Abstract
The high‐altitude environment may drive vertebrate evolution in a certain way, and vertebrates living in different altitude environments might have different energy requirements. We hypothesized that the high‐altitude environment might impose different influences on vertebrate mitochondrial genomes (mtDNA). We used selection pressure analyses and PIC (phylogenetic independent contrasts) analysis to detect the evolutionary rate of vertebrate mtDNA protein‐coding genes (PCGs) from different altitudes. The results showed that the ratio of nonsynonymous/synonymous substitutions (dN/dS) in the mtDNA PCGs was significantly higher in high‐altitude vertebrates than in low‐altitude vertebrates. The seven rapidly evolving genes were shared by the high‐altitude vertebrates, and only one positive selection gene (ND5 gene) was detected in the high‐altitude vertebrates. Our results suggest the mtDNA evolutionary rate in high‐altitude vertebrates was higher than in low‐altitude vertebrates as their evolution requires more energy in a high‐altitude environment. Our study demonstrates the high‐altitude environment (low atmospheric O2 levels) drives vertebrate evolution in mtDNA PCGs.
Keywords: evolution, low atmospheric O2 levels, mtDNA protein‐coding genes, vertebrate
We studied the influence of different altitudes on the evolution of 104 vertebrate mtDNA PCGs for the first time. Our results demonstrated that the rapid evolution and positive selection sites of mitochondrial genes are one of the important factors for vertebrates to adapt to plateau environment through this research and the high‐altitude environment drove the evolution of vertebrate mtDNA PCGs.

1. INTRODUCTION
Due to the distinctive geographical environment, high altitudes involve severe environmental conditions, such as low pressure, low oxygen, and high UV (ultraviolet) radiation. Low oxygen is one of the most important factors affecting vertebrate survival. Vertebrates adapt to hypoxia in high‐altitude habitats through changes in their physiological phenotype and gene evolution (McCracken et al., 2009; Zhu et al., 2018). High‐altitude vertebrates represent a unique scenario for studying adaptation. Therefore, studying adaptation in high‐altitude vertebrates is of great significance in allowing us to explore the adaptive evolution of organisms.
Mitochondrion energy metabolism plays an important role in the adaptation to the plateau environment by vertebrates through aerobic respiration (Das, 2006; Gershoni et al., 2009). The adenosine triphosphate (ATP) produced by mitochondrion can maintain vertebrate body temperatures and energy for activities (Sun et al., 2011). The mitochondrial genome contains thirteen protein‐coding genes (mtDNA PCG) encoding thirteen proteins involved in aerobic respiration (Chong & Mueller, 2013). To adapt to hypoxia environments, aerobic respiration in high‐altitude vertebrates may undergo natural selection to increase efficiency. Therefore, the mitochondrial evolutionary rate may be affected by the low atmospheric O2 levels at high altitudes (Scott et al., 2009).
At present, several studies using mitochondrial genome analyses have demonstrated that the thirteen animal mtDNA PCGs have different environmental adaptations based on different evolutionary rates and selection constraints, including mammals (Gu et al., 2012; Luo et al., 2008), aves (Scott et al., 2009; Zhou et al., 2014), and actinopterygii (Li et al., 2013; Wang et al., 2016). For example, the NS/S values of the ATP6, ATP8, and Cytb genes were larger (>1) in the Tibetan than the Han population (Gu et al., 2012). Similarly, the bar‐headed geese had a striking alteration in the kinetics of the cytochrome c oxidase (COX) in high‐altitude environments (Scott et al., 2009). Wang et al. (2016) discovered that Tibetan loaches accumulated more nonsynonymous mutations and exhibited rapid evolution when compared to non‐Tibetan loaches. It is possible that the mtDNA of low‐altitude vertebrates might have a lower evolutionary rate. In contrast, the mtDNA of high‐altitude vertebrates may have a faster evolutionary rate as they adapt to the low oxygen environment to maintain an efficient energy metabolism. Therefore, we tested the rate of mtDNA evolution between vertebrates living at different altitudes using the rate ratio of nonsynonymous/synonymous nucleotide substitutions (dN/dS (ω)). We aimed to determine whether there is a molecular basis for high‐altitude adaptation that is related to vertebrate mitochondrial evolutionary rates. This adaptation to low atmospheric O2 levels shows the mtDNA evolution is a critical molecular process enabling the survival of vertebrates at high altitudes and hypoxic environments driving the evolution of vertebrate mitochondrial genes.
2. MATERIALS AND METHODS
2.1. Species sample and mtDNA sequence data
Based on previous studies and the existing mitochondrial genome data of NCBI, we selected 104 vertebrate species (with habitats at varying altitudes; high‐altitude vertebrate, 52; low‐altitude vertebrate, 52). We downloaded mitochondrial genomes from the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/). Our initial criterion was to only select species with clear information available on altitude. If there was no clear altitude information, our second criteria were to include species specific to the plateau. The third criteria were to select the low‐altitude species related to the high‐altitude species. The data set covered five taxa (mammal, aves, reptile, amphibian, and actinopterygii), and the data set of each taxon was greater than or equal to 20 mitochondrial genomes. For most families investigated within each class (mammal, aves, reptile, amphibian, and actinopterygii) in this study, more than one representative of each species was selected. The species and mitochondrial genome accession numbers are listed in Appendix S1.
