Abstract
Background
Several opioids have pharmacogenomic associations impacting analgesic efficacy. However, germline pharmacogenomic testing is not routinely incorporated into supportive oncology. We hypothesized that CYP2D6 profiling would correlate with opioid prescribing and hospitalizations.
Materials and Methods
We analyzed 61,572 adult oncology patients from 2012 to 2018 for opioid exposures. CYP2D6 metabolizer phenotype (ultra‐rapid [UM], normal metabolizer [NM], intermediate [IM], or poor [PM]), the latter two of which may cause inefficacy of codeine, tramadol, and standard‐dose hydrocodone, was determined for patients genotyped for reasons unrelated to pain. The primary endpoint was number of opioid medications received during longitudinal care (IM/PMs vs. NMs). Secondary endpoint was likelihood of pain‐related hospital encounters.
Results
Most patients with cancer (n = 34,675, 56%) received multiple opioids (average 2.8 ± 1.6/patient). Hydrocodone was most commonly prescribed (62%), followed by tramadol, oxycodone, and codeine. In the CYP2D6 genotyped cohort (n = 105), IM/PMs received a similar number of opioids (3.4 ± 1.4) as NMs (3.3 ± 1.9). However, IM/PMs were significantly more likely to experience pain‐related hospital encounters compared with NMs, independent of other variables (odds ratio [OR] = 5.4; 95% confidence interval [CI], 1.2–23.6; p = .03). IM/PMs were also more likely to be treated with later‐line opioids that do not require CYP2D6 metabolism, such as morphine and hydromorphone (OR = 3.3; 95% CI, 1.1–9.8; p = .03).
Conclusion
CYP2D6 genotype may identify patients with cancer at increased risk for inadequate analgesia when treated with typical first‐line opioids like codeine, tramadol, or standard‐dose hydrocodone. Palliative care considerations are an integral part of optimal oncology care, and these findings justify prospective evaluation of preemptive genotyping as a strategy to improve oncology pain management.
Implications for Practice
Genomic variation in metabolic enzymes can predispose individuals to inefficacy when receiving opioid pain medications. Patients with intermediate and/or poor CYP2D6 metabolizer status do not adequately convert codeine, tramadol, and hydrocodone into active compounds, with resulting increased risk of inadequate analgesia. This study showed that patients with cancer frequently receive CYP2D6‐dependent opioids. However, patients with CYP2D6 intermediate and poor metabolizer status had increased numbers of pain‐related hospitalizations and more frequently required the potent non‐CYP2D6 opioids morphine and hydromorphone. This may reflect inadequate initial analgesia with the common “first‐line” CYP2D6‐metabolized opioids. Preemptive genotyping to guide opioid prescribing during cancer care may improve pain‐related patient outcomes.
Keywords: Oncology, Pain, Opioids, Pharmacogenomics, Cancer
Short abstract
Some opioids have pharmacogenomic associations that affect analgesic efficacy; however, germline pharmacogenomic testing is not routinely incorporated into supportive oncology.This article evaluates the impact of CYP2D6 genotype on pain‐related clinical outcomes in an oncology population.
Introduction
Pain is one of the most commonly reported symptoms in patients with cancer, and opioid medications are an integral part of treatment [1, 2, 3, 4]. However, patients who are prescribed opioids are at risk for adverse drug effects, inadequate pain control, or developing opioid misuse syndrome [5]. Oncology patients represent a particularly vulnerable population when treating pain given their high burden of comorbidities, polypharmacy, numerous surgeries and procedures, and extensive disease courses [2, 6]. Adequate pain control is vital for quality of life and health outcomes in patients with cancer [2, 7].
Patients can differ greatly in their interindividual responses to distinct opioid agents [8, 9]. Several opioid agents require activation by cytochrome P450 (CYP) enzymes to achieve their analgesic activities (Fig. 1) [8, 10]. Notably, codeine and tramadol are metabolized by CYP450 enzymes in phase I reactions, with key metabolites of CYP2D6 enzyme conversion having enhanced μ‐receptor potency resulting in the predominant analgesic properties of these drugs [8, 9, 10, 11, 12, 13, 14]. Hydrocodone is also metabolized by CYP2D6 to hydromorphone, a metabolite that likely mediates the primary analgesic effect; although, the relationship of this CYP2D6‐mediated conversion to pain relief is not as established [10, 13, 15, 16, 17, 18]. In contrast, although CYP2D6 also contributes to the metabolism of oxycodone, and although many studies have shown a relationship between CYP2D6 genotype/phenotype and oxymorphone concentrations after administering oxycodone, most have not shown a correlation with pain outcomes, perhaps because the majority of this agent's analgesic activity is hypothesized to result from its parent form [8, 10, 19, 20, 21, 22]. Finally, opioids such as morphine and hydromorphone are potent parent compounds themselves, which undergo further metabolism via phase II glucuronidation, with little to no impact from CYP450 genetic or drug‐based interactions [8, 11].
Figure 1.
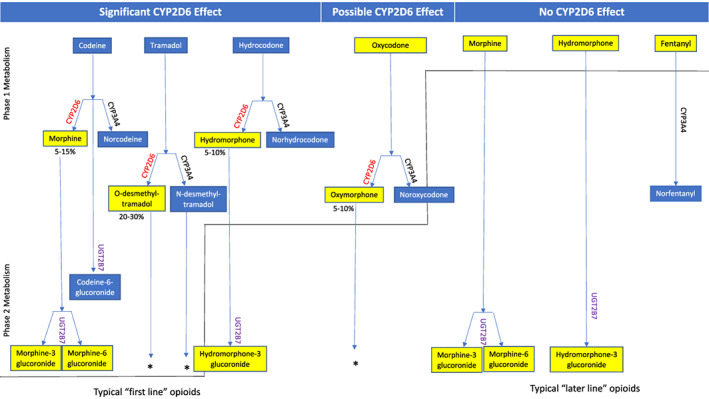
Clinical impacts of CYP2D6 metabolism on opioid analgesic effects. Significant contributors to opioid metabolism, including modification reactions performed by CYP450 enzymes (phase I metabolism) and/or conjugation reactions (phase II metabolism), are displayed. The opioid forms predominantly responsible for the majority of analgesic activities are highlighted in yellow. CYP2D6 plays an essential role in the prodrugs codeine and tramadol bioactivation to their active metabolites, morphine and O‐desmethyltramadol. CYP2D6 is also responsible for hydrocodone metabolism to the active metabolite hydromorphone, which has a 100‐fold higher affinity for μ‐opioid receptors than its parent compound and is postulated to contribute significantly to its analgesic effects. Although CYP2D6 may minimally contribute to the metabolism of oxycodone, the majority of its analgesic activity is hypothesized to result from its parent drug and CYP2D6 metabolism is not generally considered to have a meaningful impact on clinical outcomes for oxycodone [10]. Morphine and hydromorphone do not undergo phase I metabolism but undergo glucuronidation by the enzyme UGT2B7 in phase II metabolism. Because of the infrequent incidence of methadone use in the CYP2D6 genotyped cohort, methadone was omitted from this figure; however, CYP2D6 does play a minor role in this agent's metabolism as well. The information displayed was adopted and summarized from multiple references [8, 11].*Some phase II reactions are omitted from the diagram for simplicity.
