Abstract
DDP1 is a single-stranded nucleic acid binding protein of Drosophila melanogaster that associates with pericentric heterochromatin. DDP1 contains 15 consecutive KH domains and is homologous to the highly conserved vigilin proteins that, in Saccharomyces cerevisiae, are involved in the control of cell ploidy. DDP1 was identified and purified on the basis of its binding to the pyrimidine-rich C strand of the centromeric Drosophila dodeca-satellite. Here, the interaction of DDP1 with the dodeca-satellite C strand was analyzed in detail. This interaction is sequence specific. In particular, a guanine residue which is highly conserved in natural dodeca-satellite sequences was found to be essential for the efficient binding of DDP1. DDP1 binding was also found to be strongly influenced by the length and extent of secondary structure of the DNA substrate. Efficient DDP1 binding required a minimal length of about 75 to 100 nucleotides and was facilitated by the lack of secondary structure of the substrate. DDP1 also showed a significant affinity for the unstructured pyrimidine-rich strand of the most abundant centromeric Drosophila AAGAG satellite. The stoichiometry of the complexes formed with the dodeca-satellite C strand suggests that, in DDP1, the 15 consecutive KH domains are organized such that they define two nucleic acid binding surfaces. These results are discussed in the context of the possible contribution of DDP1 to heterochromatin organization and function.
DDP1 is a multi-KH-domain protein of Drosophila melanogaster that is found associated with pericentric heterochromatin (8). DDP1 contains 15 tandemly organized KH domains and is homologous to the highly conserved vigilin proteins that have been found in all eukaryotic organisms analyzed to date, from yeasts to humans (25, 29, 30, 38). Little is known about the functions of vigilins. They are up-regulated in rapidly dividing cells (29); in yeast, disruption of the corresponding gene (SCP160) results in cells with increased ploidy, suggesting a role in chromosome segregation (38). The expression of DDP1 complements a Δscp160 deletion in yeast (8). The association of DDP1 with pericentric heterochromatin also suggests a possible contribution to chromosome segregation. Vigilins could also play a role in RNA metabolism (12, 17, 19–21). They were found to bind in vitro the 3′ untranslated regions (UTRs) of the dystrophin and vitellogenin mRNAs and were proposed to be responsible for the increased stability of the latter induced by estrogens (12, 17).
Like the vigilins, DDP1 binds single-stranded nucleic acids with high affinity and specificity (8). Actually, DDP1 was identified and isolated on the basis of its interaction with the C strand of the Drosophila dodeca-satellite, a highly repeated DNA sequence which is localized to the pericentric heterochromatin on chromosome 3 in D. melanogaster (1, 5, 24). In polytene chromosomes, the distribution of DDP1 is not constrained to the regions containing dodeca-satellite sequences, being associated as well with other pericentric heterochromatin regions containing no detectable dodeca-satellite sequences (8). DDP1 is also found at some discrete sites on the euchromatic chromosome arms, colocalizing with heterochromatin protein 1 (8). The dodeca-satellite has a marked Pu-Py strand asymmetry which results in one strand, the C strand, being enriched in pyrimidines and the complementary strand, the G strand, being enriched in purines. In vitro, the dodeca-satellite can form altered DNA structures in which the G strand forms very stable intramolecular hairpins while the complementary C strand remains unstructured (13). Other centromeric satellites, such as the abundant Drosophila AAGAG satellite, show similar structural properties (6, 7, 14, 27). Formation of these altered DNA structures could therefore provide an adequate substrate for the efficient binding of DDP1 to heterochromatin. In vitro, DDP1 was found to bind the unstructured dodeca-satellite C strand but not the G strand (8, 13).
In this study, we have analyzed in vitro the interaction of DDP1 with single-stranded nucleic acids. The interaction of DDP1 with the dodeca-satellite C strand is sequence specific but is also strongly influenced by the length and extent of secondary structure of the DNA substrate. DDP1 also shows a significant affinity for the pyrimidine strand of the Drosophila AAGAG satellite.
MATERIALS AND METHODS
DNAs and RNAs.
Table 1 summarizes the different DNAs used in these experiments. Fragments 42R and 9R were obtained, respectively, from plasmids pBK6E215 and pBK6E218, which are pBluescript (Stratagene) derivatives carrying dodeca-satellite sequences inserted at the unique SpeI site (1). The dodeca-satellite fragments were released by digestion with SpeI, and the corresponding C strands were obtained as described earlier (13). All synthetic oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis before use. The DNA concentration was determined by UV spectroscopy as described previously (15). When needed, DNAs were radioactively labeled by conventional methods.
