Abstract
Climate change presents distinct ecological and physiological challenges to plants as extreme climate events become more common. Understanding how species have adapted to drought, especially ecologically important nonmodel organisms, will be crucial to elucidate potential biological pathways for drought adaptation and inform conservation strategies. To aid in genome‐to‐phenome research, a draft genome was assembled for a diploid individual of Artemisia tridentata subsp. tridentata, a threatened keystone shrub in western North America. While this taxon has few genetic resources available and genetic/genomics work has proven difficult due to genetic heterozygosity in the past, a draft genome was successfully assembled. Aquaporin (AQP) genes and their promoter sequences were mined from the draft genome to predict mechanisms regulating gene expression and generate hypotheses on key genes underpinning drought response. Fifty‐one AQP genes were fully assembled within the draft genome. Promoter and phylogenetic analyses revealed putative duplicates of A. tridentata subsp. tridentata AQPs which have experienced differentiation in promoter elements, potentially supporting novel biological pathways. Comparison with nondrought‐tolerant congener supports enrichments of AQP genes in this taxon during adaptation to drought stress. Differentiation of promoter elements revealed that paralogues of some genes have evolved to function in different pathways, highlighting these genes as potential candidates for future research and providing critical hypotheses for future genome‐to‐phenome work.
Keywords: adaptation, aquaporins, drought stress, genome mining, genome‐to‐phenome, sagebrush
A draft genome and novel mining/analysis pipeline was sued to assess Aquaporin genes in a nonmodel, ecologically significant plant species. We found that the genome of this species is enriched in Aquaporin genes and that homologs have likely diverged experienced promoter element divergence, potentially contributing to drought adaptation.

1. INTRODUCTION
Drought is a major factor determining plant survival, growth, and reproduction, as well as species distributions (Brenes‐Arguedas et al., 2009; Samarah, 2005). Drought stress has numerous deleterious effects on plants, including reduced growth (Samarah, 2005), increased production of reactive oxygen species (ROS; Cruz De Carvalho, 2008), and reduction of photosynthetic efficiency (Galmés et al., 2007). Water deficits generate hydraulic and chemical signals, particularly abscisic acid (ABA), that trigger signaling cascades and ABA‐dependent and ABA‐independent transcriptome changes (Chaves et al., 2003; Christmann et al., 2013; Lata et al., 2015; Takahashi et al., 2018). Plants respond to water deficit through mechanisms that reduce water loss, increase water uptake, and alter hydraulic conductivity (Sharp et al., 2004; Kaldenhoff et al., 2008; Hsu et al., 2021). In the long term, adaptations to dry environments come through the evolution of structural, biochemical, and physiological traits that maximize the ability to acquire and retain water (reviewed in Shinozaki & Yamaguchi‐Shinozaki, 2007; Shinozaki et al., 2003). These traits may include deep roots, thick cuticles, sunken stomata, CAM metabolism, and characteristics that reduce or enable repair of xylem embolism (Fahn, 1964; Lüttge, 2004; McElrone et al., 2007; Secchi et al., 2017).
Aquaporins (AQP) are a large family of proteins known to function in the transport of water and other molecules across cell membranes (Yakata et al., 2007; Azad et al., 2012; Uehlein et al., 2012; Reviewed in Li et al., 2014; Afzal et al., 2016). The defining characteristics of AQP proteins include having six alpha‐helices, including two conserved asparagine–proline–alanine (NPA) motifs (Mitsuoka et al., 1999; Murata et al., 2000). While the NPA motifs are generally highly conserved, there are some AQP genes that have undergone mutations of the alanine residue in the NPA motif (Ishibashi, 2006). AQPs in flowering plants comprise five subfamilies: (1) NOD26‐like intrinsic proteins (NIPs), (2) plasma membrane intrinsic proteins (PIPs), (3) small basic intrinsic proteins (SIPs), (4) tonoplast intrinsic proteins (TIPs), and (5) X intrinsic proteins (XIPs) (Danielson & Johanson, 2008). Genes from each subfamily tend to move water or other substrate depending on their NPA motifs. Some AQPs, such as NIPs, have acquired a mutation in their NPA motif, such as alanine to leucine, which confer the ability to move substrates such as urea or ammonium (reviewed in Chaumont et al., 2005; Kaldenhoff & Fischer, 2006; Maurel, 2007).
Most of the water plants take from the soil is lost to the atmosphere via transpiration (Steudle, 2001). In this water flow through the soil–plant–atmosphere continuum, AQPs play an important role in controlling the radial movement of water from the soil to the root xylem as well as water movement from the leaf xylem to evaporation sites in the mesophyll (Sade & Moshelion, 2017). Various environmental and internal changes such as in soil moisture, evaporative demand, salinity, and ABA can alter the expression and activity of AQPs and plant hydraulic conductivity (Afzal et al., 2016; Ding et al., 2015; Fang et al., 2019; Maurel et al., 2016). Through these changes in expression and activity, AQPs contribute to regulate plant water balance and maintain cellular water homeostasis, which ultimately affects plant growth, photosynthesis, and water use efficiency (Chaumont & Tyerman, 2014; Moshelion et al., 2015). As important regulators of the plant water balance, AQPs are excellent targets to increase our understanding of how plants can deal with drought stress and survive in arid environments (Shekoofa & Sinclair, 2018; Zargar et al., 2017; Zhang et al., 2019). Understanding the mechanisms that promote expression of AQPs would allow rapid identification of key AQP genes underpinning drought adaptation and provide a tool to screen natural populations to predict their abilities to cope with climate change. Such endeavors are especially important for natural habitats that are dominated by few foundational species, including the threatened sagebrush steppe ecosystem in western North America (Davies et al., 2011). Promoter sequence analyses, such as those in Lopez et al. (2013), may provide valuable information about what drives the expression of AQP genes for plant species in these habitats.
Researchers have primarily focused on studying the roles and importance of AQPs using crop and model plants; very little effort has been devoted to species occurring in natural environments. Artemisia tridentata Nutt. (sagebrush; Asteraceae) exists as a polyploid species complex with a history of hybridization (Freeman et al., 1991; McArthur et al., 1988; Taylor et al., 1964) that occupies environments with contrasting precipitation regimens and drought occurrences (Kolb & Sperry, 1999). This complex evolutionary history, in conjunction with limited resources available for research, has complicated genetic studies of this taxon. Natural variation in drought tolerance within this species may provide an excellent system to study the role of AQPs in plants’ responses to water stress and survival in arid environments (Maurel et al., 2010). This long‐lived shrub is the most ecologically important and dominant species of the steppes of northwestern North America (Karban, 2007; Leonard et al., 2000; Prevéy et al., 2010). Sagebrush steppe habitat once covered much of western North America (McArthur & Plummer, 1978; Mueggler & Stewart, 1980; Requena‐Mullor et al., 2019), though it is now threatened by various habitat disturbances (Barnard et al., 2019; Prevéy et al., 2010) and anthropogenic climate change (Richardson et al., 2017; Still & Richardson, 2015).
Anthropogenic climate change poses a great threat to many organisms across the globe. Average temperatures experienced an increase of 0.6°C within the 20th century (reviewed in Jones et al., 2001), with a potential increase in future temperatures of 0.4°C per decade (IPCC, 2001). While shifts in species distributions have been identified across latitudinal and elevational gradients (Grabherr et al., 1994; Kullman, 2002; Lloyd & Fastie, 2003; Parmesan & Yohe, 2003; Penuelas & Boada, 2003; Sanz‐Elorza et al., 2003; Sturm et al., 2001; Walther, 2003; Walther et al., 2002), with ranges of many more species expected to shift (Bakkenes et al., 2002), the rate of climate change will likely outpace the ability of many species to acclimate, adapt, or disperse (Cang et al., 2016; Jezkova & Wiens, 2016; Quintero & Wiens, 2013; Wiens, 2016). Along with rising temperatures, many areas will see changes in precipitation and aridity, leading to increased water‐related stress in sessile plants (Gao & Giorgi, 2008; Zarch et al., 2017; reviewed in Huang et al., 2017).
Given the rapid improvements in genomic tools, use of draft genomes in the study of nonmodel organisms can greatly decrease cost and help generate focused genome‐to‐phenome (G2P) hypotheses for long‐term experiments (reviewed in Wojahn et al., 2021). A draft genome can be assembled and then mined for relevant genetic information, as opposed to a more classical G2P experiment, in which genetic/transcriptomic data would be linked to an experimentally induced phenome. In this study, we explored the evolution of AQP genes that may promote drought adaptation in plants using diploid Artemisia tridentata Nutt. subsp. tridentata (2n = 2x = 18; McArthur & Sanderson, 1999). Artemisia tridentata subsp. tridentata is a nonmodel plant that can be difficult to use in genetic research, with few resources currently available, though propagated lines are in development (Barron et al., 2020). Here, we test the hypothesis that the genome of the drought‐tolerant taxon A. tridentata subsp. tridentata will be enriched with AQP genes, particularly those of the PIP and TIP subfamilies, which produce proteins key in channeling water between cells, by being located in the plasma membrane, and the tonoplast, respectively, and that differentiation of promoter sequences have driven the evolution of novel functional groups of these AQP genes underpinning biochemical pathways adapted to drought tolerance.
These hypotheses were tested by assembling a draft genome of a diploid A. tridentata subsp. tridentata (2n = 2x = 18) and mining for AQP genes. Candidate AQP genes were characterized with regard to their amino acid sequences, predicted three‐dimensional structures, promoter elements, and phylogenetic inference. AQPs mined from the draft genome were compared to those of an annual, nondrought‐tolerant congener, Artemisia annua L. (Shen et al., 2018), to determine whether the genome of A. tridentata subsp. tridentata is enriched for AQP genes, which may confer increased tolerance to drought stress.