The CYP2D6 gene is highly polymorphic, with numerous alleles described and categorized as conferring normal, reduced, or no‐function based on the activity of the enzyme they encode [23]. CYP2D6 poor metabolizers (PMs‐ two nonfunctional alleles) and intermediate metabolizers (IMs‐ one nonfunctional and one reduced function allele, or one functional and one nonfunctional allele) have decreased CYP2D6 activity [10, 23]. Compared with normal metabolizers (NMs), IM/PMs demonstrate lower concentrations of the active metabolites of codeine, tramadol, and hydrocodone with the potential for decreased analgesic effects when treated with standard dosing regimens (such as in hydrocodone/acetaminophen formations) [23, 24, 25, 26, 27, 28, 29]. In contrast, ultra‐rapid metabolizers (UMs) demonstrate increased CYP2D6 activity and may be at risk for toxicity [10].
A recent prospective study of CYP2D6‐guided opioid prescribing in patients with chronic pain demonstrated that this approach resulted in improved patient outcomes and pain control in IM/PM patients, particularly in those treated with codeine, tramadol, and hydrocodone [10, 30]. As a result of these and other data, the Clinical Pharmacogenetics Implementation Consortium (CPIC) recently published updated guidelines recommending against codeine and tramadol use in CYP2D6 PMs (and UMs), and close monitoring in codeine and tramadol‐treated IMs (and in IM and PM hydrocodone‐treated subjects) with transition to an alternative non‐CYP2D6‐metabolized analgesic if inadequate response is observed [10]. These precision medicine approaches deserve consideration for potentially informing the selection or dosing of opioids when treating patients with cancer to maximize treatment efficacy and minimize adverse drug effects. To our knowledge, no large studies have previously studied the role of CYP2D6 genotype on key palliative outcomes in a broad cancer population receiving different CYP2D6‐dependent opioids. Given the necessary and frequent role of opioids in the appropriate palliative care of patients with cancer, we first sought to characterize usual opioid prescribing patterns in a large oncology population in this study. We then hypothesized that germline CYP2D6 genotypes (available for a subset of these patients) would predict risk for inadequate analgesia when using standard opioid prescribing patterns.
Materials and Methods
Data Collection and Analysis for the Opioid Exposed Oncology Cohort
We reviewed the chronologic medication histories of patients with cancer treated at the University of Chicago Medical Center (UCMC) between December 1, 2012, and December 31, 2018, for exposure to opioids to demonstrate the prevalence of opioid medication exposure in a large oncology population (Fig. 2). Oncology patients were defined by International Classification of Diseases 9/10 diagnostic codes for neoplasm, with benign neoplasms excluded [31]. We reviewed and analyzed deidentified medical records obtained from the University of Chicago Clinical Research Data Warehouse for exposure to any of eight opioid medications (codeine, tramadol, hydrocodone, oxycodone, morphine, hydromorphone, fentanyl, and methadone), including both scheduled and as needed prescriptions and exposures. Patients were excluded if they did not have a documented opioid exposure during the study period. Relevant patient demographics were captured from the initial oncology provider encounter, including age, gender, self‐reported race and ethnicity, and tumor type.
Figure 2.
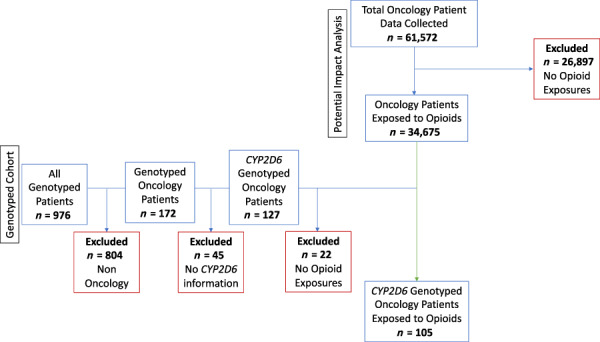
Opioid exposed oncology cohort and CYP2D6 genotyped cohort consort diagram. The chronologic medical histories of 61,572 oncology patients (as defined by ICD9/10 malignant neoplasm diagnostic codes) treated at the University of Chicago Medical Center from December 1, 2012, to December 31, 2018, were analyzed for exposure to opioid medications (codeine, tramadol, hydrocodone, oxycodone, morphine, hydromorphone, fentanyl, and methadone; all formulation types). Patients were excluded if they did not have a documented opioid exposure (n = 26,887). A total of 34,675 were included in the opioid exposed oncology cohort. Oncology patients who were genotyped as part of the 1200 Patients Project (Clinicaltrials.gov #NCT01280825) during a study time period of January 21, 2011 to December 31, 2018, and for whom CYP2D6 metabolizer status was available, were analyzed for opioid exposure. A total of 105 patients were included in the CYP2D6 genotyped cohort analysis.
Data Collection and Analysis for the CYP2D6 Genotyped Cohort
Since 2011, outpatients at UCMC have been eligible to enroll in an ongoing institutional review board–approved institutional pharmacogenomics implementation study entitled The 1200 Patients Project (ClinicalTrials.gov #NCT01280825), which evaluated the feasibility of incorporating pharmacogenomic testing into routine outpatient medical care. Eligible patients included all adults without a history of liver or kidney transplant who were receiving routine outpatient care from a selected study provider representing one of eight different primary care and subspecialty clinics including cardiology, gastroenterology, nephrology, oncology, hepatology, pulmonology, and executive health. Subjects consented to preemptive genotyping across a broad panel of variants selected for their clinical pharmacogenomic role, including CYP2D6 [32]. For CYP2D6 specifically, genotyping was performed using an Invader technology from Hologic (Marlborough, MA), complemented with copy number assessment using Taqman (ThermoFisher, Waltham, MA), with 29 variants assessed including: *1, *2, *3, *4, *4N, *6, *7, *8, *9, *10, *11, *12, *15, *17, *18, *19, *20, *21, *29, *35, *26, *40, *41, *42, *43, *44, *45, *56, *59 [33].
For our current study, we identified all opioid‐treated oncology patients (from step one, above paragraph) who had genotype results available for post‐hoc analysis because of their past participation in The 1200 Patients Project. These patients had not been genotyped for the purposes of pain prescribing, and genotypes were not routinely available to oncology physicians at the outset of oncology care and thus were not universally available prior to opioid prescriptions with the exception of one patient, whose results were viewed by one oncologist at a single patient encounter during the observation period, but with no opioids prescribed at that visit. We therefore analyzed clinical endpoints for the purposes of this study independent of knowledge of genotype, and retrospectively correlated the CYP2D6 genotype data with pain‐related outcomes.
Data Abstraction
For the CYP2D6 genotyped cohort, we defined the period of observation as starting on the date of the first oncology provider encounter and ending on the date of the last encounter type of any kind, within The 1200 Patients Project study time period of January 21, 2011, through December 31, 2018. For both the opioid exposed and CYP2D6 genotyped cohorts, we obtained patient demographics including age, gender, self‐reported race and ethnicity, and cancer diagnosis at the time of first oncology provider encounter. On the date of first opioid exposure, we recorded additional clinical information, including the clinical cancer stage (early‐stage I–III; advanced or metastatic, stage IV), the Eastern Cooperative Oncology Group performance status (if available), the number of concomitant active medication prescriptions, and the number and type of comorbidities, as defined by diagnosis code and active problem list.
