Summary
Blocking the activity of cytokines is an efficient strategy to combat inflammatory diseases. Interleukin-6 (IL-6) fulfills its pro-inflammatory properties via its soluble receptor (IL-6 trans-signaling). The selective trans-signaling inhibitor olamkicept (sgp130Fc) is currently in clinical development. We have previously shown that sgp130Fc can also efficiently block trans-signaling of the closely related cytokine IL-11, which elicits the question how selectivity for one of the two cytokines can be achieved. Using structural information, we show that the interfaces between IL-6R-gp130 and IL-11R-gp130, respectively, within the so-called site III are different between the two cytokines. Modification of an aromatic cluster around Q113 of gp130 within these interfaces allows the discrimination between IL-6 and IL-11 trans-signaling. Using recombinant sgp130Fc variants, we demonstrate that these differences can indeed be exploited to generate a truly selective IL-6 trans-signaling inhibitor. Our data highlight how the selectivity of a clinically relevant designer protein can be further improved.
Subject areas: Biochemistry, Medical biochemistry, Immunology, Cell biology
Graphical abstract

Highlights
-
•
The designer protein olamkicept/sgp130Fc is an IL-6 trans-signaling inhibitor
-
•
Our data show that sgp130Fc also efficiently blocks IL-11 trans-signaling
-
•
The signaling complexes of IL-6 and IL-11 differ in the so-called site III
-
•
Mutation of Q113 of sgp130Fc allows selectivity between the cytokines
Biochemistry; Medical biochemistry; Immunology; Cell biology
Introduction
Blocking the activity of pro-inflammatory cytokines is a well-established therapeutic strategy used to treat patients with several different inflammatory diseases. Examples are the blockade of tumor necrosis factor alpha (TNFα) in rheumatoid arthritis and psoriasis (Kalliolias and Ivashkiv, 2016) or the inhibition of interleukin-6 (IL-6) in Castleman disease, giant cell arteritis, systemic juvenile idiopathic arthritis, and also rheumatoid arthritis (Garbers et al., 2018; Jones and Jenkins, 2018). Selective inhibition of cytokines can either be achieved by antibodies against the cytokine and their receptors or by designer proteins based on soluble cytokine receptors (Lokau and Garbers, 2020).
IL-6 is a multifunctional cytokine with distinct pro- and anti-inflammatory properties (Kang et al., 2020). It can activate its target cells via the membrane-bound IL-6R (termed classic signaling) or via soluble forms of the IL-6R (sIL-6R, trans-signaling) (Rose-John, 2021). Both modes of signaling function via the formation of a gp130 homodimer, which is the signal-transducing receptor used by all IL-6 family cytokines (Garbers et al., 2012). The membrane-bound IL-6R shows an expression pattern that is restricted to hepatocytes and immune cells and thus limits the number of cells that can be activated by classic signaling, whereas IL-6 trans-signaling can activate in principle all human cells due to the ubiquitous expression of gp130 (Jones and Jenkins, 2018; Rose-John, 2018). The sIL-6R can be generated by alternative processing of the mRNA encoding the IL-6R (Lust et al., 1992). However, about 80% of the 20–80 ng/mL sIL-6R found in the human circulation originates from proteolytic cleavage of the membrane-bound IL-6R (Riethmueller et al., 2017).
Intriguingly, the trans-signaling pathway accounts for the pro-inflammatory properties of IL-6 (Garbers et al., 2018). To selectively block IL-6 trans-signaling without affecting IL-6 classic signaling, the designer protein sgp130Fc, later renamed olamkicept, has been constructed (Jostock et al., 2001). sgp130Fc consists of the extracellular part of gp130 fused to the Fc portion of a human IgG antibody. Because neither IL-6 nor the IL-6R alone have an affinity toward sgp130Fc, only the complex of IL-6/sIL-6R can be bound and thus neutralized by sgp130Fc (Jostock et al., 2001). The compound is currently undergoing clinical evaluation in patients with inflammatory bowel disease (IBD), and positive results from a prospective phase 2a trial comprising 16 patients with active IBD (Schreiber et al., 2021) and a phase 2 randomized, placebo-controlled trial in moderately to severely active ulcerative colitis with 91 treated patients (Chen et al., 2021) have recently been reported.
The cytokine that is most closely related to IL-6 is IL-11 (Garbers and Scheller, 2013). Like IL-6, IL-11 signals through a homodimer of gp130, but binds first to a unique membrane-bound IL-11R (classic signaling) that is expressed on several different cell types, e.g., in the lung, heart, intestine, spleen, and bone (Davidson et al., 1997; Putoczki and Ernst, 2010). In addition to classic signaling, IL-11 can also bind to and signal via soluble forms of the IL-11R (sIL-11R), a pathway that we have termed IL-11 trans-signaling and that is also controlled by proteolysis (Koch et al., 2021; Lokau et al., 2016, 2017a, 2017c; Sammel et al., 2019). Importantly, we have shown that sgp130Fc not only blocks IL-6 trans-signaling but also is a potent inhibitor of IL-11 trans-signaling (Lokau et al., 2016). This can be explained by the fact that both cytokines signal via a gp130 homodimer, whereas all other members of the IL-6 family use heterodimers of gp130 with another β-receptor for signaling and consequently are either not blocked by sgp130Fc at all or are blocked only at very high concentrations (Jostock et al., 2001; Scheller et al., 2005). How sgp130Fc could be modified to selectively only block IL-6 or IL-11 trans-signaling is not known so far.
A structure of an IL-6 signaling complex has been determined using X-ray crystallography (Boulanger et al., 2003; Chow et al., 2001), whereas no such complex exists for IL-11. Although it is thought that the two signaling complexes should be similar, the structures of IL-11 and the IL-11R alone suggest indeed structural differences from IL-6 (Metcalfe et al., 2020; Putoczki et al., 2014). In this study, we show that the interfaces between IL-6R-gp130 and IL-11R-gp130, within the so-called site III of the signaling complexes, are different between the two cytokines. We further demonstrate that modification of an aromatic cluster around Q113 of gp130 within this interface allows the discrimination between IL-6 and IL-11 trans-signaling and the generation of selective sgp130Fc variants.
