Abstract
Background
Over the past few years, tumor next‐generation sequencing (NGS) panels have evolved in complexity and have changed from selected gene panels with a handful of genes to larger panels with hundreds of genes, sometimes in combination with paired germline filtering and/or testing. With this move toward increasingly large NGS panels, we have rapidly outgrown the available literature supporting the utility of treatments targeting many reported gene alterations, making it challenging for oncology providers to interpret NGS results and make a therapy recommendation for their patients.
Methods
To support the oncologists at Vanderbilt‐Ingram Cancer Center (VICC) in interpreting NGS reports for patient care, we initiated two molecular tumor boards (MTBs)—a VICC‐specific institutional board for our patients and a global community MTB open to the larger oncology patient population. Core attendees include oncologists, hematologist, molecular pathologists, cancer geneticists, and cancer genetic counselors. Recommendations generated from MTB were documented in a formal report that was uploaded to our electronic health record system.
Results
As of December 2020, we have discussed over 170 patient cases from 77 unique oncology providers from VICC and its affiliate sites, and a total of 58 international patient cases by 25 unique providers from six different countries across the globe. Breast cancer and lung cancer were the most presented diagnoses.
Conclusion
In this article, we share our learning from the MTB experience and document best practices at our institution. We aim to lay a framework that allows other institutions to recreate MTBs.
Implications for Practice
With the rapid pace of molecularly driven therapies entering the oncology care spectrum, there is a need to create resources that support timely and accurate interpretation of next‐generation sequencing reports to guide treatment decision for patients. Molecular tumor boards (MTB) have been created as a response to this knowledge gap. This report shares implementation strategies and best practices from the Vanderbilt experience of creating an institutional MTB and a virtual global MTB for the larger oncology community. This report describe a reproducible framework that can be adopted to initiate MTBs at other institutions.
Keywords: Molecular tumor board, Precision oncology, Implementation, Targeted therapies, Next‐generation sequencing
Short abstract
This article describes the initiation and implementation of two molecular tumor boards (MTBs) at Vanderbilt‐Ingram Cancer Center: an institutional board for patients and a global community MTB open to the larger oncology patient population. This reproducible framework may be adopted by other institutions to recreate MTBs.
Introduction
An increasing number of molecularly driven therapies are being approved for patients with cancer. This has prompted a dramatic increase in the use of next‐generation sequencing (NGS) panel tests to identify treatment options for individual patients through currently approved drugs, off‐label drug uses, or clinical trial opportunities. Recent studies [1, 2, 3] have shown that a successfully sequenced tumor sample can identify a wide range of actionable targets based on panel size; 39%–90% of patients who received NGS testing had at least one targeted therapy option identified [4]. Furthermore, several studies have shown that patients who were treated with a biomarker‐based therapy had an increased progression‐free survival and enhanced response rate compared with those who were not [4].
As biomarker‐driven treatment options continue to grow, it must be acknowledged that interpreting NGS tests to guide treatment decisions is challenging. This is due to both a limited understanding of genomics data and the complexity of the panels themselves. Over the past few years, NGS panels have evolved from a selected handful of genes to larger panels with hundreds of genes. Additionally, although the incorporation of paired normal DNA NGS to “filter” germline from somatic variants has great advantages, newer challenges such as incidental findings have arisen [5]. Furthermore, whole exome and whole genome sequencing panels may be adapted into clinical practice in the coming years. With this move toward increasingly large NGS panels, we have rapidly outgrown the available literature supporting the utility of treatments targeting many reported gene alterations. This makes it challenging for oncology providers to meaningfully interpret NGS results and make a therapy recommendation for their patients. Studies have shown that only 30%–50% of oncologists feel confident in their knowledge of genomics, explaining NGS results to patients, and using genomic information to make treatment decisions [6].
To address this issue, several institutes, including community hospitals, have started their own molecular tumor boards (MTBs) [7, 8, 9, 10, 11] to support clinicians in interpreting NGS test results. Traditional tumor boards are focused on cancers that arise within a limited range of organs. In contrast, MTBs are often organ‐agnostic and generally comprise various experts with experience in cancer research, cancer treatment of various types, and cancer genomics. At the Vanderbilt‐Ingram Cancer Center (VICC), we have successfully initiated two MTBs—a VICC‐specific institutional MTB and a global MTB. The VICC MTB focuses on patients at VICC and other affiliate sites and specializes in discussing molecular reports from NGS as well as germline tests and provides written recommendations in a brief summary report that is uploaded to the patient's electronic health chart. The Our Cancer Genomes virtual MTB, from here on referred to as the Global MTB, is open and accessible to all oncology providers globally, thereby lending collective expertise to the entire oncology community.
Past publications on MTBs have primarily focused on patient genomics, therapy recommendations, patient outcomes, and the broader utility of services such as MTB. Herein, we focus on the challenges of implementing such a service at a large comprehensive cancer center such as VICC. The collaborative experiences of running these MTBs successfully for over 2.5 years strategically positions us to share our learnings from this implementation experience. In this report, we share implementation strategies and best practices that have enabled us to perform a seamless integration of MTB in routine care and consequently helped us enhance the reach of MTB to community oncologists and international colleagues. Through this report, we hope to put forth a reproducible framework that can be adopted to initiate MTBs at other institutions.