Categorization of Opioid Treatment Efficacy and Pain‐Related Outcomes
For both the opioid exposed and CYP2D6 cohorts, we analyzed medical histories throughout the entire study period for opioid prescriptions with start and end dates, prescription route, dose, and prescription setting (ambulatory, inpatient, postoperative, or postprocedure). For the CYP2D6 cohort, the following variables were also obtained: (a) Date and dosage of adjunct pain medications including acetaminophen, nonsteroidal anti‐inflammatory drugs (ibuprofen, naproxen, meloxicam, celecoxib, ketorolac, indomethacin), lidocaine patches, and agents prescribed primarily for the treatment of neuropathy (amitriptyline, carbamazepine, duloxetine, gabapentin, pregabalin); (b) date and dosage of CYP2D6 strong, moderate, and weak inhibitors (supplemental online Table 1) [34]; (c) dates and type of all surgeries and procedures (i.e., biopsies, resections, reconstructions, endoscopy/colonoscopy/bronchoscopy); (d) number and type of pain‐related hospital encounters including pain‐related hospitalizations, pain clinic encounters, and pain‐related procedures (including joint and epidural injections), and Emergency Department (ED) encounters for pain (as defined by problem list/diagnostic code for the encounter), including only those occurring during the study observation period of January 21, 2011–December 31, 2018; and (e) date and frequency of palliative care consults. During data extraction from the UCMC electronic medical record, the dedicated study personnel performing data collection were blinded to patients' CYP2D6 genotype/metabolizer status.
Table 1.
Demographics of the opioid exposed and CYP2D6 genotyped oncology populations
| Characteristic | Opioid exposed oncology cohort (n = 34,675) | CYP2D6 genotyped cohort (n = 105) |
|---|---|---|
| Age at first oncology provider encounter, yr | ||
| Mean | 65.3 | 61.1 |
| Median | 66.8 | 64 |
| Gender, female, n (%) | 17,930 (51.7) | 61 (58.1) |
| Race | ||
| Black, n (%) | 10,844 (31.4) | 31 (29.5) |
| White, n (%) | 21,452 (61.9) | 69 (65.7) |
| Other a , n (%) | 2,379 (6.7) | 5 (4.8) |
| Ethnicity | ||
| Not Hispanic or Latino, n (%) | 32,409 (93.5) | 34 (32.4) |
| Hispanic or Latino, n (%) | 1,644 (4.7) | 4 (3.8) |
| Unknown, n (%) | 622 (1.8) | 67 (63.8) |
| Cancer type | ||
| Breast | 3,373 (9.7) | 42 (40) |
| Central nervous system b | 333 (0.9) | 0 |
| Gastrointestinal c | 4,059 (11.7) | 33 (31.4) |
| Genitourinary d | 7,438 (21.5) | 30 (28.6) |
| Head and neck e | 1,932 (5.6) | 0 |
| Leukemia/lymphoma | 7,443 (21.5) | 0 |
| Skin/melanoma | 1,239 (3.6) | 0 |
| Thoracic | 2,972 (8.6) | 0 |
| Other f | 5,886 (16.9) | 0 |
Other includes Asian/Mid‐East Indian, American Indian/Alaska Native, Native Hawaiian/Other Pacific Islander, or Unknown.
Central nervous system includes malignancies of the brain and eye.
Gastrointestinal includes esophagogastric, colorectal, pancreas, gallbladder, small bowel malignancies, and neuroendocrine tumors of the gastrointestinal tract.
Genitourinary includes ovarian, uterine, cervical, prostate, renal, and bladder malignancies.
Head and neck includes lip, oral cavity, pharynx, and thyroid malignancies.
Other includes bone or soft tissue malignancies, neoplasm of uncertain behavior/unspecified metastatic sites, or multiple oncologic diagnoses reported.
CYP2D6 Phenotype Determination
For those with archival CYP2D6 results, metabolizer phenotypes were assigned using genotypes and copy number results. Our primary analysis was based on this CYP2D6 genotype‐predicted phenotype. Each allele was given an activity score, and the metabolizer phenotype was assigned based on the cumulative value. Scores of 0, 0.5, and 1 were assigned for each no activity, decreased activity, and normal activity allele, respectively. The phenotype was assigned based on the sum of the scores (0, PM; 0.5, IM; 1–2, NM; and > 2, ultra‐rapid metabolizer [UM]) using the phenotype classification schema that was in place during the study period [35, 36], although we acknowledge that an updated system was recently adopted that would potentially increase by a very small fraction the number of patients classified as IMs [10, 23].
A separate analysis was performed for patients prescribed a CYP2D6 inhibitor synchronously with opioid therapy to encompass phenoconversion potential. Specifically, we reviewed participants' medical histories for exposure to CYP2D6 strong, moderate, and weak inhibitors, as defined by the U.S. Food and Drug Administration (supplemental online Table 1) [34]. Patients who received a CYP2D6 inhibitor for ≥50% of the time that they received a CYP2D6‐metabolized opioid (defined as codeine, tramadol, hydrocodone) were phenoconverted by multiplying the activity score by an inhibitor factor (0.5 for weak and moderate inhibitors, 0 for strong inhibitors), as performed previously in other studies [10, 30, 34, 37, 38].
Statistical Analysis
Because of the hypothesis that different CYP2D6 phenotypes (and the resulting interindividual variability in analgesia responses) may contribute to imprecision in opioid prescribing, the study's primary endpoint was the number of different opioid medications required for pain control throughout longitudinal care, comparing IM/PM patients with NM patients. The secondary endpoint was the composite likelihood of UCMC hospitalization, pain clinic encounter/procedure, or ED visit for a pain diagnosis comparing IM/PM with NM patients. Both analyses were repeated for phenoconversion. There was no minimum amount of follow up time required. Instead, the varied observation periods between patients were specifically accounted for in the analyses, with the number of each patient's observation years included as a clinical covariate in the Poisson regression analyses. Descriptive statistics were used to summarize the demographic and clinical variables of interest. We also examined multiple clinical variables (described below) between the NM and IM/PM groups. Two‐sample t‐test or Fisher's exact tests were used to assess whether these clinical factors were significantly different between the two groups.
For the primary endpoint, Poisson regression analyses were used to evaluate the relationship between the number of different opioid medications and CYP2D6 metabolizer status and other clinical variables. Clinical covariates included number of observation years, patient age, gender, race, ethnicity, number of concomitant medications, number of comorbidities, presence of osteoarthritis and/or neuropathy, cancer type and stage, performance status, exposure to adjunct pain medications, surgeries/procedures, and palliative care consults. Univariable analysis was first used on each factor, and those with a p value < .10 from the univariable analysis were included in the multivariable model selection process. Backward selection was used to identify significant predictors of the primary endpoint in the final multivariable Poisson model. Logistic regression models were used to analyze the secondary endpoint of pain‐related hospital encounters. A similar approach was used to determine the final multivariable logistic regression model for pain‐related hospital encounters. Although the main analyses used the genotype‐assigned metabolizer status for each patient, both the primary and secondary analyses were repeated, accounting for phenoconversion. Nominal p values < .05 were considered significant.