Results
Modification of T102, Q113, and N114 does not disturb blockade of IL-6 trans-signaling by sgp130Fc
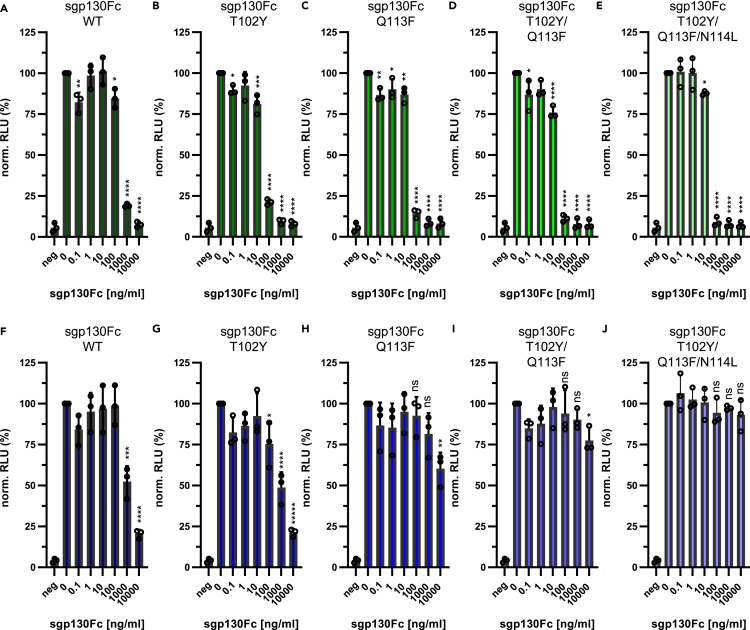
Olamkicept (sgp130Fc) blocks selectively the pro-inflammatory IL-6 trans-signaling (Figure 1A). To activate its target cells, IL-6 binds to its receptors via three distinct binding sites. The initial binding to the IL-6R is mediated through the so-called site I, whereas the subsequent binding to two gp130 molecules occurs via site II and site III (Figure 1B). A previous study showed that gp130 mutations within site II did not increase binding to IL-6/IL-6R, whereas certain mutations within site III increased affinity toward the cytokine (Tenhumberg et al., 2008). Based on a structure obtained by X-ray crystallography comprising domains 1–3 of gp130, IL-6 and domains 2–3 of the IL-6R (Boulanger et al., 2003; Chow et al., 2001), the key determinants appear to be two phenylalanine residues of the IL-6R (F153 and F187), which directly participate in the interface and are in close contact with gp130 residues, most prominently a glutamine residue (Q113, Figure 1C). Modification of gp130 via the introduction of another aromatic side chain into this cluster (Q113F) and further adjustment of neighboring residues (N114L and T102Y, Figure 1D) increased affinity to IL-6/IL-6R, resulting in sgp130Fc variants with increased efficacy (Tenhumberg et al., 2008).
Figure 1.
Modification of T102, Q113, and N114 does not disturb blockade of IL-6 trans-signaling by sgp130Fc
(A) Schematic overview of IL-6 classic and trans-signaling. IL-6 can either bind to the membrane-bound IL-6R (classic signaling) or to soluble forms of the IL-6R (trans-signaling). Both modes of signaling activate a gp130 homodimer on the target cell. sgp130Fc blocks IL-6 trans-signaling, but has no affinity to IL-6 alone. The individual domains of the receptors are shown as ellipses.
(B) Schematic overview of the IL-6 signaling complex. IL-6 has three distinct binding sites, designated as sites I, II, and III (indicated as Roman numerals in the figure panel). IL-6 binds via site I to the cytokine-binding module (CBM) of the IL-6R, which is located in domains D2 and D3. It further binds to the CBM of one gp130 molecule via site II and to the Ig-like domain of the other gp130 molecule via site III.
(C and D) Close-up of site III of (C) the IL-6/sIL-6R/sgp130Fc WT complex and (D) the IL-6/sIL-6R/sgp130Fc T102Y/Q113F/N114L complex based on the structure of the IL-6 complex (Boulanger et al., 2003; Chow et al., 2001). Amino acid residues important for the interaction are indicated (see main text for further details).
(E–I) Equal numbers of Ba/F3-gp130 cells were incubated with 5 nM Hyper-IL-6 and increasing amounts (0–10,000 ng/mL) of recombinant (E) sgp130Fc WT, (F) sgp130Fc T102Y, (G) sgp130Fc Q113F, (H) sgp130Fc T102Y/Q113F, and (I) sgp130Fc T102Y/Q113F/N114L. Cell viability was measured 48 h later. Each experiment was conducted with three technical replicates. Shown are the mean ± SD (normalized to cells treated only with Hyper-IL-6) from three independent experiments. RLU: relative light units. Experiments with Ba/F3-gp130 cells shown in this figure have been performed in parallel and therefore share the same negative control. Statistical analysis was performed using ordinary one-way ANOVA with Dunnett's multiple comparisons test (samples treated with sgp130Fc compared with the sample treated only with cytokine; ∗∗∗∗p < 0.0001).
See also Figure S1.
Because further insights into the selective modification of sgp130Fc are lacking, we first verified these previous results. For this purpose, we used Ba/F3-gp130 cells, a murine pro B cell line that proliferates in the presence of IL-6/sIL-6R and undergoes apoptosis without cytokine stimulation. We stimulated the cells with 5 nM Hyper-IL-6, which is a designer protein consisting of IL-6 fused to the sIL-6R that is capable of stimulating cells via gp130 and thus mimics IL-6 trans-signaling (Fischer et al., 1997). We then added increasing amounts (0–10,000 ng/mL) of sgp130Fc variants and determined cell viability 48 h later. In line with previous observations, sgp130Fc wild-type (WT) reduced proliferation in a dose-dependent manner (Figure 1E), and the same was true for the variants sgp130Fc T102Y (Figure 1F), sgp130Fc Q113F (Figure 1G), the double mutant sgp130Fc T102Y/Q113F (Figure 1H), and the triple mutant sgp130Fc T102Y/Q113F/N114L (Figure 1I). Because we already observed a major effect when introducing the Q113F mutation, we did not perform experiments with the N114L mutation in isolation, and therefore only analyzed N114L in the context of the triple mutant.
To not only rely on cell viability, we also determined the capacity of sgp130Fc WT and sgp130Fc T102Y/Q113F/N114L to suppress target gene induction in response to Hyper-IL-6. As shown in Figure S1A, both sgp130Fc variants successfully blocked mRNA expression of the IL-6 target gene SOCS3. In summary, these data confirm that modification of the site III interface within gp130 is possible and does not disturb blockade of IL-6 trans-signaling.
