Abstract
Among the deadliest pandemics in history, coronavirus disease 2019 (COVID-19) has wreaked havoc on human lives, economies and public health systems worldwide. To temper its effects, diagnostic methods that are simple, rapid, inexpensive, accurate, selective and sensitive continue to be necessary. In our study, we developed an electrochemical biosensing platform based on gold clusters, mercaptoethanol, the spike protein of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) antigen and bovine serum albumin-modified glassy carbon electrode able to detect the SARS-CoV-2 spike antibody. Moreover, during the detection of the SARS-CoV-2 spike antibody in spiked-real samples, the anodic signal of the produced biosensor at 0.85 V decreased as the amount of the SARS-CoV-2 spike antibody increased. Meanwhile, the recovery and relative standard deviation values for saliva and oropharyngeal swab samples were 97.73% and 3.35% and 102.43% and 4.63%, respectively. In 35 min, the biosensing platform could detect 0.03 fg/mL of the SARS-CoV-2 spike antibody in synthetic media and spiked-saliva or -oropharyngeal swab samples. The method thus issues a linear response to the SARS-CoV-2 spike antibody from 0.1 fg/mL to 10 pg/mL. The cross-reactivity studies with spike antigens of Middle East respiratory syndrome-coronavirus and influenza A and the antigen of pneumonia confirmed the excellent selectivity of the proposed method. The developed method was compared with the lateral flow immunoassay method in terms of sensitivity and it was found to be approximately 109 times more sensitive.
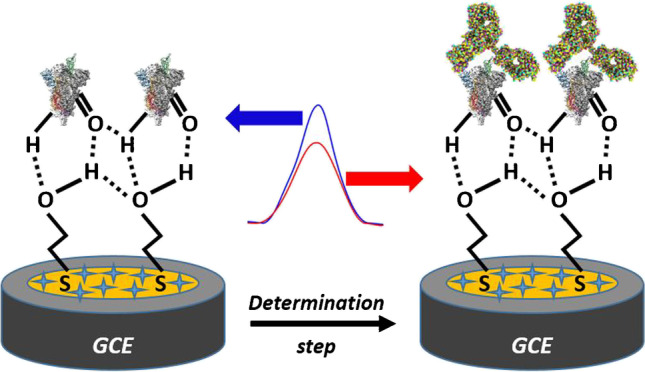
Graphical abstract
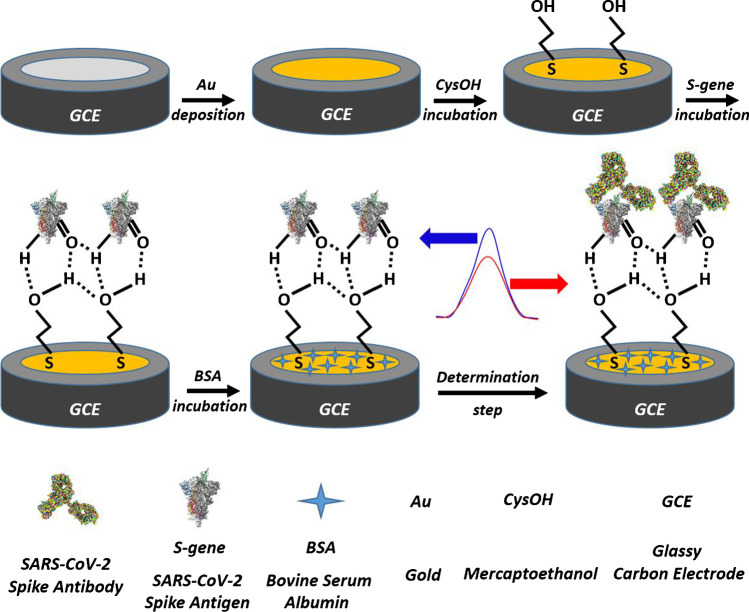
Biosensing mechanism of the platform to the SARS-CoV-2 spike antibody
Supplementary Information
The online version contains supplementary material available at 10.1007/s00216-021-03752-3.
Keywords: Biosensor, SARS-CoV-2, COVID-19, Antibody determination, Gold cluster, Square wave voltammetry
Introduction
Among the deadliest pandemics in history, novel coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) and spreading from the capital of China’s Hubei Province, has posed severe risks for human lives, public health systems and economies around the world [1–3]. To counter the pandemic’s effects, countries with advanced economies, countries with emerging markets and low-income developing countries have respectively spent $9021, $1387 and $37 billion combined as of April 2021. For example, Macao, the United States and New Zealand, as the three countries with the highest gross domestic product expenditures (GDP) in 2020, have respectively allocated 27.4%, 25.5% and 19.4% of GDP to combatting COVID-19 [4]. Despite the world’s collective efforts, as of 18 August 2021, more than 208 million cumulative cases of COVID-19 and 4.3 million deaths have been reported worldwide [5].
Coronaviruses have been divided into four subgenus: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Alpha- and betacoronaviruses are estimated to originate from mammals, especially bats, whereas gamma- and deltacoronaviruses are suspected to be transmitted by birds and pigs. Although only mild symptoms, if any, are associated with alphacoronaviruses, the effects of betacoronaviruses can be fatal [6]. Less than a week after infection, clinical signs of COVID-19 typically manifest including coughing, fever, fatigue, nasal congestion and other symptoms common to upper respiratory system infections. As observed by computed tomography, the infection can worsen with symptoms similar to pneumonia such as dyspnoea and severe chest abnormality [6, 7] and even lead to death [8, 9]. Asymptomatic individuals have also spread COVID-19 and led to the underestimation of cases [10–12]. In response, 18 vaccines with human trials and efficacy tests have shown promise for controlling COVID-19 [12, 13]. Even though effective vaccination is indeed one step to ensure efficient control of the pandemic, the need for rapid, selective and accurate methods of diagnosing COVID-19 will persist [13–15].
