Abstract
An 81-year-old immunocompetent patient with bronchiectasis and refractory Mycobacterium abscessus lung disease was treated for six months with a three-phage cocktail active against the strain. In this case study of phage to lower infectious burden, intravenous administration was safe and reduced the M. abscessus sputum load 10-fold within one month. However, after two months, M. abscessus counts increased as the patient mounted a robust IgM- and IgG-mediated neutralizing antibody response to the phages, which associated with limited therapeutic efficacy.
Nontuberculous mycobacteria (NTM)—especially Mycobacterium abscessus infections-represent emerging pathogens of increasing clinical importance1. There is an urgent need for novel treatments for NTM as outcomes are often poor, particularly with macrolide-resistant strains2–4. Bacteriophages offer an innovative therapeutic approach for difficult-to-treat infections5,6 Recently, a 15-year-old lung transplant recipient with cystic fibrosis (CF) and disseminated M. abscessus subsp. massiliense (strain GD01) was treated successfully with an intravenous (IV) mycobacteriophage cocktail 7. In the setting of an immunosuppressive regimen to prevent allograft rejection, no phage-neutralizing immune response was observed. The role of host immunity in phage therapy is uncertain, and although serum neutralization might limit phage efficacy 8–10, its extent and clinical significance in immunocompetent persons is unclear 11–14 or has not been rigorously studied 15–18.
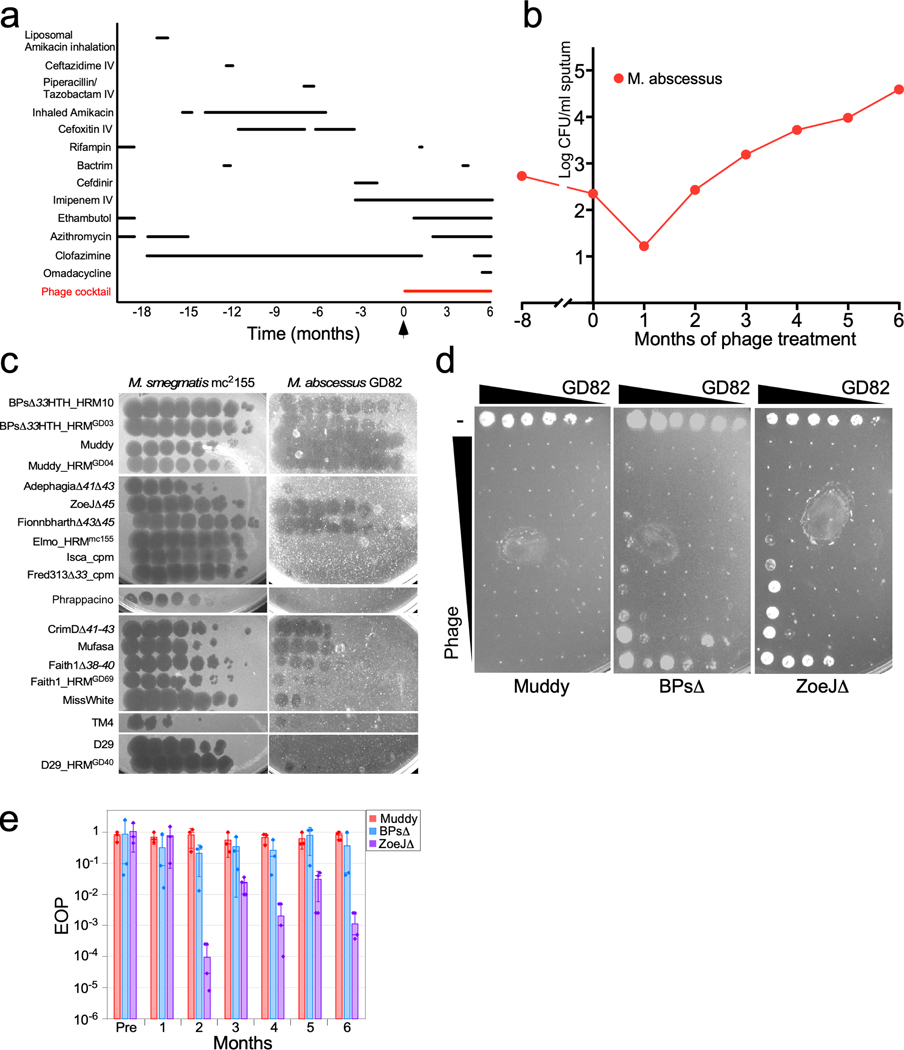
We identified an 81-year-old man with bronchiectasis and refractory NTM-lung disease as a candidate for mycobacteriophage therapy. For the prior five years he had been treated with differing antibiotic regimens for co-infection with M. abscessus and M. avium complex (MAC) (Fig. 1a), yet he remained acid-fast bacterium (AFB) sputum smear positive (Fig. 1b, Extended Data Fig. 1) with right upper lobe cavitary disease (Extended Data Fig. 2). The M. abscessus isolate, designated GD82, was a rough morphotype that subspeciated as M. abscessus subsp. massiliense. GD82 is closely related to strain GD01 from the previously mycobacteriophage-treated patient7 and is similarly resistant to a broad range of antibiotics, including macrolides (Extended Data Table 1). Screening of a mycobacteriophage library19 identified several phages with potent antimycobacterial activity against GD82 (Fig. 1c), including the same three phages (Muddy and derivatives of BPs and ZoeJ) used in the treatment of the GD01-infected patient 7. Muddy and the engineered BPsΔ33HTH_HRM10 and ZoeJΔ457 (hereafter referred to as ‘BPsΔ’ and ‘ZoeJΔ’, respectively) each were shown to effectively kill strain GD82 in vitro (Fig. 1d). Although potently bactericidal, a few surviving colonies were recovered following challenge of 108 colony forming units (CFU) of bacteria with phages; rescreening showed several patterns of resistance, but no derivative was resistant to all three phages (Extended Data Fig. 3).