Materials and Methods
Case Submission and Preparation
A Research Electronic Data Capture (REDCap) form [12] has been designed to collect patient information in a secure, online fashion (https://is.gd/vicc_hope_mtb). The form allows providers to pick a convenient presentation date. Providers are encouraged to upload redacted patient NGS reports within the case submission form. No specific protected health information (PHI) is requested other than medical record number (MRN). Institutional review board approval was obtained prior to beginning the MTB (no. 191885). Once the cases for a week are finalized, they are checked to remove any PHI not redacted, and clinical and genomic details are compiled on a slide deck. At this point, all dates are converted to MM/YYYY format, and any missing details are populated after consulting with the submitting provider. A reminder e‐mail with the list of cases to be presented is sent to the e‐mail list server along with the presenting providers on the day prior to presentation. The case submission process is similar for VICC MTB and the Global MTB.
Molecular Tumor Board Meeting
Meetings for VICC MTB typically comprise oncologists, physician scientists, hematologists, cancer geneticists, pathologists, scientists, cancer genetic counselors, clinical fellows/residents, and medical students. Meetings are held weekly in a large theatre style conference room that can hold up to 100 people. A maximum of four cases are presented at each meeting to allow sufficient opportunities for discussion. A staff scientist is present at all meetings and captures important points of discussion and recommendations. These are later transcribed into a formal recommendation report that goes through a review process, including by the practicing physician leading the session, before being finalized and uploaded to the electronic medical record (EMR) (Fig. 1). Typically, an individual with a Ph.D. in biological sciences and extensive training in precision oncology and current oncology‐related U.S. Food and Drug Administration (FDA) approvals was selected to be a staff scientist for this study. The MTB recommendations are advisory, and the final authority rests with the treating oncologist. Beginning in March 2020, the meetings were transitioned to completely virtual online meetings because of the SARS‐CoV‐2 pandemic.
Figure 1.
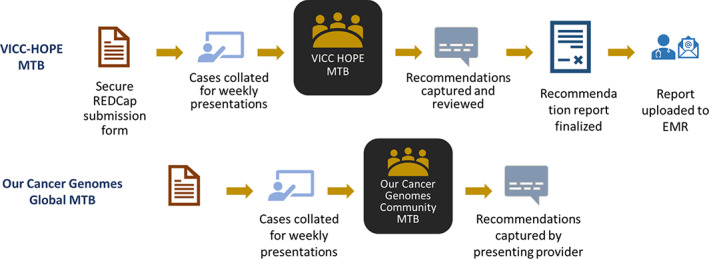
Workflow of MTB. Cases are submitted by providers using a secure REDCap form. These are collated by a staff scientist into a slide deck. A maximum of four cases are presented at each weekly meeting. For the Vanderbilt‐Ingram Cancer Center MTB, the recommendations are captured in a formal report that undergoes review before being finalized and uploaded to the EMR.Abbreviations: EMR, electronic medical record; MTB, molecular tumor board; REDCap, Research Electronic Data Capture; VICC HOPE, Vanderbilt‐Ingram Cancer Center, Hereditary & Oncology Personalized Evaluation.
Meeting attendees for Global MTB are similarly qualified but come from across the global oncology community. Virtual meetings are held monthly. Although recommendations are made during the Global MTB, these are not formalized into a report unless specifically requested by the submitting provider.
Analysis
Patient MRNs from the REDCap submission portal were extracted for all VICC patients and used for running an informatics query through the EMR to evaluate the vital status for all patients presented through December 2020. When a vital status of deceased was encountered, date of death, if available, was captured. Data pertaining to therapy recommendations was manually extracted from recommendation reports and was visualized using Microsoft Excel via bar charts and sunburst plots.
Provider Survey
A REDCap survey was created and sent to providers to assess their feedback for VICC MTB. The responses were collected and plotted on a bar chart. The survey is ongoing and can be found at https://redcap.vanderbilt.edu/surveys/?s=EME4RAEAY3.
Results
In this report, we discuss our experience with initiating MTBs at VICC. Figure 1 shows the overall workflow process for the institutional VICC MTB and Global MTB. We alternate VICC MTB meetings between Mondays and Tuesdays to increase provider engagement and opportunities to participate for clinicians with varied schedules. To encourage attendance and participation, one Continuing Medical Education (CME) and one Maintenance of Certification (MOC) credit are made available to attendees. The attendance varies from 10 to 25 attendees per session. The Global MTB meets on the fourth Monday of every month and is similarly attended. No educational credits are provided for attending Global MTB.
To date through December 2020, at the VICC MTB, we have discussed over 170 patient cases from 77 unique oncology providers from VICC and its affiliate sites. Figure 2 shows the breakdown of VICC MTB cases by diagnoses. The highest number of cases were presented for patients with lung cancer followed by breast cancer. This seems reasonable because of the abundance of molecularly driven therapies in these cancers and the integration of biomarker/protein testing into routine care. Additionally, these are also among the most common cancers in the southern states of Tennessee, Alabama, Mississippi, and Texas [13, 14]. There were several relatively rare cancers that were also presented, including mixed adrenoneuroendocrine carcinoma, malignant peripheral nerve sheath tumor, Wilms tumor, and adenoid cystic carcinoma. Patients with conditions that increase the likelihood of developing cancer, such as PIK3CA‐related overgrowth syndrome, intraductal papillary mucinous neoplasm, or a family history significant for cancers, were also presented.
