Abstract
Background: Metastases are responsible for over 70% of deaths from lung adenocarcinomas. Previous large-scale studies of LUAD mainly focused on primary diseases. We aimed to comprehensively analyze the genomic landscape of metastatic LUADs and elucidate its clinical implications in the context of precision medicine.
Methods: We performed retrospective analyses on targeted sequencing data of 3,743 primary tumors and 934 metastases from 4,480 patients with lung adenocarcinomas, and PD-L1 immunohistochemical data of 1,336 primary tumors and 252 metastases from 1,588 LUAD patients.
Results: Metastases generally manifested significantly higher mutational burdens and chromosomal instability than primary lung adenocarcinomas. Clinically actionable alterations, including ALK mutations, ALK and ROS1 fusions, and MET copy number gains, were enriched in metastases, particularly metastases to some specific organs/tissues, such as lymph nodes, liver, and brain. PD-L1 expression decreased as the approximate metastatic distance increased. Additional data of paired primary tumors and metastases to lymph nodes and brain validated patterns of actionable alterations and candidates for metastatic drivers. Two evolutionary modes of metastatic dissemination, common origins and distinct origins, were identified in both types of primary-metastasis pairs.
Conclusions: Our study showed heterogenous patterns of clinically actionable alterations, PD-L1 expressions, metastatic driver candidates, and evolutionary patterns among multiple types of metastases of lung adenocarcinomas, which may advise the planning of treatments and the identification of novel therapeutic targets.
Keywords: Genomic profiling, NSCLC, Metastasis, Therapeutic target
Introduction
Most lung adenocarcinomas (LUAD) are diagnosed at advanced stages [1], and metastasis is the major cause of LUAD-related death. Treatments for metastases, especially distant ones, have limited efficacy. Given that most previous large-scale lung cancer studies focused on primary diseases [[2], [3], [4]], a more comprehensive understanding of the tumor genomics that drives metastases is warranted, particularly in the field of targeted therapy and precision medicine.
Recently, multiple researchers investigated the genetic profile of LUAD metastasizing to a specific site or tissue. For example, a study of 41 primary LUAD-lymph node metastasis (LNM) pairs first described the heterogeneity of actionable alterations between primary tumors and metastases [5]. Another study of 73 cases of brain metastases (BRMs) from LUAD identified key metastatic drivers and validated their functions of increasing BRM incidence in xenograft mouse models [6]. However, most of these LUAD studies included only one type of metastasis and comparisons among different organs/tissues were limited. Muller et al. investigated metastatic breast cancer using a cohort of 22 patients and found heterogeneous actionable alterations among different metastatic types [7]. Whether this heterogeneous genetic pattern also applies to metastatic LUADs is still illusive and need to be investigated in large-scale patient cohorts.
In the present study, we established a discovery dataset of LUAD metastases from lymph nodes, pleura, bone, liver, and brain. The metastatic samples were collected as part of standard clinical care, and these samples were then compared with a control population of primary LUADs tumors. We aimed to study the unique molecular features among primary LUAD and various metastatic types and elucidate its potential clinical significance in directing therapeutic strategies and identifying novel targets against metastatic LUADs.
Materials and Methods
Patients and samples
We retrospectively analyzed targeted deep sequencing data of 3,743 primary tumors and 934 metastases from 4,480 LUAD patients using the GeneseeqPrime™ panel, a next-generation sequencing platform that can detect mutations, copy number alterations, and gene fusions in more than 425 cancer-associated genes. PD-L1 IHC data from 1,588 LUAD patients, including results of 1,336 primary tumors and 252 metastases, were also included in our analyses. All sequencing samples were from a clinical sequencing database, which solely included samples that had tumor contents of over 10% and have passed in-house quality control procedures to minimize the effect of DNA damage and contamination. The matched whole blood samples were used as normal controls. All 4,677 samples had at least one alteration event (e.g., somatic mutation, small insertion/deletion, copy number variation, or gene fusion). Written informed consent was collected from each patient upon sample collection. All detected alteration events were manually reviewed and reported to patients and physicians in the electronic medical record.
Library preparation and sequencing
Processes of library preparation and targeted sequencing were performed as described in Yang et al.’s study [8]. The detailed procedures were provided in the Supplementary Methods.
Single nucleotide variant and gene fusion calling
Processes of variant calling were performed as described in Yang et al.’s study [8]. The details of the analyses were provided in the Supplementary Methods.
Copy number alteration analysis
Gene-level copy number alterations were identified using FACETS [9], and the detailed procedures were provided in the Supplementary Methods.
Phylogenetic tree reconstruction
Rooted phylogenetic trees were reconstructed by the Phangorn package with the binary presence/absence matrix of alterations, including single nucleotide variants (SNVs), copy number variants (CNVs), and gene fusions, in each sample [10]. Branches were in scale of the number of alterations.