The IL-11R/gp130 interface at site III differs from the IL-6R/gp130 interface
We have previously shown that besides IL-6 trans-signaling, sgp130Fc can also efficiently block IL-11 trans-signaling via the sIL-11R (Lokau et al., 2016) (Figure 2A). It is therefore desirable to modify sgp130Fc in a way that allows the selective inhibition of either IL-6 or IL-11 trans-signaling. The IL-11 signaling complex is similar to the IL-6 complex and also characterized via three distinct bindings sites, with the only exception that IL-11 binds via site I to a unique, non-signaling IL-11R, whereas sites II and III mediate binding to the two gp130 molecules (Figure 2B). We generated a molecular model of the IL-11/IL-11R/gp130 complex based on the X-ray structure of the IL-6/IL-6R/gp130 complex and the structures of IL-11 and the IL-11R and inspected site III and especially the interface between IL-11R and gp130 (Figure 2C). Intriguingly, the IL-11R has no amino acid residues with aromatic side chains at the respective interface (Figure 2C), which is in considerable contrast to the IL-6R (Figure 1C). Accordingly, the gp130 mutations Q113F, N114L, and T102Y, which increase binding of sgp130Fc to the IL-6/IL-6R complex (Figure 1D), should not increase, but rather weaken the affinity to the respective sgp130Fc mutant (Figure 2D). A direct comparison of the IL-11/IL-11R and the IL-6/IL-6R complexes illustrates that the interface between gp130 and the cytokine receptors is strikingly different and might hold the key for the selective inhibition of one of the two trans-signaling pathways (Figure 2E).
Figure 2.
The IL-11R/gp130 interface at site III differs from the IL-6R/gp130 interface
(A) Schematic overview of IL-6 classic and trans-signaling (left) and IL-11 classic and trans-signaling (right). Both cytokines can either bind to their membrane-bound receptors (classic signaling) or to soluble forms of the receptors (trans-signaling). Both mode of signaling activate a gp130 homodimer on the target cell. sgp130Fc blocks IL-6 and IL-11 trans-signaling, but has no affinity to IL-6 or IL-11 alone. The individual domains of the receptors are shown as ellipses.
(B) Schematic overview of the IL-11 signaling complex. Like IL-6, IL-11 has three distinct binding sites, designated as sites I, II, and III (indicated as Roman numerals in the figure panel). IL-11 binds via site I to the cytokine-binding module (CBM) of the IL-11R, which is located in domains D2 and D3. It further binds to the CBM of one gp130 molecule via site II and to the Ig-like domain of the other gp130 molecule via site III.
(C and D) Close-up of site III of (C) the IL-11/sIL-11R/sgp130Fc WT complex and (D) the IL-11/sIL-11R/sgp130Fc T102Y/Q113F/N114L complex based on the structure of the IL-6 complex (Boulanger et al., 2003; Chow et al., 2001). Amino acid residues important for the interaction are indicated (see main text for further details).
(E) Overlay of the IL-6/sIL-6R/sgp130Fc WT (shown in gray, see Figure 1C for a colored version) and the IL-11/sIL-11R/sgp130Fc WT complexes (shown in the same color code as in Figure 2C) with labeling of the three residues in gp130. For labeling of all other important residues, see Figures 1C and 2C. Further details can be found in the main text and the methods section.
The Q113F mutation in sgp130Fc allows the discrimination between IL-11 and IL-6 trans-signaling
Based on our structural considerations (Figure 2), we next set out to test experimentally whether the sgp130Fc variants were indeed able to discriminate between IL-6 and IL-11 trans-signaling. For this, we used again Ba/F3-gp130 cells, but used sIL-6R + IL-6 and sIL-11R + IL-11 and not a fusion protein, as it mimics more closely the actual situation in vivo. Viability of Ba/F3-gp130 cells stimulated with 0.5 nM IL-6 and 5 nM sIL-6R was reduced in a dose-dependent manner by sgp130Fc WT (Figure 3A). Interestingly, sgp130Fc T102Y was more effective in blocking IL-6 trans-signaling compared with sgp130Fc WT, because already 100 ng/mL significantly reduced cell proliferation (Figure 3B), which was not seen for sgp130Fc WT (Figure 3A) and is in good agreement with the initial publication (Tenhumberg et al., 2008). Similarly, sgp130Fc Q113F (Figure 3C), sgp130Fc T102Y/Q113F (Figure 3D), and sgp130Fc T102Y/Q113F/N114L (Figure 3E) blocked IL-6-induced proliferation in a dose-dependent manner and were all more effective than sgp130Fc WT (Figure 3A).
Figure 3.
The Q113F mutation in sgp130Fc allows the discrimination between IL-11 and IL-6 trans-signaling
(A–E) Equal numbers of Ba/F3-gp130 cells were incubated with 0.5 nM IL-6 and 5 nM sIL-6R and increasing amounts (0–10,000 ng/mL) of recombinant (A) sgp130Fc WT, (B) sgp130Fc T102Y, (C) sgp130Fc Q113F, (D) sgp130Fc T102Y/Q113F, and (E) sgp130Fc T102Y/Q113F/N114L. Cell viability was measured 48 h later. (F–J) The experiments were performed as described for (A–E), but 0.5 nM IL-11 and 5 nM sIL-11R were used instead. Each experiment was conducted with three technical replicates. Data shown in all panels are the mean ± SD (normalized to cells treated only with cytokine and soluble receptor) from three independent experiments. RLU: relative light units. Experiments shown in (A–E) and (F–J) have been performed in parallel and therefore share the same negative control. Statistical analysis was performed using ordinary one-way ANOVA with Dunnett's multiple comparisons test (samples treated with sgp130Fc compared with the sample treated only with cytokine; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns: not significant).
See also Figure S2.
To directly compare inhibition of IL-6 and IL-11 trans-signaling by the different sgp130Fc variants, we next stimulated Ba/F3-gp130 cells with 0.5 nM IL-11 and 5 nM sIL-11R. As shown previously (Lokau et al., 2016), addition of sgp130Fc WT reduced viability in a dose-dependent manner (Figure 3F). We did not observe any difference compared with sgp130Fc WT when we used sgp130Fc T102Y (Figure 3G). However, whereas sgp130Fc Q113F showed enhanced capacity to block IL-6 trans-signaling (Figure 3C), inhibition of IL-11 trans-signaling was less effective (Figure 3H). This difference became even more apparent when the double mutant sgp130Fc T102Y/Q113F or the triple mutant sgp130Fc T102Y/Q113F/N114L were used, as the latter one showed no inhibition at all toward IL-11 trans-signaling (Figures 3I and 3J). When directly comparing the inhibitory capacity of the different sgp130Fc variants at a concentration of 10,000 ng/mL using two-way ANOVA, we detected no differences between all sgp130Fc variants with regard to IL-6 trans-signaling. Furthermore, there was no difference for sgp130Fc WT and sgp130Fc T102Y when inhibiting IL-11 trans-signaling, underlining that both proteins were efficient inhibitors of IL-11 trans-signaling. In contrast, inhibition of IL-11 trans-signaling by sgp130Fc Q113F, sgp130Fc T102Y/Q113F, and the triple mutant sgp130Fc T102Y/Q113F/N114L was significantly different from the other conditions variants (p < 0.001 for all three proteins), further confirming our notion that these sgp130Fc variants do not potently inhibit IL-11 trans-signaling.