Figure 2.
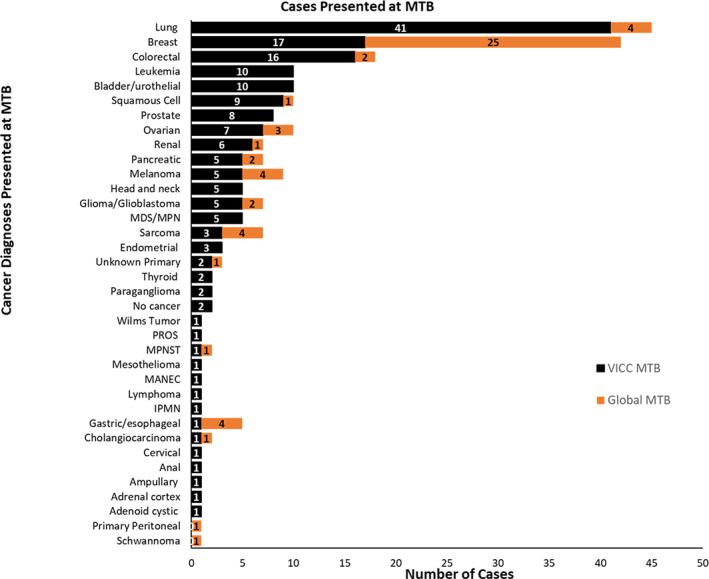
Disease Distribution for MTB cases. Several cancer diagnoses have been presented at Vanderbilt‐Ingram Cancer Center MTB (n = 177) and at the Global MTB (n = 57). If patients had multiple diagnoses, those were counted separately.Abbreviations: IPMN, intraductal papillary mucinous neoplasm; MANEC, mixed adrenoneuroendocrine carcinoma; MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; MPNST, malignant peripheral nerve sheath tumor; MTB, molecular tumor board; PROS, PIK3CA‐related overgrowth syndrome.
A total of 58 international patient cases have been presented at the Global MTB by 25 unique providers from six different countries across the globe. The disease distribution is shown in Figure 2. Breast cancer was the most commonly presented diagnosis with almost 50% of the cases involving patients with triple‐negative breast cancer subtype.
A summary of MTB recommendation types is shown in Figure 3. Recommendations made at MTB broadly focus on identifying and prioritizing all potential molecularly directed treatment options for patients. These recommendations are mostly for off‐label use of FDA‐approved therapies (n = 56) and for clinical trials (n = 59). These are either local trials or multiarm studies, such as MATCH from the National Cancer Institute [15] or TAPUR from the American Society of Clinical Oncology (ASCO) [16]. FDA‐approved therapies for indicated use were also recommended for some patients (n = 36). In the presented cohort, additional testing was requested for 108 patients. An additional somatic NGS test was deemed appropriate for 20% (n = 34) of patients to gather more information on their tumor. This is resorted to in cases requiring additional clarification, for example, in patients with multiple malignancies, tests from outdated samples (>3 years), or if the report is thought to be potentially erroneous. We performed a chart review to assess these patients and found that 9 of the 34 patients went on to receive an additional somatic test as recommended at MTB. Of the remaining, nine were either sent to hospice or were deceased less than 6 months following presentation, seven were lost to follow‐up or were receiving primary care elsewhere, one patient had no evidence of disease, and one patient was on active surveillance. The remaining seven patients did not receive any additional testing; no reason was provided by the treating oncologist for this decision. Germline testing is recommended to generally follow consensus guidelines, including either a strong family history of cancer being observed, in cases of multiple malignancies at a young age, or a close to 50% variant allele frequency (VAF) in a gene known to have pathogenic germline mutations. Forty‐four percent (n = 74) of the patients presented at VICC MTB were referred either to our Hereditary Cancer Clinic (HCC) or for germline testing to clarify somatic versus heritable variants. At times, when sufficient data are not available to warrant the use of a particular therapy or if a test panel is thought to be limiting, additional testing recommendations using a previous biopsy or circulating tumor DNA are suggested. A paired normal control, nontumor sample submitted along with the tissue biopsy in some instances can shed insight on such cases and alleviates the need to recommend an additional test.
Figure 3.
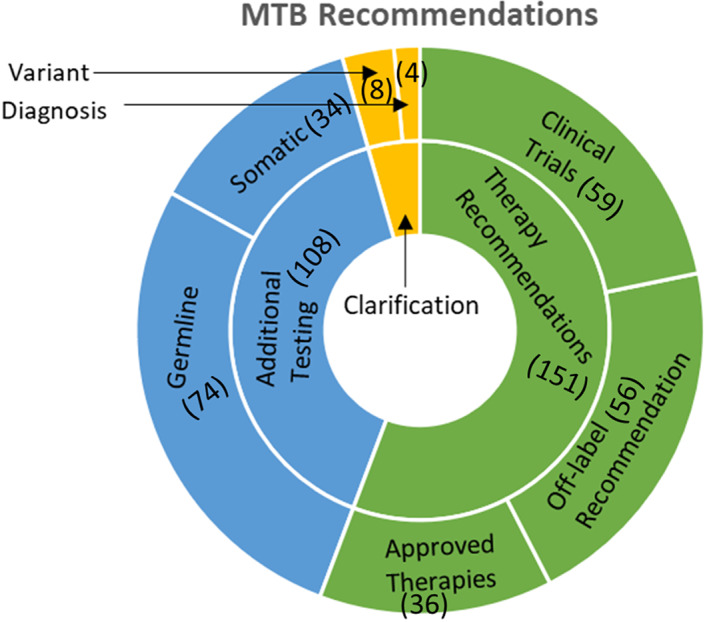
Vanderbilt‐Ingram Cancer Center MTB recommendations. The sunburst chart shows recommendations broadly classified as therapy recommendations (n = 151) and additional testing recommendations (n = 108). These were subdivided into approved therapies (n = 36), off‐label therapies (n = 56), clinical trials (n = 59), and germline (n = 74) and somatic testing (n = 34), respectively. A small fraction of cases (n = 12) were presented to obtain clarification on diagnosis (n = 4) and a reported variant (n = 8).Abbreviation: MTB, molecular tumor board.