The high‐altitude vertebrate habitat is over 2000 m, and the low‐altitude vertebrate habitat is below 1000 m. According to the habitat altitude and vertebrate phylogenetic relationships, the 104 vertebrate species were divided into 5 high‐altitude groups (high‐altitude mammal (MH), high‐altitude aves (BH), high‐altitude amphibian (AH), high‐altitude reptile (RH), and high‐altitude actinopterygii (FH)) and 5 low‐altitude groups (low‐altitude mammal (ML), low‐altitude aves (BL), low‐altitude reptile (RL), low‐altitude amphibian (AL), and low‐high actinopterygii (FL)), respectively.
2.2. Phylogenetic construction
We retrieved the 13 mtDNA PCGs of the mitochondrial genomes from the mtDNA sequence data and aligned the mtDNA PCGs using MUSCLE v3.8.31 (Edgar, 2004). We obtained fourteen sequence datasets (Appendix S1). The phylogenetic relationships of the 104 vertebrate species were determined using 13 mtDNA PCGs datasets and their Bayesian inference (BI). We used Mega 7.0 to assess the base substitution saturation in the 13 mtDNA PCGs datasets and used DAMBE 6.4 (Xia, 2001) to calculate the Iss and Iss.c for testing the substitution saturation. The optimal model (GTI +T + G) was selected in the modelfinder function of PhyloSuite software (Zhang et al., 2020) based on the greedy search algorithm and the Bayesian information criterion. The concatenated matrix was performed with four independent Markov Chain Monte Carlo (MCMC) chains for 2,000,000 generations and sampling one tree every 1000 generations.
2.3. Selection pressure analyses
We used the one ratio (M0) model and the free ratio model (model = 1) to estimate the ratio of nonsynonymous (dN) to synonymous (dS) substitutions rates (ω = dN/dS) using the CodeML program in PAML version 4 (Yang, 2007). The likelihood ratio tests (LRTs) were used to determine which mtDNA PCG free ratio model was not effective models. The free ratio model allowed different ω values for each branch on a tree. The ω values of the 13 mtDNA PCGs from the 104 species and each mtDNA PCG with their mitochondrial genomes were computed separately for the terminal branches to evaluate the selective pressure. If the dN or dS values were equal to zero, the ω values could be smaller or larger and we did not use these ω values in our analysis, and we assigned these ω values as NA data in the ω dataset. The ω values from the 5 high‐altitude groups (MH, BH, RH, AH, and FH) and 5 low‐altitude groups (ML, BL, RL, AL, and FL) were compared using one‐way ANOVA and multiple comparisons analysis. We used false discovery rates (FDR) to correct the p values. We used a Wilcoxon test to evaluate the statistical significance in the differences in ω values of 13 mtPCGs and each mtPSG between the species at different altitudes (high‐altitude vertebrate, HV; low‐altitude vertebrate, LV).
To identify the positive selection gene or rapidly evolving gene for high‐altitude adaptation, we used a branch model (one ratio (M0) model, two ratio (M2) model, and NSsites = 0) to detect each mtDNA PCGs in the 104 species. In our research, the branches with high‐altitude vertebrates were used as foreground branches, and the low‐altitude vertebrates were the background branches. Furthermore, we used a branch‐site model (one ratio (M0) model, two ratio (M2) model, and NSsites = 2) in PAML to detect each mtDNA PCG of the 104 species (foreground branches, HV; background branches, LV). We used LRTs to determine which mtDNA PCGs were positive selection genes or rapidly evolving genes. These analyses were based on the BI tree (Figure 1).
FIGURE 1.
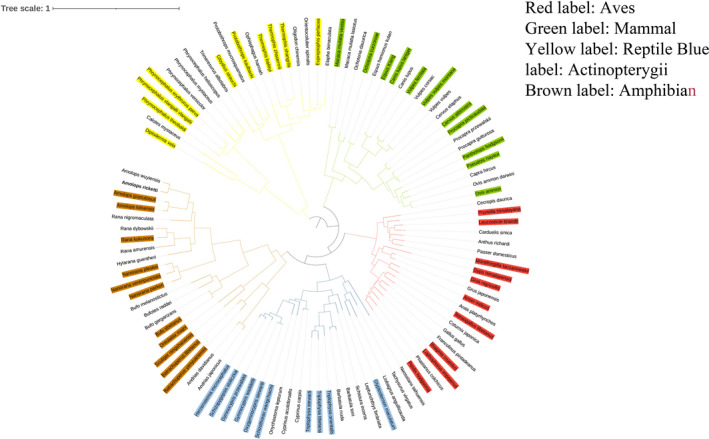
The BI phylogenetic tree of 104 vertebrates based on 13 PCGs of mitochondrial genomes (red background, high‐altitude aves; green background, high‐altitude mammal; yellow background, reptile; blue background, high‐altitude actinopterygii; brown background, high‐altitude amphibian)
2.4. Phylogenetic independent contrasts analysis
The closely related species (shared inheritance) may affect the species comparative analysis. Therefore, we used the phylogenetic independent contrasts (PIC) analysis (Felsenstein Joseph, 1985) to remove the closely related species influences and explore the relationship between altitude and the ω values of the 13 mtDNA PCGs using the ape package in R software. Firstly, we used FigTree (v1.4.3) to transform the BI tree into a binary tree. The binary tree was used as the input file in the analysis. Secondly, we classified the 104 species into two groups (high‐altitude and low‐altitude). The high‐altitude group and low‐altitude groups were coded 0 and 1, respectively. We used the ω values, 0 and 1, as the character data in the PIC analysis.