Immunohistochemistry
FFPE tissue slices were used for PD-L1 immunohistochemical testing (22C3 clone DAKO, Carpenteria, CA, USA). Blocks were sectioned at 4 μm, and tumors were classified by PD-L1 expression levels:
| Proportions of tumor cells or tumor infiltrating lymphocytes that express PD-L1 (p) | Tumor type |
| p < 1% | PD-L1 negative |
| 1% ≤ p < 50% | PD-L1 low expression |
| 50% ≤ p ≤ 100% | PD-L1 high expression |
Identification of metastasis-enriched alterations
In order to identify candidates for metastatic drivers, Fisher's exact tests were performed to compare the mutational prevalence between metastases and primary tumors. Alterations that were detected in >2% of primary tumors or >2% of metastases (at least two tumors) were included. P values were adjusted for false discovery rates (q values) independently for mutations and CNVs. Alterations with lower bound of 95% confidence interval of odds ratio (metastases to primaries) greater than 1 and q<0.2 (P<0.05 for gene fusions) were considered significantly enriched in metastases.
Selection for metastasis-private events
The method aimed to test whether candidate metastatic drivers identified from the unpaired study cohort were selected to be private in metastases in the primary-metastasis paired validation datasets. The method was adapted from the one described in the TRACERX renal study for comparisons of metastatic selection [11]. Briefly, a background null distribution of proportions of metastasis-private events was determined, based on all synonymous mutations in this study; a Binomial test was then used to compare the proportion of the metastasis-private events to that of events shared or private in the primary. The reference probability of metastasis-private events was taken from the null model, and the number of trials was based on the number of primary-metastasis pairs with the given alteration detected. All mutations and CNVs that occurred in at least 2 tumors were tested. P<0.05 was considered statistically significant; P<0.1 was considered as a trend of significance.
Statistical analysis
Comparisons between continuous data were performed using the Wilcoxon test. Comparisons of proportion between groups were conducted using the Fisher's exact test. For trend analyses of proportions, the Chi-squared trend test was employed. For the selection of metastasis-private alterations, the binomial test was employed. For multiple tests, P values were adjusted using the Benjamini-Hochberg method. A two-sided P value of less than 0.05 was considered significant unless otherwise indicated. All statistical analyses were done in R (v.4.0.2).
Results
Sample overview
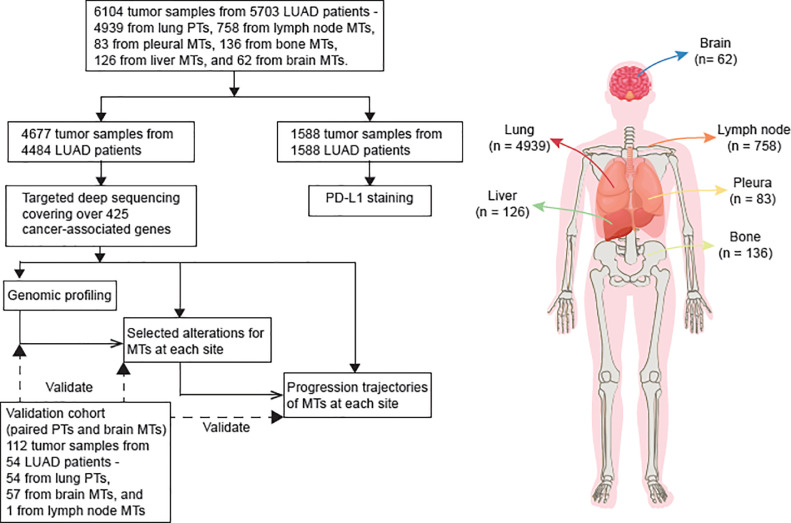
We retrospectively analyzed a total of 6,104 tumor samples, consisting of 4,939 primary tumors and 1,165 metastases, which were collected from 5,699 LUAD patients (Fig. 1, Table S1-S2). Among these samples, targeted sequencing data of 4,677 tumors, including 3,743 primary tumors and 934 metastases, were available from a clinical sequencing database. A total of 29,707 somatic non-synonymous mutations (median: 6, QTR: 3 - 8) were identified (Table S3), together with 6,267 copy number variations and 338 gene fusions (Table S4-S5). Genomic heterogeneity was observed between primary tumors and metastases, as well as among various metastatic types. An external dataset of paired primary tumor and brain metastases (BRMs) was used to validate the specific genomic patterns. Besides sequencing data, 1,588 tumors within our cohort were subjected to immunohistochemical analysis of PD-L1 expressions, with 770 being PD-L1 positive and 818 being PD-L1 negative (Table S6).
Fig. 1.
Overview of samples.
5916 tumor samples from 5387 patients with LUAD were collected. Targeted deep sequencing was performed on 4677 tumor samples, including 3743 from primary tumors and 934 from metastases. Immunohistochemistry assays were performed on 1588 tumor samples, including 1336 from primary tumors and 252 from metastases. Abbreviations: MT – metastatic tumor, PT – primary tumor, TMB – tumor mutational burden.
Genomic characteristics of primaries and metastases
To identify genomic landscape of metastases, we profiled molecular alterations in primary tumors and metastases. Three types of alterations were investigated, including gene mutations (i.e. all types of non-synonymous variants), copy-number variations (CNVs), and gene fusions (Tables S3-S5). All CNVs were adjusted with tumor purities using the FACETS tool (see Methods for more details). Only tumors with purities greater than 10% were included, and the tumor purity was comparable between primary tumors and metastases (mean: 56.2% vs. 54.9%, P = 0.77).