Results
Demographics
A total of 61,572 patients treated at UCMC during the study period were found to have an oncologic diagnosis, with the majority (56.3%, n = 34,675) receiving at least one opioid medication during their course of treatment. Of the 127 oncology patients for whom CYP2D6 genotype data were available, most (n = 105, 82.7%) had a documented opioid exposure (Fig. 2). Subject demographics, including age, self‐reported race/ethnicity, and cancer type, were similar between the total opioid exposed and CYP2D6 genotyped patient cohorts (Table 1). Patients in the CYP2D6 genotyped cohort were followed for an average of 6.1 ± 2.3 years.
Opioid Prescribing Patterns Among Oncology Patients
In the opioid exposed oncology cohort (n = 34,675), a total of 97,986 separate opioid exposures were identified. The majority (73.7%) of patients with cancer received more than one opioid type during longitudinal care. Median follow up was 426 days, with an average of 2.8 ± 1.6 different opioid medication exposures per patient (representing a median of 2.07 different opioids/year for each patient). The most common opioid prescribed was hydrocodone (n = 21,753), as detailed in Figure 3A. A total of n = 28,530 (82.3%) received one or more of the opioids for which CYP2D6 metabolism is important for analgesic activity (codeine, tramadol, and/or hydrocodone). A second analysis (Fig. 3B) subsequently excluded intravenous (IV) formulations of fentanyl, hydromorphone, and morphine to more closely approximate outpatient prescribing patterns. There was no significant difference in prevalence of different opioid types prescribed when analyzed by each demographic variable (age, race, ethnicity, and cancer type), with hydrocodone remaining the most commonly used agent within each specific group (data not shown). These findings demonstrate that a majority of oncology patients are exposed to opioids throughout their longitudinal care and that a high proportion of the prescribed agents are dependent on CYP2D6 metabolism for their analgesic activity.
Figure 3.
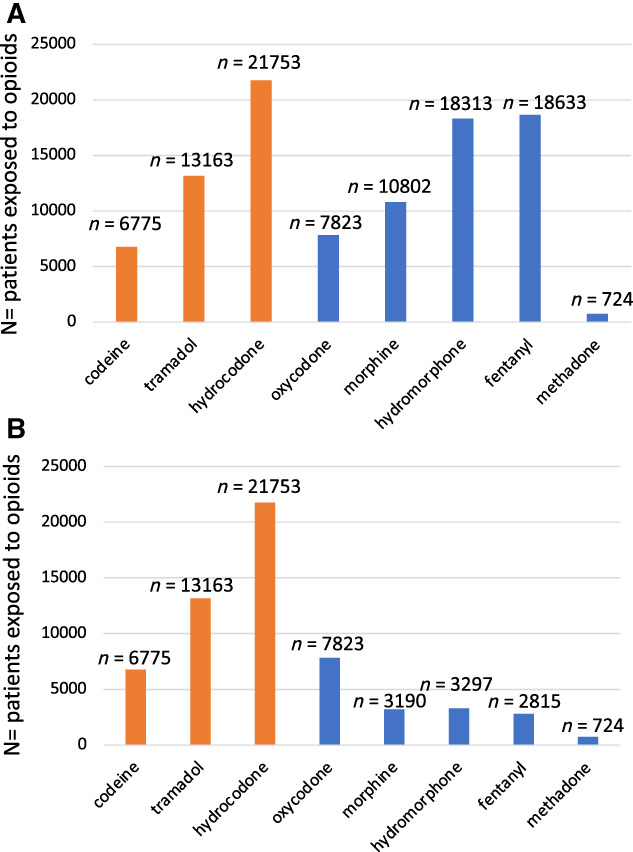
(A): Opioid prescribing patterns in a population with cancer. A total of 97,986 separate opioid drug exposures were identified in 34,675 patients with an oncologic diagnosis during the study period (December 1, 2012–December 31, 2018). The most common oral opioid prescribed was hydrocodone. CYP2D6‐metabolized opioids (codeine, tramadol, hydrocodone) are highlighted in orange. (B): Oral and transdermal opioid prescribing patterns in a population with cancer. To approximate outpatient prescribing patterns, intravenous formulations of morphine, hydromorphone, and fentanyl were excluded. A total of 59,540 separate oral (and transdermal, i.e., fentanyl) opioid drug exposures were identified in 34,675 patients with an oncologic diagnosis during the study period (December 1, 2012–December 31, 2018). CYP2D6‐metabolized opioids (codeine, tramadol, hydrocodone) are highlighted in orange.
CYP2D6 Genotype and Metabolizer Assignments
The primary analysis for the CYP2D6 genotyped cohort was based on CYP2D6 genotype‐predicted phenotypes. One genotyped patient was determined to have a UM phenotype (n = 1, 0.95%). Given that this phenotype is not correlated with the inadequate analgesic response outcomes of interest in this study (increased number of opioid exposures and composite evaluation of inadequate pain control) this patient was included for analysis within the NM phenotype group for a total of n = 91. Although the finding of only one UM patient was perhaps slightly lower than expected, this is likely because of the relatively large proportion of Black patients included in our cohort, a group with a known lower frequency of observed UM phenotypes compared with European‐descent populations (0.4% vs. 2.5%, respectively) [10].
Seven patients (6.7%) each were IM and PM, respectively, for a total of n = 14 in the combined IM/PM metabolizer group, which is consistent with expected allele frequencies previously published [23, 24]. Eight patients in the CYP2D6 genotyped cohort were found to be taking CYP2D6 inhibitors ≥50% of the time concurrently with a prescription for codeine, tramadol, and/or hydrocodone. For the secondary phenoconversion analysis described previously, these eight patients were subsequently included in the IM/PM metabolizer group (n = 22).
Clinical Factors Contribute to Opioid Use Prevalence Among CYP2D6 Genotyped Oncology Patients
Patients in both IM/PM and NM groups were exposed to a similar number of opioids throughout their longitudinal course, as demonstrated in Table 3. Prescribing of CYP2D6‐metabolized opioids such as codeine, tramadol, and hydrocodone was very common, with the majority of patients in both IM/PM and NM groups receiving at least one of these agents (13/14 or 93% vs. 74/91 or 81%, respectively). Of note, seven of the IM/PM patients (50%) were prescribed codeine, tramadol, or hydrocodone contemporaneously with a non‐CYP2D6 metabolized opioid (morphine and/or oxycodone), which could have dampened the clinical manifestations of CYP2D6 effects in these patients.
Table 3.