2. MATERIAL AND METHODS
2.1. Sampling, genome sequencing, and assembly
A diploid (2n = 2x = 18) individual of Artemisia tridentata Nutt. subsp. tridentata from a common garden in Orchard, Idaho (USA), grown from seed collected near Mountain Home, Idaho, USA (43.3371, −116.0081), was sampled for DNA extraction and genome sequencing (1C = 2.98 Gbp; Richardson et al., 2012; other individuals of this species have been found to have much larger genome sizes: 1C = 4.12–4.21 Gbp; Garcia et al., 2008). This individual, known as IDT2‐2, was grown as part of a long‐term experiment conducted by the USDA Forest Service in the Orchard common garden (Richardson & Chaney, 2018). DNA extraction was performed at Boise State University using a Qiagen Plant Mini kit per manufacturer protocol and quantified using Qubit (Thermo Fisher Scientific, Waltham, MA USA). A sample of 30 ng/μl was sent to the HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA) for sequencing. A PCR‐free 2 ×150 bp paired‐end library of 350 bp standard was constructed using the Illumina TruSeq DNA PCR‐Free LT Library Preparation Kit (cat #20015962). After construction, the library was assessed for concentration by a Qubit™ fluorometer, fragment size with an Agilent Bioanalyzer, and optimal loading concentration by qPCR.
2.2. De novo genome assembly
A whole‐genome shotgun sequencing and standard sequencing protocols were utilized to sequence the Artemisia tridentata subsp. tridentata genome. Reads were generated using the Illumina NovaSeq platform at the HudsonAlpha Institute for Biotechnology in Huntsville, Alabama (USA). Two TruSeq PCR free 400 bp insert 2 × 150 Illumina fragment libraries (176.3 × raw coverage) were generated. Prior to assembly, Illumina fragment reads were screened for PhiX contamination. Reads composed of >95% simple sequence were removed. Illumina reads <50 bp after trimming for adapter and quality <20 were removed. The final read set consists of 5,388,578,188 reads for a total of 169.9× of high‐quality Illumina bases. The genome assembly was generated by assembling the 5,388,578,188 Illumina reads (169.9× sequence coverage) using the HipMCL assembler (version 1.1‐27‐g69eb6141; Azad et al., 2018). To validate our draft genome assembly for gene mining purposes, BUSCO V.5.1.2 (Seppey et al., 2019) was used to estimate the percent of orthologous genes from the eukaryote (n = 255) and eudicots (n = 2326) db10 databases that were completely and partially assembled or missing within the draft genome. BUSCO analysis was performed using default parameters and the Augustus gene prediction algorithm (Stanke & Waack, 2003).
2.3. Identification and validation of Aquaporin genes
Scaffolds containing candidate AQP genes were identified via BLASTN search using a custom reference BLAST database containing all DNA sequences available from GenBank encoding for AQP genes (for data regarding Artemisia tridentata subsp. tridentata AQP genes used in downstream analyses, refer to Table S1). Accession numbers of top BLAST hits were recorded, as well as positions of hits along scaffolds allowing the identification of genes that were fully contained within scaffold sequences. Open reading frames (ORFs) were predicted on scaffolds identified by the BLAST analysis using the findORFs function from the “ORFik” R package (Tieldnes & Labun, 2020). The number of predicted ORFs, strands, and positions along each scaffold were recorded. To identify ORFs coding for AQP codons, ORFs were converted into amino acid sequences and BLASTP analyses were run on the online BLAST portal. Top BLAST hits for each ORF coding for AQP exons were recorded to further refine gene hypotheses (Table S1).
2.4. Predicting secondary and tertiary protein structures
AQPs are characterized by having six helices, two loops, and the presence of NPA motifs in loops (Park et al., 2010). The online TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the number of transmembrane helices (Krogh et al., 2001), and a custom R script using base R functions was used to predict number, positions and amino acid composition of NPA motifs, and number of amino acid residues (R Core Team, 2020; https://github.com/aemelton/DraftGenomeMineR). We confirmed locations of NPA motifs in loops by using output of the TMHMM analyses. These analyses were based on concatenated amino acid sequences resulting from the ORFs and BLAST analyses.
The Phyre2 online platform with default parameters (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) was used to predict and analyze AQP protein tertiary structures, functions, and locate amino acid regions where mutations would change functions (Kelley et al., 2016). This analysis was performed on AQP amino acid sequences with at least five predicted helices and two NPA motifs. Tertiary protein models were saved in pdb formats, and data on model accuracy and coverage, closest protein(s) in Phyre2 database, and locations of mutations along proteins were recorded to further validate and infer functions of AQP s in Artemisia tridentata subsp. tridentata (Table S1).
2.5. Phylogenetic analysis
The phylogenetic reconstruction based on amino acid sequences was used to (i) confirm the identities of putative AQP genes in Artemisia tridentata subsp. tridentata genome and (ii) compare them to those of Artemisia annua L. identified via the NCBI BLAST portal to identify potential candidates underpinning climate‐induced adaptations in A. tridentata subsp. tridentata, that may have evolved to function in potential new pathways relating to drought stress.
Extracted AQP amino acid sequences were aligned with AQP sequences from Artemisia annua and Arabidopsis thaliana (L.) Heynh. mined from GenBank. Arabidopsis thaliana AQPs were used as the initial queries for Artemisia annua mining from GenBank and for BLASTP identification hypotheses for A. tridentata subsp. tridentata due to high level of validation of these proteins (Quigley et al., 2001). The alignment was performed via the online mafft V.7 portal (https://mafft.cbrc.jp/alignment/server/; Katoh et al., 2019) using the E‐INS‐i algorithm. Phylogenetic reconstructions were performed via raxmlGUI 2.0.0‐beta.14 (Edler et al., 2020) using the protgamma model and 1000 rapid bootstrap replicates.
2.6. Promoter analysis
Promoter analyses were performed to investigate mechanisms triggering AQP gene expression using the approach described in Lopez et al. (2013). One and half kilobases of upstream sequence data were extracted from scaffolds containing at least 1500 bp upstream from the AQP start codon. Upstream sequences were analyzed for putative cis‐acting sequences (regulatory distal element; RDE) using the Plant Cis‐acting Regulatory DNA Elements (New PLACE) signal scan software package (https://www.dna.affrc.go.jp/PLACE/?action=newplace, Higo et al., 1999). These RDEs comprise short nucleotide motifs that occur upstream of the start codon of a gene and influence the expression of genes. Detected RDEs were sorted into biological categories which included light, abscisic acid (ABA), water stress, temperature stress, stress hormone, growth, and other stress, and targeted area of expression (e.g., leaf, aerial tissue, and roots) based on keywords for each RDE listed within the New Place database file using a custom R script (https://github.com/aemelton/DraftGenomeMineR). Due to some RDE’s having functions in multiple pathways, some RDEs could be categorized into multiple categories to account for their various functions (e.g., many drought‐responsive elements, such as DREDR1ATRD29AB, respond to both temperature and water stresses; Yamaguchi‐Shinozaki & Shinozaki, 1994). A Kruskal–Wallis test via the function kruskal.test in base R (R Core Team, 2020) followed by Dunn tests via the function dunn.test in the R package “dunn.test” V.1.3.5 (Dinno, 2017) were performed on category count data per gene to determine if there are differences in the occurrences of RDEs for any given category, indicating enrichment of RDEs in certain categories and indicating important triggers for the expression of these AQP genes.
A singular value decomposition (SVD) on the occurrence counts of RDEs for each category within upstream sequence for each gene was used to determine how AQPs may form functional groups, as proxy for biochemical pathways, with transcription potentially being promoted by similar processes. These analyses were performed using the “LinearAlgebra” and “Clustering” v.0.13.5 packages in Julia v.1.5.3 (Bezanson et al., 2017). Additionally, we performed a k‐means clustering analysis on the raw RDE category data to identify clusters of genes that have similar drivers of expression and presumably function in similar response pathways. To determine the most appropriate k for clustering, k‐means analyses were performed with k values ranging from three to 20 assessed by silhouette analysis using the “Clustering” package in Julia. The silhouette analysis aims to assess the cohesion of points within clusters and the separation of points from different clusters. This allows for the identification of the ideal number of clusters, k, by determining the k value that produces clusters with high cohesion and separation. Given that these RDEs play important roles in controlling expression, identifying clusters of AQPs based on the RDEs in principal component space (PC‐space) will allow us to potentially identify important drivers of the expression of these genes.
3. RESULTS
3.1. Draft genome assembly
In total, 3.33 million scaffolds containing3.82 million contigs were assembled. Contigs covered 97.87% of the total length of the scaffolds. These comprise 4.50 Gbp and 4.41 Gbp, for scaffolds and contigs, respectively, covering the entirety of the haploid genome (1C = 2.98 Gbp; Richardson et al., 2012). The N50 for all scaffolds and contigs were 324,094 and 409,868, respectively. The L50 for scaffolds and contigs were 3.5 and 2.6 kbp, respectively. Fifty‐nine scaffolds and 191 contigs greater than 50 kbp were assembled. BUSCO predicted 100 (39.2% of orthologues in database) and 1035 (44.5% of orthologues in database) complete orthologous genes from the eukaryote and eudicot databases, respectively. Eighty‐five (33.3% of orthologues in database) and 844 (36.3% of orthologues in database) orthologues were missing from the draft genome for the eukaryote and eudicot databases, respectively.
3.2. Identification and validation of Aquaporin genes
Eighty‐three scaffolds were identified as containing a total of 84 putative AQP genes. Of these, 50 scaffolds were predicted to contain fully assembled AQP sequences. Scaffold number 128070 contained two tandem, fully assembled putative AQP genes. Thus, a total of 51 AQPs were considered in downstream analyses (Table S1; File S1 contains scaffolds from which the AQP genes were extracted). Of these AQP genes, 11 were identified as NIPs, 21 as PIPs, three as SIPs, and 16 as TIPs based on BLASTP searches. Gene length ranged from 888 to 1658 nucleotides for the 51 AQP genes and included two to five predicted exons. Six variants of the NPA motif were found: NPA, NPS, NPV, NPT, PPA, and FPA. Relevant gene data (i.e., gene length, predicted exon number, and NPA motifs) are listed in Table S1.