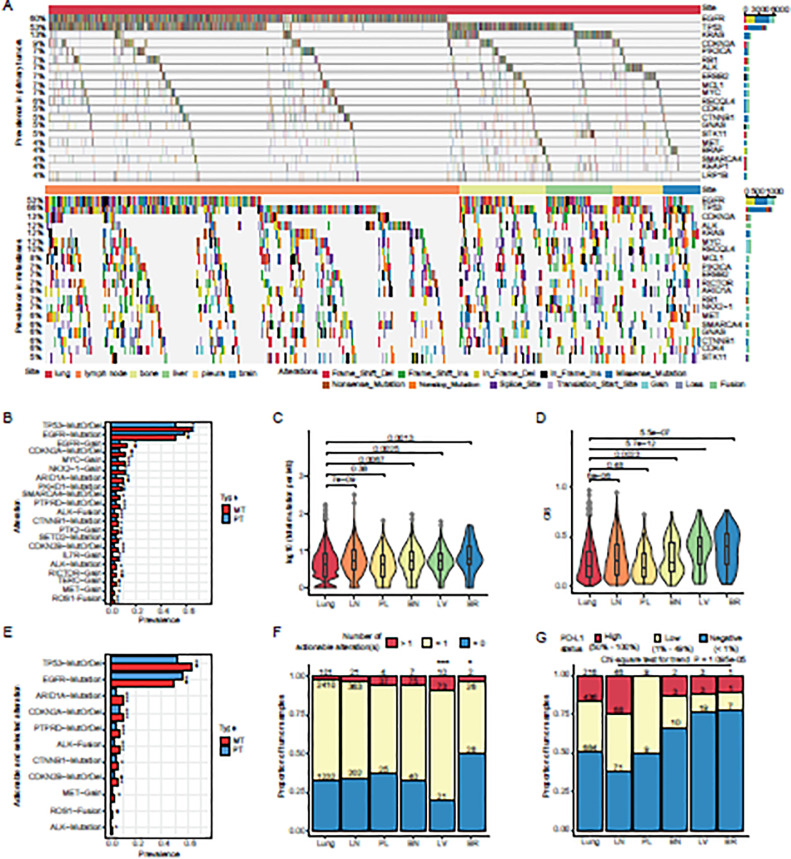
Twenty most frequently altered genes in primary tumors overlapped largely (15/20) with those in metastases (Fig. 2A). Intriguingly, among thirty most prevalent recurrent alterations in metastases, twenty showed significant difference in prevalence from those in primary tumors. Particularly, EGFR mutations exhibited a significantly lower prevalence in metastases than in primary tumors (Fig. 2B), and this trend was conserved when compared with different stages of the primary tumors (stage I, III, IV) (Fig. S1A).
Fig. 2.
Genomic profiling of primary tumors and metastases.
A). Genomic profiling of primary tumors (upper panel) and metastases (lower panel). 20 most frequently altered genes are displayed. Alterations including SNVs, InDels, CNVs, and Fusions, are denoted with different colors. B). Prevalence of alterations in primary tumors and metastases. Deep deletions and mutations of tumor suppressors were integrated and labeled as “MutOrDel”. Fisher's exact tests were performed to compare the differences of prevalence in primary tumors and metastases. P values were adjusted for multiple comparisons with Benjamini and Hochberg method. Adjusted P values (q values) smaller than 0.05 were considered of statistical significance ***q < 0.005, **q < 0.01, *q < 0.05, .q < 0.1. Twenty alterations that showed significantly different prevalence between primaries and metastases are displayed in the descending order of metastatic prevalence. C). Tumor mutational burdens of samples from primary tumors and metastases to the lymph nodes, pleura, bone, liver, and brain. D). Chromosomal instability scores of samples from primary tumors and metastases to the lymph nodes, pleura, bone, liver, and brain. E). Prevalence of actionable alterations and selected oncogenic mutations in primary tumors and metastases. Alterations were ordered by their prevalence in metastases. P values were calculated and adjusted in the same way as (B). Adjusted P values (q values) smaller than 0.05 were considered of statistical significance ***q < 0.005, **q < 0.01, *q < 0.05, .q < 0.1. F). Proportions of tumor samples that harbored more than one actionable alteration, only one actionable alteration, and no actionable alteration, from primary tumors and metastases to the lymph nodes, pleura, bone, liver, and brain. G). Proportions of tumor samples that showed high PD-L1+ compositions (50% - 100%), low PD-L1+ compositions (1% - 49%), and no PD-L1+ composition (< 1%), from primary tumors and metastases to the lymph nodes, pleura, bone, liver, and brain. Abbreviations: MT – metastatic tumor, PT – primary tumor, TMB – tumor mutational burden, CIS – chromosomal instability score, LN – lymph node, PL – pleura, BN – bone, LV – liver, BR – brain, PD-L1 – programmed death ligand 1.