We then verified our results on the level of signaling and transcription. For this, we stimulated Ba/F3-gp130 cells with the same amounts of recombinant proteins and determined phosphorylation of the transcription factor STAT3 via western blot. As shown in Figure S2A, all five sgp130Fc variants were able to block IL-6 trans-signaling-induced STAT3 phosphorylation. In contrast, only sgp130Fc WT and T102Y, but not the other three sgp130Fc variants, blocked STAT3 phosphorylation induced by IL-11 trans-signaling. Furthermore, whereas both sgp130Fc WT and sgp130Fc T102Y/Q113F/N114L blocked transcription of the STAT3 target gene SOCS3 after stimulation via IL-6 trans-signaling, only sgp130Fc WT but not sgp130Fc T102Y/Q113F/N114L blocked SOCS3 induction after IL-11 trans-signaling (Figure S2B). In conclusion, our data reveal that sgp130Fc variants containing the Q113F mutation are efficient inhibitors of IL-6 trans-signaling, but do not block IL-11 trans-signaling, thus allowing the selective blockade of one of the trans-signaling pathways.
Soluble receptors generated by proteolytic cleavage are differentially blocked by sgp130Fc variants
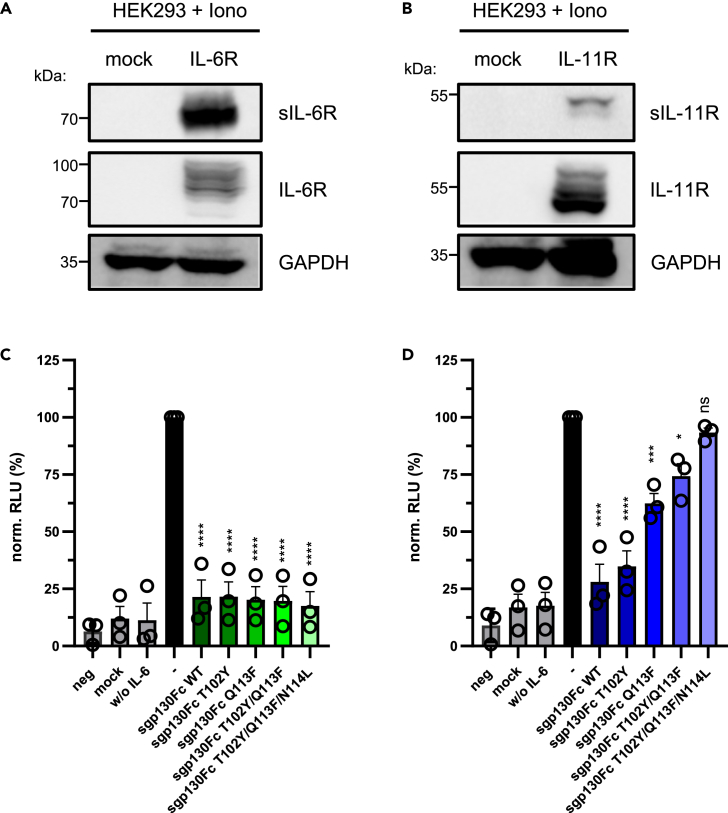
More than 80% of the sIL-6R in humans are generated by proteolytic cleavage, and one of the proteases capable of cleaving the IL-6R is ADAM10 (Riethmueller et al., 2017). Furthermore, we have previously shown that ADAM10-mediated cleavage of the IL-11R initiated IL-11 trans-signaling (Lokau et al., 2016). Therefore, we analyzed whether the observed differences in trans-signaling inhibition would also hold true for soluble receptors generated by proteolytic cleavage. For this, we transiently transfected HEK293 cells with expression plasmids encoding either IL-6R or IL-11R, activated ADAM10, and monitored proteolytic cleavage and thus release of sIL-6R and sIL-11R into the cell culture supernatant (Figures 4A and 4B). Afterward, we added IL-6 or IL-11 and the different sgp130Fc variants to the supernatant and incubated Ba/F3-gp130 cells with the mixture for 48 h. In agreement with our previous results, all sgp130Fc variants were able to block IL-6 trans-signaling (Figure 4C). Likewise, whereas sgp130Fc WT blocked IL-11 trans-signaling also, all sgp130Fc variants containing Q113F were not able to fully suppress IL-11 trans-signaling, and especially the triple mutant sgp130Fc T102Y/Q113F/N114L did not block IL-11 trans-signaling induced by a proteolysis-derived sIL-11R (Figure 4D). Thus, the identified selectivity of the sgp130Fc triple mutant also holds true for proteolysis-dependent trans-signaling events.
Figure 4.
Soluble receptors generated by proteolytic cleavage are differentially blocked by sgp130Fc variants
(A and B) HEK293 cells were transiently transfected with expression plasmids encoding either myc-tagged IL-6R or IL-11R or mock control. 48 h post-transfection, medium was replaced by new serum-free medium and cells were stimulated for 60 min with 1 μM of the ionophore ionomycin. Proteins in the cell supernatant were precipitated by trichloracetic acid and visualized by western blot. Cells were lysed and expressed receptors as well as GAPDH as loading control detected via western blot. One experiment out of three performed is shown.
(C) Equal numbers of Ba/F3-gp130 cells were incubated with conditioned supernatant derived from the experiment shown in (A) and with 5 nM IL-6. 10 μg/mL of the different recombinant sgp130Fc variants was added as indicated. Cell viability was measured 48 h later.
(D) The experiment was performed as described in (C), but conditioned supernatant derived from the experiment shown in (B) and 5 nM IL-11 were used. The experiments using Ba/F3-gp130 cells were conducted with three technical replicates. Shown in (C) and (D) are the mean ± SD (normalized to cells treated only with cytokine and soluble receptor) from three independent experiments. RLU: relative light units. Statistical analysis was performed using ordinary one-way ANOVA with Dunnett's multiple comparisons test (samples treated with sgp130Fc compared with the sample treated only with cytokine; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns: not significant).