3.3. Predicting secondary and tertiary protein structures
A total of 19 putative AQP genes met criteria for Phyre2 analyses. All genes were validated as members of the AQP gene family. Identification of TIP proteins using the Phyre2 database suggested these unidentified TIPs belong to the TIP2‐1 group. Phyre2 predicted that the greatest effects of mutations would occur at the NPA motifs. Mutations expected to affect function were identified in two NIP5‐1 proteins, which have NPS and NPV motifs (Table S1).
3.4. Phylogenetic reconstruction
Phylogenetic reconstructions of AQP amino acid sequences from Artemisia tridentata subsp. tridentata (51 protein sequences), A. annua (19 protein sequences), and Arabidopsis thaliana (35 proteins sequences) recovered four clades, each comprising a given subfamily (Figure 1; File S2 contains aligned amino acid sequences used in this analysis). One exception was the Artemisia annua gene PWA34518_1. While originally identified as NIP5‐1 by BLASTP analysis, PWA34518_1 formed a clade with members of the SIP subfamily. Several other A. annua AQPs that were not identified to the subfamily level were found to be members of the TIP subfamily within the phylogeny. Phylogenetic reconstructions for AQP genes revealed several potential duplication events for AQPs within Artemisia tridentata subsp. tridentata. These putative duplications were particularly common in the PIP and TIP subfamilies, with PIP1‐3, PIP1‐4, PIP2‐2, PIP2‐4, TIP, and TIP2‐1 comprising multiple copies (Figure 1). Genes identified as TIP genes for Artemisia tridentata subsp. tridentata formed a clade with the TIP2‐2 and TIP2‐3 genes of Arabidopsis thaliana, suggesting these unidentified TIPs are TIP2 genes. TIP2‐1 genes formed a distinct clade sister to the clade containing TIP2‐2 and TIP2‐3 genes. Therefore, the TIP genes are most likely TIP2‐2 or TIP2‐3 homologs (Figure 1). Phylogenetic analysis revealed that a PIP gene identified via BLAST as a PIP2‐1 gene is actually a PIP2‐4, as it forms a clade with the PIP2‐4 and not PIP2‐1 genes. The Artemisia tridentata subsp. tridentata NIP1‐1 gene has also experienced numerous putative duplication events, comprising five AQP genes. (Figure 1; Table S1).
FIGURE 1.
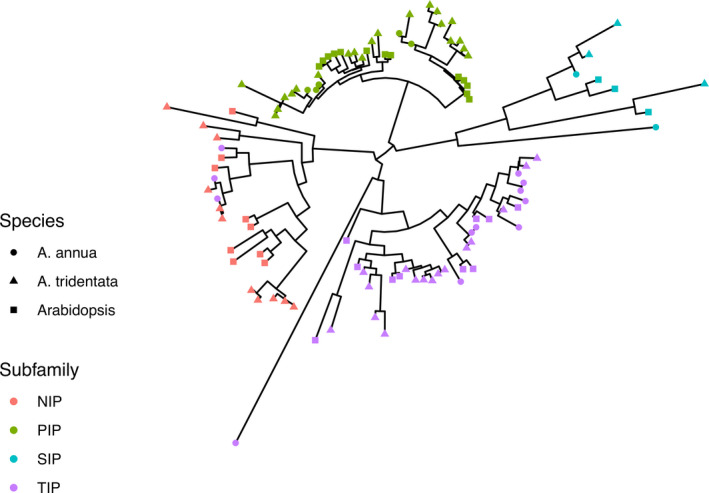
Midpoint rooted phylogeny of AQP genes from Arabidopsis, Artemisia annua, and Artemisia tridentata subsp. tridentata. Several genes previously identified as TIP genes in Artemisia annua via other methods formed a clade with the NIP genes, indicating these were previously misidentified and belong to the NIP subfamily
3.5. Promoter anal–ysis
Of the 51 AQP genes retained for further analysis, 29 were on scaffolds with at least 1.5 kbp upstream sequence to be included in promoter analyses (File S3 contains sequences extracted for this analysis). In total, 224 RDEs were identified across all upstream sequences. Light, ABA, and stress hormones, including cytokinin and auxin, were the most represented categories, with 420, 189, and 157 occurrences of classified RDEs in total across all scaffolds, respectively. The RDE “CACTFTPPCA1” was the most prominent RDE among these genes, with a total of 636 occurrences. Other highly prominent RDEs include the following: DOFCOREZM (593), CAATBOX1 (515), ARR1AT (423), and GATABOX (373).
Most RDEs play roles in tissue‐specific expression, particularly in the aerial tissue. The most prominent RDE, CACTFTPPCA1, promotes expression in leaf mesophyll. Some RDEs identified target expression in roots: NTBBF1ARROLB (56), OSE1ROOTNODULE (43), OSE2ROOTNODULE (105), and ROOTMOTIFTAPOX1 (263). These root‐specifying promoter elements were most prominent in the upstream regions of several NIP and PIP genes, particularly NIP2‐1, PIP1‐3, PIP1‐4, and PIP2‐4. One TIP gene was also found to be enriched for ROOTMOTIFTAPOX1.
Results of the Kruskal–Wallis and Dunn tests show that light category RDEs are statistically significantly enriched relative to all other categories (p‐values = .0002 and 0 for ABA and all other categories, respectively). ABA category RDEs were statistically significantly greater than all categories except light (p‐value = .0002) and stress hormone (nonstatistically significant difference), with p‐values of 0.0006 for growth, 0.0010 for other stress, and 0 for temperature and water stress. Growth category RDEs occurred statistically significantly less than light (p‐value = 0), ABA (p‐value = .0006), stress hormone (p‐value = .0114), temperature (p‐value = .0097), and water stress (p‐value = .0040) category RDEs. Other stress category RDEs occurred statistically significantly more than temperature (p‐value = .0061) and water stress (p‐value = .0024) category RDEs, and statistically significantly less than those of light, ABA, and stress hormone (p‐value = .0174) category RDEs. Stress hormone category RDEs occurred statically significantly more than temperature stress (p‐value = 0) and water stress (p‐value = 0) category RDEs. Temperature stress category RDEs did not occur statistically significantly more than RDEs of any other category.
Silhouette analysis results suggested a k of three was most appropriate to describe the clustering of gene promoter sequences in PC‐space. Clustering in PC‐space was not related to the AQP phylogeny; at least one member of each subfamily was present in each of the three clusters, with members of each gene group also being spread out in PC‐space and clusters.
SVD of RDE categories (Figure 2) showed that light has the greatest influence on AQP expression (PC1 = 72% variation explained), while a combination of ABA and water stress were the secondary drivers of AQP expression (PC2 = 7.5% of variation explained). PIP genes were most greatly influenced by PC1, primarily light category RDEs, while TIPs were primarily driven by PC2, which included ABA and water stress category RDEs. NIP gene RDEs occupied an area of PC‐space, which was influenced by both PC1 and PC2. The most highly represented genes, PIP1‐3 and TIP2‐1, each show at least one copy greatly diverging in PC‐space (Figure 2). Several copies of each of these two genes also occupy an area of PC‐space that indicates their expression is largely driven by ABA, water, and temperature stresses.
FIGURE 2.
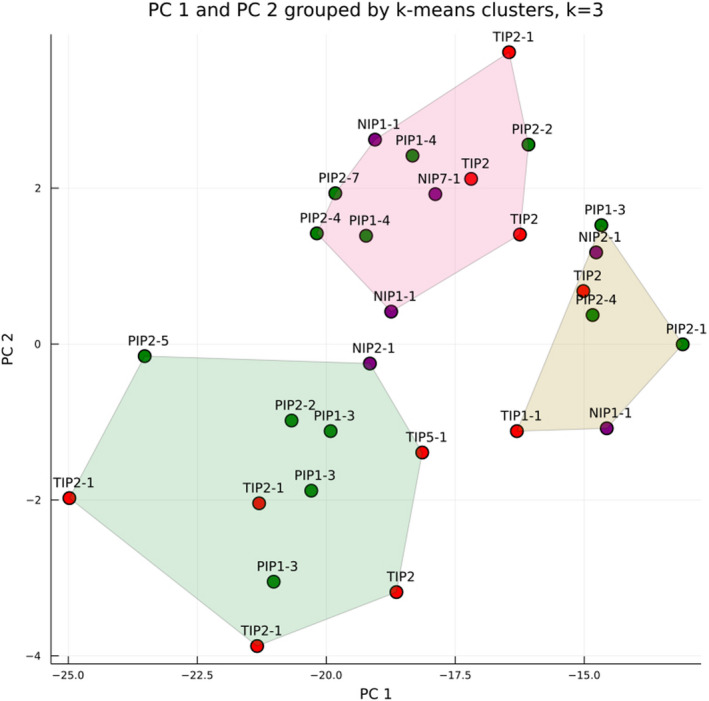
Result of SVD for RDE categories across AQP genes. Three distinct clusters were identified in the silhouette analysis, each enclosed in a convex hull. Each cluster consists of genes from multiple AQP subfamilies (NIP, purple; PIP, green; and TIP, red). PC1 explained 75% of variance, while PC2 explained 7.5% of variance
Assessing RDE enrichment per AQP gene in a phylogenetic context revealed that putative AQP paralogues have experienced some degree of differentiation (Figure 3). For example, the two PIP1‐4 genes included in promoter analyses differ in their enrichment for ABA, temperature stress, and growth categories. The PIP2‐4 clade exhibits clear differentiation in their enrichment of RDE in all categories except water stress and stress hormones. PIP1‐3 RDEs have primarily differentiated in the light and ABA categories. TIP2‐1 genes have experience clear differentiation of RDEs in all categories except the stress hormone category.