On the other hand, functional loss (mutations and/or copy number loss) of TP53, CDKN2A, ARID1A, PKHD1, SMARCA4, CTNNB1, SETD2, CDKN2B, and ALK, gains of EGFR, NKX2-1, MYC, PTK2, IL7R, RICTOR, TERC, and MET, and fusions of ALK and ROS1 were enriched in metastases (Fig. 2B). Of note, gains of EGFR were significantly more prevalent in metastases than in different stages of primaries (Fig. S1B-C). A higher proportion of EGFR mutations in metastases were accompanied with EGFR gains than those in primaries (Fig. S1D). These results suggest that EGFR gains may be more competent in driving metastasis than EGFR mutations.
Metastases generally accumulated more mutations and genomic aberrations than primary tumors, except for pleural metastases that had similar tumor mutational burden (TMB) and chromosomal instability (CIS) to primary tumors, which might be due to their early occurrence and close anatomic distance to primary lesions (Fig. 2C-D, Table S6).
Heterogeneity of actionable alterations in primaries and metastases
We next tried to compare the targetable alterations between primaries and metastases, which may provide useful guides for precision medicine. We used the OncoKB database as reference [12], which included both targets of standard-of-care treatments and those with clinical or laboratory evidence. As expected in Asian population, EGFR mutations were the most common therapeutic targets in LUAD patients, detected in 52.23% of primary tumors and 45.07% of metastases (Fig. 2E). Several druggable alterations showed significantly higher prevalence in metastases than in primary tumors, including mutations of ALK (1.50% vs. 0.61%), gains of MET (3.96% vs. 2.32%), and gene fusions of ALK and ROS1 (6.85% vs. 3.77% and 2.68% vs. 1.28%, respectively). Moreover, we examined the prevalence of oncogenic mutations that were not currently actionable but of great interest (Table S7). Among them, mutations of TP53, CTNNB1, and ARID1A were significantly enriched in metastases (63.7% vs. 53.5%, 6.5% vs. 4.8%, and 10.0% vs. 5.3%, respectively; Fig. 2E). Metastases to lymph node, pleura, and bone showed similar proportions of tumors that harbored actionable alterations (Fig. 2F). Notably, among all the metastases in our cohort, liver metastases were most likely to response to target therapies (92/104, 88.46%), and over two fifths (41/92, 44.57%) of liver metastases had multiple actionable alterations (Fig. 2F). On the contrary, brain metastases had the least proportion of actionable targets in our cohort (28/56, 50.00%) (Fig. 2F), implying the challenge to target brain metastases. Furthermore, different metastatic types tended to enrich different spectra of targetable alterations (Fig. S2). Specifically, liver metastases and primary tumors shared some frequently mutated genes, especially EGFR, whereas liver metastases were highly enriched for ALKmutations when compared with the primary tumors (6.73% vs. 0.61%) (Fig. S2). Brain and lymph node metastases, on the other hand, had lower prevalence of EGFR mutations than primary tumors (33.93% vs. 52.23% and 43.00% vs. 52.23%, respectively), but they were both enriched for ROS1 fusions (5.36% in brain metastases and 3.07% in lymph node metastases vs. 1.28% in primary tumors) (Fig. S2). In addition, lymph node metastases specifically featured ALK fusions (7.85% vs. 3.77%) (Fig. S2). Overall, these results suggest that different metastatic types may possess unique actionable mutation file, thus favoring different target therapies.
PD-L1 expression in primaries and metastasis
The activation of the programmed cell death protein 1 (PD-1)/PD-L1 axis serves as an immune escape mechanism of tumors. Blockade of PD-1/PD-L1 has been proved to be an effective cancer immunotherapy for a spectrum of malignancies. We conducted immunohistochemical assays to assess the feasibility of anti-PD-L1 immunotherapies in metastatic lung cancers (Table S8). Around half (652/1336, 48.80%) of the primary tumors were PD-L1 positive (PD-L1 content≥1%), with LNMs showing the highest PD-L1 positive proportion (114/185, 61.62%). Interestingly, the proportion of PD-L1 positive tumors decreased as the approximate metastatic distance increased (Fig. 2G), implying the increasing complexity of immune-escaping mechanisms, which is in line with the increasingly disrupted genomes in the corresponding metastatic samples (Fig. 2D). However, considering the elevated mutational burdens in metastases (Fig. 2C), especially the distant ones, these LUAD patients with relatively low PD-L1 contents may be still suitable for immunotherapies.