TABLE 1.
DNA fragments used in these experiments
| DNA fragment | Nucleotide sequence |
|---|---|
| Fragment 42Ra | |
| Fragment 12Ra | A CTAGTCCCGTAC TCTGTCCCGTAC TCTGTCCCGTAC TCCGTCTCGTAC TCTGTCCCATAT TGGTCCCGTAC TGGTCCCGCAC ATGGTCCCGAAC TGGTCCCCTAC TCCGTCCCGTAC TCGGTCCCGTAC TGATCCCGTAC TAGT |
| Oligo 9Ra | TAGTCCCGTAC TCTGTCCCGTAC TCTGTCCCGTAC TCCGTCTCGTAC TCTGTCCCATAT TGGTCCCGTAC TGGTCCCGCAC ATGGTCCCGAAC TGGTCCCCTAC |
| Oligo 4Ra | CCCGTAC TCTGTCCCGTAC TCCGTCTCGTAC TCTGTCCCATAT TGGT |
| Oligo 9Rc | (TCGGTCCCGTAC)9 |
| Oligo 6Rc | (TCGGTCCCGTAC)6 |
| Oligo 6RcTb | (TCGGTTTTGTAC)6 |
| Oligo 6RcTG1b | (TCCGTTTTGTAC)6 |
| Oligo 6RcTG2b | (TCGCTTTTGTAC)6 |
| Oligo 6RcTG3b | (TCGGTTTTCTAC)6 |
| Oligo CTCTT | (CTCTT)20 |
| Oligo VIT | CTC TAT ATC TCT ATC AAA TGA ATA AGC TGT AAT ATC ACT GAT GAT GAT AAA CTG ATC TCA ATT TCA AAC CAA ATG TAT ATT ATA CTA TTG TAA ACA ATT CAA TT |
| Oligo DYS | ACA TTT ACG AAT TAT TTT TTT AAA CTT CAG TTT TAC TGC ATT TTC ACA ACA TAT CAG ACT TCA CCA AAT ATA TGC CTT ACT ATT GTA TTA TAG TAC TGC TTT AC |
The nucleotide sequences are presented in blocks corresponding to the dodeca-satellite repeats. See reference 1 for the fragment 42R sequence.
Nucleotides substituted with respect to the dodeca-satellite C-strand consensus sequence are shown in bold.
RNAs were obtained by in vitro transcription with T7 RNA polymerase (Promega) of plasmids pbsVIT and pbsDYS, which are pBluescript derivatives carrying the double-stranded VIT and DYS sequences, respectively (Table 1), flanked by a SacI site at the 5′ end and an EcoRI site at the 3′ end. Before in vitro transcription, plasmids were linearized with HindIII. RNA products were visualized on 2% nondenaturing agarose gels, and their concentrations were determined by UV spectroscopy. Transcripts obtained in this way contained, in addition to the VIT and DYS sequences, 15 and 12 nucleotides (nt) of unrelated sequences at their 5′ and 3′ ends, respectively.
Proteins.
DDP1 was either purified from SL2 nuclear extracts or obtained as a recombinant by expression in Escherichia coli cells (8). Both proteins behaved indistinctly in electrophoretic mobility shift assay (EMSA) experiments (8). 1/2DDP1 was obtained as a recombinant by the expression in E. coli of the first 651 amino acids of DDP1 by use of the PET29-b expression vector (Novagen). 1/2DDP1 was produced as a fusion protein carrying a C-terminal His6 tag and purified as DDP1 (8).
EMSA experiments.
EMSA experiments were performed as described previously (8) with ∼0.2 ng of radioactively labeled DNA and, when necessary, an excess of competitor. Competition experiments were always performed at a protein concentration providing 75 to 97% binding in the absence of any added competitor. When binding to fragment 42R was studied, complexes were resolved on native 4% instead of 5% polyacrylamide gels. For high-resolution analysis, the protein-DNA complexes were subjected to electrophoresis through 40-cm-long native 5% polyacrylamide gels for 12 h at 150 V. Autoradiographs were recorded on HyperFilm (Amersham) and analyzed quantitatively on a Molecular Dynamics laser densitometer. The percent competition was expressed as the percent binding observed in the presence of competitor DNA relative to the percent binding obtained in the absence of competitor DNA.