FIGURE 3.
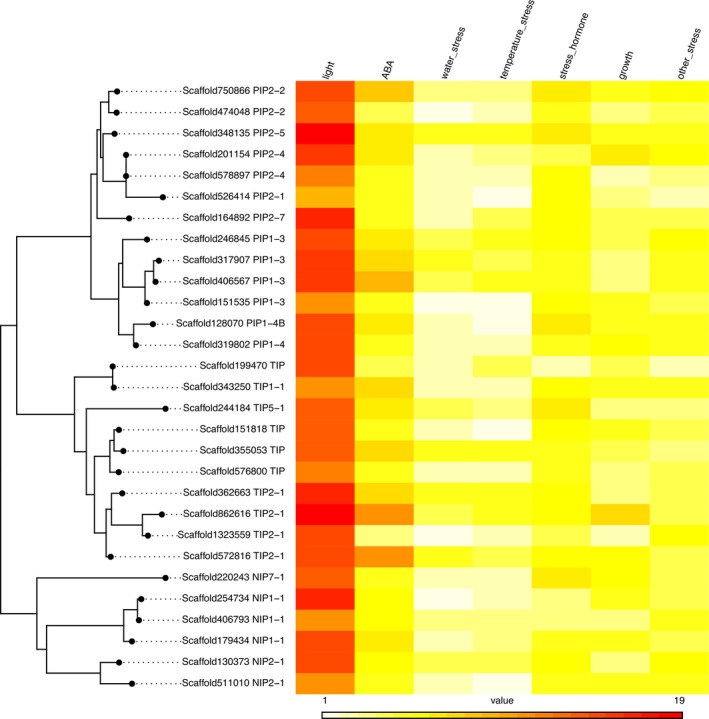
Heatmap of RDE categories per scaffold along with phylogenetic context for each gene. The value within each cell is equal to the number of occurrences of RDEs for a given category within the promoter sequences of each gene. Per Kruskal–Wallis and Dunn test results, the light category of RDEs was significantly enriched relative to all other categories. ABA category RDEs were significantly enriched relative to all other categories except light. Temperature category RDEs were the least prevalent and did not occur significantly more than those of other categories
4. DISCUSSION
4.1. Providing a draft genome to investigate drought tolerance
A first draft genome for Artemisia tridentata, a keystone species of western North America, was successfully assembled for use in genome mining and drought adaptation research. The individual used for this study was previously estimated to have a diploid genome size of 5.95 Gbp (6.09 pg; Richardson et al., 2012). Our draft genome assembly is at least four times that of Artemisia annua (1.38–1.78 Gb; Liu et al., 2018; Shen et al., 2018), while maintaining the same number of chromosomes (2n = 2x = 18). Draft genome assemblies for both species represent essentially complete haploid genomes (Shen et al., 2018). The draft genome assembled for A. tridentata subsp. tridentata was assembled without filtering of homologous scaffolds or assembling haploid genome sets. This does not affect the results of the analyses described here, as no homologous scaffolds containing AQPs were found. The BUSCO analysis demonstrated that this genome assembly was of sufficient quality to be mined for genes, as nearly two‐thirds of eudicot orthologues (63.7%) were partially or fully recovered. While sufficient for gene mining, genes were undoubtedly missed in our analyses due to the incontiguous nature of the draft assembly.
The methods described in this study provide a clear methodological pathway to validate genes within draft genome assemblies. While not all the 84 putative AQP genes were fully assembled within scaffolds and were thus excluded from downstream analyses, we were able to validate to a high degree the identity of 51 of the fully assembled putative AQPs. Given the results of the BUSCO analysis, it is likely that around 30% of AQP genes in the genome are left to be discovered. Our combination of BLAST, phylogenetic reconstruction, and prediction of tertiary structure allowed for high confidence validation of many of the putative Artemisia tridentata subsp. tridentata AQP genes (Figure 1; Table S1). While not all genes were validated using all methods, we can still be confident that they fall within the AQP gene family. Many of these genes were validated to at least subfamily and clade level (e.g., PIP1‐1; Table S1).
4.2. Enrichment of AQPs in a drought‐tolerant species
Our results indicate that the genome of drought‐tolerant Artemisia tridentata subsp. tridentata has at least 51 AQPs (Table S1). This number of AQPs is relatively high when compared to other species. Based on genome‐wide identification, Deshmukh et al. (2016) reported the number of AQPs in 31 species, ranging from 23 in the moss Physcomitrella patens to 72 in Glycine max (soybeans). Within the list presented in this review (Deshmukh et al., 2016), only four species have more AQPs than A. tridentata subsp. tridentata. Notably, this species has a much higher number of AQPs than its congener A. annua, which has 19 AQPs. This difference in gene enrichment could be due to whole‐genome duplication followed by diploidization and adaptation to more arid climates of western North America. The average genome size of Artemisia species in the North American group of the Tridentatae subgenus is approximately twice that of other Artemisia species (Garcia et al., 2008; Pellicer et al., 2010). The North American group of Artemisia subgenus Tridendatae likely diverged from the Asian clade approximately 10.8 mya (± 1.5 my; Sanz et al., 2011) after a vicariance event from Asia to North America via the Bering Land bridge. Given the divergence times, difference in genome sizes, and different climatic regimes occupied by the two species (Figure S1 highlights the difference in occupied climatic niches of these two species), it is likely that genomic processes, such as polyploidization followed by diploidization, and adaptation to a drought‐prone and arid environment have driven the evolution of the AQP gene family in the genome of A. tridentata subsp. tridentata.
The PIP and TIP subfamilies were the most enriched in the Artemisia tridentata subsp. tridentata genome, relative to the A. annua genome. Putative duplications of AQPs within the Artemisia tridentata subsp. tridentata genome seem to largely fall within the PIP and TIP subfamilies (Figure 1). These two subfamilies are the primary AQPs that function in water transport across the plasmalemma and tonoplast (Bienert et al., 2018; Siefritz et al., 2002; Song et al., 2016). The PIPs and TIPs identified in the Artemisia tridentata subsp. tridentata genome almost exclusively have NPA motifs, though one FPA and five PPA motifs were identified in proteins of the PIP subfamily (PIP1‐3, PIP2‐1, PIP2‐2; Table S1). The NPA to PPA mutation has been reported in Linum usitatissimum, though otherwise appears to be a rare change (Shivaraj et al., 2017). The conservation of these motifs is essential for the movement of water across membranes, as modifications of the NPA motifs lead to changes in the specificity, including nitrogenous compounds transported by NIPs. The asparagine residue in the NPA is highly conserved, as it provides helix cap stability and functions in cation exclusion (Wree et al., 2011). Therefore, it is likely that the AQPs with FPA and PPA motifs have experienced some degree of functional differentiation, though these have not been assessed. NIP5‐1 contains NPS and NPV motifs that would likely alter the function of the gene, per Phyre2 analysis. While these are deviations from the typical NPA motif, AQP genes with variations of the NPA motif and confirmed function in water transport have been identified in other organisms (Ishibashi, 2006).
4.3. Light drives AQP expression
The most enriched RDE category presented here was “light,” occurring statistically significantly more than RDEs of any other category, with the GATABOX element being the most common. This RDE functions in chlorophyll a/b binding in aerial tissue. The observation of light‐responsive elements in Artemisia tridentata subsp. tridentata AQPs is consistent with several studies that have reported upregulation of AQPs in response to light (Baaziz et al., 2012; Cochard et al., 2007; Lopez et al., 2013). Moreover, these increases in AQPs expression were correlated to increases in leaf hydraulic conductivity, presumably adjusting water supply to transpirational demand (Baaziz et al., 2012; Lopez et al., 2013). However, light effects on aquaporin expression and leaf hydraulic conductivity vary between species. For example, Juglans regia showed a fourfold increase in hydraulic conductivity with light, while the increase was minimal upon illumination of Salix alba and Quercus rubra (Baaziz et al., 2012; Rockwell et al., 2011). Given the large complement of AQPs with light‐responsive elements in Artemisia tridentata subsp. tridentata (Figure 3), it would be interesting to determine how its hydraulic conductivity response to light compares to that of other species. ABA‐responsive elements and the MYCCONSENSUSAT RDE were also highly enriched. The latter has functions in both cold responses associated with CBF/DREB1 genes and drought responses associated with dehydration‐responsive gene, rd22, genes (Liu et al., 2015). This is likely an important promoter element for Artemisia tridentata subsp. tridentata, which experiences long periods of drought during the summer, and cold to freezing temperatures during the winter (Kolb & Sperry, 1999; Lambrecht et al., 2007).
The presence of many tissue‐specific RDEs indicates that many of the identified AQPs are expressed primarily in aerial tissues. Since many RDEs in these AQPs have roles in light response, their expression in these tissues would be expected. Also, the RDE analysis suggests special expression in the leaf mesophyll, a tissue enriched in chloroplasts where photosynthesis occurs. Artemisia tridentata has a compact mesophyll, with air spaces mainly limited to the substomatal cavity (Downs & Black, 1999). Tight packing of cells may increase the proportion of transcellular water transport over the apoplastic pathway (Maurel & Prado, 2017). Under this scenario, high AQP expression in the mesophyll and other leaf parenchyma cells may be particularly important for regulating leaf water balance. Apart from water, AQPs in the mesophyll may facilitate the movement of other molecules across membranes, including CO2, which could contribute to increase mesophyll conductance to this gas during photosynthesis (Carriquí et al., 2019; Flexas, Bota, et al., 2006; Flexas, Ribas‐Carbó, et al., 2006; Singh et al., 2020). Overall, the presence of leaf‐specific RDE in several of the identified PIPs and TIPs are congruent with observations made in other species showing high expression of these subfamilies in leaves (Heinen et al., 2009). We also found some PIPs and NIPs with promoter elements indicative of root localization. Unfortunately, no SIP genes had sufficient upstream sequence to meet criteria for inclusion in promoter analyses.