Opioid medication exposures in CYP2D6 genotyped (n = 105) population with phenoconversion analysis
| Opioid medication | CYP2D6 genotyped cohort IM/PM | CYP2D6 genotyped cohort NM | CYP2D6 genotyped cohort ‐ IM/PM phenoconversion | CYP2D6 genotyped cohort – NM phenoconversion |
|---|---|---|---|---|
| (n = 14) | (n = 91) | (n = 22) | (n = 83) | |
| Codeine n (%) a , b | 8 (57) | 37 (41) | 13 (59) | 33 (40) |
| Tramadol n (%) a , b | 3 (21) | 30 (33) | 5 (23) | 28 (34) |
| Hydrocodone n (%) a , b | 8 (57) | 57 (63) | 12 (55) | 52 (63) |
| Oxycodone n (%) a , b | 4 (29) | 22 (24) | 7 (32) | 19 (23) |
| Morphine n (%) a , b | 7 (50) | 29 (32) | 11 (50) | 25 (30) |
| Hydromorphone n (%) a , b | 8 (57) | 52 (57) | 12 (55) | 47 (57) |
| Fentanyl n (%) a , b | 11 (79) | 68 (75) | 17 (77) | 62 (75) |
| Methadone n (%) a , b | 0 (0.0) | 2 (2.2) | 1 (4.5) | 1 (1.2) |
| The average number of distinct opioid exposures per patient | ||||
| Mean ± SE c | 3.5 ± 1.4 | 3.3 ± 1.9 | 3.6 ± 1.7 | 3.2 ± 1.9 |
| Patients exposed to morphine and/or hydromorphone n (%) c | 12 (85.7) | 55 (60.4) | 17 (77.3) | 42 (50.6) |
| Odds ratio of morphine and/or hydromorphone exposure, IM/PM vs. NM, c (95% CI) | 3.9 (0.83–18.6), p = .08 | 3.3 (1.12–9.8), p = .03 | ||
These data are categorized by distinct opioid medications.
The same patient might be represented in multiple fields within a column if exposed to multiple opioid agents during the study period.
These data are categorized by distinct patients.
Abbreviations: CI, confidence interval; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; SE, standard of error.
As oncology patients frequently undergo diagnostic and therapeutic procedures during their treatment, with opioids being a mainstay of management in these settings, we crucially included the presence (or lack thereof) of surgery and/or a procedure as a clinical covariate for predicting opioid exposure (Table 2). The receipt of a surgery/procedure was expectedly predictive of exposure to a greater number of opioids (p = .003), particularly in the setting of IV opioids (85.9% of IV opioids exposures were associated with surgery/procedure) and, to a lesser extent, oral opioids (50.2% of oral opioids were prescribed in conjunction with a surgery/procedure). In addition, patients who were prescribed additional adjunct pain medications during treatment (i.e., ibuprofen, naproxen, meloxicam, celecoxib, ketorolac, indomethacin, lidocaine patches, amitriptyline, carbamazepine, duloxetine, gabapentin, pregabalin) also had more opioid exposures throughout their longitudinal course (p = .0008). The associations of these two clinical factors with patients' increasing exposure to opioid medications remained similar when repeated with the exclusion of IV fentanyl and also when the Poisson regression was repeated for the phenoconversion population.
Table 2.
Characteristics of CYP2D6‐genotyped study population (n = 105)
| Characteristic | NM | IM/PM | p value |
|---|---|---|---|
| n = 91 | n = 14 | ||
| Age at first encounter, years | |||
| Mean | 58.8 | 61.5 | .43 |
| Median | 55.5 | 64 | |
| Gender, female, n (%) | 50 (54.9) | 11 (78.6) | .15 |
| Race | |||
| Black, n (%) | 24 (26.4) | 7 (50) | .23 |
| White, n (%) | 62 (68.1) | 7 (50) | |
| Other, n (%) | 5 (5.5) | 0 | |
| Ethnicity | |||
| Not Hispanic or Latino, n (%) | 30 (33.0) | 4 (28.6) | .87 |
| Hispanic or Latino, n (%) | 4 (4.4) | 0 | |
| Unknown, n (%) | 57 (62.6) | 10 (71.4) | |
| Cancer type | |||
| Breast | 34 (37.4) | 8 (57.1) | .36 |
| Gastrointestinal a | 29 (31.9) | 4 (28.6) | |
| Genitourinary b | 28 (30.8) | 2 (14.3) | |
| Cancer stage at 1st opioid prescription | |||
| I–III, n (%) | 49 (53.9) | 12 (85.7) | .10 |
| IV, n (%) | 38 (41.8) | 2 (14.3) | |
| n/a c , n (%) | 4 (4.4) | 0 (0) | |
| Diagnosis of osteoarthritis d , n (%) | 36 (39.6) | 4 (28.6) | .56 |
| Diagnosis of neuropathy d , n (%) | 28 (30.8) | 5 (35.7) | .76 |
| Number of comorbid medical conditions | |||
| Mean ± SE d | 3.58 ± 0.43 | 2.57 ± 0.60 | .18 |
| Number of total medication prescriptions | |||
| Mean ± SE d | 7.80 ± 0.46 | 6.50 ± 0.81 | .29 |
| Number of surgeries/procedures | |||
| Mean ± SE e | 3.41 ± 0.31 | 3.64 ± 0.44 | .67 |
A two‐sample t‐test was used to compare continuous variables, and Fisher's exact test was used to compare categorical variables between NM and IM/PM groups.
Gastrointestinal includes colorectal, pancreas, and small bowel malignancies.
Genitourinary includes prostate, renal, and bladder malignancies.
Cancer stage was not known at the time of the first opioid prescription.
Assessment of diagnoses was made at the time point of the date of the first opioid prescription.
Surgeries and procedures performed during the entire study observation period of January 21, 2011, to December 31, 2018, were included. Examples of surgeries and procedures include resections, reconstructions, biopsies, endoscopies, colonoscopies, and bronchoscopies.
Abbreviations: IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; SE, standard of error.
Correlation of CYP2D6 Genotype with Efficacy of Pain Control and Incidence of Pain‐Related Hospital Encounters
The vast majority of IM/PM patients (10/14, or 71.4%) experienced a pain‐related hospitalization, ED visit, and/or pain clinic consultation for a pain‐related procedure. This finding was in stark contrast to NM patients, who required these in only one third of cases (30/91, or 33%). Half of the IM/PM patients who required a pain‐related hospital encounter were prescribed one of the CYP2D6‐activated opioids (codeine, tramadol, hydrocodone) as an outpatient before their pain‐related event.
The majority of pain‐related encounters in both IM/PM and NM patients included pain clinic visits, pain‐related procedures (e.g., injections), and/or ED visits (75%). However, as a surrogate for capturing the potential burden of inadequate pain control on health care utilization, for patients with hospitalizations we examined hospital length of stay (arguably the most costly of the three types of included encounters). In this exploratory analysis (limited by the number of IM/PM patients), we did not find a significant difference in hospital length of stay between NM and IM/PM patients (median 3.5 days [range 1–15] vs. 1.0 days [1, 2, 3], p = .07). Also, the average time between first opioid exposure and first pain‐related hospitalization was variable and did not appear explanatory (190.2 ± 357.2 days for NMs vs. 429.5 ± 324.6 for IM/PMs).