Although real-time polymerase chain reaction (RT-PCR) [16–23] is the most prominent technique among the many methods of diagnosing COVID-19 to date, the methods based on enzyme-linked immunosorbent assay (ELISA) [24], lateral flow assay (LFA) [25], lateral flow immunoassay (LFIA) [26–32], UV–visible spectroscopy [33], clustered regularly interspaced short palindromic repeats (CRISPR) [34–36], loop-mediated isothermal amplification (LAMP) [37–40], haematological parameters [41], computed tomography (CT) imaging [42], plasmonic sensors [43, 44] and electrochemical biosensors [45–60] stand at the fore given their advantages such as simplicity, rapidity, sensitivity and accuracy. Among those techniques, RT-PCR is the one most commonly used due to its standardisation, good sensitivity and selectivity. Even so, RT-PCR is also expensive, labour-intensive and time-consuming, as well as requires experienced personnel, remains exclusive to laboratory-based medical institutions [3, 17, 45, 47, 48] and, even worse, has a high false-negative ratio (i.e. 20–67%) depending on the time since infection [61, 62]. Indeed, Wang et al. [63] investigated the performance of six commercial RT-PCR diagnostic kits for COVID-19 and found that all six kits could detect a large amount of the RNA of SARS-CoV-2 and thus, sometimes issued false-negative results. Meanwhile, ELISA-, LFA-, LFIA- and UV–visible spectroscopy-based methods [24–33] are simple, inexpensive, user-friendly and rapid, despite their low sensitivity and thus frequent false-negative results. Methods based on plasmonic sensors, CRISPR and LAMP techniques [34–40, 43, 44] are also affordable and highly sensitive; however, they too require experienced personnel and entail labour-intensive experimental procedures. Beyond those methods, haematological investigation and CT imaging are not appropriate for on-site analysis or the early and certain diagnosis of COVID-19 [43, 64].
By contrast, electrochemical biosensing methods are not only simple, rapid, cost-effective, robust and highly sensitive and selective for diagnosing COVID-19, as demonstrated in various strategies, but also detect the whole virus, the antibody produced in the body and their specific fragments and proteins [65–69]. Antigen-based electrochemical methods based on viral detection (i.e. RNA) requiring 7 h and 29 h for sensor preparation and 40 min and 3 h for measurement provide LODs of 6900 copy/mL [45] and 200 copy/mL [50], respectively. Both antigen- [47–49, 51–56, 59, 60] and antibody-based electrochemical methods [3, 46, 48, 58] use either a spike protein [3, 46, 47, 49, 51, 52, 55, 56, 58–60] or nucleocapsid protein [46, 48, 53, 54] to diagnose COVID-19. Whereas antigen-based methods using proteins offer sensor preparation times ranging from 5 to 73 h and measurement times between 30 s and 45 min, antibody-based ones require between 3 and 72 h to prepare and offer measurement times between 15 min and 1 h.
Using those methods, Seo et al. [47], Rahmati et al. [51], Mavrikou et al. [55], Eissa et al. [53], Hashemi et al. [59] and Liv et al. [60] were able to detect (LODs) 1 fg/mL and 100 fg/mL of the spike protein in synthetic and clinical media, 0.04 fg/mL and 1 fg/mL of the spike protein, 0.8 pg/mL of the nucleocapsid protein in synthetic media, 1.68 × 10−22 μg/mL in biological media and 1 ag/mL of the spike protein in synthetic media, saliva and oropharyngeal swab samples. By comparison, Raziq et al. [54] and Mahari et al. [56] detected 15 fM of the nucleocapsid protein in nasopharyngeal swab samples and 90 fM of the spike protein in saliva samples, respectively. Vadlamani et al. [49] and Mojsoska et al. [52], however, calculated LODs of 0.1 μg/mL and 20 μg/mL for the spike protein in synthetic media, respectively, and the latter found an LOD of 5.5 × 105 PFU/mL in plaque assay media. Although Rashed et al. [58], who studied the spike antibody, and Torrente-Rodríguez et al. [48], who developed a method to detect the spike and nucleocapsid antibodies as well as antigen nucleocapsid protein, did not provide results regarding LODs or dynamic range, Fabiani et al. [46] studied both the spike and nucleocapsid antibodies in synthetic media and found LODs of 19 ng/mL and 8 ng/mL, respectively. Last, in our previous work [3], using gold clusters, cysteamine, glutaraldehyde and the SARS-CoV-2 spike antigen-modified glassy carbon electrode (GCE), we found an LOD of 0.01 ag/mL for the spike antibody in synthetic media.
Against that background, we here report a novel biosensor platform based on gold clusters (Au), mercaptoethanol (CysOH), the SARS-CoV-2 spike antigen protein (S-gene) and bovine serum albumin (BSA)-modified GCE (BSA/S-gene/CysOH/Au/GCE) for determining the SARS-CoV-2 spike antibody in synthetic and spiked-real samples.
