8439–8446. doi: 10.1021/bi00447a025. [DOI] [PubMed] [Google Scholar]
- 5.Oka M., McCormick D.B. Complete purification and general characterization of FAD synthetase from rat liver. J. Biol. Chem. 1987;262:7418–7422. [PubMed] [Google Scholar]
- 6.Efimov I., Kuusk V., Zhang X., McIntire W.S. Proposed steady-state kinetic mechanism for Corynebacterium ammoniagenes FAD synthetase produced by Escherichia coli. Biochemistry. 1998;37:9716–9723. doi: 10.1021/bi972817j. [DOI] [PubMed] [Google Scholar]
- 7.Yamada Y., Merrill A.H., Jr., McCormick D.B. Probable reaction mechanisms of flavokinase and FAD synthetase from rat liver. Arch. Biochem. Biophys. 1990;278:125–130. doi: 10.1016/0003-9861(90)90240-y. [DOI] [PubMed] [Google Scholar]
- 8.Huerta C., Borek D., Machius M., Grishin N.V., Zhang H. Structure and mechanism of a eukaryotic FMN adenylyl transferase. J. Mol. Biol. 2009;389:388–400. doi: 10.1016/j.jmb.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes S.Y., Scholle M.D., D'Souza M., Bernal A., Baev M.V., Farrell M., Kurnasov O.V., Daugherty M.D., Mseeh F., Polanuyer B.M., Campbell J.W., Anantha S., Shatalin K.Y., Chowdhury S.A., Fonstein M.Y., Osterman A.L. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagihara T., Fujio T., Aisaka K. Cloning of FAD synthetase gene from Corynebacterium ammoniagenes and its application to FAD and FMN production. Appl. Microbiol. Biotechnol. 1995;42:724–729. doi: 10.1007/BF00171952. [DOI] [PubMed] [Google Scholar]
- 11.Mack M., van Loon A.P., Hohmann H.P. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 1998;180:950–955. doi: 10.1128/jb.180.4.950-955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarebout G., Villers C., Leclercq R. Macrolide resistance gene mreA of Streptococcus agalactiae encodes a flavokinase. Antimicrob. Agents Chemother. 2001;45:2280–2286. doi: 10.1128/AAC.45.8.2280-2286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solovieva I.M., Kreneva R.A., Leak D.J., Perumov D.A. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology. 1999;145(Pt 1):67–73. doi: 10.1099/13500872-145-1-67. [DOI] [PubMed] [Google Scholar]
- 14.Santos M.A., Jimenez A., Revuelta J.L. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:28618–28624. doi: 10.1074/jbc.M004621200. [DOI] [PubMed] [Google Scholar]
- 15.Kashchenko V.E., Shavlovskii G.M. Purification and properties of the riboflavin kinase of the yeast Pichia guilliermondii. Biokhimiia. 1976;41:376–383. [PubMed] [Google Scholar]
- 16.McCormick D.B., Oka M., Bowers-Komro D.M., Yamada Y., Hartman H.A. Purification and properties of FAD synthetase from liver. Methods Enzymol. 1997;280:407–413. doi: 10.1016/s0076-6879(97)80132-2. [DOI] [PubMed] [Google Scholar]
- 17.Schrecker A.W., Kornberg A. Reversible enzymatic synthesis of flavin-adenine dinucleotide. J. Biol. Chem. 1950;182:795–803. [PubMed] [Google Scholar]
- 18.Abbas C.A., Sibirny A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 2011;75:321–360. doi: 10.1128/MMBR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu M., Repetto B., Glerum D.M., Tzagoloff A. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:264–271. doi: 10.1128/mcb.15.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brizio C., Galluccio M., Wait R., Torchetti E.M., Bafunno V., Accardi R., Gianazza E., Indiveri C., Barile M. Over-expression in Escherichia coli and characterization of two recombinant isoforms of human FAD synthetase. Biochem. Biophys. Res. Commun. 2006;344:1008–1016. doi: 10.1016/j.bbrc.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Sandoval F.J., Zhang Y., Roje S. Flavin nucleotide metabolism in plants: monofunctional enzymes synthesize fad in plastids. J. Biol. Chem. 2008;283:30890–30900. doi: 10.1074/jbc.M803416200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torchetti E.M., Brizio C., Colella M., Galluccio M., Giancaspero T.A., Indiveri C., Roberti M., Barile M. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion. 2010;10:263–273. doi: 10.1016/j.mito.2009.12.149. [DOI] [PubMed] [Google Scholar]