The logistic regression model identified CYP2D6 metabolizer status, Black race, and exposure to adjunct pain medications as each being independently associated with an increased need for pain‐related hospitalizations and/or procedures (Fig. 4). Notably, in the multivariable regression model, IM/PM patients were five times more likely than NMs to require a pain‐related procedure/hospital encounter (odds ratio [OR], 5.4; confidence interval [CI], 1.2–23.6; p = .03). Black race (OR, 3.1; CI, 1.2–8.4; p = .03) and exposure to adjunct pain medications (OR, 16.3; CI, 3.0–87.7; p = .001) were other independent predictors of pain‐related hospitalization/emergency visit. Given the clinical importance of considering phenoconversion (which appropriately captures the “effective” metabolizer status), these analyses were repeated in the larger group (n = 22) of phenoconverted IM/PM subjects. The results were importantly reproduced and accentuated for all three significant factors predictive of increased risk for pain‐related hospitalization, ED visit, or pain procedure: IM/PM vs. NM (OR, 5.5; CI, 1.6–19.2; p = .007); Black vs. White race (OR, 3.5; CI, 1.3–9.6; p = .02); and exposure to adjunct pain medications (OR, 18.7; CI, 3.4–101.4; p = .0007).
Figure 4.
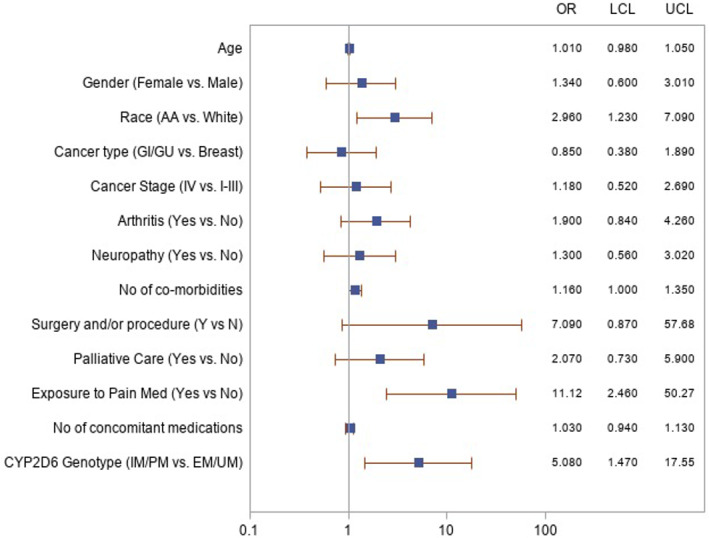
Forest plot demonstrating independent risk factors for pain‐related hospital encounters in CYP2D6‐genotyped cohort. Pain‐related hospital encounters during the study period (January 21, 2011–December 31, 2018) were defined as a hospitalization, emergency department visit, or pain clinic visit/procedure for a pain‐related diagnosis (as defined by problem list/diagnostic code for the encounter). The relationship between the likelihood of pain‐related hospital encounters and important demographic and clinical variables was examined using univariable logistic regression models. The corresponding model results are graphically presented. The following three factors were identified as significant predictors of pain‐related hospital encounters through the multivariable logistic regression model selection process: additional exposure to adjunct pain medications, Black race, and IM/PM CYP2D6 genotypes. LCL and UCL are defined.Abbreviations: EM, extensive metabolizer; GI, gastrointestinal; GU, genitourinary; IM, intermediate metabolizer; LCL, lower confidence limit; OR, odds ratio; PM, poor metabolizer; UCL, upper confidence limit; UM, ultra‐rapid metabolizer.
Finally, although the majority (13/14, or 93%) of IM/PM patients received a CYP2D6‐metabolized opioid such as codeine, tramadol, or hydrocodone, prescribing patterns demonstrated that these patients also more frequently received treatment with CYP2D6‐independent opioids such as morphine and/or hydromorphone. IM/PM patients were more likely than NM patients to be exposed to morphine and/or hydromorphone during their longitudinal treatment course (12/14 IM/PM patients or 85.7% vs. 60.4% NMs; OR 3.9, 95% CI, 0.8–18.6; p = .08). Six of the 12 (50%) of these exposures occurred during a pain‐related hospital encounter. Most of these patients (four of the six, 67%) had an active outpatient prescription for codeine, tramadol, or hydrocodone at the time of the pain‐related hospital encounter, suggesting failure of their prescribed CYP2D6‐metabolized medication. The association of increased exposure to morphine/hydromorphone in the IM/PM population was significant when accounting for phenoconversion (77.3% vs. 50.6% with OR, 3.3; CI 1.1–9.8; p = .03), as displayed in Table 3.
Discussion
A high prevalence of opioid exposures among oncology patients was found in our study, in which patients were noted to receive numerous different types of opioid medications throughout their longitudinal course of care. Hydrocodone and tramadol, both dependent on CYP2D6 activation, were the most frequently used oral opioid medications, regardless of cancer type or demographic factors (race, age). When analyzed by CYP2D6 status, IM/PM patients were more likely to have a pain‐related hospital encounter than NM patients despite active outpatient prescriptions for CYP2D6‐metabolized opioids, suggesting that IM/PMs may have experienced insufficient analgesia with the commonly used CYP2D6‐dependent opioids. In addition, an increased proportion of IM/PM patients were subsequently found to be exposed to less commonly used but more potent CYP2D6‐independent opioids such as morphine and/or hydromorphone. These data suggest that initial inadequate pain control may have precipitated a pain‐related hospital encounter in which patients were exposed to later‐line opioids not dependent on CYP2D6 activation. A pattern of prescribing tramadol, hydrocodone, or codeine‐containing medications as “first choice” pain control drugs (as recommended by the World Health Organization Cancer Pain Management Ladder [39]) or using opioid “trial and error” may thus be inadequate or potentially harmful in CYP2D6 IM/PM patients. Prospective studies will be necessary to confirm these important findings for cancer care.
Despite some prior investigations in small cohorts or examining single opioids, convincing evidence of the pharmacogenomic role for CYP2D6 in guiding pain prescribing in the oncology setting has not been previously established [19, 40, 41, 42]. Smith et al. recently demonstrated the successful implementation of CYP2D6‐guided opioid therapy (using both genotype and phenoconversion) in the primary care setting [30]. Their results showed improved pain control in the population postulated to benefit most from this strategy (IM/PMs initially prescribed a CYP2D6‐bioactivated opioid such as codeine, tramadol, or hydrocodone), with improved patient outcomes including pain control. Our findings now extend these important implications to the oncology treatment setting. Despite the relatively modest size of our genotyped cohort in this study, our genotyped cohort represents to our knowledge the largest collection of CYP2D6‐genotyped patients with cancer receiving multiple different CYP2D6‐dependent opioids analyzed for pain outcomes. Our findings were reproduced and strengthened when considering CYP2D6 phenoconversion, which extended the analysis to an even larger group of functional IM/PM patients. This step is likely important, given that concomitant use of CYP2D6 inhibitors and opioids is common in cancer care, highlighting the potentially important role of considering both drug‐drug interactions as well as drug‐gene interactions in perpetuating the clinical variability of opioid responses.