Materials and methods
Chemicals and equipment
The spike proteins of SARS-CoV-2 (2019-nCoV) (spike S1-his recombinant protein, verified by HPLC, Cat: 40591-V08H), Middle East respiratory syndrome-coronavirus (MERS-CoV) (S1 Subunit, aa 1–725, His Tag, Cat: 40069-V08B1), Influenza A (H1N1 Hemagglutinin/H0A protein, Cat: 11055-VNAB) and the spike antibody protein of SARS-CoV-2 (Chimeric MAb Cat: 40150-D00) were purchased from Sino Biological Inc. and the native extract of Streptococcus Pneumoniae antigen was supplied from Native Antigen Company. Gold solution (1000 mg/L Au in 2 M HCl, traceable to NIST SRM, Merck 170216), 2-mercaptoethanol (≥ 99.0%, Sigma-Aldrich M6250), bovine serum albumin (BSA, ≥ 98%, Sigma-Aldrich 05470), phosphate-buffered saline (Tablet, Sigma-Aldrich P4417, 0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride, pH 7.2–7.6, at 25 °C) and all the other chemicals were used as analytical reagent grade.
Vitrosens RapidFor™ SARS-CoV-2 Rapid IgG/IgM test kits were used for comparative study.
Eppendorf protein LoBind tubes were used for the preparation and storage of all spike antigen and antibody solutions in 0.01 M (pH 7.5) of PBS solution to prevent sticking of proteins to the walls of their containers, while high-density polyethylene bottles or falcon tubes were used to store the other solutions prepared in ultrapure water.
Milli-Q Direct 8 system was used to produce ultrapure water. A Metrohm Autolab PGSTAT 128 N potentiostat–galvanostat system consisting of chemically modified glassy carbon electrode (Au clusters capped with CysOH, S-gene and BSA modified GCE–BSA/S-gene/CysOH/Au/GCE, GCE: BASi MF-2012, 3.0 mm diameter) as the working electrode, Ag/AgCl/3 M NaCl (BASi MF-2052 RE-5B) as a reference electrode and platinum wire (BASi MW-1032, 7.5 cm) as a counter electrode was used for voltammetrically determining the SARS-CoV-2 spike antibody in synthetic and spiked-real samples.
A Mettler Toledo Seven Easy pH meter with InLab Routine Pro-ISM combined pH electrode and a thermostatic circulator (Thermo Haake DC 10 K20) was used for the preparation of buffer solutions.
A FEI Quanta FEG 250 environmental scanning electron microscope (SEM) and a FEI Quanta 250 XFLASH 5030 energy-dispersive X-ray spectroscopy (EDX) were used for the characterisation of the prepared electrodes.
Preparation of the biosensing platform
First, we sequentially polished the surface of the GCE with 6.0 µm and 1.0 µm of diamond and 0.05 µm of aluminium oxide suspension on velvet felt, washed it with ultrapure water and thereafter exposed it to ultrasonic waves in an ethanol–ultrapure water mixture (1:1, v/v) and in ultrapure water for 3 min each. The gold clusters formed on the GCE’s surface in the same way as in our previous study [3]. Onto Au/GCE’s surface was deposited 25 μL of 20 mM CysOH for 60 min to prepare CysOH/Au/GCE, after which the thiol sites of CysOH were chemisorbed and hydroxyl terminals were placed facing the outside of the electrode. Next, 10 μL of 10 μg/mL S-gene was dropped on the surface of CysOH/Au/GCE for 45 min to provide an interaction between –OH groups of CysOH and –C = O and –H groups of S-gene, as shown in Fig. 1. Free spaces of the obtained electrode, namely S-gene/CysOH/Au/GCE, were blocked with 2% BSA for 20 min to obtain BSA/S-gene/CysOH/Au/GCE. All incubations were performed at room temperature, and the biosensing platform was stored at 4 °C until further use.
Fig. 1.
Procedures for preparing BSA/S-gene/CysOH/Au/GCE and voltammetric measurement for the detection of the SARS-CoV-2 spike antibody
The procedures for preparing BSA/S-gene/CysOH/Au/GCE and voltammetric measurement for the detection of the SARS-CoV-2 spike antibody also appear in Fig. 1.
Voltammetric measurement procedure
Measurements using square wave voltammetry (SWV) and cyclic voltammetry (CV) were applied as in our previous work [3]. Briefly, SWV was performed with a potential range of 0.1–1.4 V with 10 Hz of frequency, 20 mV of pulse amplitude, 5 mV of step potential and an interval time of 0.1 s. Meanwhile, CV scans had the same potential range, with 3 mV of step amplitude and a scan rate of 100 mV/s. An appropriate amount of the SARS-CoV-2 spike antibody or real spiked-sample in 0.01 M (pH 7.5) of PBS solution with a final volume of 10 mL was used. The oxidation peak belonging to BSA/S-gene/CysOH/Au/GCE at 0.85 V decreased with the proportional amount of the SARS-CoV-2 spike antibody, and that signal was used for determining the SARS-CoV-2 spike antibody. Measurements were carried out at 21 ± 3 °C and in 45 ± 15% relative humidity.
LFIA measurement procedure
Measurements were performed according to the manufacturer’s instructions. Briefly, 10 μL of the spiked-saliva sample was dropped onto the sample well of the test card using the plastic dropper and thereafter added 80 μL of sample diluent to the sample well. Finally, the images were interpreted for IgG.