Figure 1. Development and efficacy of a phage cocktail for M. abscessus treatment.
a, Administration timeline of anti-mycobacterial drugs and phage cocktail. b, Quantitative M. abscessus counts in expectorated sputum (log CFU mL−1) pre- and post-treatment; CFU counts at each timepoint were derived from plating in triplicate. c, Identifying therapeutically useful phages for M. abscessus GD82. Ten-fold serial dilutions (left to right) of phages (as shown) were plated on M. smegmatis mc2155 and M. abscessus GD82 as indicated. See Supplementary Materials for further details. Assays are representative of at least two replicates. d, Phage killing of M. abscessus GD82. A M. abscessus GD82 culture was 10-fold serially diluted (most concentrated samples contain ~107 CFU) and incubated without phage (−) or with 10-fold serial dilutions of phage (uppermost phage rows contain: Muddy, 1010 PFU; BPsΔ, 109 PFU; ZoeJΔ, 109 PFU), for 24 hrs at 37° C and plated onto solid media. e, Phage sensitivities of post-treatment M. abscessus isolates, demonstrating emergence of ZoeJ resistance. Efficiencies of plaquing (EOP) are shown for Muddy (red), BPsΔ (blue) and ZoeJΔ (purple) on M. abscessus isolated 1–6 months post treatment, relative to M. smegmatis (Pre is the pre-treatment M. abscessus GD82). Bar heights and error bars are mean ± s.d. from three replicate experiments (shown as points).
A three-phage cocktail was prepared from high titer, highly purified preparations and shown to be endotoxin free and sterile, as described previously7. Regulatory and institutional approval for IV mycobacteriophage administration was obtained from the Food and Drug Administration and the Johns Hopkins Institutional Review Board. While continuing multidrug antibiotic therapy for NTM, the three-phage cocktail was initiated at a dose of 1×109 plaque forming units (PFU) IV twice daily (Fig. 1a). To assess safety and efficacy, regular clinical, laboratory assessments and chest CT scans were performed (Extended Data Figs. 2, 4). M. abscessus and MAC burden were calculated monthly as mean CFU mL−1 of expectorated sputum (Fig. 1b, Extended Data Fig. 1). During treatment, M. abscessus was assessed for changes in antibiotic susceptibility (Extended Data Table 1) and for acquisition of phage resistance (Fig. 1e).
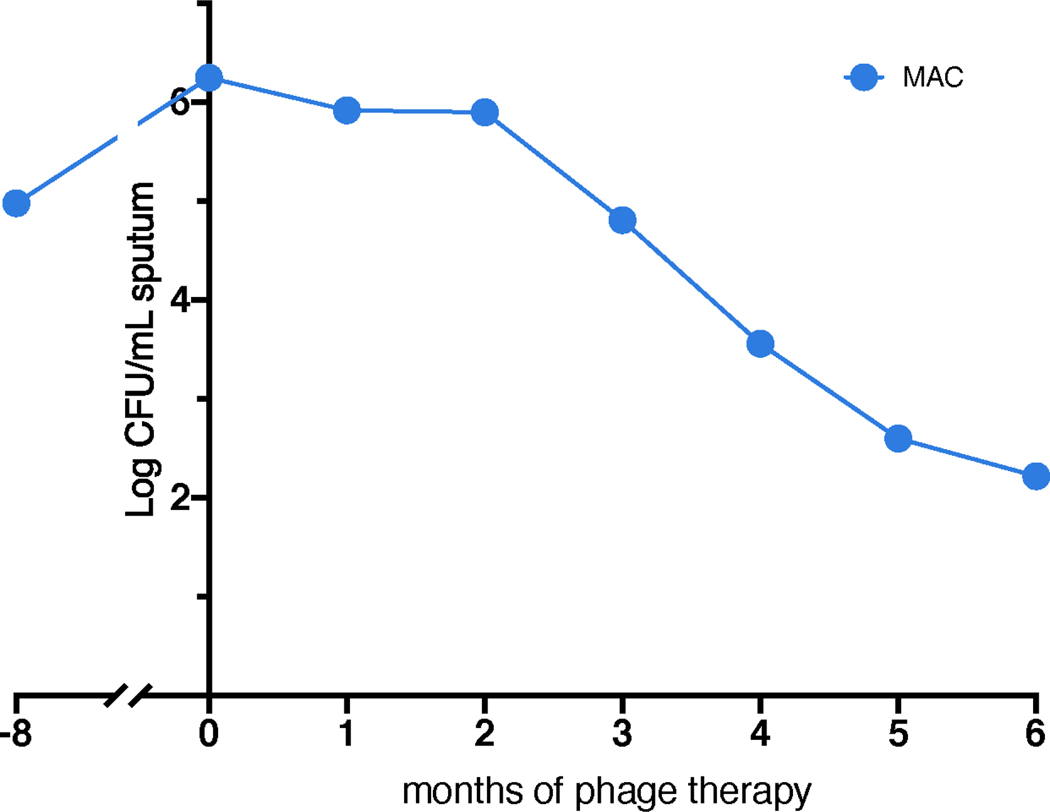
During the six-month phage treatment period, no serious adverse events were observed. Phage treatment was not associated with observable changes in respiratory or systemic symptoms, or evident changes in various clinical indicators and chest imaging (Extended Data Figs. 2, 4). There were no interruptions in phage treatment, as during safety monitoring there were no clinically significant changes in blood cell counts or differentials, liver or kidney function, and inflammatory markers (Extended Data Fig 4). On serial chest CT scans, the majority of chronic airway abnormalities appeared stable, with the exception of mildly increased right upper lobe cavitation and focal nodularity. Changes in companion antibiotics (Fig. 1a) were motivated by antibiotic-related side effects and efforts to improve anti-mycobacterial activity against MAC (Extended Data Fig. 1), against which the phages are not active.
Prior to phage treatment, M. abscessus was present at 2.4 log CFU ml−1 in sputum and decreased to 1.2 log CFU mL−1 one-month post-phage initiation (Fig. 1b). However, in subsequent months, bacterial counts rebounded such that six months-post phage initiation, M. abscessus was 2.2 log CFU ml−1 higher than pre-treatment levels (Fig. 1b). Post-treatment M. abscessus isolates remain fully sensitive to phages Muddy and BPsΔ, although there was intermittent resistance to ZoeJΔ (Fig. 1e). The full sensitivity to Muddy and BPsΔ indicates that phage resistance is not the primary cause of the increase in sputum bacterial count (Fig. 1b), although the acquisition of ZoeJΔresistance suggests that even given extensive structural lung disease, with IV administration the phages reached their target and imposed selective pressure. The antibiotic resistance profiles did not change post-treatment (Extended Data Table 1).