During cancer care, the need for ED visits/hospitalizations and pain consults (with related procedures) to address pain directly not only impacts quality of life but leads to increased health care utilization and associated costs [43, 44]. Despite the fact that most IM/PM patients in our study had early‐stage cancers, we found that these patients were five times more likely than NM patients to have a pain‐related hospital encounter throughout their clinical course, even when controlled for additional clinical variables. In this study, self‐reported Black race was also independently associated with an increased risk of pain‐related hospital encounters, supporting results found in prior studies [45, 46].
Our study had several limitations. First, our findings describe a single institution experience, and our analyses were retrospective in nature. However, we used a diverse patient population first to establish opioid prescribing patterns, and our CYP2D6‐based findings (although based on a relatively small number of IM/PM genotypic individuals) were recapitulated when considering both CYP2D6 genotype‐conferred metabolizer status and when considering phenoconversion (representing a larger group), and were directionally consistent across two different pain‐related phenotypes (increased pain‐related encounters; increased requirement for CYP2D6‐independent opioids), thus significantly decreasing the possibility that these results were found by chance alone. Although CYP2D6 enzymatic activities and opioid pharmacokinetic parameters were not directly analyzed, the use of CYP2D6 genotype to reliably predict phenotype is well‐established [10, 23, 47]. It is nonetheless acknowledged that although CPIC does include hydrocodone prescribing guidance in its latest recommendations (albeit as limited evidence and “optional”), given the strong data demonstrating the role of CYP2D6 genotype/phenotype in impacting hydrocodone‐related pain outcomes demonstrated by Smith et al. [30], and consistent with current ongoing large‐scale National Institutes of Health‐sponsored prospective trials that include CYP2D6‐guided adjustment for hydrocodone, tramadol, and codeine (clinicaltrials.gov #NCT04445792), we included hydrocodone in our analysis but the role of hydrocodone/CYP2D6 effects deserves ongoing validation. Separately, although pain scores were not available in our analysis, our findings on the role of genotype on pain‐related clinical outcomes were on‐par with several clinical factors that sensibly correlated with pain outcomes, and CYP2D6 genotype was independently associated even after controlling for these clinical factors in multivariable analyses. To prevent potential coding bias given the retrospective nature of this study, the two independent phenotype coders were blinded to CYP2D6 metabolizer status at the time of clinical data abstraction. Another limitation of our study is that CYP3A4 status, which impacts hydrocodone metabolism, was not included in our analysis. Other pharmacogenes, including COMT and the pharmacodynamic OPRM1 were also not analyzed because consensus guidelines have not established a meaningful clinical impact on pain outcomes for these genes [10]. If further evidence justifies the consideration of these additional genes or other pharmacodynamic variables [48], future studies could consider composite models of biological factors (including CYP2D6) on opioid‐related outcomes. One such consideration, organ dysfunction, which may have effects on opioid metabolism and could affect response, was not explicitly evaluated in our study; instead, patients' comorbidities at the time of first opioid prescription were recorded and were not found to be predictive of either the primary or secondary outcomes. Cancer progression and refractory disease is also a well‐known mechanism of worsening cancer pain; whereas cancer stage was included as an independent factor and not found to be significant, future studies should explore this relationship further. Lastly, information regarding costs of pain‐related health care utilization were not available for this analysis but could be important to explore in future studies.
Conclusion
Our findings suggest that CYP2D6 IM/PM patients may not be adequately palliated by commonly used front‐line opioid agents including codeine, tramadol, and hydrocodone, and such patients have an increased risk of experiencing hospitalization or pain‐related encounters and requiring non‐CYP2D6 activated opioids for pain control. These results provide support for the prospective consideration of preemptive CYP2D6 genotyping to assist in personalized opioid prescribing among oncology patients.
Author Contributions
Conception/design: Natalie Reizine, Peter H. O'Donnell
Provision of study material or patients: Keith Danahey, Emily Schierer, Merisa Middlestadt, Jenna Ludwig, Xander M.R. van Wijk, Kiang‐Teck J. Yeo, Mark Ratain, Peter H. O'Donnell
Collection and/or assembly of data: Natalie Reizine, Keith Danahey, Emily Schierer, Ping Liu, Merisa Middlestadt, Jenna Ludwig
Data analysis and interpretation: Natalie Reizine, Ping Liu, Tien M. Truong, Xander M.R. van Wijk, Kiang‐Teck J. Yeo, Peter H. O'Donnell
Manuscript writing: Natalie Reizine, Peter H. O'Donnell
Final approval of manuscript: Natalie Reizine, Keith Danahey, Emily Schierer Ping Liu, Merisa Middlestadt, Jenna Ludwig, Tien M. Truong, Xander M.R. van Wijk, Kiang‐Teck J. Yeo, Monica Malec, Mark Ratain, Peter H. ODonnell
Disclosures
Kiang‐Teck J. Yeo: Truvian Sciences, Roche Diagnostics (C/A, H, SAB), Truvian Sciences (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Table S1 FDA Classification of CYP2D6 Inhibitors
Acknowledgments
This work was supported by the National Institutes for Health [R01 HG009938‐01A1 to P.H.O, 5T32GM007019‐41 through the University of Chicago Committee on Clinical Pharmacology and Pharmacogenomics to N. R.]; the University of Chicago Comprehensive Cancer Center; The William F. O'Connor Foundation; and the Benjamin McAllister Research Fellowship Award [to N. R.].