Sample preparation
Saliva and oropharyngeal swab samples were collected from six healthy individuals and treated by following a far easier approach than in our previous work [3]. After 10 fg of the SARS-CoV-2 spike antibody was added to half of the samples for each 5 μL, the spiked- and nonspiked-samples (i.e. the other half of the samples) were analysed to determine the SARS-CoV-2 spike antibody via external calibration by depositing 5 μL of the spiked- or nonspiked-sample on the surface of BSA/S-gene/CysOH/Au/GCE without any preprocessing.
1 ng/mL, 10 ng/mL, 100 ng/mL, 1 μg/mL, 10 μg/mL and 100 μg/mL of the SARS-CoV-2 spike antibody (as a final concentration for each) were added to the saliva samples and analysed by LFIA.
Results and discussion
Characterisation of the electrodes
We examined the surfaces of the electrodes after each modification with CV, scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDX). At first, CV measurements for the GCE, Au/GCE, CysOH/Au/GCE, S-gene/CysOH/Au/GCE and BSA/S-gene/CysOH/Au/GCE electrodes were recorded in 1 mM of K3[Fe(CN)6], 1 mM of K4[Fe(CN)6] and 0.1 M of KCl, as shown in Fig. 2. The peak heights of [Fe(CN)6]3− and [Fe(CN)6]4− couple significantly increased due to the improved electron transfer rate resulting from the conductivity of Au after the modification of gold clusters on the bare GCE (Fig. 2b). The peak heights belonging to the redox couple decreased after the incubation of CysOH on the surface of Au/GCE (Fig. 2c) due to the electrostatic repulsion forces between the [Fe(CN)6]3− and [Fe(CN)6]4− and the unpaired electrons of oxygen from CysOH. With S-gene attached on CysOH/Au/GCE, the peak heights increased (Fig. 2d), which could be attributed to both/either the interaction and attraction between the hydrogen atoms of the amine form in the structure of S-gene and the redox couple and/or the increasing effective surface area. Because BSA is a kind of protein, the peak heights of [Fe(CN)6]3− and [Fe(CN)6]4− couple continued to increase after BSA’s modification on S-gene/CysOH/Au/GCE (Fig. 2e). BSA’s immobilisation was pivotal to determining the SARS-CoV-2 spike antibody, because the non-use of BSA reduced the peak intensities and distorted their symmetry.
Fig. 2.

Cyclic voltammograms of (a) GCE, (b) Au/GCE, (c) CysOH/Au/GCE, (d) S-gene/CysOH/Au/GCE and (e) BSA/S-gene/CysOH/Au/GCE in 1 mM of K3[Fe(CN)6], 1 mM of K4[Fe(CN)6] and 0.1 M of KCl with a scan rate of 50 mV/s
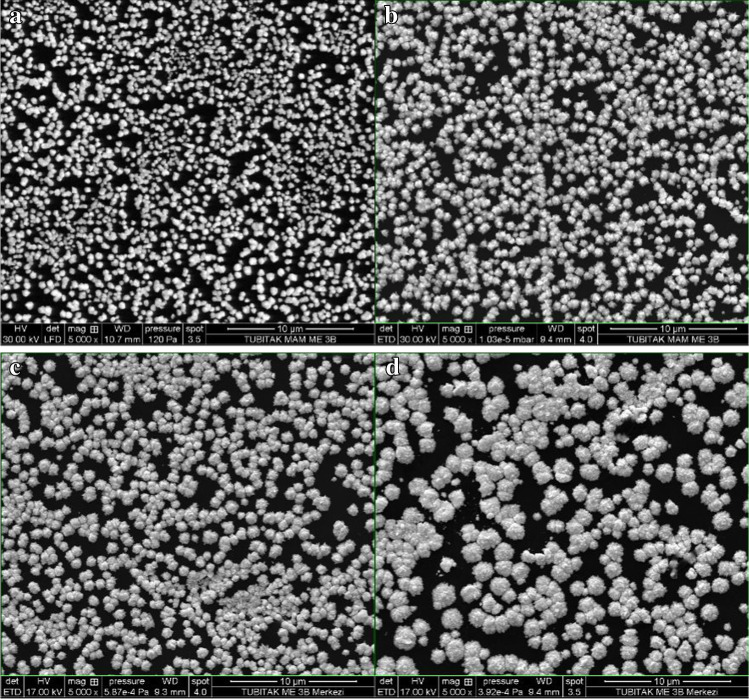
SEM images and EDX spectra for the electrodes (i.e. Au/GCE, CysOH/Au/GCE, S-gene/CysOH/Au/GCE and BSA/S-gene/CysOH/Au/GCE) were recorded to identify the surface state and the elemental composition, as shown in Fig. 3 and Fig. S1, respectively. With a surface of carbon and oxygen, GCEs are known to appear smooth in SEM images [3]. In our study, gold clusters formed on the surface of the GCE with roundish structures, and nearly half of the electrode surface was covered by Au clusters (51.60%), as illustrated in Fig. 3A and Fig. S1A, respectively. As immobilisation occurred on the surface of the gold clusters, the size of the clusters increased with each subsequent modification, as depicted in Fig. 3A–D, which resulted in lowering the Au content on the surface of the electrodes (Fig. S1A–D). Moreover, the colour of the gold clusters turned from white to grey, which we attributed to reduced conductivity. CysOH was chemisorbed from sulphur sites by gold clusters, and oxygen content increased from 0.74 to 4.56% in the EDX spectrum due to hydroxyl terminals on the surface of CysOH/Au/GCE (Fig. S1B). After S-gene had interacted with the hydroxyl terminals of CysOH, the amount of oxygen at the surface of CysOH/Au/GCE decreased from 4.56 to 2.11%, and the nitrogen peaks originating from S-gene appeared as shown in Fig. S1C. Upon BSA’s immobilisation on S-gene/CysOH/Au/GCE, the increasing amount of nitrogen and oxygen added to a trace amount of sulphur on the surface suggested that the protein-like structures had attached to the surface (Fig. S1D).