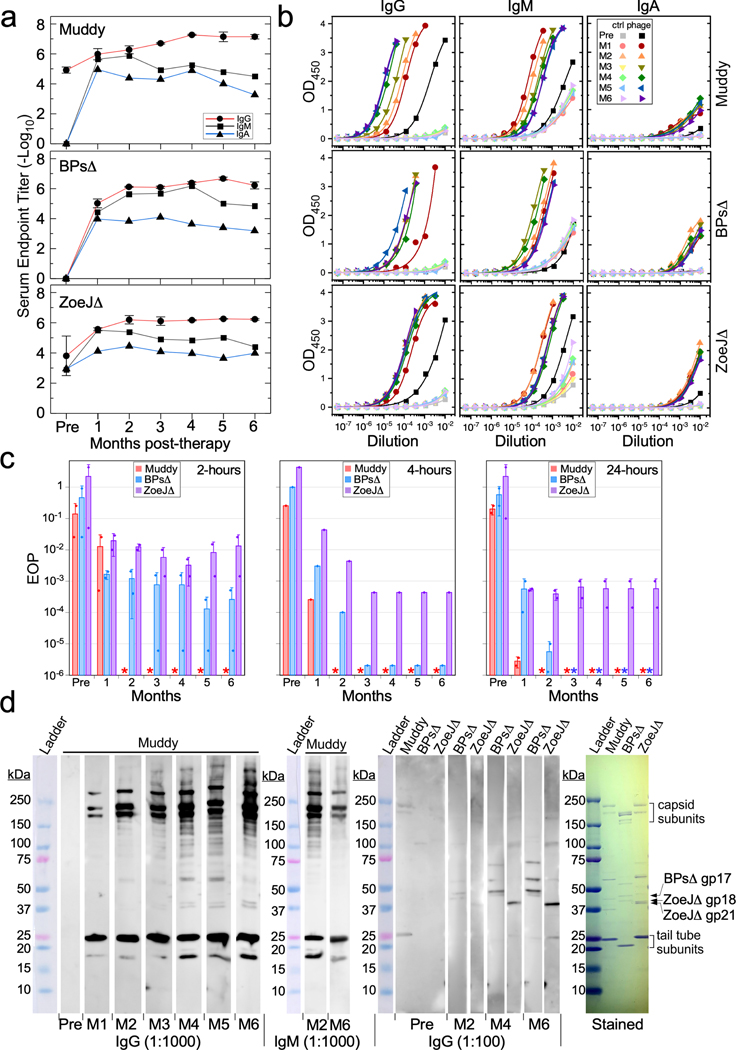
Pre-phage treatment, the patient had little or no antibody response to phage BPsΔ, and weak to moderate antibody binding to Muddy and ZoeJΔ proteins primarily IgG-mediated (Fig. 2a, b). Interestingly, the pre-treatment serum did not significantly impair infectivity of any of the phages, as there was only very mild phage neutralization (see Methods) (Fig. 2c). However, one month after starting phage treatment, we observed increases in IgG, IgM, and IgA antibody responses to all three phages (Fig. 2a, b). For BPsΔ and ZoeJΔ, the IgG response increased for 2–3 months and then stabilized (Fig. 2a, b), whereas for Muddy it continued to increase for several months; by month-4, IgG antibodies to each of the phages exceeded end-point titers of 1:1,000,000 dilution (Fig. 2a, b). IgM responses generally mirrored IgG activity early in treatment but diminished at later times (Fig. 2a, b); IgA responses were generally weaker than IgM or IgG.
Figure 2. Phage-neutralizing immune responses.
a, ELISA endpoint titers of IgG, IgM, and IgA antibodies in pre-phage serum (Pre) and sera 1–6 months (M1 – M6) post-phage therapy, for phages Muddy (top panel), BPsΔ (middle panel), and ZoeJΔ (bottom panel). Values are mean ± s.d. from two technical replicates. b, ELISA curves and logistic fits for IgG, IgM, and IgA responses to Muddy (top panels), BPsΔ (middle panels), and ZoeJΔ (bottom panels). Control (ctrl) samples without immobilized phage, to measure non-specific antibody binding to wells, are shown in light-colored symbols; additional controls are shown in Figure S1. Fitted curves display whole responses not wholly captured by end point determinations. c, Phage neutralization assays, in which sera were incubated with phage Muddy (red), BPsΔ (blue) and ZoeJΔ (purple) lysates for two, four and twenty-four hours, then 10-fold serially diluted and plated on lawns of M. smegmatis. The data are presented as EOP relative to a no-serum phage control. The pre-phage (Pre) serum is slightly neutralizing for Muddy and ZoeJΔ. Sera from months 1–6 post-treatment are shown, with potent serum neutralization of phage observed as early as 1 month. The limit of detection is approximately 103 PFU/ml (corresponding to EOP ~10−6), and asterisks indicate titers below the detection limit. Raw data are shown in Figure S4. Bar heights and error bars are mean ± s.d. from two replicate experiments (shown as points). d, Western blot analysis of serum responses to phages Muddy, BPsΔ, and ZoeJΔ, using either 1:100 or 1:1000 serum dilutions (as indicated) and detection with IgG or IgM-specific secondary antibodies. Pre-phage (Pre) serum shows weak responses using a 1:100 serum dilution, but there is a strong IgG-mediated recognition of Muddy that increases from month-1 (M1) to month-6 (M6) post-treatment, especially against the capsid and major tail subunits. Recognition of BPsΔ and ZoeJΔ is weaker but readily detected using a 1:100 serum dilution. Protein molecular weight ladder with sizes shown in kDa. Coomassie-stained gel (right) shows the structural proteins of phages Muddy, BPsΔ, and ZoeJΔ, with the abundant capsid and tail tube subunits indicated. The BPsΔ gp17, ZoeJΔ gp18, and ZoeJΔ gp21 minor tail proteins recognized by IgG antibodies were identified by mass spectrometry. IgG data is representative of at least two replicate experiments and unprocessed images are shown in Figure S5.
The antibody responses one-month post-treatment had substantial anti-phage neutralization activity and reduced phage infectivity by 2–3 orders of magnitude following two hours incubation of serum with phage lysates, with infectivity further reduced on longer incubations (Fig. 2c). At two months and beyond post-treatment, the serum was potently neutralizing for Muddy, reducing infectivity by at least six orders of magnitude after just 2 hours of incubation (Fig. 2c). Serum responses to ZoeJΔ and BPsΔ reduced infectivity by more than three orders of magnitude after 2 hours of incubation, but more so at longer incubation times (Fig. 2c). In general, these phage neutralization profiles reflect the ELISA titers at one month and beyond.