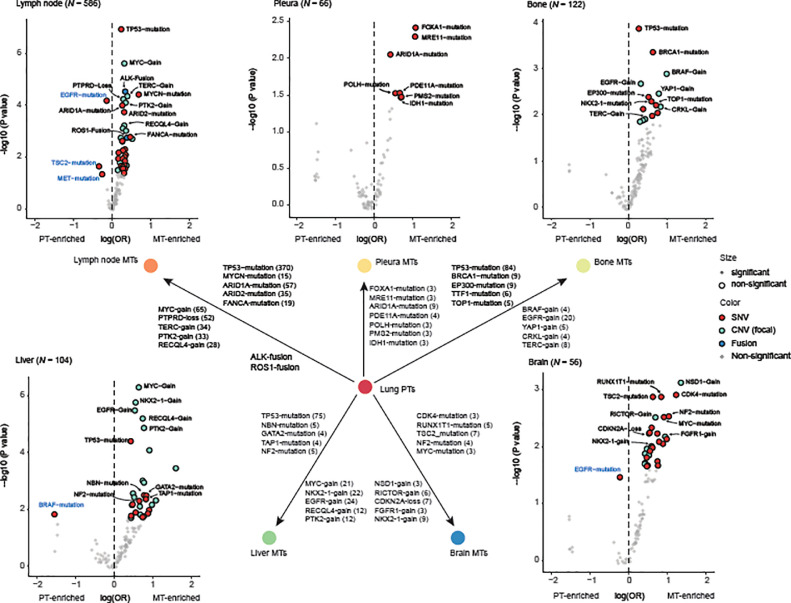
Identification of metastatic drivers for different organs
To identify drivers for metastatic progression, we investigated alterations that showed elevated frequencies at each metastatic site (Table S9). In LNMs, mutations of TP53, MYCN, ARID1A, and ARID2 were significantly enriched (q value=1.04*10−5, 1.92*10−3,2.24*10−3, and 3.72*10−3, respectively). Gains of MYC, TERC, PTK2, and RECQL4 (q value=6.00*10−5, 4.71*10−4, 4.79*10−4, and 3.18*10−3, respectively) and loss of PTPRD (q value=4.71*10−4) also showed significant higher prevalence in LNMs than that in primary tumors. In addition, of 586 LNMs we detected gene fusions of ALK and ROS1 in 46 (7.85%) and 19 (3.25%) cases, respectively, which is significantly higher than that in primary tumors (ALK: 141/3743, 3.77%, q value=5.46*10−5; ROS1: 48/3743, 1.28%, q value=1.64*10−3).
Fewer alterations were enriched in pleural metastases and only 7 of them showed statistical significance (P<0.05), including mutations of FOXA1, MRE11, ARID1A, PDE11A, POLH, PMS2, and IDH1 (P=0.00395, 0.00459, 0.00889, 0.0279, 0.0311, 0.0332, and 0.0353, respectively). This suggested that pleural metastases may diverge less from primary tumors, as also supported by the similarity of TMB and CIS between them (Fig. 2C-D).
Mutations of TP53 and gains of EGFR were enriched in both bone and liver metastases, indicating their roles in driving metastasis. While unique feature alterations of each type may define different adaptabilities of primary dispersions to microenvironments at bones and liver. Bone metastases featured mutations of TP53, BRCA1, EP300, NKX2-1, and TOP1 (q = 0.0176, 0.0306, 0.137, 0.137, and 0.137, respectively), and gains of BRAF, EGFR, YAP1, CRKL, and TERC (q = 0.0281, 0.0281, 0.0281, 0.0385, and 0.0537, respectively), whereas liver metastases featured mutations of TP53, NBN, GATA2, TAP1, and NF2 (q = 0.00230, 0.0942, 0.0942, 0.0942, and 0.0942, respectively), and gains of MYC, NKX2-1, EGFR, RECQL4, and PTK2 (q = 1.42*10−5, 2.47*10−5, 3.24*10−5, 4.70*10−5, and 8.87*10−5, respectively). Of note, if more stringent cutoff for statistical significance was employed, for example q < 0.05, only mutations of TP53 and gains of five genes remained significantly enriched in liver metastases, implying that the migration to the liver may be empowered mainly by copy number variations.
In BRMs, our data revealed several novel candidates for metastatic drivers, such as mutations of TSC2 and NF2 (q = 0.0402 and 0.0502, respectively) in addition to some known drivers for neural system carcinomas, such as mutations of CDK4, RUNX1T1, and MYC, gains of RICTOR, FGFR1, and NKX2-1, and loss of CDKN2A (Fig. 3, Table S8). In sum, the site-specific patterns of candidate metastatic drivers suggest that LUADs dispersed with different genomic determinants in adaption to complex microenvironments at different organs/tissues.
Fig. 3.
Metastasis-enriched alterations.
The odds ratio and p value for the comparison of prevalence in primary tumors and metastases of each alteration are shown. P values were adjusted for false discovery rates (q values). Each scatter denotes an alteration. Alteration scatters with P < 0.05 in pleural metastases or q < 0.2 at other metastatic sites are colored and enlarged. Five mutations with the smallest q values, five copy number variations with the smallest q values, and all gene fusions with P < 0.05 were labeled black and displayed in the schematic diagram at the center. Alterations that were enriched in primaries were additionally labeled blue. Pleural metastases showed a total of only 6 alterations that had P < 0.05. Abbreviations: MT – metastatic tumor, PT – primary tumor, OR – odds ratio, SNV – single nucleotide variation, CNV – copy number variation.