Fig. 3.
SEM images of a Au/GCE, b CysOH/Au/GCE, c S-gene/CysOH/Au/GCE and d BSA/S-gene/CysOH/Au/GCE (SEM analysis: 20 kV voltage, 4.0 spot value, ETD detector)
CV, SEM and EDX measurements, agreeing with each other and the literature [3, 69], demonstrated that the various electrode modifications had been effectively performed for the voltammetric determination of the SARS-CoV-2 spike antibody in synthetic and spiked-real samples.
Cyclic voltammetric characteristics of the system
The cyclic voltammetric behaviour of the biosensor and the added SARS-CoV-2 spike antibody were examined in 0.01 M (pH 7.5) of PBS solution. The oxidation peak of the biosensor at 0.85 V decreased with the addition of the SARS-CoV-2 spike antibody (Fig. S2), and the irreversibility of the oxidation reaction could have resulted from the partly negative sites on the biosensor’s surface after anodic scan and the repulsion forces between these negative sites and the phosphate buffer species (i.e. H2PO4− and HPO42−), hence the diminished electron transfer rate [3]. Because the cathodic peak at 0.35 V had not changed with the proportional amount of the SARS-CoV-2 spike antibody, that peak was not taken into account for determining the SARS-CoV-2 spike antibody.
Optimisation studies
Significant parameters affecting the biosensor’s performance, including the concentration of CysOH and S-gene, and the binding time of CysOH, S-gene, BSA and the SARS-CoV-2 spike antibody, were optimised to 20 mM, 10 μg/mL, 60 min, 45 min, 20 min and 30 min, respectively, in the presence of 1 fg/mL of the SARS-CoV-2 spike antibody (Fig. S3). The measurements were taken in 0.01 M (pH 7.5) of PBS solution because the pH of bodily fluids is generally neutral.
Method validation
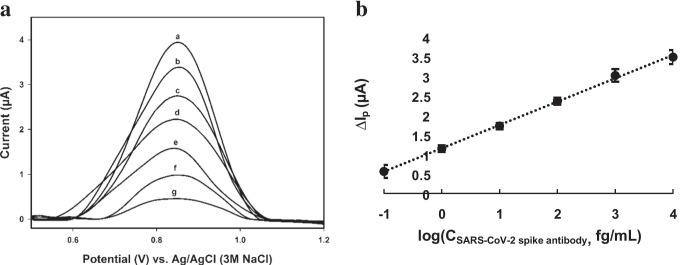
The electrochemical oxidation reaction of the biosensor and the signal’s decrease after the SARS-CoV-2 spike antibody was added are associated with the oxidation of heteroatoms, including hydroxyl, on the surface of the SARS-CoV-2 spike antigen and the formation of a large immuno-complex that hinders electron transfer, respectively [3, 67]. Figure 4 shows the related SWV voltammograms and calibration curve for determining the SARS-CoV-2 spike antibody. ΔIp, the y-axis of the calibration curve, was calculated by subtracting the signal of the added SARS-CoV-2 spike antibody from the signal of the produced biosensor, BSA/S-gene/CysOH/Au/GCE. The method has an LOD (i.e. from blank signal) of 0.03 fg/mL of the SARS-CoV-2 spike antibody and has an analytical range of 0.1 fg/mL to 10 pg/mL in 0.01 M (pH 7.5) of PBS solution.
Fig. 4.
(a) The obtained square wave voltammograms and (b) the calibration curve using BSA/S-gene/CysOH/Au/GCE in 0.01 M (pH 7.5) of PBS solution. (a) 0.01 M (pH 7.5) of PBS solution, (b) + 0.1 fg/mL, (c) + 1 fg/mL, (d) + 10 fg/mL, (e) + 100 fg/mL, (f) + 1000 fg/mL and (g) + 10 pg/mL of the SARS-CoV-2 spike antibody. n = 3 for each concentration
The MERS-CoV spike protein (i.e. M-S-gene), influenza A spike protein (i.e. InfA-S-gene) and the Streptococcus pneumoniae antigen (i.e. Pneu) were used to examine the produced biosensor’s selectivity by separately immobilising them on CysOH/Au/GCE and blocking them with BSA. The produced platforms were denoted as BSA/M-S-gene/CysOH/Au/GCE, BSA/InfA-S-gene/CysOH/Au/GCE and BSA/Pneu/CysOH/Au/GCE, respectively. As a consequence, the fabricated platforms showed no significant response to 1 fg/mL of the SARS-CoV-2 spike antibody (Fig. S4). The interference effects of various anions, enzymes and compounds that could be present in saliva were investigated in the presence of 1 fg/mL of the SARS-CoV-2 spike antibody with a criterion to mark a 5% variation in the peak height for evaluation (Table 1). Those results suggest the good selectivity of the proposed method.
Table 1.