4.4. Differentiation of the drivers of AQP expression
Comparative analyses provided evidence for RDE content differentiation among homologs of some AQP genes, particularly PIP1‐3 and TIP2‐1. These genes are most highly represented in the RDE analyses, with four copies each, and each had at least one copy that took on novel functions relative to the other copies. Copies of these genes, and genes with multiple copies, do not occupy the same PC‐space as members of their homologs (Figure 2). In some cases, a copy of a gene may occupy quite distant space from other gene copies. This indicates that promoter sequence differentiation, to varying degrees, has occurred in these genes. Given the results of SVD and phylogenetic comparisons, the PIP1‐3 and TIP2‐1 genes appear to be strong candidates for future research as drought tolerance genes in A. tridentata subsp. tridentata. Both genes occur in multiple copies within the genome, and they have experienced RDE category differentiation (Figures 1, 2, 3). These genes also have copies that have diverged in promoter sequences to likely play a greater role in ABA and drought stress pathways (Figure 2). So, while these genes may share similar amino acid sequences with their respective homologs, they do exhibit differentiation RDE composition, suggesting differences in biological pathways that would drive their expression.
4.5. Perspectives
Overall, this research provides genomic resources and valuable hypotheses for further work on Artemisia tridentata subsp. tridentata. While our methods do not replace larger G2P experiments, they do offer a more rapid and cheaper method to acquire and analyze data that will generate testable G2P hypotheses, which can lead to more focused and efficient experiments. We will be able to test such hypotheses thanks to the development of a novel in vitro method of propagation developed specifically for A‐ tridentata subsp. tridentata (Barron et al., 2020). We are currently in the process of generating several individual lines, which will be used in generating more higher quality draft genomes, assembling a phased diploid genome for this taxon, and to conduct genotype‐by‐environment experiments.
5. CONCLUSIONS
We see clear evidence that the genome of the drought‐tolerant taxon, Artemisia tridentata subsp. tridentata, contains far more AQP genes, particularly PIPs and TIPs, than the genome of a nondrought‐tolerant congeneric species, A. annua. This is likely due to in situ processes within the genomes of Artemisia tridentata and the North American Artemisia clade. This research has also led to important hypotheses generation for future research, particularly that PIP1‐3 and TIP2‐1 genes could potentially confer increased drought adaptation in this foundational, keystone species.
CONFLICT OF INTEREST
The authors have no conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS
Anthony E. Melton: Conceptualization (equal); Data curation (equal); Formal analysis (lead); Investigation (lead); Methodology (equal); Validation (lead); Visualization (lead); Writing‐original draft (lead); Writing‐review & editing (equal). James D. Beck: Formal analysis (supporting); Writing‐review & editing (supporting). Stephanie J. Galla: Visualization (supporting); Writing‐review & editing (supporting). Jerry W. Jenkins: Data curation (supporting). Lori Handley: Data curation (supporting). Min Kim: Data curation (supporting). Jane Grimwood: Data curation (supporting). Jeremy Schmutz: Data curation (supporting). Bryce A. Richardson: Data curation (supporting); Writing‐review & editing (supporting). Marcelo Serpe: Formal analysis (supporting); Methodology (supporting); Writing‐review & editing (supporting). Stephen Novak: Methodology (supporting); Writing‐review & editing (supporting). Sven Buerki: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Funding acquisition (lead); Investigation (equal); Methodology (equal); Project administration (equal); Writing‐review & editing (equal).
Supporting information
Fig S1
File S1
File S2
File S3
Table S1
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to the members of the GEM3 project, especially Jennifer Forbey and Denise Pfeifer at Boise State University, Andy Kliskey and Rick Schumaker at NSF Idaho EPSCoR for their support and comment on earlier versions of the manuscript. We are also grateful to the students from the VIP Genome 2 Phenome course at Boise State University who helped testing our bioinformatic pipeline, especially Carlos D. Dumaguit and Walker Morales. The findings and conclusions in this publication are those of the author(s) and should not be construed to represent any official USDA or U.S. Government determination or policy. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Melton, A. E. , Beck, J. , Galla, S. J. , Jenkins, J. , Handley, L. , Kim, M. , Grimwood, J. , Schmutz, J. , Richardson, B. A. , Serpe, M. , Novak, S. , & Buerki, S. (2021). A draft genome provides hypotheses on drought tolerance in a keystone plant species in Western North America threatened by climate change. Ecology and Evolution, 11, 15417–15429. 10.1002/ece3.8245
Funding information
This research was made possible by the NSF Idaho EPSCoR Program and by the NSF under award number OIA‐1757324.
Contributor Information
Anthony E. Melton, Email: anthonymelton@boisestate.edu.
Sven Buerki, Email: svenbuerki@boisestate.edu.
DATA AVAILABILITY STATEMENT
Scripts used in this research have been deposited at https://github.com/aemelton/DraftGenomeMineR. The raw reads (accessions SRR14309371, SRR14309372, and SRR14309373) and draft genome assembly (accession JAHAUY000000000) have been submitted to NCBI as SAMN18747788 under PRJNA722258. Supplementary documents, including scaffolds and alignment fasta, have been submitted as part of the SI.
REFERENCES
- Afzal, Z. , Howton, T. C. , Sun, Y. , & Mukhtar, M. S. (2016). The roles of aquaporins in plant stress responses. Journal of Developmental Biology, 4(1), 1–22. 10.3390/jdb4010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad, A. , Pavlopoulos, G. A. , Ouzounis, C. A. , Kyrpides, N. C. , & Buluç, A. (2018). HipMCL: A high‐performance parallel implementation of the Markov clustering algorithm for large‐scale networks. Nucleic Acids Research, 46(6), e33. 10.1093/nar/gkx1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad, A. K. , Yoshikawa, N. , Ishikawa, T. , Sawa, Y. , & Shibata, H. (2012). Substitution of a single amino acid residue in the aromatic/arginine selectivity filter alters the transport profiles of tonoplast aquaporin homologs. Biochimica Et Biophysica Acta – Biomembranes, 1818(1), 1–11. 10.1016/j.bbamem.2011.09.014 [DOI] [PubMed] [Google Scholar]
- Bakkenes, M. , Alkemade, J. R. M. , Ihle, F. , Leemans, R. , & Latour, J. B. (2002). Assessing effects of forecasted climate change on the diversity and distribution of European higher plants for 2050. Global Change Biology, 8(4), 390–407. 10.1046/j.1354-1013.2001.00467.x [DOI] [Google Scholar]
- Barnard, D. M. , Germino, M. J. , Arkle, R. S. , Bradford, J. B. , Duniway, M. C. , Pilliod, D. S. , Pyke, D. A. , Shriver, R. K. , & Welty, J. L. (2019). Soil characteristics are associated with gradients of big sagebrush canopy structure after disturbance. Ecosphere, 10(6), e02780. 10.1002/ecs2.2780 [DOI] [Google Scholar]
- Barron, R. , Martinez, P. , Serpe, M. , & Buerki, S. (2020). Development of an in vitro method of propagation for Artemisia tridentata subsp. tridentata to support genome sequencing andgenotype‐by‐environment research. Plants, 66, 1717. 10.3390/plants9121717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Baaziz, K. , Lopez, D. , Rabot, A. , Combes, D. , Gousset, A. , Bouzid, S. , Cochard, H. , Sakr, S. , & Venisse, J.‐S. (2012). Light‐mediated Kleaf induction and contribution of both the PIP1s and PIP2s aquaporins in five tree species: Walnut (Juglans regia) case study. Tree Physiology, 32(4), 423–434. 10.1093/treephys/tps022 [DOI] [PubMed] [Google Scholar]
- Bezanson, J. , Edelman, A. , Karpinski, S. , & Shah, V. B. (2017). Julia: A fresh approach to numerical computing. SIAM Review, 59(1), 65–98. 10.1137/141000671 [DOI] [Google Scholar]
- Bienert, M. D. , Diehn, T. A. , Richet, N. , Chaumont, F. , & Bienert, G. P. (2018). Heterotetramerization of plant PIP1 and PIP2 aquaporins is an evolutionary ancient feature to guide PIP1 plasma membrane localization and function. Frontiers in Plant Science, 9, 1–15. 10.3389/fpls.2018.00382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenes‐Arguedas, T. , Coley, P. D. , & Kursar, T. A. (2009). Pests vs. drought as determinants of plant distribution along a tropical rainfall gradient. Ecology, 90(7), 1751–1761. 10.1890/08-1271.1 [DOI] [PubMed] [Google Scholar]