3. RESULTS
3.1. Phylogenetic analyses
We used 13 mtDNA PCGs datasets of the 104 vertebrates to conduct the phylogenetic analysis. The BI phylogenetic analyses of the concatenated datasets yielded consistent topological relationships between vertebrates with high bootstrap support values and Bayesian posterior probabilities. The BI tree is divided into 5 large branches: the mammal, aves, reptile, amphibian, and actinopterygii (Figure 1). Our results are consistent with previous studies (Townsend et al., 2008), and this was credible for the following analyses.
3.2. Free ratio model analysis
The ω values (13 mtDNA PCGs and each mtDNA PCG of 104 vertebrates) were estimated using CodeML within the PAML package. The ω values <1 indicate a purifying selection, the ω values = 1 indicate a neutral selection, and the ω values >1 indicate a positive selection. We found that the ω values were lower than 1 for the 13 mtDNA PCGs or each mtDNA PCG within the 104 vertebrates (provided in Appendix S2). On the other hand, we found that the ATP8 gene experienced positive selection in the Phrynocephalus theobaldi (ω = 1.209; group RH) and Protobothrops mucrosquamatus lineage (ω = 1.132; group RL); the COX1 gene (ω = 1.373) and ND5 gene (ω = 1.609) experienced positive selection in the Cyprinus acutidorsalis lineage (group FL); and the ND6 gene experienced positive selection in the Barbatula toni lineage (ω = 1.522; Group FH) and Anser indicus lineage (ω = 1.998; Group BH).
The Wilcoxon test results showed that the ω values of the COX2, COX3, Cytb, and ND2 genes were higher in the HV than the LV (Figure 2). The other mtDNA PCGs and the 13 mtDNA PCGs were not higher in the HV than the LV (p > .05). In addition, the one‐way ANOVA and multiple comparison analysis showed the 13 mtDNA PCGs and each mtDNA PCG were not significantly different between the 5 high‐altitude groups or the 5 low‐altitude groups (FDR p > .05). The results indicated that the evolutionary rates were not significantly different in the 13 mtDNA PCGs and within each mtDNA PCG in the high‐altitude vertebrates or the low‐altitude vertebrates.
FIGURE 2.
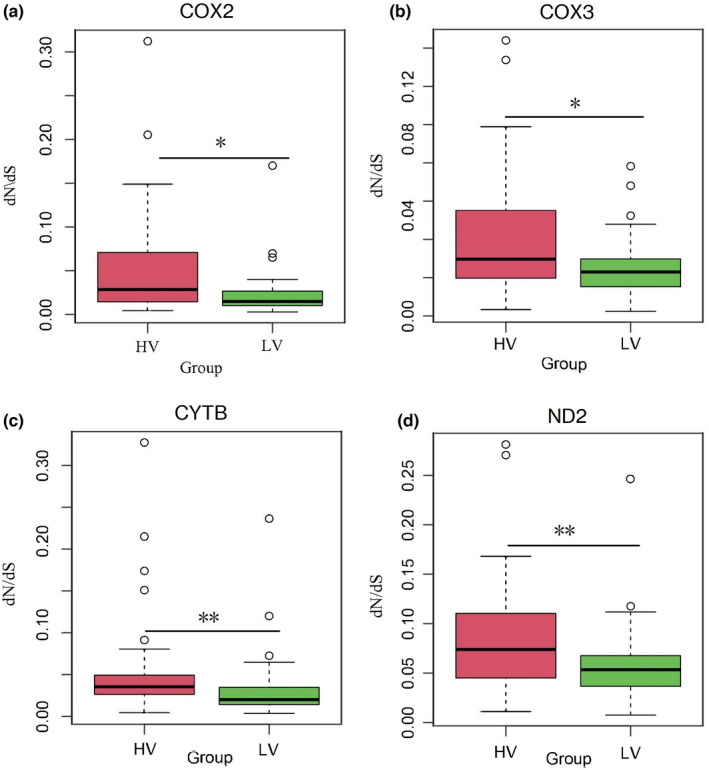
Comparisons of ω values among vertebrates of different altitudes based on 13 mtDNA PCGs (*p < .05; **p < .01)
3.3. Positive selection analysis
We used the branch model and branch‐site model analyses to test our hypothesis. The branch model analysis identified seven rapid evolutionary genes (LRTs, p < .05) in the HV, that is, the ND1 gene (HV: ω values, 0.0351; LV: ω values, 0.0274), ND2 gene (HV: ω values, 0.0735; LV: ω values, 0.0561), ND4 gene (HV: ω values, 0.0533; LV: ω values, 0.0437), COX2 gene (HV: ω values, 0.0306; LV: ω values, 0.0216), COX3 gene (HV: ω values, 0.0287; LV: ω values, 0.0215), ATP6 gene (HV: ω values, 0.0524; LV: ω values, 0.0393), and Cytb gene (HV: ω values, 0.0376; LV: ω values, 0.0255) (Table 1). The results confirmed the effectiveness of the Wilcoxon test results. We used a branch‐site model to detect the positive site of each mtDNA PCG in the HV. We found that only one positive selection gene (ND5 gene, corresponding to site 247 and 524) was detected in the HV using the branch‐site model analysis (Table 2).