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
References
- 1. Nahin RL. Estimates of pain prevalence and severity in adults: United States, 2012. J Pain 2015;16:769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Page R, Blanchard E. Opioids and cancer pain: Patients' needs and access challenges. J Oncol Pract 2019;15:229–231. [DOI] [PubMed] [Google Scholar]
- 3. van den Beuken‐van Everdingen MH, Hochstenbach LM, Joosten EA et al. Update on prevalence of pain in patients with cancer: Systematic review and meta‐analysis. J Pain Symptom Manage 2016;51:1070–1090.e9. [DOI] [PubMed] [Google Scholar]
- 4. Paice JA. Managing pain in patients and survivors: Challenges within the United States opioid crisis. J Natl Compr Canc Netw 2019;17:595–598. [DOI] [PubMed] [Google Scholar]
- 5. Benyamin R, Trescot AM, Datta S et al. Opioid complications and side effects. Pain Physician 2008;11(suppl 2):S105–120. [PubMed] [Google Scholar]
- 6. Lee JS, Hu HM, Edelman AL et al. New persistent opioid use among patients with cancer after curative‐intent surgery. J Clin Oncol 2017;35:4042–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennett M, Paice JA, Wallace M. Pain and opioids in cancer care: Benefits, risks, and alternatives. Am Soc Clin Oncol Educ Book 2017;37:705–713. [DOI] [PubMed] [Google Scholar]
- 8. Smith HS. Opioid metabolism. Mayo Clin Proc 2009;84:613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith DM, Weitzel KW, Cavallari LH et al. Clinical application of pharmacogenetics in pain management. Per Med 2018;15:117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crews KR, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6, OPRM1, and COMT genotype and select opioid therapy. Clin Pharmacol Ther 2021. [epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Overholser BR, Foster DR. Opioid pharmacokinetic drug‐drug interactions. Am J Manag Care 2011;17(suppl 11):S276–287. [PubMed] [Google Scholar]
- 12. Gong L, Stamer UM, Tzvetkov MV et al. PharmGKB summary: Tramadol pathway. Pharmacogenet Genomics 2014;24:374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DePriest AZ, Puet BL, Holt AC et al. Metabolism and disposition of prescription opioids: A review. Forensic Sci Rev 2015;27:115–145. [PubMed] [Google Scholar]
- 14. Gasche Y, Daali Y, Fathi M et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med 2014;351:2827–2831. [DOI] [PubMed] [Google Scholar]
- 15. Otton SV, Schadel M, Cheung SW et al. CYP2D6 phenotype determines the metabolic conversion of hydrocodone to hydromorphone. Clin Pharmacol Ther 1993;54:463–472. [DOI] [PubMed] [Google Scholar]
- 16. Stauble ME, et al. Hydrocodone in postoperative personalized pain management: Pro‐drug or drug? Clin Chim Acta 2014;429:26–29. [DOI] [PubMed] [Google Scholar]
- 17. Linares OA, Fudin J, Daly AL et al. Individualized hydrocodone therapy based on phenotype, pharmacogenetics, and pharmacokinetic dosing. Clin J Pain 2015;31:1026–1035. [DOI] [PubMed] [Google Scholar]
- 18. Linares OA, Tod M, Daly AL et al. Feasibility and utility of the individualized hydrocodone therapy based on phenotype, pharmacogenetics, and pharmacokinetic dosing. Clin J Pain 2016;32:1106–1107. [DOI] [PubMed] [Google Scholar]
- 19. Andreassen TN, Eftedal I, Klepstad P et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross‐sectional multicentre study. Eur J Clin Pharmacol 2012;68:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zwisler ST, Enggaard TP, Noehr‐Jensen L et al. The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharmacol Toxicol 2009;104:335–344. [DOI] [PubMed] [Google Scholar]
- 21. Samer CF, Daali Y, Wagner M et al. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharmacol 2010;160:919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zwisler ST, Enggaard TP, Mikkelsen S et al. Impact of the CYP2D6 genotype on post‐operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand 2010;54:232–240. [DOI] [PubMed] [Google Scholar]
- 23. Caudle KE, et al. Standardizing CYP2D6 genotype to phenotype translation: Consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 2020;13:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaedigk A, Sangkuhl K, Whirl‐Carrillo M et al. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 2017;19:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaedigk A, Dinh JC, Jeong H et al. Ten years' experience with the CYP2D6 activity score: A perspective on future investigations to improve clinical predictions for precision therapeutics. J Pers Med 2018;8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stamer UM, Musshoff F, Kobilay M et al. Concentrations of tramadol and O‐desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther 2007;82:41–47. [DOI] [PubMed] [Google Scholar]
- 27. St Sauver JL, Olson JE, Roger VL et al. CYP2D6 phenotypes are associated with adverse outcomes related to opioid medications. Pharmgenomics Pers Med 2017;10:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fanelli A, Palazzo C, Balzani E et al. An explorative study of CYP2D6's polymorphism in a sample of chronic pain patients. Pain Med 2020;21:1010–1017. [DOI] [PubMed] [Google Scholar]
- 29. Dagostino C, Allegri M, Napolioni V et al. CYP2D6 genotype can help to predict effectiveness and safety during opioid treatment for chronic low back pain: Results from a retrospective study in an Italian cohort. Pharmgenomics Pers Med 2018;11:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith DM, Weitzel KW, Elsey AR et al. CYP2D6‐guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: A pragmatic clinical trial. Genet Med 2019;21:1842–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ICD‐9‐CM to ICD‐10‐CM based on FY2014 ICD‐9‐CM codes ‐ Reportable Neoplasms. Available at https://seer.cancer.gov/tools/conversion/2014/ICD9CM_to_ICD10CM_2014CF.pdf. Accessed April 1, 2020.
- 32. O'Donnell PH, Bush A, Spitz J et al. The 1200 Patients Project: Creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 2012;92:446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leung EKY, Agolini E, Pei X et al. Validation of an extensive CYP2D6 assay panel based on Invader and TaqMan copy number assays. J Appl Lab Med 2019;1:471–482. [DOI] [PubMed] [Google Scholar]
- 34. Drug development and drug interactions : Table of substrates, inhibitors and inducers. U.S. Food and Drug Administration Web site, 2020. Available from: https://www.fda.gov/drugs/drug‐interactions‐labeling/drug‐development‐and‐drug‐interactions‐table‐substrates‐inhibitors‐and‐inducers. Accessed April 1, 2020.
- 35. Gaedigk A, Simon SD, Pearce RE et al. The CYP2D6 activity score: Translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 2008;83:234–242. [DOI] [PubMed] [Google Scholar]
- 36. Crews KR, Gaedik A, Dunnenberger HM et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 2014;95:376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Borges S, Desta Z, Jin Y et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 2010;50:450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shah RR, Smith RL. Addressing phenoconversion: The Achilles' heel of personalized medicine. Br J Clin Pharmacol 2015;79:222–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reidenberg MM. Pain control and the world health organization analgesic ladder. JAMA 1996;275:835. [DOI] [PubMed] [Google Scholar]
- 40. Patel JN, et al. Pain management using clinical pharmacy assessments with and without pharmacogenomics in an oncology palliative medicine clinic. JCO Oncol Pract 2020;16:e166–e174. [DOI] [PubMed] [Google Scholar]
- 41. Klepstad P, Fladvad T, Skorpen F et al. Influence from genetic variability on opioid use for cancer pain: A European genetic association study of 2294 cancer pain patients. Pain 2011;152:1139–1145. [DOI] [PubMed] [Google Scholar]
- 42. Kekic A, Seetharam M, Singh P et al. Integrating pharmacogenomics panel testing for supportive care medications in patients with solid tumors. Journal of Clinical Oncology 2020;38:e24114. [Google Scholar]
- 43. Whitney RL, Bell JF, Tancredi DJ et al. Unplanned hospitalization among individuals with cancer in the year after diagnosis. J Oncol Pract 2019;15:e20–e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Whitney RL, Bell JF, Tancredi DJ et al. Hospitalization rates and predictors of rehospitalization among individuals with advanced cancer in the year after diagnosis. J Clin Oncol 2017;35:3610–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manzano JG, Luo R, Elting LS et al. Patterns and predictors of unplanned hospitalization in a population‐based cohort of elderly patients with GI cancer. J Clin Oncol 2014;32:3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iqbal J, Ginsburg O, Rochon PA et al. Differences in breast cancer stage at diagnosis and cancer‐specific survival by race and ethnicity in the United States. JAMA 2015;313:165–173. [DOI] [PubMed] [Google Scholar]
- 47. Fang H, Liu X, Ramírez et al. Establishment of CYP2D6 reference samples by multiple validated genotyping platforms. Pharmacogenomics J 2014;14:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lopes GS, Bielinski SJ, Moyer AM et al. Sex differences in associations between CYP2D6 phenotypes and response to opioid analgesics. Pharmgenomics Pers Med 2020;13:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Table S1 FDA Classification of CYP2D6 Inhibitors