Western blot analysis shows strong IgG antibody recognition of the Muddy tail tube (~25 kDa) and interlinked capsid subunits (175 – 210 kDa) with increasing strength over the treatment course (Fig. 2d). Bindings to less-abundant proteins are evident in the higher titer samples, likely including minor tail proteins to which antibody interactions may block infection. IgM binding responses to Muddy are qualitatively similar to IgG binding but as in the ELISA data (Fig. 2a, b), decrease from 2 months to 6 months post-treatment. Both IgG and IgM reactions to BPsΔ proteins are weaker, partly reflecting low antibody titers (Fig. 2a), and primarily recognize proteins less abundant than the tail tube and capsid proteins, such as BPsΔ gp17, a minor tail protein identified by mass spectrometry (Fig. 2d). These profiles suggest independent immune reactions to these phages rather than cross-reactivity of the Muddy antibodies with BPsΔ or ZoeJΔ; the most similar proteins shared by these phages are the tail tube subunits (~40% identity), a prominent target of Muddy-specific antibodies, but not BPsΔ− or ZoeJΔ–specific antibodies (Fig. 2d). IgG reactivity to ZoeJΔ appears weaker than to both Muddy and BPsΔ, and targets less abundant structural proteins, most notably the ZoeJΔ gp18 and gp21 minor tail proteins as identified by mass spectrometry (Fig. 2d), which may be critical for host recognition. Taken together with the ELISA data, these antibody responses are consistent with the neutralization data and suggest that a prominent antibody response limits the effectiveness of phage treatment for M. abscessus. After six months of phage treatment a decision was made to discontinue phage therapy using intravenous administration due to lack of efficacy. Multidrug antibiotic therapy for NTM lung disease was continued.
The course and outcomes of this phage treatment are in noted contrast to the case reported previously, although the two M. abscessus subsp. masilliense strains are very closely-related20, the same phage cocktail was used for treatment, and the dose and regimen were identical. While there are other differences between these cases—including age and CF status—the most striking difference is the robust and neutralizing immune response observed in this immunocompetent patient that was not seen in the immunosuppressed post-lung transplant CF patient 7. The immune response and its phage neutralizing activity (Fig. 2a–c) temporally correlate with the increasing M. abscessus bacterial load (Fig. 1b) suggesting strongly that the immune reaction is a substantial impediment to phage-mediated bacterial killing. Phage susceptibility assays show that treatment failure does not result from the emergence of phage resistance (Fig. 1e). The pre-treatment immune reactions to the phages indicate the patient has been previously exposed to these or related phages that cross-react immunologically; however, it is striking that the pre-existing responses were not phage-neutralizing (Fig. 2c) and only weakly detected by Western blot (Fig. 2d). This suggests that in other cases considered for phage therapy, pre-treatment antibody responses in the absence of neutralization assays should not be interpreted as contraindicative.
Although phage therapy of NTM infections is a promising innovation, clinical experience with these agents remains in its infancy. For this patient, IV administration was pursued due to anatomic concerns regarding his cavitary lung disease potentially limiting nebulized delivery. A cocktail of three phages was chosen to avoid phage-resistance, and no post-treatment isolates were resistant to all three phages. Although clinical circumstances including antibiotic changes and active MAC co-infection cannot be excluded from contributing factors, the strong antibody-mediated phage neutralization is a likely cause of treatment failure. This is noted in contrast to the prior successful management of an immunosuppressed post-lung transplant recipient treated with the same IV mycobacteriophage cocktail, where serum phage neutralizing activity was not observed—at least up to nine months post-therapy initiation7. Immunosuppression used to prevent allograft rejection post-transplant may thus serve to avert emergence of anti-phage immune responses and preserve phage efficacy. These observations also suggest that if extended IV administration is desirable, it may be preferable to administer the phages serially or in a non-continuous fashion so as to avoid neutralization of all phages at once. It also emphasizes the benefits of understanding immune cross-reactivity among different potentially therapeutically useful phages.
Despite the failure of IV mycobacteriophage therapy to cure refractory M. abscessus lung disease in this case, there are important lessons for future clinical use of phage therapy, and differing phage treatment approaches may be required for immunocompetent and immunocompromised populations. Attention to host immune status and rigorous inclusion of immunologic parameters in the design of future phage therapy will be required to best determine the promise and pitfalls of this therapeutic strategy.
Methods
Regulatory and ethical approvals.
Regulatory approval for the use of mycobacteriophage was obtained from the United States Food and Drug Administration as Single Patient Expanded Access Investigational New Drug (IND) application #19847. This protocol was also approved by the Johns Hopkins University School of Medicine Institutional Review Board, and written informed consent was obtained prior to protocol initiation. The protocol principal investigator was responsible for safety monitoring and oversight of potential adverse events.
Mycobacteriophage administration.
A 5 mL bacteriophage solution containing Muddy, BPsΔ and ZoeJΔ diluted in normal saline at a dose of 109 PFU for each phage was injected intravenously twice daily via slow push.
Safety monitoring.
The first two lifetime doses of phage were administered in a monitored medical setting. Safety blood draws to monitor blood counts, electrolytes, liver and kidney function and inflammatory markers were performed weekly for the first month, then at least every two weeks for the second month, then at least monthly thereafter for the duration of phage therapy. Serum was collected monthly to assess for anti-phage immune responses.
Bacterial strains.
M. smegmatis mc2155 is a laboratory strain and was grown as previously described21, M. abscessus GD82 and GD82-M1 through -M6 are clinical isolates from Baltimore, Maryland. M. abscessus strains were grown in Middlebrook 7H9 media with OADC and 1 mM CaCl2 for 4–5 days, shaking, at 37° C. For plaque assays, M. abscessus cultures were sonicated briefly in a cup-horn sonicator (Q-sonica 500) at 30% amplitude with 15 sec on and 10 sec off until visibly dispersed, as in7; this does not adversely affect viability.
Mycobacterial enumeration in sputum.
Monthly spot expectorated sputa were processed and decontaminated using standard techniques (Snap n’ Digest™), per the manufacturer’s instructions. Processed sputum was plated in serial dilutions onto 7H11 plates in triplicate, and incubated for three weeks at 37° C. The plates were examined regularly for contamination, and colonies were counted when they were easily visible. The counts from triplicate measurements were used to calculate the mean colony forming units (CFU) mL−1 of sputum on days 3 and 5 (M. abscessus), and days 14 and 21 (MAC).