TABLE 1.
The rapidly evolving gene on vertebrates 13 mtDNA PCGs through branch model
| Gene | 2ΔlnL | p‐value | The ω values of high‐altitude vertebrates (HV) | The ω values of low‐altitude vertebrates (LV) |
|---|---|---|---|---|
| ATP6 | 7.976 | .004 | 0.0524 | 0.0393 |
| ATP8 | 0.098 | .754 | 0.1530 | 0.1611 |
| ND1 | 7.763 | .005 | 0.0351 | 0.0274 |
| ND2 | 13.558 | .0002 | 0.0735 | 0.0561 |
| ND3 | 0.472 | .492 | 0.0543 | 0.0496 |
| ND4 | 8.084 | .004 | 0.0533 | 0.0437 |
| ND4L | 0.048 | .826 | 0.0395 | 0.0409 |
| ND5 | 0.132 | .890 | 0.0603 | 0.0571 |
| ND6 | 0.109 | .740 | 0.0556 | 0.0581 |
| COX1 | 0.029 | .863 | 0.0107 | 0.0104 |
| COX2 | 9.366 | .002 | 0.0306 | 0.0216 |
| COX3 | 6.302 | .012 | 0.0287 | 0.0215 |
| Cytb | 22.069 | 2.63E−06 | 0.0376 | 0.0255 |
TABLE 2.
Positive selection on 13 mtDNA PCGs of mammal, aves, reptile, amphibian, and actinopterygii through branch‐site model
| Models | Parameter estimates | p‐value | Positively selected sites (BEB analysis) | ||||
|---|---|---|---|---|---|---|---|
| Site class | 0 | 1 | 2a | 2b | |||
| Null model | Proportion | 0.94622 | 0.01479 | 0.03838 | 0.00060 | 247 F 0.952* | |
| Background ω | 0.05529 | 1.00000 | 0.05529 | 1.00000 | 3.86E−14 |
524 M 0.991** |
|
| Foreground ω | 0.05529 | 1.00000 | 1.00000 | 1.00000 | |||
| Model A | Proportion | 0.98147 | 0.01853 | 0.00000 | 0.00000 | ||
| Background ω | 0.05720 | 1.00000 | 0.05720 | 1.00000 | |||
| Foreground ω | 0.05720 | 1.00000 | 20.80002 | 20.80002 | |||
*represents the significant level; PPs of Bayes Empirical Bayes (BEB) analysis with p > .95 was regarded as candidates for selection (*> 0.95, **> 0.99)
3.4. Phylogenetic independent contrasts analysis
We used a PIC analysis to overcome the potential phylogenetic biases, for example, variation in the branch lengths and any phylogenetic inertia. The PIC showed a significant decreasing trend in the dN/dS ratio with elevation reduction (R 2 = 0.057, p = .014; Figure 3). We identified the evolutionary rate of the 13 mtDNA PCGs were faster in the high‐altitude vertebrates when compared with low‐altitude vertebrates.
FIGURE 3.
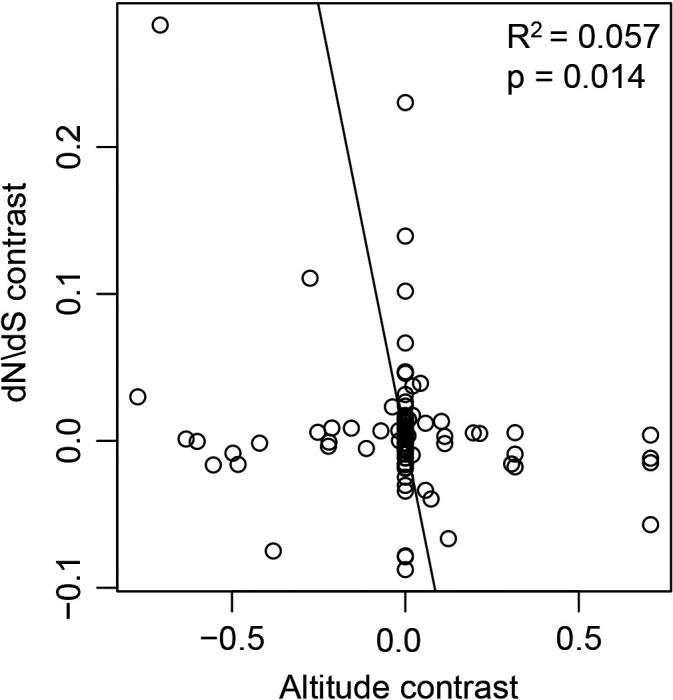
PIC analysis between different altitude and ω values of 104 vertebrates mitochondrial 13 mtDNA PCGs
4. DISCUSSION
This research focused on the adaptation of vertebrate mtDNA PCGs to high‐altitude environments. In high‐altitude environments, vertebrates must maintain normal energy production under hypoxic pressure (Qiu et al., 2012; Wang et al., 2011). Therefore, high‐altitude vertebrates require more energy than low‐altitude vertebrates. Mitochondria (as the energy metabolism center of cells) provide energy for the animals through oxidative phosphorylation (Maes et al., 2004). Therefore, vertebrate mitochondria play important roles in high‐altitude adaptation. Previous research has mainly focused on one kind or family of vertebrates, such as Tibetans (Chen et al., 2020), Tibetan pigs (Ma et al., 2019), Antilopinae (Hassanin et al., 2009), Galliform birds (Zhou et al., 2014), and Schizothoracine fishes (Li et al., 2013). However, previous research has not focused on different altitudes when assessing the five vertebrate taxa. Therefore, we studied the evolutionary rate of 13 mtDNA PCGs within vertebrates at different altitudes. Our research revealed a consistent role of vertebrate mitochondria in high‐altitude adaptation.