Validation of site-specific patterns using a primary-metastasis matched cohort
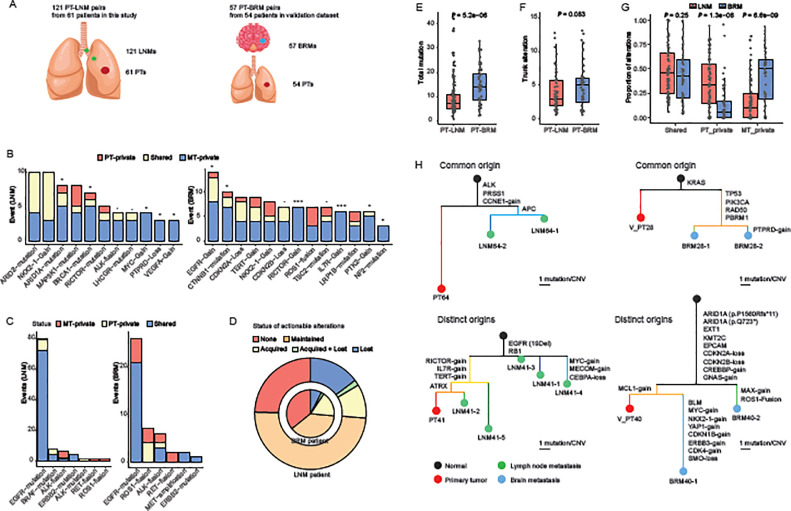
To validate the patterns of metastatic drivers and therapeutic targets, we acquired an independent cohort of 57 BRMs with 54 paired LUADs. We also sorted data of 121 LNMs that were paired with 61 primaries from our dataset, which allowed for comparisons between LNM-PT and BRM-PT pairs (Fig. 4A, Table S10-S11).
Fig. 4.
Evolutionary analyses of primary tumor-lymph node metastasis and primary tumor-brain metastasis pairs.
A). Descriptions of PT-LNM pairs in this study and PT-BRM pairs in the validation dataset. B). Numbers of metastasis-enriched alterations (q values < 0.2 in the enrichment analyses) that were PT-private, shared, and MT-private. Binomial tests were performed to determine whether an alteration was selected to be MT-private rather than in random paths of silent mutations. Alterations that showed more than two MT-private events are displayed. C). Numbers of actionable alterations that were PT-private, shared, and MT-private in LNM-PT (left panel) and BRM-PT (right panel) datasets. D). Proportions of patients that had no actionable alteration (none), maintained concordant actionable alterations in primaries and metastases (maintained), acquired new actionable alterations in metastases (acquired), or lost actionable alterations in metastases (lost). Inner circle represents patients with BRMs. Outer circle represents patients with LNMs. E). Total mutations of PT-LNM pairs and PT-BRM pairs. F). Trunk alterations of PT-LNM pairs and PT-BRM pairs. G). Proportions of alterations that were PT-private, shared, and MT-private. H). Phylogenetic trees of example cases with multiple metastases showing common origins. Colors of nodes denote different types of samples – black for virtual normal controls, red for PTs, green for LNMs, and blue for BRMs. Branch lengths are in the scale of alteration numbers. Scale bars for the branch length are shown at the bottom right of each phylogenetic tree.
Abbreviations: PT – primary tumor, LNM – lymph node metastasis, BRM – brain metastasis, MT – metastatic tumor, SNV – single nucleotide variation, CNV – copy number variation.
We first investigated candidate metastatic drivers, which were identified as significantly enriched in metastases from population-based analyses. Driver alterations that confer selective advantages during metastasis are expected to manifest elevated frequencies or be private in metastatic lesions. With the assumption that synonymous mutations were neutral for metastatic selection, we examined whether candidate drivers for LNMs and BRMs described above had higher probabilities to be private in respective metastases, using the two LNM/BRM-PT matched validation datasets. Our results revealed that mutations of ARID1A and BRCA1, gains of MYC and VEGFA, and loss of PTPRD were significantly selected for LNM-private events, with mutations of LHCGR and gene fusions of ALK showing a trend. While mutations of CTNNB1 and NF2, and gains of EGFR, RICTOR, IL7R, and PTK2 were selected for BRM-private events, with mutations of TSC2 and loss of CDKN2B showing a trend (Fig. 4B, Fig. S2).
We then evaluated therapeutic targets that may guide options for treatment. EGFR mutations were the most prevalent targetable alterations and showed high primary-metastasis concordance in both datasets, with rates of 72/80 (90.00%) in LNM-PT pairs and 21/26 (80.77%) in BRM-PT pairs. Other than that, BRM-PT pairs had a notable proportion of metastasis-private EGFR mutations (5/26, 19.23%) and no primary-private EGFR mutations, compared to respective proportions of 1/80 (1.25%) and 7/80 (8.75%) in LNM-PT pairs. Other common targets included mutations of ERBB2 and fusions of ALK, RET, and ROS1. In both LNM-PT and BRM-PT dataset, all ERBB2 mutations were shared within paired primaries and metastases whereas all RET fusions were private in metastases (Fig. 4C).
At the patient level, a higher proportion of LUAD patients with BRMs harbored no actionable alteration than those with LNMs 19/54 (35.19%) vs. 15/61 (24.59%). However, more BRMs acquired new therapeutic targets that were absent from paired primaries, which was seen in 10/54 (18.52%) of the patients, compared to a ratio of 7/61 (11.48%) in patients with LNMs. Moreover, only 5/54 (9.26%) of the patients lost actionable targets in their BRMs. The ratio almost doubled in patients with LNMs (10/61, 16.39%) (Fig. 4D). These findings correspond to the site-specific patterns of actionable alterations identified in the population-based analyses, suggesting adapted strategies for clinical trial designing and/or treatment planning against different types of metastases.