MIC determination.
Recovered clinical isolates of M. abscessus GD82 and GD82-M1 through -M6 underwent MIC determination to a standard panel of antibiotics using RAPMYCO Sensititre™ plates (TREK Diagnostic Systems), per the manufacturer’s instructions. Colony counts of the starting mycobacterial inocula were confirmed by plating onto 7H11 to ensure the adequacy of the inoculum for drug susceptibility interpretation. The lowest concentration of drug that did not show visible growth was recorded as the MIC to the respective drug. For trimethoprim/sulfamethoxazole, MIC was defined as 80% growth inhibition. For clarithromycin, extended 14-day incubation was utilized to assess for inducible clarithromycin resistance.
Phage susceptibility screening.
Phage susceptibility profiles were completed using a standard plaque assay21, and efficiencies of plaquing (EOP) were determined by spotting 10fold serial dilutions on M. smegmatis mc2155 and M. abscessus GD82 isolates, as described previously21. Phages from the collection at the University of Pittsburgh were propagated on M. smegmatis and tested on both strains.
Mycobacteriophage cocktail preparation.
Phages were grown on M. smegmatis mc2155 using solid media and recovered by collection of top agar. Top agar was centrifuged to remove debris and cells, yielding lysates with titers of >1 × 1011 PFU ml−1. Lysates were filtered using 0.22 μm filters and then concentrated into a pellet using centrifugation (28000 rpm x 1 hour). The phage pellet was resuspended in phage buffer (68 mM NaCl, 10 mM Tris HCl pH 7.5, 10 mM MgSO4, 10 mM CaCl2). Cesium chloride (CsCl) was added to a density of 1.5 g cm−3 (4.1 M) to this concentrate, subjected to equilibrium density gradient centrifugation for 16 h, the visible phage band collected (~1.5 ml), centrifuged similarly again, and stored at 4 °C; this yielded 1–2 ml of phage with titers of 1011–1013 PFU ml−1. For cocktail preparation, 1 ml of each phage sample was dialyzed against 1 L of Ringers Solution (Oxoid) four times for a minimum of 4 hours each. Undetectable levels of endotoxin were seen in any dialyzed sample using the EndoZyme II (Hyglos GmbH) assay. Sterility of each sample was confirmed by Accugen, Inc. Samples were combined to form a three-phage cocktail, each at 1×1011 PFU ml−1. Cocktail batches were prepared monthly.
Phage neutralization assays.
A 1:10 dilution of each serum sample was incubated with each of the three phages used in the cocktail separately. Specifically, 10 μl of serum was added to 90 μl of phage buffer (10 mM Tris-HCl (pH 7.5), 10 mM MgSO4, and 68 mM NaCl) containing ~1×1010 PFU of phage. A no serum control was also included. After incubating the serum-phage mixture at room temperature for 2, 4 and 24 hours, 10-fold serial dilutions were made and 3 μl of each dilution was spotted onto top agar overlays containing M. smegmatis mc2155. Plates were incubated at 37° C for 24 hours. Efficiencies of Plaquing (EOPs) were calculated by the ratio of titer for each sample to titer of the no serum phage control.
Isolation of phage-resistant mutants.
Phage resistant mutants were isolated by mixing 1ml of 1×108 CFU of M. abscessus GD82 and 1×109 PFU of each phage used in the cocktail, separately, in liquid culture and a mixture of them all together. After 5 days of incubation at 37° C shaking, 0.1 ml of the mixture was plated onto solid media and incubated for 5 days at 37° C. The number of surviving colonies were <10 with Muddy, <10 for BPsΔ33HTH_HRM10, <1000 for ZoeJΔ45, and <50 with all three phages. Surviving colonies were streaked three times onto solid media and then tested for phage susceptibility compared to M. abscessus GD82 and M. smegmatis mc2155. Of four Muddy survivors tested, one was fully resistant to Muddy and the others were fully sensitive. Four ZoeJΔ45 survivors were tested and again only one was fully resistant to ZoeJΔ45, the others were fully sensitive. No BPsΔ33HTH_HRM10 survivors were tested. Four survivors from the three-phage challenge were found to only be fully resistant to ZoeJΔ45 and fully sensitive to both Muddy and BPsΔ33HTH_HRM10.
ELISA.
Enzyme-linked immunosorbant assays (ELISAs) were performed by coating wells of EIA microplates (Corning CLS3590) with 100 μl of either pure coating buffer (carbonate-bicarbonate with pH = 9.6, Sigma C3041) or phage (Muddy, ZoeJΔ45, or BPsΔ33HTH_HRM10) diluted to 5 × 109 pfu ml−1 in coating buffer. The coated wells were incubated at 4° C for ~20 hours, then washed 5 times with 200 μl PBS (Sigma packets P3813 when resuspended in DI water contain: 10 mM phosphate, 138 mM NaCl, 2.7 mM KCl, pH 7.4) + 0.05% Tween 20 (PBST) and blocked with 300 μl PBST + 3% milk for ~20 hours at 4°C. The blocking buffer was then decanted and the wells filled with 100 μl of heat inactivated, diluted patient serum (initial dilution of 1:100, followed by 11 subsequent 3X serial dilutions in blocking buffer) and incubated at 4°C for ~20 hours, such that each serum is incubated in four sets (coated with no phage, Muddy, ZoeJΔ45, or BPsΔ33HTH_HRM10 as described above) of 12 wells each. The sera were removed and the wells washed five times with PBST, then incubated with 100 μl of secondary antibody (diluted 1:10,000 into PBST) for 1 hour in the dark at room temperature. Secondary antibodies include: 1) Goat Anti-Human IgG Fc (HRP) pre-adsorbed (Abcam ab98624), 2) Goat Anti-Human IgA alpha chain (HRP) pre-adsorbed (Abcam ab98558), and 3) Goat Anti-Human IgM mu chain (HRP) pre-adsorbed (Abcam ab98549). Finally, the secondary antibody was removed, and the wells were washed twice with PBST and three times with PBS before adding 100 μl of TMB substrate (Sigma T0440) and incubating in the dark. The reaction was allowed to develop for 8 minutes, then stopped with the addition of 100 μl of 2N H2SO4. The plates were quantified by taking the difference between the absorbance at 450nm (signal) and at 570 nm (background). The results were plotted against the dilution and fit with a logistic curve. For each serum tested, the endpoint titer was determined by calculating the ratio of the logistic curve fits for the phage-coated wells to the control (no phage) wells. The greatest dilution at which this ratio was greater than 4 defined the endpoint titer. Reciprocal serum endpoint titers were calculated as log10(1/endpoint titer).