4.1. Ratio of nonsynonymous/synonymous nucleotide substitutions
The ratio of nonsynonymous/synonymous nucleotide substitutions (i.e., dN/dS ratio or ω value) has been widely used to measure the selection pressure intensity of the protein‐coding genes (Andrieux & Arenales, 2014; Xia et al., 2019). In our research, the ω values of the 13 mtDNA PCGs and each mtDNA PCG in 104 vertebrates were generally lower than 1 (Appendix S2). The results indicated that the PCGs of the vertebrate mtDNA were under purifying selection at the different altitudes. Previous research has demonstrated that the mtDNA PCGs of Mustelidae (Wei et al., 2020), Tibetan loaches (Wang et al., 2016), and Orthoptera insects (Chang et al., 2020) were under purifying selection during their evolution. This relates to the highly conserved mitochondrial DNA protein‐coding genes (Wolstenholme, 1992). Purifying selection can delete deleterious substitutions to maintain the normal function of the protein‐coding genes. The ω values of 13 mtDNA PCGs and each mtDNA PCG were generally high among vertebrates in high altitudes. This result was consistent with previous research (Wang et al., 2016; Xu et al., 2007) and indicates that high‐altitude environments affect the evolutionary rate of vertebrate mitochondrial DNA protein‐coding genes.
Furthermore, our research also investigated the signatures of positive selection in the ATP8 gene experienced positive selection in the linage Phrynocephalus theobaldi and Protobothrops mucrosquamatus; the COX1 gene and ND5 gene experienced positive selection in the Cyprinus acutidorsalis lineage; and the ND6 gene experienced positive selection in the Barbatula toni lineage and Anser indicus lineage. These evolutionary events showed that the habitat of some low‐altitude vertebrates and high altitudes can affect the evolution rate of mitochondria. For example, the Cyprinus acutidorsalis is a specific brackish water species (Liu et al., 2004), and the special brackish water environment may drive the positive selection in the Cyprinus acutidorsalis mtPCGs. Therefore, to verify the effect of altitude on the evolutionary rate of vertebrate mitochondria, we used the Wilcoxon test (based on the ω value of the terminal branches), a branch model, a branch‐site model, and PIC analysis.
4.2. Selection pressure comparison
We used the Wilcoxon test, a branch model, and a branch‐site model to detect any rapidly evolving genes and positive selection genes shared among the five high‐altitude taxa. We used the Wilcoxon test and one‐way ANOVA analysis (ω values of 13 mtDNA PCGs and each mtDNA PCG in the terminal branches) to detect the evolutionary rate variations between the vertebrates at different altitudes and the evolutionary rate differences between different taxonomies at the same altitude (i.e., the evolutionary rate differences among mammal, aves, reptile, amphibian, and actinopterygii at high or low altitudes), respectively. The ω values (13 mtPCGs and each PCG) showed no significant difference between the five taxonomies at high‐ or low‐altitude environments. This may be due to the similar selection pressures (oxygen levels). The ω values of the COX2 gene, COX3 gene, ND2 gene, and Cytb gene suggested they were all rapidly evolving in the high‐altitude vertebrates. These results show the high‐altitude environment (hypoxia) affects the evolutionary rate of the mitochondria in vertebrates, and the partial gene evolutionary rate in HV was faster than LV (Cheviron & Brumfield, 2012; Storz & Scott, 2019; Storz et al., 2010).
The rapidly evolving genes of the ND1 gene, ND2 gene, ND4 gene, COX2 gene, COX3 gene, ATP6 gene, and Cytb gene were detected in the high‐altitude vertebrates using the branch model. The results of the branch model contained the Wilcoxon test results. These results indicated that the rapidly evolving genes of the high‐altitude vertebrates might go through similar evolutionary patterns (Li et al., 2013; Wang et al., 2016). The high‐altitude environment drives the evolution of vertebrate mitochondrial genes (Ghezzi & Zeviani, 2012; Lenaz et al., 2006; Qingqing et al., 2018).