The interference studies for the detection of the SARS-CoV-2 spike antibody using BSA/S-gene/CysOH/Au/GCE. Conditions: 1 fg/mL of the SARS-CoV-2 spike antibody, 0.01 M (pH 7.5) of PBS solution
| Interference | Tolerable amounta (unit/mL) | Tolerable ratiob |
|---|---|---|
| α-amylase | 200 | - |
| Lipase | 30 | - |
| Na+, K+, Mg2+ | - | 750 |
| Ca2+ | - | 600 |
| H2PO4−, HPO42−, HCO3− | - | 350 |
| Urea, ammonia | - | 250 |
aTolerable amount is the enzyme concentration directly in the assay medium. bTolerable ratio is how many times more than 1 fg/mL (SARS-CoV-2 spike antibody) of the interference does not change the peak height by more than 5%
RSD% values were calculated to be 7.55%, 3.79% and 5.23% for 1 fg/mL, 100 fg/mL and 10 pg/mL of the SARS-CoV-2 spike antibody, respectively. Those results indicate that the method also has good reproducibility. The stability and robustness studies were performed by storing the biosensor in an argon atmosphere by measuring the peak height at the end of six consecutive 5 day periods at 4 °C, 25 °C and 37 °C, respectively (Figs. S5 and S6). No significant difference emerged between the results when the sensor was stored at 4 °C versus 25 °C. By contrast, on day 30, the signal had preserved at least 84.5% of the signal from day 1 even when stored at 37 °C. Those results indicate the exceptional stability and robustness of BSA/S-gene/CysOH/Au/GCE as well.
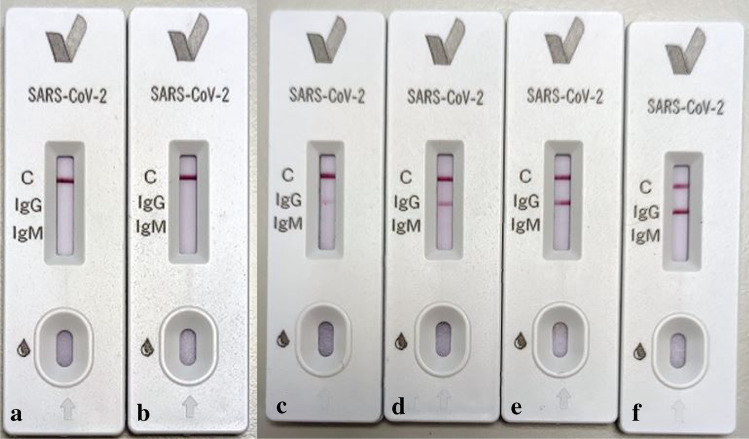
LFIA method was compared with the proposed method in terms of sensitivity with different amount of the SARS-CoV-2 spike antibody as shown in Fig. 5. It was crucial to indicate that the LFIA method responded as a faint line at 100 ng/mL of the SARS-CoV-2 spike antibody and gave a negative response at lower concentrations (i.e. 1 and 10 ng/mL). The results clearly depicted that the developed biosensor was 109 times more sensitive than the LFIA method. Since patient individuals may have different virus levels and therefore different antibody levels, it was concluded that the developed method could be successfully used after the onset of symptoms and early diagnosis of COVID-19.
Fig. 5.
Image obtained from LFIA cassettes at different concentration levels of the SARS-CoV-2 spike antibody. (a) 1 ng/mL, (b) 10 ng/mL, (c) 100 ng/mL, (d) 1 μg/mL, (e) 10 μg/mL and (f) 100 μg/mL of the SARS-CoV-2 spike antibody
Sample application
The method was used to determine the amount of the SARS-CoV-2 spike antibody in spiked-saliva and -oropharyngeal swab samples, and the recovery and relative standard deviation values obtained were 97.73% and 3.35% and 102.43% and 4.63%, respectively. The voltammograms and the results for those samples appear in Fig. S7, Fig. S8 and Table 2. The results thus show that the method additionally provides good accuracy. These results obtained with the external calibration also depict that the developed method is not affected by the sample matrix.
Table 2.
The results of spiked-saliva and -oropharyngeal swab samples (n = 6)
| Sample | Added amount of SARS-CoV-2 spike antibody (fg/mL) |
SARS-CoV-2 spike antibody found (mean ± standard deviation; fg/mL) |
Recovery (%) (mean ± standard deviation) |
Relative standard deviation (%) |
|---|---|---|---|---|
| Saliva | 1 | 0.98 ± 0.03 | 97.73 ± 3.27 | 3.35 |
| Oropharyngeal swab | 1 | 1.02 ± 0.05 | 102.43 ± 4.74 | 4.63 |
Conclusion
In our study, a rapid (approx. ~ 35 min), inexpensive, sensitive, selective biosensing platform was developed for the voltammetric determination of the SARS-CoV-2 spike antibody in spiked-saliva and -oropharyngeal swab samples. The developed biosensor has the shortest preparation time among prominent electrochemical biosensing methods based on antigen- or antibody-protein reported in the literature [3, 46–49, 51–56, 58–60] and an analysis time comparable to that of other antibody protein-based electrochemical methods [3, 46, 58], albeit far shorter than RT-PCR methods [16–23] as well as less expensive. Using saliva and oropharyngeal swab samples without any preprocessing instead of time-consuming blood and serum samples requiring different processes affords the proposed method relative simplicity. At the same time, the results of cross-reactivity and interference studies revealed its good selectivity, while the results of spiked-saliva and -oropharyngeal swab samples revealed its relative accuracy. In addition, the proposed method has much better sensitivity than LFIA-based antibody tests. Moving forward, BSA/S-gene/CysOH/Au/GCE could be easily fabricated and provided as a ready-to-use kit on a commercial scale by using disposable screen-printed electrodes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was performed in the Electrochemistry Laboratory of TUBITAK UME. The authors would like to thank Sevgi Gülyüz at TUBITAK MAM-Materials Institute for SEM and EDX measurements and Tanıl Kocagöz for supplying the LFIA kits.