- Cang, F. A. , Wilson, A. A. , & Wiens, J. J. (2016). Climate change is projected to outpace rates of niche change in grasses. Biology Letters, 12(9), 20160368. 10.1098/rsbl.2016.0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriquí, M. , Douthe, C. , Molins, A. , & Flexas, J. (2019). Leaf anatomy does not explain apparent short‐term responses of mesophyll conductance to light and CO 2 in tobacco. Physiologia Plantarum, 165(3), 604–618. 10.1111/ppl.12755 [DOI] [PubMed] [Google Scholar]
- Chaumont, F. , Moshelion, M. , & Daniels, M. J. (2005). Regulation of plant aquaporin activity. Biology of the Cell, 97(10), 749–764. 10.1042/bc20040133 [DOI] [PubMed] [Google Scholar]
- Chaumont, F. , & Tyerman, S. D. (2014). Aquaporins: Highly regulated channels controlling plant water relations. Plant Physiology, 164(4), 1600–1618. 10.1104/pp.113.233791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves, M. M. , Maroco, J. P. , & Pereira, J. S. (2003). Understanding plant responses to drought ‐ From genes to the whole plant. Functional Plant Biology, 30(3), 239–264. 10.1071/FP02076 [DOI] [PubMed] [Google Scholar]
- Christmann, A. , Grill, E. , & Huang, J. (2013). Hydraulic signals in long‐distance signaling. Current Opinion in Plant Biology, 16(3), 293–300. 10.1016/j.pbi.2013.02.011 [DOI] [PubMed] [Google Scholar]
- Cochard, H. , Venisse, J.‐S. , Barigah, T. S. , Brunel, N. , Herbette, S. , Guilliot, A. , Tyree, M. T. , & Sakr, S. (2007). Putative role of aquaporins in variable hydraulic conductance of leaves in response to light. Plant Physiology, 143(1), 122–133. 10.1104/pp.106.090092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusoe, M. R. , Alameldin, H. F. , Awad, S. , Boucher, E. , Caldwell, A. , Cartwright, R. , Charbonneau, A. , Constantinides, B. , Edvenson, G. , Fay, S. , Fenton, J. , Fenzl, T. , Fish, J. , Garcia‐Gutierrez, L. , Garland, P. , Gluck, J. , González, I. , Guermond, S. , Guo, J. , … Brown, C. T. (2015). The khmer software package: Enabling efficient nucleotide sequence analysis. F1000Research, 4, 1–12. 10.12688/f1000research.6924.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz De Carvalho, M. H. (2008). Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signaling and Behavior, 3(3), 156–165. 10.4161/psb.3.3.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson, J. Å. H. , & Johanson, U. (2008). Unexpected complexity of the Aquaporin gene family in the moss Physcomitrella patens . BMC Plant Biology, 8, 1–16. 10.1186/1471-2229-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, K. W. , Boyd, C. S. , Beck, J. L. , Bates, J. D. , Svejcar, T. J. , & Gregg, M. A. (2011). Saving the sagebrush sea: An ecosystem conservation plan for big sagebrush plant communities. Biological Conservation, 144(11), 2573–2584. 10.1016/j.biocon.2011.07.016 [DOI] [Google Scholar]
- Deshmukh, R. K. , Sonah, H. , & Bélanger, R. R. (2016). Plant aquaporins: Genome‐wide identification, transcriptomics, proteomics, and advanced analytical tools. Frontiers in Plant Science, 7, 1–14. 10.3389/fpls.2016.01896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L. , Gao, C. , Li, Y. , Li, Y. , Zhu, Y. , Xu, G. , Shen, Q. , Kaldenhoff, R. , Kai, L. , & Guo, S. (2015). The enhanced drought tolerance of rice plants under ammonium is related to aquaporin (AQP). Plant Science 234, 14–21. 10.1016/j.plantsci.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Dinno, A. (2017). dunn.test: Dunn's test of multiple comparisons using rank sums. R package version 1.3.5. https://CRAN.R‐project.org/package=dunn.test [Google Scholar]
- Downs, J. L. , & Black, R. A. (1999). Leaf surface characteristics and gas exchange in Artemisia tridentata subspecies wyomingensis and tridentata . In McArthur, E. D. , Ostler, W. K. , & Wambolt, C. L. (Eds.), Proceedings shrubland ecotones (pp. 108–112). USDA, Forest Service, RMRS‐P‐11. [Google Scholar]
- Edler, D. , Klein, J. , Antonelli, A. , & Silvestro, D. (2020). raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution, 12(2), 373–377. 10.1111/2041-210X.13512 [DOI] [Google Scholar]
- Fahn, A. (1964). Some anatomical adaptations of desert plants. Phytomorphology, 4, 93–101. [Google Scholar]
- Fang, L. , Abdelhakim, L. O. A. , Hegelund, J. N. , Li, S. , Liu, J. , Peng, X. , Li, X. , Wei, Z. , & Liu, F. (2019). ABA‐mediated regulation of leaf and root hydraulic conductance in tomato grown at elevated CO2 is associated with altered gene expression of aquaporins. Horticulture Research, 6(1), 104. 10.1038/s41438-019-0187-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexas, J. , Bota, J. , Galmés, J. , Medrano, H. , & Ribas‐Carbó, M. (2006). Keeping a positive carbon balance under adverse conditions: Responses of photosynthesis and respiration to water stress. Physiologia Plantarum, 127(3), 343–352. 10.1111/j.1399-3054.2006.00621.x [DOI] [Google Scholar]
- Flexas, J. , Ribas‐Carbó, M. , Hanson, D. T. , Bota, J. , Otto, B. , Cifre, J. , & Kaldenhoff, R. (2006). Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant Journal, 48(3), 427–439. 10.1111/j.1365-313X.2006.02879.x [DOI] [PubMed] [Google Scholar]
- Freeman, D. C. , Turner, W. A. , McArthur, E. D. , & Graham, J. H. (1991). Characterization of a narrow hybrid zone between two subspecies of big sagebrush (Artemisia tridentata: Asteraceae). American Journal of Botany, 78(6), 805–815. [PubMed] [Google Scholar]
- Galmés, J. , Medrano, H. , & Flexas, J. (2007). Photosynthetic limitations in response to water stress and recovery in Mediterranean plants with different growth forms. New Phytologist, 175(1), 81–93. 10.1111/j.1469-8137.2007.02087.x [DOI] [PubMed] [Google Scholar]
- Gao, X. , & Giorgi, F. (2008). Increased aridity in the Mediterranean region under greenhouse gas forcing estimated from high resolution simulations with a regional climate model. Global and Planetary Change, 62(3–4), 195–209. 10.1016/j.gloplacha.2008.02.002 [DOI] [Google Scholar]
- Garcia, S. , Canela, M. Á. , Garnatje, T. , Mcarthur, E. D. , Pellicer, J. , Sanderson, S. C. , & Vallès, J. (2008). Evolutionary and ecological implications of genome size in the North American endemic sagebrushes and allies (Artemisia, Asteraceae). Biological Journal of the Linnean Society, 94(3), 631–649. 10.1111/j.1095-8312.2008.01001.x [DOI] [Google Scholar]
- Grabherr, G. , Gottfried, M. , & Pauli, H. (1994). Climate effects on mountain plants [7]. Nature, 369(6480), 448. 10.1038/369448a0 [DOI] [PubMed] [Google Scholar]
- Heinen, R. B. , Ye, Q. , & Chaumont, F. (2009). Role of aquaporins in leaf physiology. Journal of Experimental Botany, 60(11), 2971–2985. 10.1093/jxb/erp171 [DOI] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. , & Korenaga, T. (1999). Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research, 27(1), 297–300. 10.1093/nar/27.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P. K. , Dubeaux, G. , Takahashi, Y. , & Schroeder, J. I. (2021). Signaling mechanisms in abscisic acid‐mediated stomatal closure. The Plant Journal, 105(2), 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Li, Y. , Fu, C. , Chen, F. , Fu, Q. , Dai, A. , Shinoda, M. , Ma, Z. , Guo, W. , Li, Z. , Zhang, L. , Liu, Y. , Yu, H. , He, Y. , Xie, Y. , Guan, X. , Ji, M. , Lin, L. , Wang, S. , … Wang, G. (2017). Dryland climate change: Recent progress and challenges. Reviews of Geophysics, 55(3), 719–778. 10.1002/2016RG000550 [DOI] [Google Scholar]
- Intergovernmental Panel on Climate Change . (2001). Climate change 2001: IPCC third assessment report. IPCC Secretariat. [Google Scholar]
- Ishibashi, K. (2006). Aquaporin subfamily with unusual NPA boxes. Biochimica Et Biophysica Acta – Biomembranes, 1758(8), 989–993. 10.1016/j.bbamem.2006.02.024 [DOI] [PubMed] [Google Scholar]
- Jezkova, T. , & Wiens, J. J. (2016). Rates of change in climatic niches in plant and animal populations are much slower than projected climate change. Proceedings of the Royal Society B: Biological Sciences, 283(1843), 20162104. 10.1098/rspb.2016.2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P. D. , Osborn, T. J. , & Briffa, K. R. (2001). The evolution of climate over the last millennium. Science, 292(5517), 662–667. 10.1126/science.1059126 [DOI] [PubMed] [Google Scholar]
- Kaldenhoff, R. , & Fischer, M. (2006). Functional aquaporin diversity in plants. Biochimica Et Biophysica Acta – Biomembranes, 1758(8), 1134–1141. 10.1016/j.bbamem.2006.03.012 [DOI] [PubMed] [Google Scholar]
- Kaldenhoff, R. , Ribas‐Carbo, M. , Sans, J. F. , Lovisolo, C. , Heckwolf, M. , & Uehlein, N. (2008). Aquaporins and plant water balance. Plant, Cell and Environment, 31(5), 658–666. 10.1111/j.1365-3040.2008.01792.x [DOI] [PubMed] [Google Scholar]
- Karban, R. (2007). Associational resistance for mule’s ears with sagebrush neighbors. Plant Ecology, 191(2), 295–303. 10.1007/s11258-006-9243-z [DOI] [Google Scholar]