We detected only one positive selection gene (ND5 gene) using the branch‐site model. The ND5 gene is involved in the regulation of electron transfer in the respiratory chain, which provides necessary energy for vertebrate activity (Brandt, 2006; Javadov et al., 2021; Sousa et al., 2018). We suggest that the ND5 gene is responsible for high‐altitude adaptation in vertebrate. These results suggested that high‐altitude vertebrates probably employ the same genic toolkit to adapt to the extreme environment.
4.3. Phylogenetic independent contrasts analysis
A PIC analysis can remove any influence by phylogenetic inertia, and we identified a significant relationship between the ω values of vertebrate mtDNA PCGs and habitat altitude using a regression analysis (R 2 = 0.057, p = .014). The ω values were negatively related to habitat altitude (the ω value decreases as the altitude decreased). This result was consistent with our previous results using the Wilcoxon test, branch model, and branch‐site model. These results show the evolution of vertebrate mtDNA PCGs is driven by high‐altitude environments.
5. CONCLUSION
In summary, we investigated the evolution of vertebrate mtDNA PCGs at different altitudes. The ω value for the 13 mtDNA PCGs was higher in high‐altitude vertebrates than in the low‐altitude vertebrates as analyzed by the Wilcoxon test, branch model, branch‐site model, and PIC analysis. Although mitochondrial evolution rates are not consistent across different vertebrate taxonomies, the ND1 gene, ND2 gene, ND4 gene, COX2 gene, COX3 gene, ATP6 gene, and Cytb gene are rapidly evolving genes that are shared among high‐altitude vertebrates. Moreover, the high‐altitude vertebrates possess one positive selection gene. The rapid evolution and positive selection sites of the mitochondrial genes are important factors in vertebrate adaptation to the plateau environment, and high‐altitude environments drive the evolution of vertebrate mtDNA PCGs.
CONFLICT OF INTEREST
No potential conflict of interest was reported by the authors.
AUTHOR CONTRIBUTIONS
Xibao Wang: Data curation (equal); Formal analysis (equal); Project administration (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Shengyang Zhou: Formal analysis (equal). Xiaoyang Wu: Project administration (supporting). Qinguo Wei: Project administration (supporting). Yongquan Shang: Formal analysis (supporting). Guolei Sun: Formal analysis (supporting). Xuesong Mei: Formal analysis (supporting). Yuehuan Dong: Formal analysis (supporting). Weilai Sha: Project administration (supporting). Honghai Zhang: Funding acquisition (lead); Project administration (equal).
Supporting information
Appendix S1
Appendix S2
ACKNOWLEDGMENTS
This work was supported by the Special Fund for Forest Scientific Research in the Public Welfare (201404420), the National Natural Science Foundation of China (31872242, 32070405, 32001228, 32000291, 31900311), and the China Postdoctoral Science Foundation (2019M661878).
Wang, X. , Zhou, S. , Wu, X. , Wei, Q. , Shang, Y. , Sun, G. , Mei, X. , Dong, Y. , Sha, W. , & Zhang, H. (2021). High‐altitude adaptation in vertebrates as revealed by mitochondrial genome analyses. Ecology and Evolution, 11, 15077–15084. 10.1002/ece3.8189
Wang and Zhou shared first authorship.
DATA AVAILABILITY STATEMENT
All the mitochondria genome sequences used in this study were accessed through the GenBank database using the accession numbers and DOI accession numbers in Appendix S1.
REFERENCES
- Andrieux, L. O. , & Arenales, D. T. (2014). Whole‐genome identification of neutrally evolving pseudogenes using the evolutionary measure dN/dS. Methods in Molecular Biology, 1167, 75. [DOI] [PubMed] [Google Scholar]
- Brandt, U. (2006). Energy converting NADH: quinone oxidoreductase (complex I). Annual Review of Biochemistry, 75, 69–92. 10.1146/annurev.biochem.75.103004.142539 [DOI] [PubMed] [Google Scholar]
- Chang, H. , Qiu, Z. , Yuan, H. , Wang, X. , Li, X. , Sun, H. , Guo, X. , Lu, Y. , Feng, X. , Majid, M. , & Huang, Y. (2020). Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Molecular Phylogenetics and Evolution, 145, 106734. 10.1016/j.ympev.2020.106734 [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Gong, L. , Liu, X. , Chen, X. , Yang, S. , & Luo, Y. (2020). Mitochondrial DNA genomes revealed different patterns of high‐altitude adaptation in high‐altitude Tajiks compared with Tibetans and Sherpas. Scientific Reports, 10(1), 1–9. 10.1038/s41598-020-67519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron, Z. A. , & Brumfield, R. T. (2012). Genomic insights into adaptation to high‐altitude environments. Heredity, 108(4), 354–361. 10.1038/hdy.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, R. A. , & Mueller, R. L. (2013). Low metabolic rates in salamanders are correlated with weak selective constraints on mitochondrial genes. Evolution: International Journal of Organic Evolution, 67(3), 894–899. 10.1111/j.1558-5646.2012.01830.x [DOI] [PubMed] [Google Scholar]