In an attempt to collect vital statistical data on the patients presented at VICC MTB, an informatics query was performed on the patient charts on March 1, 2021. One hundred forty‐five patients were used for this query; the remaining 25 patients were from affiliate sites and were not included in the analysis. Fifty patients had a confirmed date of death, and the days alive after MTB presentation were calculated and plotted. For the remaining patients, the last known date on which the patient was alive was used to calculate confirmed days alive after MTB presentation (Fig. 4).
Figure 4.
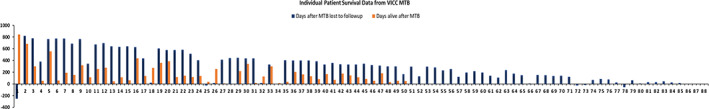
Individual deceased patient data from VICC MTB. Patient level data was assessed via Epic query on March 1, 2021. Orange bars represent patients that were found to be deceased at the time of the Epic query (March 1, 2021). The number of days alive after presentation at MTB was plotted as the length of the bars. Black bars represent patients were found to be alive (to the best of our knowledge), and the number of days patients lived after MTB presentation before being lost to follow‐up was plotted as length. Negative values imply that the patient transferred care elsewhere or was lost to follow‐up before presentation at MTB.Abbreviations: MTB, molecular tumor board; VICC, Vanderbilt‐Ingram Cancer Center.
Feedback is continually being gathered from presenting oncology providers through an ongoing survey. The initial decision to conduct MTB on a rotating schedule (i.e., alternating between Mondays and Tuesdays) was also based on feedback from the providers. A total of 19 providers have taken the survey so far. Seventy‐seven percent (n = 14) of providers reported discussing the MTB findings with their patients, and 55% (n = 10) of providers reported informing their patient prior to MTB presentation. Twenty‐six percent (n = 5) of providers presented exclusively to discuss a germline finding from an NGS test, whereas 74% (n = 14) reported that they presented to discuss the use of off‐label therapies. Thirty‐one percent (n = 6) of the case presentations were a result of a personalized e‐mail prompt regarding potentially actionable mutations. There was also a request from some providers to make the process of recommending patients even more straightforward by linking it directly to the EMR, specifically via an Epic message alert. To accommodate these providers, we do currently allow case submissions via a direct EMR alert to the staff coordinating MTB scheduling. A mock case presentation and patient report can be seen in Figure 5.
Figure 5.
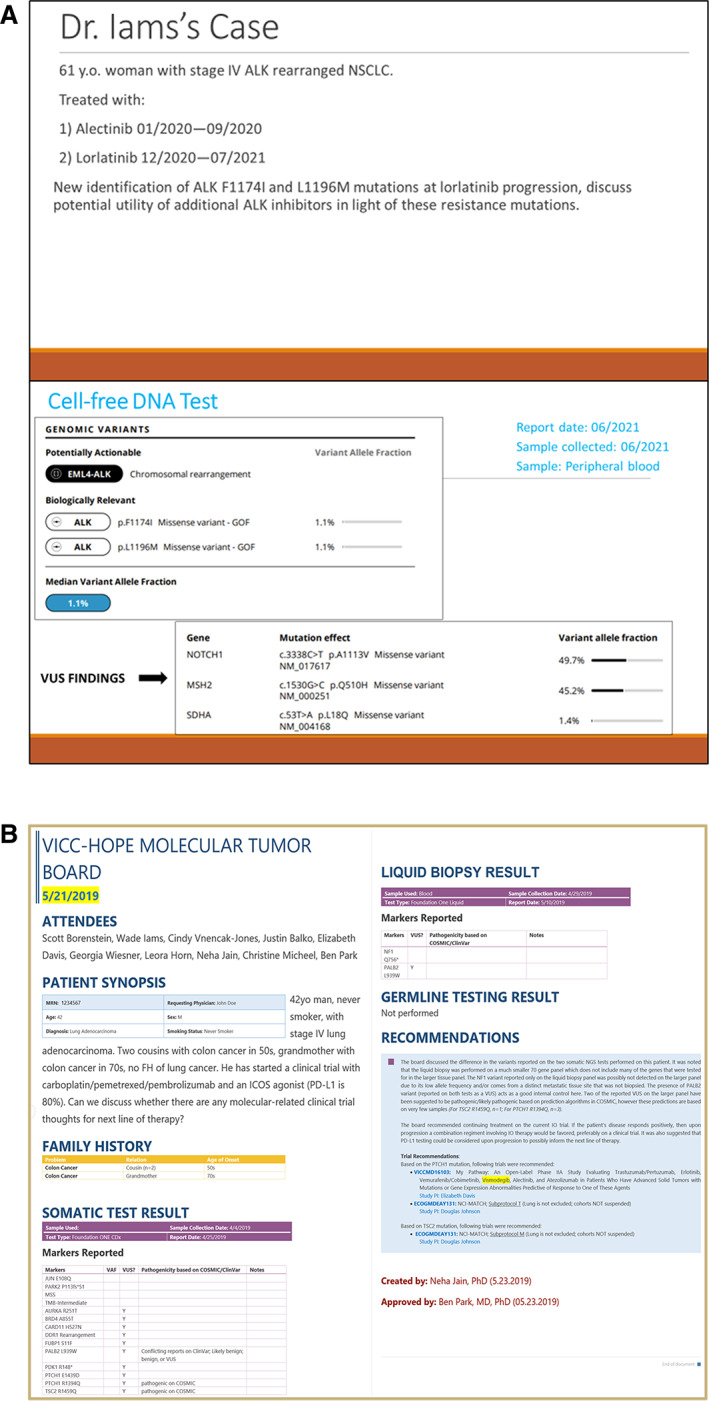
Screenshots from MTB. (A): Slides from an MTB presentation. (B): A deidentified patient report.Abbreviations: FH, family history; ICOS, inducible costimulator of T cells; GOF, gain of function; NSCLC, non‐small cell lung cancer; VAF, variant allele frequency; VUS, variant of unknown significance.