Discussion
The inhibition of pro-inflammatory cytokines is a well-established therapeutic strategy to combat inflammatory diseases, including rheumatoid arthritis and IBD. Consequently, antibodies and designer proteins targeting these cytokines are among the top-selling biopharmaceutical products (Walsh, 2018). Of note, further improvement of therapeutics is warranted, not only to reduce adverse effects but also to move further in the direction of precision medicine (Dugger et al., 2018).
IL-6 has been identified more than three decades ago, and the monoclonal antibody tocilizumab that blocks the IL-6R is approved in more than 100 countries worldwide (Kishimoto, 2010). The notion that the activities of IL-6 can be divided into classic and trans-signaling and that IL-6 trans-signaling is responsible for its pro-inflammatory properties led to the development of sgp130Fc/olamkicept, which selectively blocks trans-signaling (Jones et al., 2011). This is believed to be superior compared with the global blockade of IL-6, which compromises the hepatic acute-phase response and results in bacterial infections in patients treated with tocilizumab (Lang et al., 2012), whereas treatment with sgp130Fc does not influence the innate immune response against pathogens (Hoge et al., 2013). Indeed, first clinical trials in patients with IBD reported encouraging results (Chen et al., 2021; Schreiber et al., 2021).
IL-11 is the only other cytokine that signals through a homodimer of gp130, and consequently we have shown that IL-11 trans-signaling via the IL-11/sIL-11R complex is also blocked by sgp130Fc (Lokau et al., 2016). Although the exact roles of IL-11 trans-signaling in vivo have not been elucidated so far, it is without doubt that injection of sgp130Fc in human patients and in mice blocks them, and that some of the reported beneficial activities of sgp130Fc might be due to reduced IL-11 trans-signaling. In this study, we identify a region within the interfaces of the cytokine/receptor complexes that is different between IL-6 and IL-11. We find that an aromatic cluster around Q113 of gp130 holds the key to discriminate between IL-6 and IL-11 trans-signaling and that this knowledge can indeed be used to modify sgp130Fc in a way that it does not interfere with IL-11 trans-signaling. Our results are in good agreement with a very recent study that also analyzed the T102Y/Q113F/N114L mutations (Heise et al., 2021). Importantly, the mutation Q113F has a stronger effect than the T102Y mutation in isolation, whereas the combination of both already largely prevents the inhibition of IL-11 trans-signaling. The best discrimination between IL-6 and IL-11 trans-signaling, however, was achieved with the triple mutant T102Y/Q113F/N114L. We did not analyze the N114L mutation in isolation, because its localization at the border of the interface suggests that it would not play a major role in this context. Indeed, the difference between sgp130Fc T102Y/Q113F and sgp130Fc T102Y/Q113F/N114L was rather mild, showing that the major amino acid residue that is responsible for the discriminatory effect is Q113. However, we cannot rule out that N114L in isolation would also show a discriminatory effect with regard to the inhibition of IL-11 trans-signaling. Interestingly, the gp130 mutation R281Q mutation in a patient with craniosynostosis has been described, which leads to a selective loss of IL-11 signaling (Schwerd et al., 2020). It is tempting to speculate that the introduction of the mutations described in this study into membrane-bound gp130 would result in a similar effect.
The identification of a truly selective IL-6 trans-signaling inhibitor that has no affinity toward IL-11/sIL-11R will not only help to uncover the physiological and pathophysiological roles of IL-11 trans-signaling in vivo but also will pave the way to design selective IL-11 trans-signaling inhibitors that do not block IL-6 trans-signaling. These insights can also be used to generate designer proteins that selectively and with high affinity bind and block other pro-inflammatory cytokines, which represent potent next-generation therapeutics to treat inflammatory diseases.
Limitations of the study
Our results show how the selectivity of the therapeutically relevant designer protein sgp130Fc can be further improved. Although our in vitro data conclusively demonstrate that IL-6 trans-signaling, but not IL-11 trans-signaling, is blocked by the mutated sgp130Fc proteins, a confirmation in vivo has to be achieved in further studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-myc | Cell Signaling | Clone 71D10, Cat #2278 |
| Anti-GAPDH | Cell Signaling | Clone 14C10, Cat #2118 |
| Anti-pSTAT3 (pTyr705) | Cell Signaling | Clone D3A7, Cat#9145 |
| Anti-STAT3 | Cell Signaling | Clone 124H6, Cat#9139 |
| Mouse-POD | Cell Signaling | Cat#7076 |
| Rabbit-POD | Cell Signaling | Cat#7074 |
| Bacterial and virus strains | ||
| E. coli DH5α | Invitrogen | Cat#18265-017 |
| Chemicals, peptides, and recombinant proteins | ||
| HyperIL-6 | (Fischer et al., 1997) | N/A |
| IL-6 | (Mackiewicz et al., 1992) | N/A |
| IL-11 | (Lokau et al., 2017b) | N/A |
| sgp130Fc WT | (Jostock et al., 2001) | N/A |
| sgp130Fc T102Y | Conaris Research AG | N/A |
| sgp130Fc Q113F | Conaris Research AG | N/A |
| sgp130Fc T102Y/Q113F | Conaris Research AG | N/A |
| sgp130Fc T102Y/Q113F/N114L | Conaris Research AG | N/A |
| sIL-6R | R&D Systems | Cat# 227-SR-025/CF |
| sIL-11R | R&D Systems | Cat# 8895-MR-050 |
| Turbofect | Thermo Fisher Scientific | Cat# R0532 |
| Ionomycin | Thermo Fisher Scientific | Cat# I24222 |
| Tris-HCl | Carl Roth | Cat# 9090.3 |
| Tris Base | Carl Roth | Cat# AE15.1 |
| NaCl | Carl Roth | Cat# 9357.2 |
| Methanol | Carl Roth | Cat# 4627.5 |
| Tween-20 | Carl Roth | Cat# 9127.1 |
| TEMED | Carl Roth | Cat# 2367.1 |
| Protease Inhibitor Cocktail | Sigma Aldrich | Cat# P8340 |
| Acrylamide (Rotiphorese Gel 30) | Carl Roth | Cat# 3029.1 |
| bovine serum albumin | Carl Roth | Cat# T844.3 |
| Trichloracetic acid | Carl Roth | Cat# 8789.1 |
| SDS | Carl Roth | Cat# 2326.2 |
| β-mercaptoethanol | Carl Roth | Cat# 4227.3 |
| EDTA | Carl Roth | Cat# 8043.2 |
| Glycin | Carl Roth | Cat# 3908.3 |
| Critical commercial assays | ||
| Cell Titer Blue Cell Viability Assay | Promega | Cat# G8080 |
| SYBR Green Mastermix | Thermo Fisher Scientific | Cat# 4309155 |
| Experimental models: Cell lines | ||
| Ba/F3-gp130 | (Gearing et al., 1994) | N/A |
| HEK293 | ATCC | RRID:CVCL_0045 |
| Oligonucleotides | ||
| mSOCS3 F GCTCCAAAAGCGAGTACCAGC |
Sigma-Aldrich | N/A |
| mSOCS3 R AGTAGAATCCGCTCTCCTGCAG |
Sigma-Aldrich | N/A |
| mGAPDH F AAGGTCATCCCAGAGCTGAA |
Sigma-Aldrich | N/A |
| mGAPDH R CTGCTTCACCACCTTCTTGA |
Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | ThermoFisher Scientific | N/A |
| pcDNA3.1-Myc-hIL-6R | (Lokau et al., 2017c) | NA |
| pcDNA3.1-Myc-hIL-11R | (Lokau et al., 2017c) | N/A |
| Software and algorithms | ||
| UCSF Chimera 1.13.1 | (Pettersen et al., 2004) | RRID:SCR_004097 |
| Image Studio Lite Ver 5.2 | Li-COR Biosciences | RRID:SCR_013715 |
| GraphPad Prism 8 | GraphPad Software Inc | RRID:SCR_002798 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christoph Garbers (christoph.garbers@med.ovgu.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cultivation of Ba/F3-gp130 and HEK293 cells
The cells were cultured in DMEM supplemented with 10% FCS, 60 mg/L penicillin, 100 mg/mL streptomycin in a standard incubator at 37°C with 5% CO2 and a water-saturated atmosphere. Ba/F3-gp130 cells were additionally cultured with 10 ng/mL Hyper-IL-6. The sex of HEK293 cells is female, the sex of Ba/F3 cells is unknown. The cell lines were obtained from official repositories, and therefore no additional cell authentication was performed.