- Katoh, K. , Rozewicki, J. , & Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20(4), 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L. A. , Mezulis, S. , Yates, C. M. , Wass, M. N. , & Sternberg, M. J. (2016). Trabajo práctico No 13. Varianzas en función de variable independiente categórica. Nature Protocols, 10(6), 845–858. 10.1038/nprot.2015-053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb, K. J. , & Sperry, J. S. (1999). Differences in drought adaptation between subspecies of sagebrush (Artemisia tridentata). Ecology, 80(7), 2373–2384. 10.2307/176917 [DOI] [Google Scholar]
- Krogh, A. , Larsson, B. , Von Heijne, G. , & Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305(3), 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kullman, L. (2002). Rapid recent range‐margin rise of tree and shrub species in the Swedish Scandes. Journal of Ecology, 90(1), 68–77. 10.1046/j.0022-0477.2001.00630.x [DOI] [Google Scholar]
- Lambrecht, S. C. , Shattuck, A. K. , & Loik, M. E. (2007). Combined drought and episodic freezing effects on seedlings of low‐ and high‐elevation subspecies of sagebrush (Artemisia tridentata). Physiologia Plantarum, 130(2), 207–217. 10.1111/j.1399-3054.2007.00904.x [DOI] [Google Scholar]
- Lata, C. , Muthamilarasan, M. , & Prasad, M. (2015). Drought stress responses and signal transduction in plants. In Pandey, G. (Ed.), Elucidation of abiotic stress signaling in plants: Functional genomics perspectives (Vol. 2). Springer‐Verlag. 10.1007/978-1-4939-2540-7 [DOI] [Google Scholar]
- Leonard, K. M. , Reese, K. P. , & Connelly, J. W. (2000). Distribution, movements and habitats of sage grouse Centrocercus urophasianus on the upper snake river plain of Idaho: Changes from the 1950s to the 1990s. Wildlife Biology, 6(4), 265–270. 10.2981/wlb.2000.025 [DOI] [Google Scholar]
- Li, G. , Santoni, V. , & Maurel, C. (2014). Plant aquaporins: Roles in plant physiology. Biochimica Et Biophysica Acta ‐ General Subjects, 1840(5), 1574–1582. 10.1016/j.bbagen.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Wang, F. , Yu, G. , Zhang, X. , Jia, C. , Qin, J. , & Pan, H. (2015). Functional analysis of the maize C‐Repeat/DRE motif‐binding transcription factor CBF3 promoter in response to abiotic stress. International Journal of Molecular Sciences, 16(6), 12131–12146. 10.3390/ijms160612131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Guo, S. , Xu, J. , Zhang, Y. , Dong, L. , Xiao, S. , Bai, R. , Liao, B. , Su, H. E. , Cheng, R. , & Chen, S. (2018). Genome size estimation of Chinese cultured Artemisia annua L. Journal of Plant Biology and Crop Research, 1(1), 1002. 10.33582/2637-7721/1002 [DOI] [Google Scholar]
- Lloyd, A. H. , & Fastie, C. L. (2003). Recent changes in treeline forest distribution and structure in interior Alaska. Ecoscience, 10(2), 176–185. 10.1080/11956860.2003.11682765 [DOI] [Google Scholar]
- Lopez, D. , Venisse, J.‐S. , Fumanal, B. , Chaumont, F. , Guillot, E. , Daniels, M. J. , Cochard, H. , Julien, J.‐L. , & Gousset‐Dupont, A. (2013). Aquaporins and leaf hydraulics: Poplar sheds new light. Plant and Cell Physiology, 54(12), 1963–1975. 10.1093/pcp/pct135 [DOI] [PubMed] [Google Scholar]
- Lüttge, U. (2004). Ecophysiology of Crassulacean Acid Metabolism (CAM). Annals of Botany, 93(6), 629–652. 10.1093/aob/mch087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, C. (2007). Plant aquaporins: Novel functions and regulation properties. FEBS Letters, 581(12), 2227–2236. 10.1016/j.febslet.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Maurel, C. , & Prado, K. (2017). Aquaporins and leaf water relations. Plant aquaporins (pp. 155–165). Springer. [Google Scholar]
- Maurel, C. , Simonneau, T. , & Sutka, M. (2010). The significance of roots as hydraulic rheostats. Journal of Experimental Botany, 61(12), 3191–3198. [DOI] [PubMed] [Google Scholar]
- Maurel, C. , Verdoucq, L. , & Rodrigues, O. (2016). Aquaporins and plant transpiration. Plant Cell and Environment, 39(11), 2580–2587. 10.1111/pce.12814 [DOI] [PubMed] [Google Scholar]
- McArthur, D. E. , & Plummer, P. A. (1978). Biogeography and management of native wetern shrubs: A case study, section tridentatae of Artemisia . Great Basin Naturalist Memoirs, 2, 229–243. [Google Scholar]
- Mcarthur, E. D. , & Sanderson, S. C. (1999). Cytogeography and chromosome evolution of subgenus Tridentatae of Artemisia (Asteraceae). American Journal of Botany, 86(12), 1754–1775. 10.2307/2656673 [DOI] [PubMed] [Google Scholar]
- Mcarthur, E. D. , Welch, B. L. , & Sanderson, S. C. (1988). Natural and artificial hybridization between big sagebrush (Artemisia tridentata) subspecies. Journal of Heredity, 79(4), 268–276. 10.1093/oxfordjournals.jhered.a110508 [DOI] [Google Scholar]
- McElrone, A. J. , Bichler, J. , Pockman, W. T. , Addington, R. N. , Linder, C. R. , & Jackson, R. B. (2007). Aquaporin‐mediated changes in hydraulic conductivity of deep tree roots accessed via caves. Plant, Cell and Environment, 30(11), 1411–1421. 10.1111/j.1365-3040.2007.01714.x [DOI] [PubMed] [Google Scholar]
- Mitsuoka, K. , Murata, K. , Walz, T. , Hirai, T. , Agre, P. , Heymann, J. B. , Engel, A. , & Fujiyoshi, Y. (1999). The structure of aquaporin‐1 at 4.5‐Å resolution reveals short α‐ helices in the center of the monomer. Journal of Structural Biology, 128(1), 34–43. 10.1006/jsbi.1999.4177 [DOI] [PubMed] [Google Scholar]
- Moshelion, M. , Halperin, O. , Wallach, R. , Oren, R. , & Way, D. A. (2015). Role of aquaporins in determining transpiration and photosynthesis in water‐stressed plants: Crop water‐use efficiency, growth and yield. Plant, Cell and Environment, 38(9), 1785–1793. 10.1111/pce.12410 [DOI] [PubMed] [Google Scholar]
- Mueggler, W. F. , & Stewart, W. L. (1980). Grassland and shrubland habitat types of western Montana. USDA Forest Service. 10.5962/bhl.title.100640 [DOI] [Google Scholar]
- Murata, K. , Mitsuoka, K. , Hirai, T. , Walz, T. , Agre, P. , Heymann, J. B. , Engel, A. , & Fujiyoshi, Y. (2000). Structural determinants of water permeation through aquaporin‐1. Nature, 407(6804), 599–605. 10.1038/35036519 [DOI] [PubMed] [Google Scholar]
- Park, W. , Scheffler, B. E. , Bauer, P. J. , & Campbell, B. T. (2010). Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biology, 10, 142. 10.1186/1471-2229-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan, C. , & Yohe, G. (2003). A globally coherent fingerprint of climate change. Nature, 421, 37–42. [DOI] [PubMed] [Google Scholar]
- Pellicer, J. , Garcia, S. , Canela, M. Á. , Garnatje, T. , Korobkov, A. A. , Twibell, J. D. , & Vallès, J. (2010). Genome size dynamics in Artemisia L. (Asteraceae): Following the track of polyploidy. Plant Biology, 12(5), 820–830. 10.1111/j.1438-8677.2009.00268.x [DOI] [PubMed] [Google Scholar]
- Penuelas, J. , & Boada, M. (2003). A global change‐induced biome shift in the Montseny mountains (NE Spain). Global Change Biology, 9(2), 131–140. 10.1046/j.1365-2486.2003.00566.x [DOI] [Google Scholar]
- Prevéy, J. S. , Germino, M. J. , Huntly, N. J. , & Inouye, R. S. (2010). Exotic plants increase and native plants decrease with loss of foundation species in sagebrush steppe. Plant Ecology, 207(1), 39–51. 10.1007/s11258-009-9652-x [DOI] [Google Scholar]
- Quigley, F. , Rosenberg, J. M. , Shachar‐Hill, Y. , & Bohnert, H. J. (2001). From genome to function: the Arabidopsis aquaporins. Genome Biology, 3(1), 1–17. 10.1186/gb-2001-3-1-research0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, I. , & Wiens, J. J. (2013). Rates of projected climate change dramatically exceed past rates of climatic niche evolution among vertebrate species. Ecology Letters, 16(8), 1095–1103. 10.1111/ele.12144 [DOI] [PubMed] [Google Scholar]
- R Core Team . (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing; https://www.R‐project.org/ [Google Scholar]
- Requena‐Mullor, J. M. , Maguire, K. C. , Shinneman, D. J. , & Caughlin, T. T. (2019). Integrating anthropogenic factors into regional‐scale species distribution models—A novel application in the imperiled sagebrush biome. Global Change Biology, 25(11), 3844–3858. 10.1111/gcb.14728 [DOI] [PubMed] [Google Scholar]
- Richardson, B. A. , & Chaney, L. (2018). Climate‐based seed transfer of a widespread shrub: population shifts, restoration strategies, and the trailing edge. Ecological Applications, 28(8), 2165–2174. 10.1002/eap.1804 [DOI] [PubMed] [Google Scholar]
- Richardson, B. A. , Chaney, L. , Shaw, N. L. , & Still, S. M. (2017). Will phenotypic plasticity affecting flowering phenology keep pace with climate change? Global Change Biology, 23(6), 2499–2508. 10.1111/gcb.13532 [DOI] [PubMed] [Google Scholar]
- Richardson, B. A. , Page, J. T. , Bajgain, P. , Sanderson, S. C. , & Udall, J. A. (2012). Deep sequencing of amplicons reveals widespread intraspecific hybridization and multiple origins of polyploidy in big sagebrush (Artemisia tridentata; Asteraceae). American Journal of Botany, 99(12), 1962–1975. 10.3732/ajb.1200373 [DOI] [PubMed] [Google Scholar]