Another emerging component of tumor testing is the use of RNA NGS (RNA seq) to uncover fusions and/or structural variants that would not be discovered with traditional tumor DNA NGS. Brief summaries from two example cases that highlight the usefulness of RNA seq are presented below. The first case was presented to guide the next line of therapy, and the second case was presented to clarify the diagnosis.
Case 1
A 42‐year‐old man with no history of smoking was diagnosed with stage IV poorly differentiated, non‐small cell carcinoma. At diagnosis, his primary lung tumor was analyzed with a 324‐gene NGS panel, revealing a JUN E108Q mutation, PARK2 P113fs*51 mutation, and a tumor mutational burden (TMB) of nine mutations per megabase. He was treated on a clinical trial with platinum doublet, PD‐1 inhibitor, and an inducible costimulator of T cells agonist for 11 months. Because of central nervous system metastases, he was then treated with two lines of standard‐of‐care chemotherapy. Upon progression on third‐line systemic therapy, his tumor was again sequenced. NGS sequencing using a 648‐gene panel revealed copy number losses in CDKN2A, CDKN2B, and MTAP and a TMB of 1.6 mutations per megabase. Furthermore, whole transcriptome RNA sequencing revealed a CD74‐ROS1 chromosomal rearrangement that made the patient eligible for ROS1‐targeted therapies. The patient has had a partial response and ongoing disease control for 6 months as of this writing.
Case 2
A 3‐year‐old boy presented with rapid onset of upper extremity flaccid paralysis and an inability to walk. Imaging showed large brain and spinal cord lesions and extensive leptomeningeal disease. Biopsy revealed poorly differentiated carcinoma but no definitive cancer type. Because of progressive symptoms and no definitive diagnosis, the patient was emergently started on chemotherapy (etoposide, cytoxan, vincristine, and HD methotrexate). The patient responded favorably to his chemotherapy. An NGS report from a 648‐gene panel and whole transcriptome RNA sequencing showed an NFIA/CBFA2T3 fusion in the biopsy specimen. The evidence from molecular testing and previous case reports [17, 18] suggested that this tumor was a central nervous system erythroid leukemia with sarcomatous presentation. This confirmation of diagnosis allowed modification of the chemotherapeutic regimen to a protocol more suited for erythroid leukemia and therefore was critical to the patient's treatment. This case has been recently described in detail [19].
Discussion
Recently, the FDA Web site showed that ~33% of new drug approvals between January 1, 2021, and March 31, 2021, were based on the presence of a biomarker [20]—a trend that has continued for several years. This highlights the role of patient‐specific biomarkers in cancer care and emphasizes the need for developing resources that are available to oncologists for assisting in clinical decision making when it comes to biomarker‐driven therapies. Keeping this vision in mind, we have initiated VICC MTB so that VICC patient reports can be discussed and the appropriate treatment plan can be determined. Going a step further, we have also deployed an open and accessible monthly virtual Global MTB that invites oncology cases from across the global community.
There are 51 National Cancer Institute–designated comprehensive cancer centers [21] in the U.S. and 2,368 oncology practices according to the 2017 ASCO survey [22]. This puts cancer centers such as VICC in a strategic position in terms of sharing lessons learned and lending oncology expertise to community oncology practices. With the southern states of the country having the largest share of oncology practices (33%) and unfortunately also accounting for 38% of new cancer diagnoses, this is an area of critical need [22].
Creating a streamlined process for case submission, report generation, and report upload to the EMR allows us to increase the reach and adoption of MTB across various centers, divisions, and departments. As a result, we have seen increasing participation from neuro‐oncology, pediatric oncology, hematology, surgical oncology, Veterans Affairs, and our HCC. We have adopted REDCap for the online submission of reports because of its easy accessibility, security, and multiple customization features [12] but then also supplemented the process with human review for safeguarding PHI. Although providers are encouraged to submit redacted reports from their patients’ NGS tests, we periodically find a small percentage of reports containing PHI, which is manually removed while preparing slides. This stresses the need for manual review of cases so that PHI is not accidentally exposed to a wide audience that may be in attendance for MTB.