Author contribution
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by L. Liv, M. Yener, G. Çoban and Ş. A. Can. The first draft of the manuscript was written by L. Liv.
Funding
This study was supported by G3EK-E1-04-I project of TUBITAK UME.
Data availability
Yes, our manuscript has data included as electronic supplementary material.
Code availability
SigmaPlot 12.0, MS Powerpoint 2013, MS Excel 2013.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent of publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Informed consent
Signed informed consent was obtained from the participants who gave the saliva and oropharyngeal swab samples.
Source of biological material
Saliva and oropharyngeal swab samples were collected from six healthy individuals with their own requests. The samples were anonymized.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liv L. Electrochemical immunosensor platform based on gold-clusters, cysteamine and glutaraldehyde modified electrode for diagnosing COVID-19. Microchem J. 2021;168:106445. doi: 10.1016/j.microc.2021.106445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.IMF (2021) Fiscal monitor database of country fiscal measures in response to the COVID-19 pandemic. https://www.imf.org/en/Topics/imf-and-covid19/Fiscal-Policies-Database-in-Response-to-COVID-19. Accessed 14 June 2021.
- 5.WHO (2021) WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/. Accessed 1 June 2021.
- 6.Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12:e7423. doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenji M, Katsushi K, Alexander Z, Gerardo C. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiura H, Kobayashi T, Miyama T, Suzuki A, Jung S, Hayashi K, Kinoshita R, Yang Y, Yuan B, Akhmetzhanov AR, et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int J Infect Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JH, Marks F, Clemens JD. Looking beyond COVID-19 vaccine phase 3 trials. Nat Med. 2021;27:205–211. doi: 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 14.Pokhrel P, Hu C, Mao H. Detecting the coronavirus (COVID-19) ACS Sensors. 2020;5:2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 16.Yang P, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virology. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH, Fung AY, Ng AC, Zou Z, Tsoi H, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–e320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SW, Cornforth DM, Dushoff J, Weitz JS. The time scale of asymptomatic transmission affects estimates of epidemic potential in the COVID-19 outbreak. Epidemics. 2020;31:1–18. doi: 10.1016/j.epidem.2020.100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Kasteren PB, van der Veer B, van den Brink S, Wijsman L, de Jonge J, van den Brandt A, Molenkamp R, Reusken CBEM, Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M, Chen S, Huang B, Zhong JM, Su H, Chen YJ, Cao Q, Ma L, He J, Li X, et al. Pathological findings in the testes of COVID-19 patients: clinical implications. Eur Urol Focus. 2020;6:1124–1129. doi: 10.1016/j.euf.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visseaux B, Le Hingrat Q, Collin G, Bouzid D, Lebourgeois S, Le Pluart D, Deconinck L, Lescure F, Lucet J, Bouadma L, et al. Evaluation of the QIAstat-Dx respiratory SARS-CoV-2 panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J Clin Microbiol. 2020;58:e00630–e720. doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams ER, Anand R, Andersson MI, Auckland K, Baillie JK, Barnes E, Beer S, Bell JI, Berry T, Bibi S, et al (2020) Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. medRxiv:04.15.20066407. 10.1101/2020.04.15.20066407
- 25.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, He S, Wang X, Yan Y, Liu J, Wu S, Liu S, Lei Y, Chen M, Li L, et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4:1150–1158. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- 27.Andryukov BG. Six decades of lateral flow immunoassay: from determining metabolic markers to diagnosing COVID-19. AIMS Microbiol. 2020;6:280–304. doi: 10.3934/microbiol.2020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C, Wen T, Shi FJ, Zeng XY, Jiao YJ. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng L, Li Y, Liu J, Guo L, Wang Z, Xu X, Song S, Hao C, Liu L, Xin M, et al. Rapid, ultrasensitive and highly specific biosensor for the diagnosis of SARS-CoV-2 in clinical blood samples. Mater Chem Front. 2020;4:2000–2005. doi: 10.1039/d0qm00294a. [DOI] [Google Scholar]
- 30.Chen Z, Zhang Z, Zhai X, Li Y, Lin L, Zhao H, Bian L, Li P, Yu L, Wu Y, et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Zhang Q, Hou P, Chen M, Hui W, Vermorken A, Luo Z, Li H, Li Q, Cui Y. Gold magnetic nanoparticle conjugate-based lateral flow assay for the detection of IgM class antibodies related to torch infections. Int J Mol Med. 2015;36:1319–1326. doi: 10.3892/ijmm.2015.2333. [DOI] [PubMed] [Google Scholar]
- 32.Wen T, Huang C, Shi FJ, Zeng XY, Lu T, Ding SN, Jiao YJ. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst. 2020;145:5345–5352. doi: 10.1039/D0AN00629G. [DOI] [PubMed] [Google Scholar]