- 23.Bafunno V., Giancaspero T.A., Brizio C., Bufano D., Passarella S., Boles E., Barile M. Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 2004;279:95–102. doi: 10.1074/jbc.M308230200. [DOI] [PubMed] [Google Scholar]
- 24.Barile M., Brizio C., Valenti D., De Virgilio C., Passarella S. The riboflavin/FAD cycle in rat liver mitochondria. Eur. J. Biochem. 2000;267:4888–4900. doi: 10.1046/j.1432-1327.2000.01552.x. [DOI] [PubMed] [Google Scholar]
- 25.Barile M., Passarella S., Bertoldi A., Quagliariello E. Flavin adenine dinucleotide synthesis in isolated rat liver mitochondria caused by imported flavin mononucleotide. Arch. Biochem. Biophys. 1993;305:442–447. doi: 10.1006/abbi.1993.1444. [DOI] [PubMed] [Google Scholar]
- 26.Deluca C., Kaplan N.O. Flavin adenine dinucleotide synthesis in animal tissues. Biochim. Biophys. Acta. 1958;30:6–11. doi: 10.1016/0006-3002(58)90233-6. [DOI] [PubMed] [Google Scholar]
- 27.Pallotta M.L., Brizio C., Fratianni A., De Virgilio C., Barile M., Passarella S. Saccharomyces cerevisiae mitochondria can synthesise FMN and FAD from externally added riboflavin and export them to the extramitochondrial phase. FEBS Lett. 1998;428:245–249. doi: 10.1016/s0014-5793(98)00544-4. [DOI] [PubMed] [Google Scholar]
- 28.Galluccio M., Brizio C., Torchetti E.M., Ferranti P., Gianazza E., Indiveri C., Barile M. Over-expression in Escherichia coli, purification and characterization of isoform 2 of human FAD synthetase. Protein Expr. Purif. 2007;52:175–181. doi: 10.1016/j.pep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Torchetti E.M., Bonomi F., Galluccio M., Gianazza E., Giancaspero T.A., Iametti S., Indiveri C., Barile M. Human FAD synthase (isoform 2): a component of the machinery that delivers FAD to apo-flavoproteins. FEBS J. 2011;278:4434–4449. doi: 10.1111/j.1742-4658.2011.08368.x. [DOI] [PubMed] [Google Scholar]
- 30.Leulliot N., Blondeau K., Keller J., Ulryck N., Quevillon-Cheruel S., van Tilbeurgh H. Crystal structure of yeast FAD synthetase (Fad1) in complex with FAD. J. Mol. Biol. 2012;398:641–646. doi: 10.1016/j.jmb.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 31.Macheroux P., Kappes B., Ealick S.E. Flavogenomics-a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 32.Rajeswari S.R., Jonnalagadda V.S., Jonnalagadda S. Purification and characterization of flavokinase from Neurospora crassa. Indian J. Biochem. Biophys. 1999;36:137–142. [PubMed] [Google Scholar]
- 33.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Louis Jeune C., Andrade Navarro M.A., Perez Iratxeta C. Prediction of protein secondary structure from circular dichorism using theoretically derived spectra. Proteins. 2011;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Cabo P., Ros S., Palau F. Flavin adenine dinucleotide rescues the phenotype of frataxin deficiency. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leone P., Galluccio M., Barbiroli A., Eberini I., Tolomeo M., Vrenna F., Gianazza E., Iametti S., Bonomi F., Indiveri C., Barile M. Bacterial production, characterization and protein modeling of a novel monofunctional isoform of FAD synthetase in humans-anemergency protein? Molecules. 2018;23:116. doi: 10.3390/molecules23010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arilla-Luna S., Serrano A., Medina M. Specific features for the competent binding of substrates at the FMN adenylyl transferase site of FAD Synthase from Corynebacteriumammoniagenes. Int. J. Mol. Sci. 2019;20(20):5083. doi: 10.3390/ijms20205083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lans I., Carbonell E.A., Rodriguez K.P., Ainsa J.A., Medina M., Cossio P. Insilico discovery and biological validation of ligands of FAD synthase, a promising new antimicrobial target. Plos Computat Biol. 2020;16(8) doi: 10.1371/journal.pcbi.1007898. [DOI] [PMC free article] [PubMed] [Google Scholar]