Participation in MTB is encouraged by providing CME and MOC credit to attendees. Such incentives can be great motivators for busy clinicians. Furthermore, regular personalized e‐mail alerts to oncologists also drive participation in MTB. In our experience, MTB has mainly been advertised through colleagues. Providers who have previously trained at VICC are also encouraged to continue their relationship and use this useful resource at their current facility. In the future, integrating the EMR system to be able to receive structured data from NGS tests and create personalized alerts when detecting certain “high‐value” mutations could reduce the current manual burden of e‐mailing providers to potentially discuss cases with actionable findings.
The recommendations from the VICC MTB are manually captured in a report for documentation purposes and also to maintain a record that can be helpful in case of transfer of care. This report is directly uploaded to the patient's chart, providing easy access to providers. These reports are tagged with “VICC molecular” keyword to make them easily searchable from within the EMR. Although creating a formal report is a labor‐intensive process, formal documentation has high value since it can help achieve authorization or approval from insurance in case of off‐label drug use or, as case study–level data, a therapy recommendation based on limited evidence [23].
It has been shown that 5%–10% of cancers are related to inherited genetic mutations [24]. Synchronizing our MTB efforts and working with our HCC at VICC has allowed us to recommend surveillance plans for at‐risk individuals and their family members, thereby allowing for early detection and intervention. Patients with no cancer diagnoses but a very strong family history of cancers and/or a concerning germline test result have been presented at MTB. Forty‐four percent of the patients presented at VICC MTB were referred either to HCC or for germline testing to clarify results from tumor NGS tests.
Patients were also presented when a subclonal population potentially indicating clonal hematopoiesis of indeterminate potential (CHIP) [25] was observed. CHIP, a recently described entity, is sometimes incidentally detected by NGS tests and can be a precursor to hematologic malignancies and cardiovascular disease [26]. Individuals in whom CHIP was identified (n = 5, 3%) and discussed in MTB have been referred to a CHIP clinic recently begun at Vanderbilt University Medical Center. This clinic helps make specific therapy recommendations or modifications based on the presence of CHIP, often identified on an NGS report.
The HCC and CHIP clinics point to an underexplored application of MTBs as a preventive measure by leveraging genomic expertise to design personalized surveillance plans for at‐risk individuals. In the near future, MTB use may witness a rapid expansion with the increased number of direct‐to‐consumer genetic testing kits in the market, which has been predicted to grow by ~15% in the 2020–2026 period [27]. In the future, resources could be developed that allow patients to directly interact with an online system that supports upload of their genetic test and allows them to receive expert guidance.
In the past 2.5 years, we have had NGS reports presented from various vendors and have witnessed the challenges of not receiving complete data on these reports. It is noteworthy that more genomic data do not always translate into greater insight when it comes to treatment options. NGS data can be of varying breadth, and apart from differences in the actual genes tested, there are also differences in the level of reporting. VAFs, which can be a helpful indicator in identifying if a variant is germline or part of a small subclonal population, is typically generated for all reported variants, but information related to VAFs is not typically shared by all vendors. Furthermore, processing a nontumor, normal matched sample along with the actual tumor specimen can be extremely helpful in increasing confidence in the somatic status of a mutation, but this service is currently only offered by a few vendors. Even so, germline mutations that are detected in these NGS tests are not uniformly reported. For example, our current vendor only reports germline pathogenic results if recommended by the American College of Medical Genetics list of 59 genes for incidental reporting [28]. Importantly, germline filtering is not equated to true germline testing, and alterations such as large rearrangements in the germline will not be detected by most NGS vendors using paired normal DNA for germline filtering purposes. These issues highlight the complexity in interpreting reports uniformly and may require additional tests to elucidate the full genomic picture in an individual patient. RNA seq with whole transcriptome analysis, for example, has the potential to uncover fusions that can be otherwise missed in most DNA NGS reports. The two cases presented in this report highlight the role of RNA seq in deciphering diagnoses as well as guiding the next line of treatment.
We used a provider survey to obtain feedback on the MTB workflow and in the process discovered that although 77% of providers reported discussing the MTB findings with their patients (n = 14), only 55% (n = 10) of providers reported informing their patient prior to MTB presentation. This could be to avoid understandable patient disappointment if no actionable targets are present. Although a large number of genes are present on NGS panels, not all of them have related targeted therapies. The problem posed by variants of unknown significance can make interpretation difficult and may not be helpful in subsequent decision support for guiding the next line of treatments in addition to increasing financial burden and anxiety for patients. The survey is ongoing, and we hope to gather more insights and continue to improve the experience of NGS report interpretation for oncologists.
Cancer is a heterogenous group of diseases, and by combining our expertise and knowledge to discuss genomic reports, we hope to present novel avenues for optimal patient care. We call upon the entire oncology community to participate in our Global MTB and encourage the creation of MTBs at local institutions so that additional patients can benefit from the combined expertise of multiple institutions.