RESULTS AND DISCUSSION
Efficient binding of DDP1 to the dodeca-satellite C strand depends strongly on the length and lack of secondary structure of the DNA fragments.
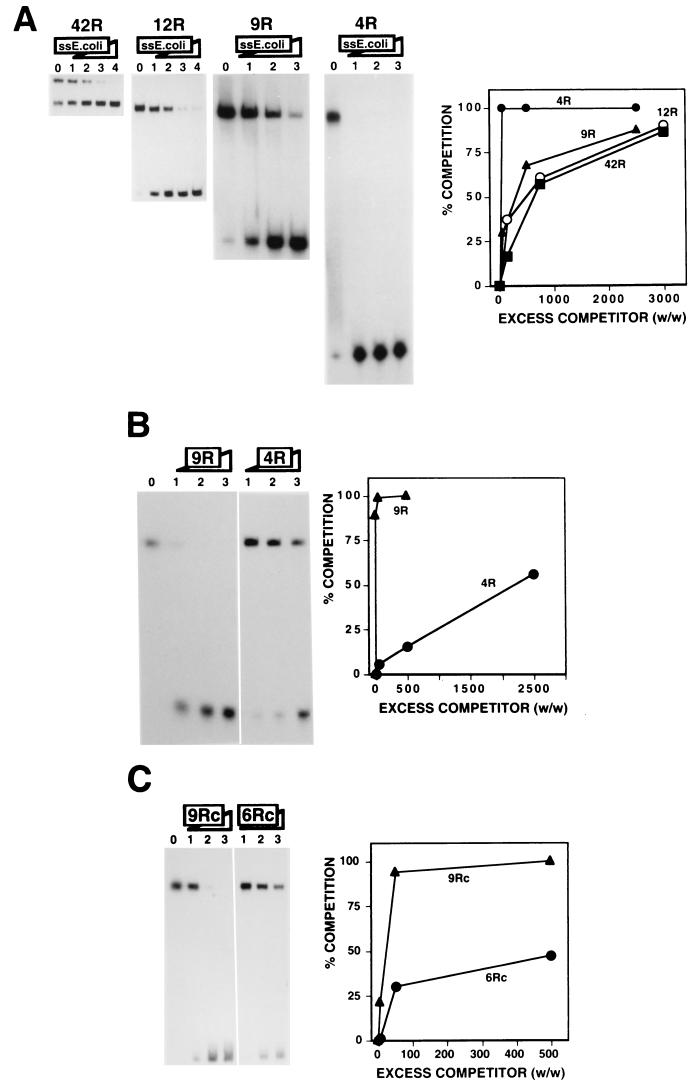
The affinity of DDP1 for the dodeca-satellite C strand depends strongly on the length of the DNA fragments; efficient interaction requires relatively long binding sites. As shown in Fig. 1, the affinity of DDP1 for the dodeca-satellite C strand decreases sharply for fragments shorter than about 70 nt. Fragments 42R and 12R correspond to the C strand of naturally occurring dodeca-satellite DNA fragments containing 42 and 12 repeats, respectively. Oligo 9R and oligo 4R, which were derived from fragment 12R, carry nine and four repeats, respectively (Table 1). The efficiency of the interaction of DDP1 with these C-strand fragments was determined through competition experiments. As judged from the excess of a nonspecific heat-denatured single-stranded E. coli DNA competitor required to obtain efficient competition, the affinities of DDP1 for fragment 42R, fragment 12R, and oligo 9R are very similar, but the affinity for oligo 4R is much lower (Fig. 1A). For oligo 4R, the binding of DDP1 is completely abolished in the presence of a 50-fold excess (wt/wt) of nonspecific competitor (Fig. 1A, 4R, lane 1), while for the longer fragments, significant binding is still detected in the presence of a 2,500- to 3,000-fold excess (Fig. 1A, 42R, 12R, and 9R, lanes 3). The lower affinity detected for oligo 4R was corroborated when the binding of DDP1 to oligo 9R was competed by oligo 9R itself or oligo 4R (Fig. 1B). A similar reduced affinity was observed for DNA fragments also containing four repeats but spanning different regions of fragment 12R (data not shown), indicating that this reduced affinity is not directly associated with the different nucleotide sequences of the fragments used. Consistent with this interpretation, similar results were obtained when the binding of DDP1 to DNA fragments carrying nine (oligo 9Rc) and six (oligo 6Rc) repeats of the C-strand consensus sequence (TCGGTCCCGTAC) was analyzed (Fig. 1C). These synthetic oligonucleotides differ in length but not in nucleotide sequence (Table 1). The affinity of DDP1 for oligo 9Rc is higher than that for oligo 6Rc, as judged from competition experiments in which the binding of DDP1 to oligo 9Rc was competed by oligo 9Rc and oligo 6Rc (Fig. 1C). Similarly, single-stranded E. coli DNA competed the binding of DDP1 to oligo 6Rc more efficiently than to oligo 9Rc (data not shown).