Method details
Production of recombinant proteins
Hyper-IL-6, a fusion protein consisting of IL-6 and the sIL-6R, IL-6, and IL-11 have been produced in house as described previously (Fischer et al., 1997; Lokau et al., 2017b; Schroers et al., 2005).
Molecular modeling of the cytokine/cytokine receptor complexes
The crystal structure of the IL-6/IL-6R/gp130 complex (pdb: 1P9M) (Boulanger et al., 2003) served as template for the model of the IL-11/IL-11R/gp130 complex. The crystal structures of IL-11 and IL-11R (Metcalfe et al., 2020) were superimposed by a least squares fit algorithm onto the IL-6 and IL-6R structure in the IL-6/IL-6R/gp130 complex, respectively. Only regular secondary structure elements were considered for the superposition by the fitting algorithm. Please see also the procedure described in (Tenhumberg et al., 2008). Visualization of the structures and in silico point mutations were performed with UCSF chimera (Pettersen et al., 2004).
All amino acids residues in this manuscript are numbered according to the full length proteins including their signal peptides. In contrast, the sequences in the pdb files which form the basis of the molecular models of the cytokine complexes do not include the signal peptides and thus have a different numbering. These numbers correspond to the structures as follows:
gp130: T102 = T80, Q113 = Q91, N114 = N92; IL-6R: F153 = F134, F187 = F168; IL-6: W184 = W157; IL-11R: L154 = L132, D209 = D187; IL-11: W168 = W168. Please note that L154 of the IL-11R is not resolved in the respective structure (pdb: 6O4P).
Viability of Ba/F3-gp130 cells and detection of STAT3 phosphorylation
To analyze the response of Ba/F3-gp130 cells to the different trans-signaling pathways, 5000 cells per well were seeded into 96 well plates. The cells were treated with 5nM Hyper-IL-6, 0.5 nM IL-6 and 5 nM sIL-6R, or 0.5 nM IL-11 and 5 nM sIL-11R in the presence of increasing amounts of the sgp130Fc variants (0–10,000 ng/mL) for 48 h. Alternatively, cells were treated with 50% conditioned supernatant from ionomycin treated HEK cells and 5 nM IL-6 or IL-11 for 48 h. The Viability of the cells was assessed using the CellTiter Blue Cell Viability Assay (Promega, Karlsruhe, Germany) according to manufacturers' instructions. Fluorescence intensity was measured on a CLARIOstar plate reader (BMG Labtech, Ortenberg, Germany) and normalized to the value measured at the starting point. Furthermore, data were normalized to the viability without addition of sgp130Fc in order to be able to compare different trans-signaling inducers. All measurements were performed in triplicates.
To determine STAT3 phosphorylation, Ba/F3-gp130 cells were washed 3 times with PBS and then serum starved for 2.5 h. 5 nM sIL6R/sIL-11R and 0.5 nM IL-6/IL-11 were incubated with 1 μg/mL sgp130Fc variant for 20 min. Then, cells were stimulated with that mixture for 15 min, collected by centrifugation and boiled in 2.5x reducing Lämmli-buffer before being subjected to SDS-PAGE and western blotting.
Shedding assay in HEK293 cells, cell lysis and precipitation of supernatants
HEK293 cells were transfected with expression plasmids encoding myc-tagged IL-6R or IL-11R, or a mock control. 48 h after transfection, cells were stimulated with 1 μM ionomycin for 1 h in serum free medium. Cells and supernatant were harvested and the supernatants were filtered through a 0.2 μm filter. The proteins in the supernatant were precipitated using trichloracetic acid (TCA). For that, supernatants were mixed with equal amounts of 20% TCA and incubated for 20 min on ice. The precipitated proteins were collected by centrifugation at 18,000 x G for 20 min at 4°C and then washed with 350 μL ice-cold acetone for 20 min on ice. After a final centrifugation step at 18,000 x G for 20 min at 4°C the acetone was removed and the precipitate allowed to dry at room temperature over night. Finally, the pellets were boiled in 2.5 x reducing Lämmli buffer and analyzed by western blot.
Cells were lysed in lysis buffer [10 mM Tris-HCl pH 7.5, 150 mM NaCl, 50 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, USA) and analyzed via western blot.
Western blot
Cell lysates and precipitated supernatants were separated by SDS-PAGE and transferred either onto a nitrocellulose membrane (for detection of (p)STAT3) or on a PVDF membrane. Membranes were blocked with 5% BSA in TBS (10 mm Tris-HCl, pH 7.6, 150 mm NaCl) for 1 h at room temperature. Afterward, membranes were probed with primary antibodies at 4°C over night. Then, the membranes were washed 3 times with TBST-T (10 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.05% Tween) and incubated with appropriate secondary antibodies either conjugated to IRDye (for detection of (p)STAT3) or horseradish peroxidase for 1 h at room temperature. The membranes were again washed once with TBS-T and 3 times with TBS and the signals were detected using the ChemiDocMP (Bio-Rad, Hercules, USA). For detection of horseradish peroxidase signals, the ECL Prime Western blotting detection reagent (GE Healthcare, Chicago, USA) was used according to the manufacturer's instructions. For detection of the loading control GAPDH the membrane was cut horizontally with a scalpel at the appropriate molecular weight in order to allow detection of multiple proteins.