We further compared the evolution of LNMs and BRMs inferred from paired data. BRM-PT pairs accumulated significantly more mutations than LNM-PT pairs (P = 5.2*10−6) (Fig. 4E). Mutations that were located at the trunk of the BRM-PT phylogeny, in other words, shared by all tumor samples of each patient with BRMs, showed the trend to exceed those in patients with LNMs (P = 0.083). (Fig. 4F). These together indicated that BRMs occurred at later stages of primary progression than LNMs. Nonetheless, BRM-PT pairs exhibited higher proportions of metastasis-private mutations than those of LNM-PT pairs (P = 6.6*10−9), implying faster accumulate mutations in BRMs (Fig. 4G).
Lastly, we unraveled two modes of metastatic evolution in both LNM-PT and BRM-PT pairs. Case P64 of the LNM-PT dataset and case P28 of the BRM-PT dataset had all metastases clustered in the same phylogenetic clades, which were defined as of common origins. While LNM-PT case P41 and BRM-PT case P40 showed metastases distributed at different clades, which were classified as of distinct origins (Fig. 4H). Different evolutionary modes indicated the heterogeneity within each type of metastases.
Discussion
Metastatic lung cancers were diseases of great heterogeneity. Here we report comprehensive genomic landscape of metastases by analyses of over 6000 tumor samples under real-world clinical settings. With a driver event-enriched bespoke panel that covers over 425 genes, we were able to systemically assess mutations, copy number variations, gene fusions, overall tumor mutational burdens, chromosomal instability, therapeutic targets, and site-enriched alterations. Besides, we analyzed PD-L1 expressions in a separate portion of primaries and metastases. ‘
A previous important large-scale study revealed that tumor mutational burden is site specific and that brain metastases harbored the highest mutational burden [13]. Our observations supported the conclusions. The mean TMB in brain metastases in our cohort was 10.1 mutations/Mb, greater than those of primaries (mean: 6.8) and metastases at any other site (mean: 6.7 – 8.7). Our results further revealed that metastases accumulated more actionable targets and may develop more complicated mechanisms of immune escape as the approximate distance of migration increased. Specifically, lymph node metastases enriched ALK and ROS1 fusions; liver metastases enriched ALK mutations; while brain metastases enriched ROS1 fusions. The heterogeneity of actionable alterations suggested different feasibility of specific target therapies among different metastasis types, leaving implications for future designs of clinical trials. Taken together, the heterogeneity of actionable alterations, PD-L1 expressions, and mutational burdens may add to the complexity of treatment planning for metastatic LUADs, highlighting the significance of comprehensive genomic analyses and clinical evaluations.
Targeted therapies, such as tyrosine kinase inhibitors, were known to influence the frequencies of certain driver events. In our cohort, in patients who received prior targeted therapies, metastases showed similar prevalence of EGFR mutations (OR = 0.82, P = 0.19) and higher prevalence of ALK fusions (OR = 2.17, P = 4.67e-03). In comparison, patients without prior targeted therapies had significantly decreased EGFR mutation prevalence (OR = 0.62, P = 1.98e-07) and less elevated ALK fusion prevalence (OR = 1.61, P = 2.43e-02, Fig. S3A-D). However, among all the other metastasis-enrichment we identified (Fig. 2B, E), none was specific to the targeted therapy-treated group (Fig. S3A-D), suggesting that prior targeted therapy status had limited influence on our findings of candidates for metastatic drivers.
The finding that EGFR mutations may be losing during lung cancer metastasis was, to some extent, biologically unexpected. However, we found that multiple smaller lung cancer cohorts, all of which included matched primary tumors and metastases, had reported the depletion of EGFR mutations in metastases [[14], [15], [16], [17]]. Different methods were used in these studies for the detection of EGFR mutations, including high-resolution melting method, ARMS methods, direct sequencing, and heteroduplex analysis, excluding the possibility of methodological error. The depletion rate could be as high as 50% [16], suggesting that EGFR mutations may not be as important in driving metastasis as in carcinogenesis.
The correlation of the PD-L1 expression with the mutational profile is a context of great interest to both laboratories and clinics. We found that primary tumors and metastases with similar levels of PD-L1 expressions may not significantly differ in mutational burden or chromosomal instability (Fig. S4B-C). However, with only a small portion of samples (9.3%, 148/1588, Fig. S4A) having both PD-L1 and sequencing data, the investigation may be under-representative. Further investigations with larger sample sizes were needed.
Furthermore, we identified metastasis-enriched events at the lymph nodes, pleura, bone, liver, and brain, each spreading a spectrum of alterations. Shih et al. identified and validated gains of MYC, YAP1, and MMP13 as drivers for brain metastases [6]. However, none of them was featured due to low occurrence in BRMs in this study (MYC gain: 3; YAP1 gain: 1), which may be due to the population differences.
We validated alterations that were significantly enriched in LNMs and BRMs by analyses of LNM-PT and BRM-PT pairs. Specifically, we found that mutations of TSC2 and NF2 were selected in BRMs, which may be novel drivers that have not been reported. Furthermore, phylogeny analyses distinguished two types of dissemination origins. Metastases of common origin may be derived from the same dispersion of the primary tumor; while those of distinct origins were likely seeded by multiple rounds of primary tumor dispersions, which may imply a more active status of the primary tumor. The evolutionary heterogeneity within each metastasis type may therefore be of clinical significance in assessing the malignancy of metastatic diseases.