Western blotting.
Phage samples for western blotting were prepared by centrifuging 2 × 1011 pfu of phage at 14,000 rpm for 30 minutes, then resuspending the pellet in 75 μl of 20 mM dithiothreitol (DTT) with vigorous vortexing. The capsids were ruptured by adding 2 μl of 0.5M Ethylenediaminetetraacetic acid (EDTA, pH 8), mixing, and completing two cycles of incubation at 75°C (for 1 minute) followed by vigorous vortexing. The released genomic DNA was digested by mixing in 4 μL of 1M MgSO4 and 2 μL of 1 mg mL−1 DNase I, and incubating at 30°C for 20 minutes. Finally, phage proteins were denatured by adding 28 μL of 4X SDS loading dye, and boiling at 95°C for 2.5 minutes. From this sample, 10 μl was loaded into each well of large format 4–20% SDS PAGE gradient gel (~2 × 1010 pfu per well). After separating the proteins with 120 V over 20 hours, part of the gel was excised and used for staining (Imperial protein stain, ThermoFisher 24615) to confirm equal loading across wells. For the remaining lanes, proteins were transferred to PVDF membrane using semi-dry transfer and a discontinuous Tris/CAPS transfer buffer (anode buffer: 60mM Tris, 40mM CAPS, pH 9.6; 15% MeOH; cathode buffer: 60mM Tris, 40mM CAPS, pH 9.6; 0.1% SDS) at 25V for 75 minutes. The membrane was then briefly rinsed in TBS (25 mM Tris-HCl pH 7.5, 300 mM NaCl), blocked for three hours at room temperature in TBS + 3% milk, and incubated overnight in heat-inactivated patient serum diluted 1:1,000 or 1:100 in TBST + 3% milk (TBS with 0.05% Tween 20). The serum was removed and the membrane washed extensively in TBST then incubated for 45 minutes at room temperature with secondary antibody diluted 1:12,500 in TBST + 3% milk (secondary antibodies used were Goat Anti-Human IgG Fc (HRP) pre-adsorbed (Abcam ab98624 and Goat Anti-Human IgM mu chain (HRP) pre-adsorbed (Abcam ab98549). Finally, the blot was washed exhaustively in TBST, rinsed once in TBS, then incubated for 5 minutes with ECL substrate (SuperSignal WestPico PLUS Chemiluminescent Substrate, ThermoFisher 34579) and imaged with an Amersham Imager 600 (GE Healthcare).
Protein identification by mass spectrometry.
Following gel electrophoresis, protein bands which were both visible on the Coomassie-stained gel and identified as antibody targets by Western blots were excised and submitted to the Biomedical Mass Spectrometry Center at University of Pittsburgh. Gel slices were trypsin digested and the resulting peptides analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). Peptides were reassembled into proteins, searched using MASCOT, and visualized with Scaffold software.
Quantification, statistics, and reproducibility.
No statistical method was used to predetermine sample size. No data were excluded from the analyses.Technical replicates were performed as described in the figure legends. From these replicates, the mean and standard deviation (s.d.) were determined using Microsoft Excel. Fixed-baseline logistic curve fitting and subsequent analyses for ELISA data were performed using OriginLab (version 9.85).
Extended Data
Extended Data Figure 1. Determination of MAC sputum counts.
Quantitative MAC counts in expectorated sputum (log CFU mL−1) pre- and post-treatment. The observed decline in MAC treatment during phage therapy is likely attributable to the addition of anti-MAC antibiotics, azithromycin and ethambutol, to the multidrug regimen. MAC colony morphology appeared smooth and opaque.
Extended Data Figure 2. Chest CT scans during therapy.
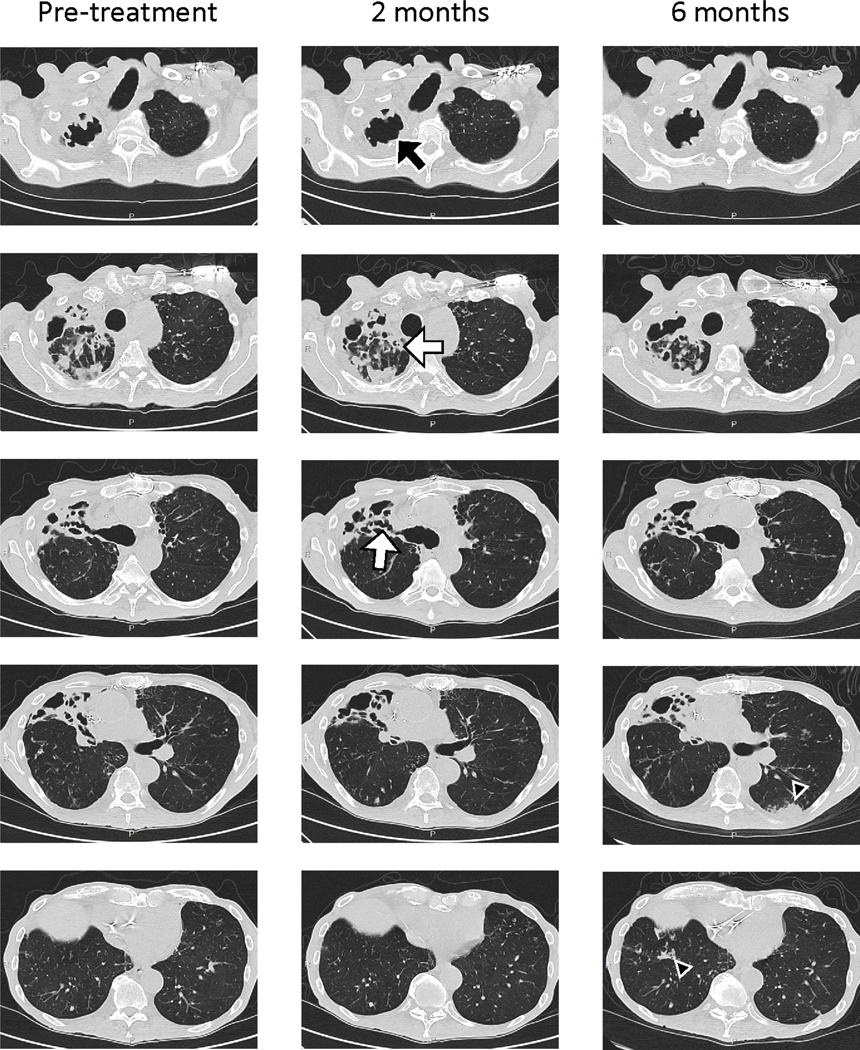
Non-contrast high resolution chest CT scans were performed immediately prior to phage treatment, and repeated after phage treatment initiation at two- and six-months. The majority of chronic airway changes such as bronchiectasis and bronchial wall thickening (white arrows) were stable throughout phage therapy. However, there was slightly increased cavitation in the right upper lobe (black arrows) noted at two months post-phage treatment, and interval development of peripheral left lower lobe consolidations and right lower lobe nodularity (black arrow heads) at six months post-phage treatment.