Conclusion
As NGS testing and FDA indications expand for increasingly complex, biomarker‐driven treatment options for patients with cancer, it is imperative that clinical decision support is provided to treating oncologists and other specialists to identify potential treatment based on molecular biomarkers. The MTBs at VICC are an example of a multidisciplinary team approach for interpreting NGS results and developing a personalized approach to therapy. As new therapies are developed, MTBs can quickly integrate new treatments into everyday care. Leveraging this expertise to our global MTB is meeting the urgent need for the growing population of patients with cancer by providing support to regional and international oncologists.
Author Contributions
Conception/design: Neha M. Jain, Ben Ho Park, Wade Iams
Provision of study material or patients: Lauren Schmalz, Christopher Cann, Travis Osterman, Thomas Stricker, Justin M. Balko
Collection and/or assembly of data: Neha M. Jain, Lauren Schmalz, Christopher Cann, Adara Holland, Wade Iams
Data analysis and interpretation: Neha M. Jain, Travis Osterman, Katie Lang, Georgia L. Wiesner, Tuya Pal, Christine Lovly, Christine Micheel, Justin M. Balko, Douglas B. Johnson, Ben Ho Park, Wade Iams
Manuscript writing: Neha M. Jain, Tuya Pal, Douglas B. Johnson, Ben Ho Park, Wade Iams
Final approval of manuscript: Neha M. Jain, Lauren Schmalz, Christopher Cann, Adara Holland, Travis Osterman, Katie Lang, Georgia L. Wiesner, Tuya Pal, Christine Lovly, Thomas Stricker, Christine Micheel, Justin M. Balko, Douglas B. Johnson, Ben Ho Park, Wade Iams
Disclosures
Neha M. Jain: GE Healthcare (RF); Travis Osterman: eHealth, AstraZeneca, Outcomes Insights, Biodesix, MD Outlook, GenomOncology, Cota Healthcare, Flagship Biosciences (C/A), GE Healthcare, Microsoft, IBM Watson Health (RF), Infostratix (OI); Christine Lovly: Pfizer, Novartis, AstraZeneca, Genoptix, Sequenom, Ariad, Takeda, Blueprints Medicine, Cepheid, Foundation Medicine, Roche, Achilles Therapeutics, Genentech, Syros, Amgen, EMD‐Serono, Eli Lilly & Co. (C/A), Xcovery, AstraZeneca, Novartis (RF); Christine Micheel: Roche, Genentech (C/A), GenomOncology (RF); Justin M. Balko: Genentech/Roche, Bristol‐Myers Squibb, Incyte Corporation (RF), Novartis (C/A), patent regarding immunotherapy biomarkers in cancer (IP); Douglas B. Johnson: Array, Bristol‐Myers Squibb, Catalyst, Iovance, Jansen, Merck, Novartis, Oncosec (C/A), Bristol‐Myers Squibb, Incyte (RF); Ben Ho Park: Jackson Labs, Sermonix, EQRx, Hologics (C/A), Celcuity Inc. (SAB, OI), Horizon Discovery Ltd, The Johns Hopkins University (IP); Wade Iams: Genentech, Jazz Pharma, G1 Therapeutics, Clinical Care Options, Curio Science, Chardan, Outcomes Insights, Defined Health (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Acknowledgments
The authors acknowledge all oncologists from VICC, its affiliate sites, and oncologists from other institutions, who contributed cases to the MTBs. These include but are not limited to Saint Thomas— Midtown Hospital, Nashville, TN; Baptist Memorial Hospital, Columbus, MS; The University of Mississippi Medical Center Jackson, MS; Johns Hopkins Medical Institute, Baltimore, MD; Allegheny Health Network, Pittsburgh, PA; Woodlands Medical Specialists, Pensacola, FL; Virginia Commonwealth University Health, Richmond, VA; University of Wisconsin‐Madison, Madison, WI; White Plains Hospital, White Plains, NY; Institutul Oncologic Prof. Dr.I. Chiricuta, Cluj‐Napoca, Romania; Tan Tock Seng Hospital, Singapore (Novena); Medic Primar Oncologie, Bucharest, Romania; and Hospital Beatriz Ângelo, Loures, Portugal. L.S. is currently affiliated with Wake Forest Baptist Comprehensive Cancer Center, Winston‐Salem, NC. W.I. was supported by a National Comprehensive Cancer Network Young Investigator Award. B.H.P. acknowledges support from the Susan G. Komen Foundation, the Breast Cancer Research Foundation, the Canney Foundation, the Marcie and Ellen Foundation, Amy and Barry Baker, and Stephen Kandel from the SAGE Patient Advocates. Funding for this project included support from the Vanderbilt‐Ingram Cancer Center's National Cancer Institute Cancer Center Support Grant (P30CA068485) and from philanthropic sources, including a grant from the Nashville Wine Auction, and the Kent & Laurie Parker Family Foundation.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
Disclosures of potential conflicts of interest may be found at the end of this article.