- Rockwell, F. E. , Holbrook, N. M. , & Zwieniecki, M. A. (2011). Hydraulic conductivity of red oak (Quercus rubra L.) leaf tissue does not respond to light. Plant, Cell and Environment, 34(4), 565–579. 10.1111/j.1365-3040.2011.02263.x [DOI] [PubMed] [Google Scholar]
- Sade, N. , & Moshelion, M. (2017). Plant aquaporins and abiotic stress. In Chaumont, F. , & Tyerman, S. D. (Eds). Signaling and communication in plants. Plant aquaporins: From transport to signaling (pp. 185–206). Springer International Publishing. [Google Scholar]
- Samarah, N. H. (2005). Effects of drought stress on growth and yield of barley. Agronomy for Sustainable Development, 25(1), 145–149. 10.1051/agro:2004064 [DOI] [Google Scholar]
- Sanz, M. , Schneeweiss, G. M. , Vilatersana, R. , & Vallès, J. (2011). Temporal origins and diversification of Artemisia and allies (Anthemideae, Asteraceae). Collectanea Botanica, 30, 7–15. 10.3989/collectbot.2011.v30.001 [DOI] [Google Scholar]
- Sanz‐Elorza, M. , Dana, E. D. , González, A. , & Sobrino, E. (2003). Changes in the high‐mountain vegetation of the central Iberian Peninsula as a probable sign of global warming. Annals of Botany, 92(2), 273–280. 10.1093/aob/mcg130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchi, F. , Pagliarani, C. , & Zwieniecki, M. A. (2017). The functional role of xylem parenchyma cells and aquaporins during recovery from severe water stress. Plant Cell and Environment, 40(6), 858–871. 10.1111/pce.12831 [DOI] [PubMed] [Google Scholar]
- Seppey, M. , Manni, M. , & Zdobnov, E. M. (2019). BUSCO: Assessing genome assembly and annotation completeness. In Kollmar, M. (Eds.), Gene prediction. Methods in molecular biology (Vol. 1962). Humana. [DOI] [PubMed] [Google Scholar]
- Sharp, R. E. , Poroyko, V. , Hejlek, L. G. , Spollen, W. G. , Springer, G. K. , Bohnert, H. J. , & Nguyen, H. T. (2004). Root growth maintenance during water deficits: Physiology to functional genomics. Journal of Experimental Botany, 55(407), 2343–2351. 10.1093/jxb/erh276 [DOI] [PubMed] [Google Scholar]
- Shekoofa, A. , & Sinclair, T. (2018). Aquaporin activity to improve crop drought tolerance. Cells, 7(9), 123. 10.3390/cells7090123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q. , Zhang, L. , Liao, Z. , Wang, S. , Yan, T. , Shi, P. U. , Liu, M. , Fu, X. , Pan, Q. , Wang, Y. , Lv, Z. , Lu, X. U. , Zhang, F. , Jiang, W. , Ma, Y. , Chen, M. , Hao, X. , Li, L. , Tang, Y. , … Tang, K. (2018). The genome of Artemisia annua provides insight into the evolution of asteraceae family and artemisinin biosynthesis. Molecular Plant, 11(6), 776–788. 10.1016/j.molp.2018.03.015 [DOI] [PubMed] [Google Scholar]
- Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany, 58(2), 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Shinozaki, K. , Yamaguchi‐Shinozaki, K. , & Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology, 6(5), 410–417. 10.1016/S1369-5266(03)00092-X [DOI] [PubMed] [Google Scholar]
- Shivaraj, S. M. , Deshmukh, R. K. , Rai, R. , Belanger, R. , Agrawal, P. K. , & Dash, P. K. (2017). Genome‐wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Scientific Reports 7, 46137. 10.1038/srep46137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefritz, F. , Tyree, M. T. , Lovisolo, C. , Schubert, A. , & Kaldenhoff, R. (2002). PIP1 plasma membrane aquaporins in tobacco: From cellular effects to function in plants. The Plant Cell, 14(4), 869–876. 10.1105/tpc.000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. K. , Deshmukh, R. , Muthamilarasan, M. , Rani, R. , & Prasad, M. (2020). Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiology and Biochemistry, 149, 178–189. 10.1016/j.plaphy.2020.02.009 [DOI] [PubMed] [Google Scholar]
- Song, L. I. , Nguyen, N. A. , Deshmukh, R. K. , Patil, G. B. , Prince, S. J. , Valliyodan, B. , Mutava, R. , Pike, S. M. , Gassmann, W. , & Nguyen, H. T. (2016). Soybean tip gene family analysis and characterization of GmTIP1;5 and GmTIP2;5 water transport activity. Frontiers in Plant Science, 7, 1–15. 10.3389/fpls.2016.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke, M. , & Waack, S. (2003). Gene prediction with a hidden‐markov model and a new intron submodel. Bioinformatics, 19(2), ii215–ii225. 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- Steudle, E. (2001). The cohesion‐tension mechanism and the acquisition of water by plant roots. Annual Review of Plant Physiology and Plant Molecular Biology, 52(1), 847–875. 10.1146/annurev.arplant.52.1.847 [DOI] [PubMed] [Google Scholar]
- Still, S. M. , & Richardson, B. A. (2015). Projections of contemporary and future climate niche for wyoming big sagebrush (Artemisia tridentata subsp. wyomingensis): A guide for restoration. Natural Areas Journal, 35(1), 30–43. 10.3375/043.035.0106 [DOI] [Google Scholar]
- Sturm, M. , Racine, C. , & Tape, K. (2001). Increasing shrub abundance in the Arctic. Nature, 411(6837), 546–547. 10.1038/35079180 [DOI] [PubMed] [Google Scholar]
- Takahashi, F. , Kuromori, T. , Sato, H. , & Shinozaki, K. (2018). Regulatory gene networks in drought stress responses and resistance in plants. In Iwaya‐Inoue, M. , Sakurai, M. , & Uemura, M. (Eds). Advances in experimental medicine and biology. Survival strategies in extreme cold and desiccation: Adaptation mechanisms and their applications (pp. 189–214). Springer Singapore. [DOI] [PubMed] [Google Scholar]
- Taylor, R. L. , Marchand, L. S. , & Crompton, C. W. (1964). Cytological observations on the Artemisia tridentata (Compositae) complex in British Columbia. Canadian Journal of Botany, 6(1), 42–45. [Google Scholar]
- Tieldnes, H. , & Labun, K. (2020). ORFik: Open Reading Frames in Genomics. R Package version 1.10.2. https://github.com/Roleren/ORFik [Google Scholar]
- Uehlein, N. , Sperling, H. , Heckwolf, M. , & Kaldenhoff, R. (2012). The Arabidopsis aquaporin PIP1;2 rules cellular CO2 uptake. Plant, Cell and Environment, 35(6), 1077–1083. 10.1111/j.1365-3040.2011.02473.x [DOI] [PubMed] [Google Scholar]
- Walther, G. (2003). Plants in a warmer world. Perspectives in Plant Ecology, Evolution and Systematics, 6(3), 169–185. 10.1078/1433-8319-00076 [DOI] [Google Scholar]
- Walther, G. , Post, E. , Convey, P. , Menzel, A. , Parmesank, C. , Beebee, T. J. C. , & Bairlein, F. (2002). Ecological response to recent climate change. Nature, 416, 389–395. [DOI] [PubMed] [Google Scholar]
- Wiens, J. J. (2016). Climate‐related local extinctions are already widespread among plant and animal species. PLoS Biology, 14(12), 1–18. 10.1371/journal.pbio.2001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojahn, J. M. A. , Galla, S. J. , Melton, A. E. , & Buerki, S. (2021). G2PMineR: A genome to phenome literature review approach. Genes, 12(2), 293. 10.3390/genes12020293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wree, D. , Wu, B. , Zeuthen, T. , & Beitz, E. (2011). Requirement for asparagine in the aquaporin NPA sequence signature motifs for cation exclusion. FEBS Journal, 278(5), 740–748. 10.1111/j.1742-4658.2010.07993.x [DOI] [PubMed] [Google Scholar]
- Yakata, K. , Hiroaki, Y. , Ishibashi, K. , Sohara, E. , Sasaki, S. , Mitsuoka, K. , & Fujiyoshi, Y. (2007). Aquaporin‐11 containing a divergent NPA motif has normal water channel activity. Biochimica Et Biophysica Acta – Biomembranes, 1768(3), 688–693. 10.1016/j.bbamem.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. , & Shinozaki, K. (1994). A novel cis‐acting element in an Arabidopsis gene is involved in responsiveness to drought, low‐temperature, or high‐salt stress. The Plant Cell, 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarch, M. A. A. , Sivakumar, B. , Malekinezhad, H. , & Sharma, A. (2017). Future aridity under conditions of global climate change. Journal of Hydrology, 554, 451–469. 10.1016/j.jhydrol.2017.08.043 [DOI] [Google Scholar]
- Zargar, S. M. , Nagar, P. , Deshmukh, R. , Nazir, M. , Wani, A. A. , Masoodi, K. Z. , Agrawal, G. K. , & Rakwal, R. (2017). Aquaporins as potential drought tolerance inducing proteins: Towards instigating stress tolerance. Journal of Proteomics, 169, 233–238. 10.1016/j.jprot.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Feng, M. , Chen, W. , Zhou, X. , Lu, J. , Wang, Y. , Li, Y. , Jiang, C.‐Z. , Gan, S.‐S. , Ma, N. , & Gao, J. (2019). In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nature Plants, 5(3), 290–299. 10.1038/s41477-019-0376-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
File S1
File S2
File S3
Table S1
Supplementary Material
Data Availability Statement
Scripts used in this research have been deposited at https://github.com/aemelton/DraftGenomeMineR. The raw reads (accessions SRR14309371, SRR14309372, and SRR14309373) and draft genome assembly (accession JAHAUY000000000) have been submitted to NCBI as SAMN18747788 under PRJNA722258. Supplementary documents, including scaffolds and alignment fasta, have been submitted as part of the SI.