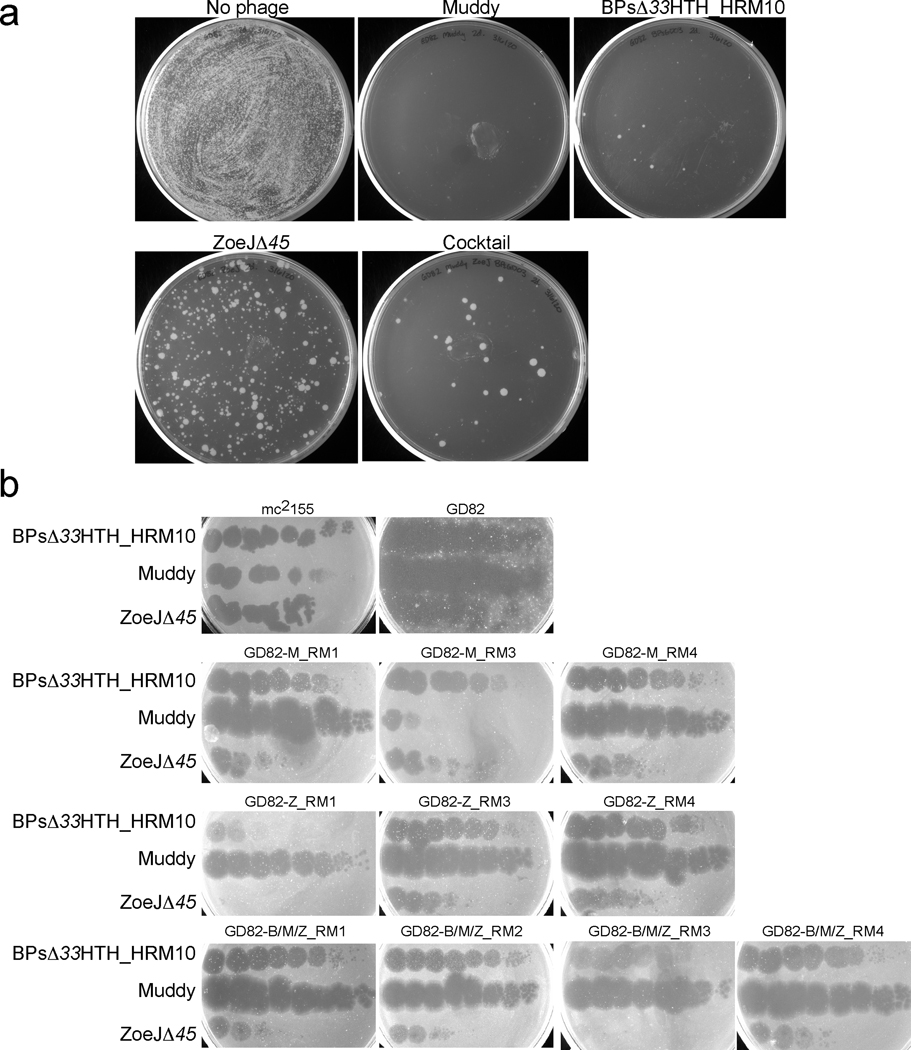
Extended Data Figure 3. Analysis of phage resistance in vitro.
a, 1 ml of 2 × 108 CFU GD82 was mixed with 2 × 109 PFU of each phage used in the cocktail or with all three phages, and incubated at 37° C with shaking. After 48 hours, 0.1 ml of the mix was spread on solid media and incubated for 5 days at 37° C. b, Four surviving colonies of each phage challenge were grown and retested for phage sensitivity by spotting 10-fold serial dilutions of phages as indicated. GD82-M_RM2 and GD82-Z_RM2 could not be propagated and were not tested further. BPsΔ33HTH_HRM10 RMs were not tested.
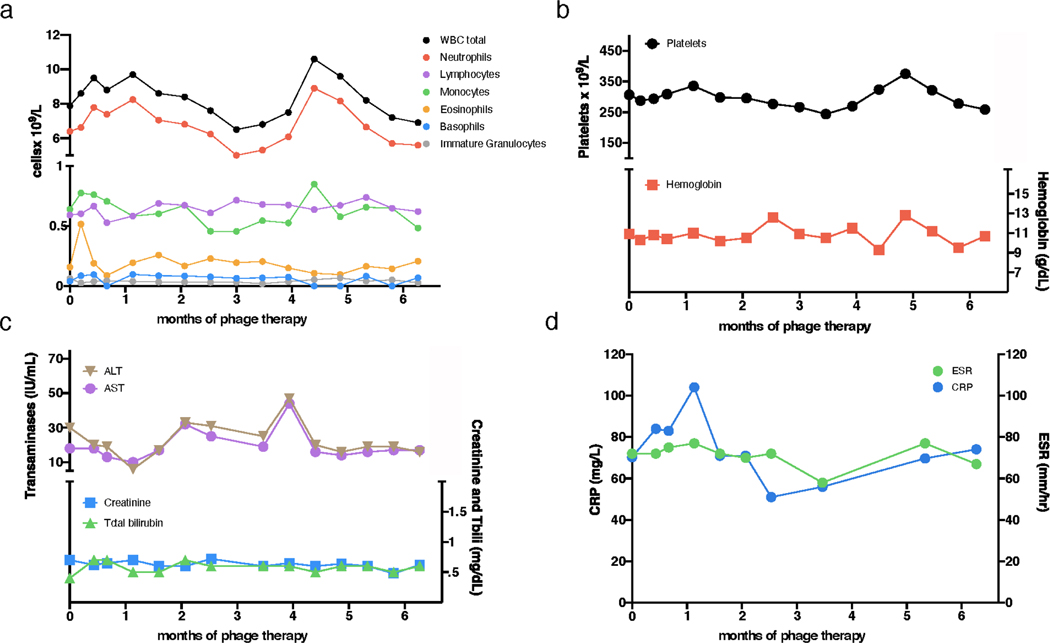
Extended Data Figure 4. Clinical indicators during phage treatment.