- Das, J. (2006). The role of mitochondrial respiration in physiological and evolutionary adaptation. BioEssays, 28(9), 890–901. 10.1002/bies.20463 [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with improved accuracy and speed. Paper presented at the Proceedings. 2004 IEEE Computational Systems Bioinformatics Conference, 2004. CSB 2004. [Google Scholar]
- Felsenstein, J. (1985). Phylogenies and the comparative method. The American Naturalist, 125, 1–15. 10.1086/284325 [DOI] [PubMed] [Google Scholar]
- Gershoni, M. , Templeton, A. R. , & Mishmar, D. (2009). Mitochondrial bioenergetics as a major motive force of speciation. BioEssays, 31(6), 642–650. 10.1002/bies.200800139 [DOI] [PubMed] [Google Scholar]
- Ghezzi, D. , & Zeviani, M. (2012). Assembly factors of human mitochondrial respiratory chain complexes: Physiology and pathophysiology. Mitochondrial Oxidative Phosphorylation, 748, 65–106. 10.1007/978-1-4614-3573-0_4 [DOI] [PubMed] [Google Scholar]
- Gu, M. , Dong, X. , Shi, L. , Shi, L. , Lin, K. , Huang, X. , & Chu, J. (2012). Differences in mtDNA whole sequence between Tibetan and Han populations suggesting adaptive selection to high altitude. Gene, 496(1), 37–44. 10.1016/j.gene.2011.12.016 [DOI] [PubMed] [Google Scholar]
- Hassanin, A. , Ropiquet, A. , Couloux, A. , & Cruaud, C. (2009). Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). Journal of Molecular Evolution, 68(4), 293–310. 10.1007/s00239-009-9208-7 [DOI] [PubMed] [Google Scholar]
- Javadov, S. , Jang, S. , Chapa‐Dubocq, X. R. , Khuchua, Z. , & Camara, A. K. (2021). Mitochondrial respiratory supercomplexes in mammalian cells: Structural versus functional role. Journal of Molecular Medicine, 99(1), 57–73. 10.1007/s00109-020-02004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaz, G. , Baracca, A. , Fato, R. , Genova, M. L. , & Solaini, G. (2006). Mitochondrial Complex I: structure, function, and implications in neurodegeneration. Italian Journal of Biochemistry, 55(3–4), 232–253. [PubMed] [Google Scholar]
- Li, Y. , Ren, Z. , Shedlock, A. M. , Wu, J. , Sang, L. , Tersing, T. , Hasegawa, M. , Yonezawa, T. , & Zhong, Y. (2013). High altitude adaptation of the schizothoracine fishes (Cyprinidae) revealed by the mitochondrial genome analyses. Gene, 517(2), 169–178. 10.1016/j.gene.2012.12.096 [DOI] [PubMed] [Google Scholar]
- Liu, L. G. , Zhao, J. , Cui, M. , & Chen, X. L. (2004). Karyotype studies of Cyprinus acutidorsalis . Journal of South China Normal University (Natural ence). 1(1), 108–111. https://mall.cnki.net/magazine/Article/HNSF200401021.htm [Google Scholar]
- Luo, Y. , Gao, W. , Gao, Y. , Tang, S. , Huang, Q. , Tan, X. , Chen, J. , & Huang, T. (2008). Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion, 8(5–6), 352–357. 10.1016/j.mito.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Ma, Y.‐F. , Han, X.‐M. , Huang, C.‐P. , Zhong, L. I. , Adeola, A. C. , Irwin, D. M. , Xie, H.‐B. , & Zhang, Y.‐P. (2019). Population genomics analysis revealed origin and high‐altitude adaptation of Tibetan pigs. Scientific Reports, 9(1), 1–11. 10.1038/s41598-019-47711-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes, D. , Collins, D. , Declercq, L. , Foyouzi‐Yousseffi, R. , Gan, D. , Mammone, T. , Pelle, E. , Marenus, K. , & Gedeon, H. (2004). Improving cellular function through modulation of energy metabolism. International Journal of Cosmetic Ence, 26(5), 268–269. 10.1111/j.1467-2494.2004.00230_4.x [DOI] [Google Scholar]
- McCracken, K. G. , Johnson, K. P. , Kuhner, M. K. , Trucco, J. , Valqui, T. H. , Wilson, R. E. , & Peters, J. L. (2009). Gene flow in the face of countervailing selection: Adaptation to high‐altitude hypoxia in the betaA hemoglobin subunit of yellow‐billed pintails in the Andes. Molecular Biology & Evolution, 26(4), 815. [DOI] [PubMed] [Google Scholar]
- Qingqing, W. , Wenkang, L. U. , Jianke, Y. , Jiang, L. , Zhang, Q. , Kan, X. , & Yang, X. (2018). Comparative transcriptomics in three Passerida species provides insights into the evolution of avian mitochondrial complex I. Comparative Biochemistry & Physiology Part D Genomics & Proteomics, 28, 27–36. 10.1016/j.cbd.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Qiu, Q. , Zhang, G. , Ma, T. , Qian, W. , Wang, J. , Ye, Z. , Cao, C. , Hu, Q. , Kim, J. , Larkin, D. M. , Auvil, L. , Capitanu, B. , Ma, J. , Lewin, H. A. , Qian, X. , Lang, Y. , Zhou, R. , Wang, L. , Wang, K. , … Liu, J. (2012). The yak genome and adaptation to life at high altitude. Nature Genetics, 44(8), 946–949. 10.1038/ng.2343 [DOI] [PubMed] [Google Scholar]