- 33.Karakuş E, Erdemir E, Demirbilek N, Liv L. Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor. Anal Chim Acta. 2021;1182:338939. doi: 10.1016/j.aca.2021.338939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM, Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng Q, Wan W, Ma X, Liu J, Yang G. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull. 2020;65:1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z, Tian D, Liu Y, Lin Z, Lyon CJ, Lai W, Fusco D, Drouin A, Yin X, Hu T, et al. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chow FW, Chan TT, Tam TR, Zhao S, Yao W, Fung J, Cheng FK, Lo GC, Chu S, Aw-Yong KL, et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int J Mol Sci. 2020;21:5380. doi: 10.3390/ijms21155380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang M, Pan W, Arasthfer A, Fang W, Ling L, Fang H, Daneshnia F, Yu J, Liao W, Pei H, et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front Cell Infect Microbiol. 2020;10:331. doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan C, Cui J, Huang L, Du B, Chen L, Xue G, Li S, Zhang W, Zhao L, Sun Y, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infec. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang T, Wang YC, Shen CF, Cheng CM. Point-of-care RNA-based diagnostic device for COVID-19. Diagnostics. 2020;10:165. doi: 10.3390/diagnostics10030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan BE, Chong VCL, Chan SSW, Lim GH, Lim KGE, Tan GB, Mucheli SS, Kuperan P, Ong KH. Hematologic parameters in patients with COVID-19 infection. Am J Hematol. 2020;95:E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 42.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 44.Ahmadivand A, Gerislioglu B, Ramezani Z, Kaushik A, Manickam P, Ghoreishi SA. Functionalized terahertz plasmonic metasensors: femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens Bioelectron. 2021;177:112971. doi: 10.1016/j.bios.2021.112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alafeef M, Dighe K, Moitra P, Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabiani L, Saroglia M, Galatà G, De Santis R, Fillo S, Luca V, Faggioni G, D’Amore N, Regalbuto E, Salvatori P, et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens Bioelectron. 2020;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo G, Lee G, Kim MJ, Baek S, Choi M, Ku KB, Lee C, Jun S, Park D, Kim HG, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 48.Torrente-Rodríguez RM, Lukas H, Tu J, Min J, Yang Y, Xu C, Rossiter HB, Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1–18. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vadlamani BS, Uppal T, Verma SC, Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors. 2020;20:5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Liu F, Xie W, Zhou TC, OuYang J, Jin L, Li H, Zhao CY, Zhang L, Wei J, et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens Actuators B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahmati Z, Roushani M, Hosseini H, Choobin H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Microchim Acta. 2021;188:105. doi: 10.1007/s00604-021-04762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mojsoska B, Larsen S, Olsen DA, Madsen JS, Brandslund I, Alatraktchi FA. Rapid SARS-CoV-2 detection using electrochemical immunosensor. Sensors. 2021;21:390. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eissa S, Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal Chem. 2021;93:1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 54.Raziq A, Kidakova A, Boroznjak R, Reut J, Öpik A, Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mavrikou S, Moschopoulou G, Tsekouras V, Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahari S, Roberts A, Shahdeo D, Gandhi S (2020) eCovSens-Ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv:2020.04.24.059204. 10.1101/2020.04.24.059204
- 57.Tripathy S, Singh SG. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans Indian Natl Acad Eng. 2020;5:205–209. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rashed MZ, Kopechek JA, Priddy MC, Hamorsky KT, Palmer KE, Mittal N, Valdez J, Flynn J, Williams SJ. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashemi SA, Behbahan NGG, Bahrani S, Mousavi SM, Gholami A, Ramakrishna S, Firoozsani M, Moghadami M, Lankarani KB, Omidifar N. Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens Bioelectron. 2021;171:112731. doi: 10.1016/j.bios.2020.112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liv L, Çoban G, Nakiboğlu N, Kocagöz T. A rapid, ultrasensitive voltammetric biosensor for determining SARS-CoV-2 spike protein in real samples. Biosens Bioelectron. 2021;192:113497. doi: 10.1016/j.bios.2021.113497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Yao H, Xu X, Zhang P, Zhang M, Shao J, Xiao Y, Wang H. Limits of detection of 6 approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2) Clin Chem. 2020;66:977–979. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol. 2020;30:4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan MS, Misra SK, Dighe K, Wang Z, Schwartz-Duval AS, Sar D, Pan D. Electrically-receptive and thermally responsive paper-based sensor chip for rapid detection of bacterial cells. Biosens Bioelectron. 2018;110:132–140. doi: 10.1016/j.bios.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 66.Khan MS, Dighe K, Wang Z, Srivastava I, Schwartz-Duval AS, Misra SK, Pan D. Electrochemical-digital immunosensor with enhanced sensitivity for detecting human salivary glucocorticoid hormone. Analyst. 2019;144:1448–1457. doi: 10.1039/C8AN02085J. [DOI] [PubMed] [Google Scholar]
- 67.Liu F, Choi KS, Park TJ, Lee SY, Seo TS. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011;5:123–128. doi: 10.1007/s13206-011-5204-2. [DOI] [Google Scholar]
- 68.Wang R, Xue C, Gao M, Qi H, Zhang C. Ultratrace voltammetric method for the detection of DNA sequence related to human immunodeficiency virus type 1. Microchim Acta. 2011;172:291–297. doi: 10.1007/s00604-010-0490-2. [DOI] [Google Scholar]
- 69.Layqah LA, Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yes, our manuscript has data included as electronic supplementary material.
SigmaPlot 12.0, MS Powerpoint 2013, MS Excel 2013.