Safety assessments during phage treatment included regular monitoring of complete blood counts with differential, comprehensive metabolic panels, and inflammatory markers. During six-months of intravenous phage treatment there were no clinically significant differences observed in a, white blood cell (WBC) total counts and differentials, b, hemoglobin and platelets, c, liver function (alanine transaminase, ALT; aspartate transaminase, AST; and total bilirubin) or kidney function (creatinine), and d, C-reactive protein (CRP) and erythrocyte sedimentation rate (ESR).
Extended Data Table 1.
Antibiotic susceptibility of M. abscessus isolates
| Months relative to phage treatment initiation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Antibiotic 1 | −8 | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
|
| ||||||||
| T rimethoprim/sulfamethoxazole | >8/152 | >8/152 | >8/152 | >8/152 | >8/152 | >8/152 | >8/152 | >8/152 |
| Ciprofloxacin | >4 | >4 | >4 | >4 | >4 | >4 | >4 | >4 |
| Moxifloxacin | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
| Cefoxitin | 64 | 64 | 64 | 64 | 64 | 32 | 64 | 64 |
| Amikacin | 32 | 32 | 32 | 32 | 32 | 32 | 16 | 32 |
| Doxycycline | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Tigecycline | 4 | 2 | 4 | 4 | 4 | 4 | 4 | 4 |
| Linezolid | 32 | 32 | 32 | 32 | 32 | >32 | >32 | >32 |
| Imipenem | 64 | 64 | 64 | 64 | >64 | 64 | >64 | >64 |
| Cefepime | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| Amoxicillin/clavulanate | >64/32 | >64/32 | >64/32 | >64/32 | >64/32 | >64/32 | >64/32 | >64/32 |
| Ceftriaxone | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| Minocycline | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
| Tobramycin | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Clarithromycin (3 days) | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Clarithromycin (14 days) | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
Antibiotic minimum inhibitory concentration (MIC) determination s (in μg/mL) active against M. abscessus GD82 were determined pre-phage therapy initiation and monthly during phage therapy using a standard antibiotic panel for rapidly growing mycobacteria (Sensititre RAPMYCO ™).
Supplementary Material
Acknowledgements
We thank Debora Jacobs-Sera and Haley G. Aull for phage cocktail preparation, Rebecca Garlena and Daniel Russell at the University of Pittsburgh for DNA sequencing and analysis, and Meghan Ramsay, Chris Lippincott, Jonathan Zenilman, Kelly Dooley and Elisa Ignatius at John Hopkins University School of Medicine for their clinical discussions regarding the care of this patient. This work was funded by Cystic Fibrosis Foundation grant # HATFUL19GO, National Institutes of Health grant GM131729, Howard Hughes Medical Institute grant GT12053, and by the Fowler Fund for Phage Research, to GFH, as well as grants from the NHLBI/NIH K08 HL139994 and the Burroughs Wellcome Fund Career Award for Medical Scientists to KAC. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank Chip Schooley, Eric Rubin, and Bill Bishai for comments on the manuscript.
J.A.N. has received consulting fees from AstraZeneca. G.F.H. is a compensated consultant for Janssen Inc. K.A.C. has received consulting fees from Insmed Inc, HillRom and Merck.
Footnotes
Competing Interests
Other authors declare no competing interests.
Data Availability
GD82, GD01A and GD01B sequence data is available under accession numbers JADWXT000000000m, CP035923 and CP035924, respectively. All phage information can be found at www.phagesdb.org. Sequence information for Muddy, BPs and ZoeJ are available under accession numbers KF024728, EU568876 and KJ510412, respectively. ELISA raw data is available in supplementary excel file “Raw Data Fig2ab”.
References:
- 1.Johansen MD, Herrmann JL & Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol 18, 392–407 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Griffith DE et al. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med 175, 367–416 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Choi H. et al. Clinical Characteristics and Treatment Outcomes of Patients with Macrolide-Resistant Mycobacterium massiliense Lung Disease. Antimicrob. Agents Chemother 61, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi H. et al. Clinical characteristics and treatment outcomes of patients with acquired macrolide-resistant Mycobacterium abscessus lung disease. Antimicrob. Agents Chemother 61, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kortright KE, Chan BK, Koff JL & Turner PE Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 25, 219–232 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Salmond GPC & Fineran PC A century of the phage: Past, present and future. Nat. Rev. Microbiol 13, 777–786 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Dedrick RM et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med 25, 730–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerne NK & Avegno P. The development of the phage-inactivating properties of serum during the course of specific immunization of an animal: reversible and irreversible inactivation. J. Immunol 76, 200–8 (1956). [PubMed] [Google Scholar]
- 9.Roach DR et al. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 22, 38–47.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Hodyra-Stefaniak K. et al. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci. Rep 5, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Górski A. et al. Phage as a Modulator of Immune Responses. Practical Implications for Phage Therapy. Adv. Virus Res 83, 41–71 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Aslam S. et al. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect. Dis 7, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Łusiak-Szelachowska M. et al. Phage neutralization by sera of patients receiving phage therapy. Viral Immunol.27, 295–304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamme C. ANTIBODIES AGAINST STAPHYLOCOCCAL BACTERIOPHAGES IN HUMAN SERA: I. Assay of antibodies in healthy individuals and in patients with staphylococcal infections. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol 81 B, 741–748 (1973). [PubMed] [Google Scholar]
- 15.Aslam S. et al. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am. J. Transplant 19, 2631–2639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schooley RT et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother 61, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jault P. et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis 19, 35–45 (2019). [DOI] [PubMed] [Google Scholar]
- 18.El Haddad L, Harb CP, Gebara MA, Stibich MA & Chemaly RF A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin. Infect. Dis 69, 167–178 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Hatfull GF Actinobacteriophages: Genomics, Dynamics, and Applications. Annu. Rev. Virol 7, 37–61 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedrick RM et al. Mycobacterium abscessus strain morphotype determines phage susceptibility, the repertoire of therapeutically useful phages, and phage resistance. MBio in revision (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.