RNA isolation and quantitative PCR
Ba/F3-gp130 cells were washed 3 times with PBS and serum starved for 2.5 h. 5 nM Hyper-IL-6 or 5nM sIL-6R/sIL-11R plus 5 nM IL-6/IL-11 were incubated with 1 μg/mL sgp130Fc variant for 20 min. Then, the cells were stimulated with the mixture for 1 h. Cells were harvested and the RNA was isolated using the NuceloSpin RNA Kit (Macherey-Nagel, Düren, Germany) according to manufacturers' instructions. The RNA concentration was measured on a ScanDrop2 (Analytik Jena, Jena, Germany) and 2 μg RNA were reversely transcribed into cDNA to a final volume of 20 μL using 5 μM Oligo-dT primer and the RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, USA) according to manufacturers' instructions. For quantitative PCR, cDNA transcribed from 50ng RNA was analyzed using the SYBR Green Mastermix (Thermo Fisher Scientific, Waltham, USA) in a total volume of 10μL according to manufacturers' instructions. The PCR was performed on a QuantStudio 6 (Thermo Fisher Scientific, Waltham, USA). The samples were initially incubated at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The measurements were performed in triplicates. SOCS3 expression was normalized to GAPDH and the relative gene expression levels were calculated using the 2−ΔΔCt method.
Quantification and statistical analysis
Quantification
Densiometric analysis of western blot images was performed using the Image Studio Lite 5.2 software (LI-COR Biosciences, Lincoln, USA). Data are presented as ratio of pSTAT/STAT3 signal intensity.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). In order to determine the inhibition capacity of the individual sgp130Fc variants, ordinary one-way ANOVA with Dunnett's multiple comparisons test was used (samples treated with sgp130Fc compared to the sample treated only with cytokine). Further details about the values that were considered significant are given in the respective figure legends. In order to compare the different sgp130Fc variants with each other at a fixed concentration, we used a two-way ANOVA with multiple comparisons combined with the Tukey post-hoc test.
Acknowledgments
This work was funded by grants from the Deutsche Forschungsgemeinschaft, Bonn, Germany (SFB877 projects A10 and A14 to C.G.).
Author contributions
C.G. conceived the project and designed the experiments. J.L. performed the experiments and analyzed the data. Y.G. advised on and performed the statistical analysis. J.G. helped with molecular modeling. C.G. wrote the manuscript. All other authors contributed to the final version of the manuscript.
Declaration of interests
C.G. has received a research grant from Corvidia Therapeutics (Waltham, MA, USA) and has acted as a consultant for AbbVie. All other authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103309.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Boulanger M.J., Chow D.C., Brevnova E.E., Garcia K.C. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- Chen B., Zhang S., Wang B., Chen H., Li Y., Cao Q., Zhong J., Xie M., Ran Z., Tang T. DOP01 Efficacy and safety of the IL-6 trans-signalling inhibitor olamkicept: a phase 2 randomized, placebo-controlled trial in moderately to severely active Ulcerative Colitis. J. Crohn's Colitis. 2021;15:S041–S042. [Google Scholar]
- Chow D., He X., Snow A.L., Rose-John S., Garcia K.C. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–2155. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- Davidson A.J., Freeman S.A., Crosier K.E., Wood C.R., Crosier P.S. Expression of murine interleukin 11 and its receptor alpha-chain in adult and embryonic tissues. Stem Cells. 1997;15:119–124. doi: 10.1002/stem.150119. [DOI] [PubMed] [Google Scholar]
- Dugger S.A., Platt A., Goldstein D.B. Drug development in the era of precision medicine. Nat. Rev. Drug Discov. 2018;17:183–196. doi: 10.1038/nrd.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Goldschmitt J., Peschel C., Brakenhoff J.P., Kallen K.J., Wollmer A., Grötzinger J., Rose-John S. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 1997;15:142–145. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- Garbers C., Heink S., Korn T., Rose-John S. Interleukin-6: designing specific therapeutics for a complex cytokine. Nat. Rev. Drug Discov. 2018;17:395–412. doi: 10.1038/nrd.2018.45. [DOI] [PubMed] [Google Scholar]
- Garbers C., Hermanns H., Schaper F., Müller-Newen G., Grötzinger J., Rose-John S., Scheller J. Plasticity and cross-talk of Interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012;23:85–97. doi: 10.1016/j.cytogfr.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Garbers C., Scheller J. Interleukin-6 and interleukin-11: same same but different. Biol. Chem. 2013;394:1145–1161. doi: 10.1515/hsz-2013-0166. [DOI] [PubMed] [Google Scholar]
- Gearing A.J., Beckett P., Christodoulou M., Churchill M., Clements J., Davidson A.H., Drummond A.H., Galloway W.A., Gilbert R., Gordon J.L. Processing of tumour necrosis factor-alpha precursor by metalloproteinases. Nature. 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- Heise D., Derrac Soria A., Hansen S., Dambietz C., Akbarzadeh M., Berg A.F., Waetzig G.H., Jones S.A., Dvorsky R., Ahmadian M.R. Selective inhibition of IL-6 trans-signaling by a miniaturized, optimized chimeric soluble gp130 inhibits TH17 cell expansion. Sci. Signal. 2021;14 doi: 10.1126/scisignal.abc3480. [DOI] [PubMed] [Google Scholar]
- Hoge J., Yan I., Jänner N., Schumacher V., Chalaris A., Steinmetz O.M., Engel D.R., Scheller J., Rose-John S., Mittrücker H.-W.W. IL-6 controls the innate immune response against Listeria monocytogenes via classical IL-6 signaling. J. Immunol. 2013;190:703–711. doi: 10.4049/jimmunol.1201044. [DOI] [PubMed] [Google Scholar]
- Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018;18:773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- Jones S.A., Scheller J., Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 2011;121:3375–3383. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostock T., Müllberg J., Özbek S., Atreya R., Blinn G., Voltz N., Fischer M., Neurath M.F., Rose-John S. Soluble gp130 is the natural inhibitor of soluble IL-6R transsignaling responses. Eur. J. Biochem. 2001;268:160–167. doi: 10.1046/j.1432-1327.2001.01867.x. [DOI] [PubMed] [Google Scholar]