References
- 1. Johnson DB, Dahlman KH, Knol J et al. Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next‐generation sequencing panel. The Oncologist 2014;19:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tan O, Shrestha R, Cunich M et al. Application of next‐generation sequencing to improve cancer management: A review of the clinical effectiveness and cost‐effectiveness. Clin Genet 2018;93:533–544. [DOI] [PubMed] [Google Scholar]
- 3. Meric‐Bernstam F, Brusco L, Shaw K et al. Feasibility of large‐scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol 2015;33;2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwaederle M, Zhao M, Lee JJ et al. Association of biomarker‐based treatment strategies with response rates and progression‐free survival in refractory malignant neoplasms: A meta‐analysis. JAMA Oncol 2016;2:1452–1459. [DOI] [PubMed] [Google Scholar]
- 5. Reid S, Pal T. Update on multi‐gene panel testing and communication of genetic test results. Breast J 2020;26:1513–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gray SW, Hicks‐Courant K, Cronin A et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol 2014;32:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kato S, Kim KH, Lim HJ et al. Real‐world data from a molecular tumor board demonstrates improved outcomes with a precision N‐of‐One strategy. Nat Commun 2020;11:4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luchini C, Lawlor RT, Milella M et al. Molecular tumor boards in clinical practice. Trends Cancer 2020;6:738–744. [DOI] [PubMed] [Google Scholar]
- 9. VanderWalde A, Grothey A, Vaena D et al. Establishment of a molecular tumor board (MTB) and uptake of recommendations in a community setting. J Pers Med 2020;10:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koopman B, Groen HJM, Ligtenberg MJL et al. Multicenter comparison of molecular tumor boards in The Netherlands: Definition, composition, methods, and targeted therapy recommendations. The Oncologist 2021;26:e1347–e1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harada S, Arend R, Dai Q et al. Implementation and utilization of the molecular tumor board to guide precision medicine. Oncotarget 2017;8:57845–57854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris PA, Taylor R, Thielke R et al. Research Electronic Data Capture (REDCap) ‐ A metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Statistics Center American Cancer Society Web site. Available at http://cancerstatisticscenter.cancer.org/. Accessed June 1, 2021.
- 14. Common cancer types National Cancer Institute Web site. Available at https://www.cancer.gov/types/common-cancers. Accessed June 1, 2021.
- 15. Targeted Therapy Directed by Genetic Testing in Treating Patients with Advanced Refractory Solid Tumors, Lymphomas, or Multiple Myeloma (the MATCH Screening Trial) ClinicalTrials.gov identifier NCT02465060. Bethesda, MD: U.S. National Library of Medicine, June 8, 2015. Available at https://clinicaltrials.gov/ct2/show/NCT02465060. Accessed June 1, 2021.
- 16. TAPUR: Testing the Use of Food and Drug Administration (FDA) Approved Drugs That Target a Specific Abnormality in a Tumor Gene in People with Advanced Stage Cancer (TAPUR) ClinicalTrials.gov identifier NCT02693535. Bethesda, MD: U.S. National Library of Medicine, February 26, 2016. Available at https://clinicaltrials.gov/ct2/show/NCT02693535. Accessed June 1, 2021.
- 17. Micci F, Thorsen J, Panagopoulos I et al. High‐throughput sequencing identifies an NFIA/CBFA2T3 fusion gene in acute erythroid leukemia with t(1;16)(p31;q24). Leukemia 2013;27:980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu H, Guinipero TL, Schieffer KM et al. De novo primary central nervous system pure erythroid leukemia/sarcoma with t(1;16)(p31;q24) NFIA/CBFA2T3 translocation. Haematologica 2020;105:e194–e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linnik Y, Pastakia D, Dryden I et al. Primary central nervous system erythroid sarcoma with NFIA‐CBFA2T3 translocation: A rare but distinct clinicopathologic entity. Am J Hematol 2020;95:E299–E301. [DOI] [PubMed] [Google Scholar]
- 20.Center for Drug Evaluation and Research. Hematology/Oncology (Cancer) Approvals & Safety Notifications. U.S. Food and Drug Administration, 2021. Available at https://www.fda.gov/drugs/resources‐information‐approved‐drugs/oncology‐cancer‐hematologic‐malignancies‐approval‐notifications. Accessed June 1, 2021.
- 21.NCI‐designated cancer centers. National Cancer Institute Web site. Available at https://www.cancer.gov/research/infrastructure/cancer-centers. Accessed June 1, 2021.
- 22. Kirkwood MK, Hanley A, Bruinooge SS et al. The state of oncology practice in America, 2018: Results of the ASCO Practice Census Survey. J Oncol Pract 2018;14:e412–e420. [DOI] [PubMed] [Google Scholar]
- 23. Tafe LJ, Gorlov IP, de Abreu FB et al. Implementation of a molecular tumor board: The impact on treatment decisions for 35 patients evaluated at Dartmouth‐Hitchcock Medical Center. The Oncologist 2015;20:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The genetics of cancer. National Cancer Institute Web site. Available at https://www.cancer.gov/about-cancer/causes-prevention/genetics. Accessed June 1, 2021.
- 25. Steensma DP, Bejar R, Jaiswal S et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papa V, Marracino L, Fortini F et al. Translating evidence from clonal hematopoiesis to cardiovascular disease: A systematic review. J Clin Med 2020;9:2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Global direct‐to‐consumer (DTC) genetic testing market to expand at about 14.9% CAGR during 2020–2026. News release. New York: Zion Market Research. Available at https://apnews.com/press‐release/marketers‐media/business‐corporate‐news‐north‐america‐products‐and‐services‐genetic‐testing‐2a8ee087dccb8c1efdf078596b341dbc. Accessed June 1, 2021.
- 28.ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. National Center for Biotechnology Information Web site. Available at https://www.ncbi.nlm.nih.gov/clinvar/docs/acmg/. Accessed June 1, 2021.