- Scott, G. R. , Egginton, S. , Richards, J. G. , & Milsom, W. K. (2009). Evolution of muscle phenotype for extreme high altitude flight in the bar‐headed goose. Proceedings of the Royal Society B: Biological Sciences, 276(1673), 3645–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, J. S. , D'Imprima, E. , & Vonck, J. (2018). Mitochondrial Respiratory Chain Complexes. In: Harris, J. , & Boekema, E. (Eds.), Membrane Protein Complexes: Structure and Function. Subcellular Biochemistry (87, pp. 167–227). 10.1007/978-981-10-7757-9_7 [DOI] [PubMed] [Google Scholar]
- Storz, J. F. , & Scott, G. R. (2019). Life ascending: Mechanism and process in physiological adaptation to high‐altitude hypoxia. Annual Review of Ecology, Evolution, and Systematics, 50, 503–526. 10.1146/annurev-ecolsys-110218-025014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, J. F. , Scott, G. R. , & Cheviron, Z. A. (2010). Phenotypic plasticity and genetic adaptation to high‐altitude hypoxia in vertebrates. Journal of Experimental Biology, 213(24), 4125–4136. 10.1242/jeb.048181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.‐B. , Shen, Y.‐Y. , Irwin, D. M. , & Zhang, Y.‐P. (2011). Evaluating the roles of energetic functional constraints on teleost mitochondrial‐encoded protein evolution. Molecular Biology and Evolution, 28(1), 39–44. 10.1093/molbev/msq256 [DOI] [PubMed] [Google Scholar]
- Townsend, J. P. , López‐Giráldez, F. , & Friedman, R. (2008). The phylogenetic informativeness of nucleotide and amino acid sequences for reconstructing the vertebrate tree. Journal of Molecular Evolution, 67(5), 437–447. 10.1007/s00239-008-9142-0 [DOI] [PubMed] [Google Scholar]
- Wang, H. , Long, R. , Liang, J. B. , Guo, X. , Ding, L. , & Shang, Z. (2011). Comparison of nitrogen metabolism in yak (Bos grunniens) and indigenous cattle (Bos taurus) on the Qinghai‐Tibetan Plateau. Asian‐Australasian Journal of Animal Sciences, 24(6), 766–773. 10.5713/ajas.2011.10350 [DOI] [Google Scholar]
- Wang, Y. , Shen, Y. , Feng, C. , Zhao, K. , Song, Z. , Zhang, Y. , Yang, L. , & He, S. (2016). Mitogenomic perspectives on the origin of Tibetan loaches and their adaptation to high altitude. Scientific Reports, 6(1), 1–10. 10.1038/srep29690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Q. , Zhang, H. , Wu, X. , & Sha, W. (2020). The selective constraints of ecological specialization in mustelidae on mitochondrial genomes. Mammal Research, 65(1), 85–92. 10.1007/s13364-019-00461-2. [DOI] [Google Scholar]
- Wolstenholme, D. R. (1992). Animal mitochondrial DNA: Structure and evolution. International Review of Cytology (141, pp. 173–216). 10.1016/S0074-7696(08)62066-5 [DOI] [PubMed] [Google Scholar]
- Xia, T. , Zhang, H. , Zhang, L. , Yang, X. , & Zhao, C. (2019). Comparative and evolutionary analysis of the reptilian hedgehog gene family (Shh, Dhh, and Ihh). PeerJ, 7(2), e7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X. (2001). DAMBE: Software package for data analysis in molecular biology and evolution. Journal of Heredity, 92(4), 371–373. 10.1093/jhered/92.4.371 [DOI] [PubMed] [Google Scholar]
- Xu, S. , Luosang, J. , Hua, S. , He, J. , Ciren, A. , Wang, W. , Tong, X. , Liang, Y. U. , Wang, J. , & Zheng, X. (2007). High altitude adaptation and phylogenetic analysis of tibetan horse based on the mitochondrial genome. Journal of Genetics and Genomics, 34(8), 720–729. 10.1016/S1673-8527(07)60081-2 [DOI] [PubMed] [Google Scholar]
- Yang, Z. (2007). PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Gao, F. , Jakovlić, I. , Zou, H. , Zhang, J. , Li, W. X. , & Wang, G. T. (2020). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20(1), 348–355. 10.1111/1755-0998.13096 [DOI] [PubMed] [Google Scholar]
- Zhou, T. , Shen, X. , Irwin, D. M. , Shen, Y. , & Zhang, Y. (2014). Mitogenomic analyses propose positive selection in mitochondrial genes for high‐altitude adaptation in galliform birds. Mitochondrion, 18, 70–75. 10.1016/j.mito.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Guan, Y. , Qu, Y. , David, G. , Song, G. , & Lei, F. (2018). Elevational divergence in the great tit complex revealed by major hemoglobin genes. Current Zoology, 64(4), 455–464. 10.1093/cz/zox042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Appendix S2
Data Availability Statement
All the mitochondria genome sequences used in this study were accessed through the GenBank database using the accession numbers and DOI accession numbers in Appendix S1.