- Kalliolias G.D., Ivashkiv L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016;12:49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Narazaki M., Metwally H., Kishimoto T. Historical overview of the interleukin-6 family cytokine. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T. IL-6: from its discovery to clinical applications. Int. Immunol. 2010;22:347–352. doi: 10.1093/intimm/dxq030. [DOI] [PubMed] [Google Scholar]
- Koch L., Kespohl B., Agthe M., Schumertl T., Düsterhöft S., Lemberg M.K., Lokau J., Garbers C. Interleukin-11 (IL-11) receptor cleavage by the rhomboid protease RHBDL2 induces IL-11 trans-signaling. FASEB J. 2021;35 doi: 10.1096/fj.202002087R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V.R., Englbrecht M., Rech J., Nusslein H., Manger K., Schuch F., Tony H.P., Fleck M., Manger B., Schett G. Risk of infections in rheumatoid arthritis patients treated with tocilizumab. Rheumatology. 2012;51:852–857. doi: 10.1093/rheumatology/ker223. [DOI] [PubMed] [Google Scholar]
- Lokau J., Agthe M., Flynn C.M., Garbers C. Proteolytic control of interleukin-11 and interleukin-6 biology. Biochim. Biophys. Acta. 2017;1864:2105–2117. doi: 10.1016/j.bbamcr.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Lokau J., Garbers C. Biological functions and therapeutic opportunities of soluble cytokine receptors. Cytokine Growth Factor Rev. 2020;55:94–108. doi: 10.1016/j.cytogfr.2020.04.003. [DOI] [PubMed] [Google Scholar]
- Lokau J., Göttert S., Arnold P., Düsterhöft S., Massa Lopez D., Grötzinger J., Garbers C. The SNP rs4252548 (R112H) which is associated with reduced human height compromises the stability of IL-11. Biochim. Biophys. Acta. 2017;1865:496–506. doi: 10.1016/j.bbamcr.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Lokau J., Nitz R., Agthe M., Monhasery N., Aparicio-Siegmund S., Schumacher N., Wolf J., Möller-Hackbarth K., Waetzig G.H., Grötzinger J. Proteolytic cleavage governs interleukin-11 trans-signaling. Cell Rep. 2016;14:1761–1773. doi: 10.1016/j.celrep.2016.01.053. [DOI] [PubMed] [Google Scholar]
- Lokau J., Wandel M., Garbers C. Enhancing interleukin-6 and interleukin-11 receptor cleavage. Int. J. Biochem. Cell Biol. 2017;85:6–14. doi: 10.1016/j.biocel.2017.01.014. [DOI] [PubMed] [Google Scholar]
- Lust J., Donovan K., Kline M., Greipp P., Kyle R., Maihle N. Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine. 1992;4:96–100. doi: 10.1016/1043-4666(92)90043-q. [DOI] [PubMed] [Google Scholar]
- Mackiewicz A., Schooltink H., Heinrich P.C., Rose-John S. Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J. Immunol. 1992;149:2021–2027. Baltimore, Md : 1950. [PubMed] [Google Scholar]
- Metcalfe R.D., Aizel K., Zlatic C.O., Nguyen P.M., Morton C.J., Lio D.S., Cheng H.C., Dobson R.C.J., Parker M.W., Gooley P.R. The structure of the extracellular domains of human interleukin 11alpha receptor reveals mechanisms of cytokine engagement. J. Biol. Chem. 2020;295:8285–8301. doi: 10.1074/jbc.RA119.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Putoczki T., Ernst M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J. Leukoc. Biol. 2010;88:1109–1117. doi: 10.1189/jlb.0410226. [DOI] [PubMed] [Google Scholar]
- Putoczki T.L., Dobson R.C.J., Griffin M.D.W. The structure of human interleukin-11 reveals receptor-binding site features and structural differences from interleukin-6. Acta Crystallogr. D. Biol. Crystallogr. 2014;70:2277–2285. doi: 10.1107/S1399004714012267. [DOI] [PubMed] [Google Scholar]
- Riethmueller S., Somasundaram P., Ehlers J.C., Hung C.-W.W., Flynn C.M., Lokau J., Agthe M., Düsterhöft S., Zhu Y., Grötzinger J. Proteolytic origin of the soluble human IL-6R in vivo and a decisive role of N-glycosylation. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-John S. Interleukin-6 family cytokines. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028415. a028415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-John S. Therapeutic targeting of IL-6 trans-signaling. Cytokine. 2021;144:155577. doi: 10.1016/j.cyto.2021.155577. [DOI] [PubMed] [Google Scholar]
- Sammel M., Peters F., Lokau J., Scharfenberg F., Werny L., Linder S., Garbers C., Rose-John S., Becker-Pauly C. Differences in shedding of the interleukin-11 receptor by the proteases ADAM9, ADAM10, ADAM17, meprin alpha, meprin beta and MT1-MMP. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20153677. 3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Schuster B., Hölscher C., Yoshimoto T., Rose-John S. No inhibition of IL-27 signaling by soluble gp130. Biochem. Biophys. Res. Commun. 2005;326:724–728. doi: 10.1016/j.bbrc.2004.11.098. [DOI] [PubMed] [Google Scholar]
- Schreiber S., Aden K., Bernardes J.P., Conrad C., Tran F., Hoper H., Volk V., Mishra N., Blase J.I., Nikolaus S. Therapeutic IL-6 trans-signalling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology. 2021;160:2354–2366.e11. doi: 10.1053/j.gastro.2021.02.062. [DOI] [PubMed] [Google Scholar]
- Schroers A., Hecht O., Kallen K.J., Pachta M., Rose-John S., Grötzinger J. Dynamics of the gp130 cytokine complex: a model for assembly on the cellular membrane. Protein Sci. 2005;14:783–790. doi: 10.1110/ps.041117105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerd T., Krause F., Twigg S.R.F., Aschenbrenner D., Chen Y.H., Borgmeyer U., Muller M., Manrique S., Schumacher N., Wall S.A. A variant in IL6ST with a selective IL-11 signaling defect in human and mouse. Bone Res. 2020;8:24. doi: 10.1038/s41413-020-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhumberg S., Waetzig G.H., Chalaris A., Rabe B., Seegert D., Scheller J., Rose-John S., Grotzinger J. Structure-guided optimization of the interleukin-6 trans-signaling antagonist sgp130. J. Biol. Chem. 2008;283:27200–27207. doi: 10.1074/jbc.M803694200. [DOI] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018;36:1136–1145. doi: 10.1038/nbt.4305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.