One limitation of this study lies in that only a small portion of metastases studied had paired primary samples. This prevented us from drawing definitive conclusions on genomic heterogeneity between primaries and metastases, as there is substantial heterogeneity among primaries that match different types of metastases. Over 80% of our primary samples with clear clinical information were from Stage IV lesions though, the heterogeneity between advanced tumors and earlier stage ones may still affect the comparisons between primaries and metastases. For example, we could not exclude the possibility that the higher number of therapeutic targets in liver metastases was inherited from paired primary tumors that were more actionable than others. Another limitation is the incompleteness of the medical records. It would be interesting to know how the smoking history and/or treatment influences the patterns of actionable alterations and metastatic evolution. We will continue to clean and standardize clinical information from the medical records of this dataset. With more informative annotations, analyses with more clinically relevant stratifications were possible.
In conclusion, our large-scale study of metastatic LUAD refined the genomic landscape and highlighted heterogeneity both among metastasis types and within each metastasis type. Our findings thus were of significance for both further genomic studies and clinical practice of metastatic diseases.
Ethics approval and patient consent
Written informed consent was obtained from each patient upon sample collection according to the protocols approved by the ethics committee of each hospital.
Consent for publication
All authors have approved the manuscript and agreed to the submission to and publication in Neoplasia.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Funding
None declared.
Author Contributions
D.L. - Data curation, formal analysis, investigation, resources, and writing original draft; Y.H. - Data curation, formal analysis, investigation, resources, and writing original draft; L.C. - Data curation, formal analysis, investigation, resources, and draft editing; M.W. - Methodology, formal analysis, visualization, writing original draft, and draft editing; H. B. - Methodology, investigation, supervision, and draft editing; Y.X. - Investigation and draft editing; Y.W. - Formal analysis, visualization, and draft editing; S.W. - Formal analysis and draft editing; X.W. – Investigation, supervision, and draft editing; Y.S. (Yang Shao) – Investigation, supervision, and draft editing; W.Z. - Data curation, resources, and draft editing; G.L. - Data curation, resources, and draft editing; S.H. - Conceptualization, investigation, supervision, and draft editing; T.Z. - Conceptualization, investigation, supervision, and draft editing; Y.S. (Yunfei Shi) - Conceptualization, investigation, supervision, and draft editing.
Declaration of Competing Interest
M.W., H.B., Y.X., Y.W., S.W., X.W., and Y.S. (Yang Shao) are employees of Nanjing Geneseeq Technology Inc. All remaining authors have declared no conflict of interests.
Acknowledgements
We thank Ran Cao and Xia Peng for providing technical and administrative supports for this project.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.10.001.
Contributor Information
Shan Huang, Email: Shan_huang2020@126.com.
Tao Zhang, Email: zhangt10@126.com.
Yunfei Shi, Email: km-syf@163.com.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayekar MK, Bivona TG. Current Landscape of Targeted Therapy in Lung Cancer. Clin Pharmacol Ther. 2017;102(5):757–764. doi: 10.1002/cpt.810. [DOI] [PubMed] [Google Scholar]
- 4.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim EY, Cho EN, Park HS, Kim A, Hong JY, Lim S. Genetic heterogeneity of actionable genes between primary and metastatic tumor in lung adenocarcinoma. BMC Cancer. 2016;16:27. doi: 10.1186/s12885-016-2049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shih DJH, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet. 2020;52(4):371–377. doi: 10.1038/s41588-020-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller KE, Marotti JD, de Abreu FB, Peterson JD, Miller TW, Chamberlin MD. Targeted next-generation sequencing detects a high frequency of potentially actionable mutations in metastatic breast cancers. Exp Mol Pathol. 2016;100(3):421–425. doi: 10.1016/j.yexmp.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018;24(13):3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 9.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell. 2018;173(3) doi: 10.1016/j.cell.2018.03.057. 581-94 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein MK, Pandey M, Xiu J, Tae H, Swensen J, Mittal S. Tumor Mutational Burden Is Site Specific in Non–Small-Cell Lung Cancer and Is Highest in Lung Adenocarcinoma Brain Metastases. JCO Precis Oncol. 2019;(3):1–13. doi: 10.1200/PO.18.00376. [DOI] [PubMed] [Google Scholar]
- 14.Gow CH, Chang YL, Hsu YC, Tsai MF, Wu CT, Yu CJ. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20(4):696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Holmes-Tisch AJ, Cho EY, Shim YM, Kim J, Kim HS. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4(7):809–815. doi: 10.1097/JTO.0b013e3181a94af4. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZY, Zhong WZ, Zhang XC, Su J, Yang XN, Chen ZH. EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist. 2012;17(7):978–985. doi: 10.1634/theoncologist.2011-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherwood J, Dearden S, Ratcliffe M, Walker J. Mutation status concordance between primary lesions and metastatic sites of advanced non-small-cell lung cancer and the impact of mutation testing methodologies: a literature review. J Exp Clin Cancer Res. 2015;34:92. doi: 10.1186/s13046-015-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.