FIG. 1.
Efficient binding of DDP1 to the dodeca-satellite C strand depends on the length of the DNA substrate. (A) The binding of DDP1 to dodeca-satellite C-strand DNA fragments of different lengths is shown as a function of increasing amounts of heat-denatured single-stranded (ss) E. coli DNA. The excess quantities (weight to weight) of competitor used were as follows: panels 42R and 12R, 0 (lanes 0), 150 (lanes 1), 750 (lanes 2), 3,000 (lanes 3), and 9,000 (lanes 4); and panels 9R and 4R, 0 (lanes 0), 50 (lanes 1), 500 (lanes 2), and 2,500 (lanes 3). Quantitative analysis of the results is shown on the right for fragment 42R, fragment 12R, oligo 9R, and oligo 4R. (B) The binding of DDP1 to oligo 9R is shown as a function of increasing amounts of oligo 9R (panel 9R) and oligo 4R (panel 4R). The excess quantities (weight to weight) of competitor used were as follows: panel 9R, 5 (lane 1), 50 (lane 2), and 500 (lane 3); and panel 4R, 50 (lane 1), 500 (lane 2), and 2,500 (lane 3). Lane 0 shows the binding obtained in the absence of any added competitor. Quantitative analysis of the results is shown on the right for oligo 9R and oligo 4R. (C) The binding of DDP1 to oligo 9Rc is shown as a function of increasing excess quantities (weight to weight) of oligo 9Rc (panel 9Rc) and oligo 6Rc (panel 6Rc): 0 (lane 0), 5 (lanes 1), 50 (lanes 2), and 500 (lanes 3). Quantitative analysis of the results is shown on the right for oligo 9Rc and oligo 6Rc. See Table 1 for a description of the DNA fragments used in these experiments.
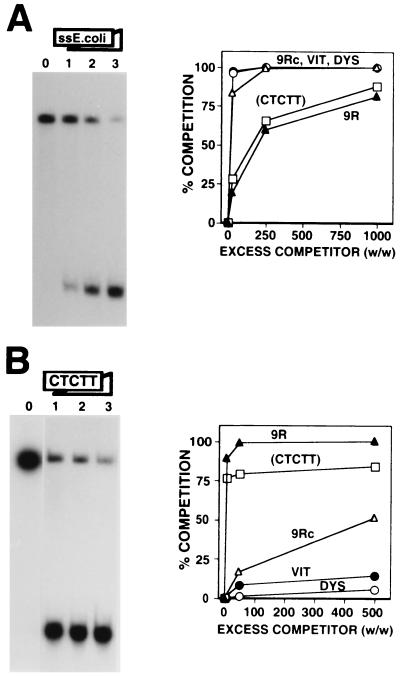
FIG. 4.
DDP1 binds the pyrimidine-rich strand of the Drosophila AAGAG satellite. (A) The binding of DDP1 to oligo CTCTT is shown as a function of increasing excess quantities (weight to weight) of single-stranded (ss) E. coli DNA: 0 (lane 0), 25 (lane 1), 250 (lane 2), and 1,000 (lane 3). (B) The binding of DDP1 to oligo 9R is shown as a function of increasing excess quantities (weight to weight) of oligo CTCTT: 0 (lane 0), 5 (lane 1), 50 (lane 2), and 500 (lane 3). Quantitative analysis of the results is shown to the right of each panel for oligo CTCTT. The results obtained with oligo 9R (▴), oligo 9Rc (▵), oligo DYS (○), and oligo VIT (●), taken from Fig. 2 and 3, are included for comparison. See Table 1 for a description of the DNA fragments used in these experiments.
Altogether, these results indicate that the binding of DDP1 to the dodeca-satellite C strand requires a minimum length of between six repeats (72 nt) and nine repeats (104 to 108 nt). The affinity of DDP1 for DNA fragments above this length threshold does not increase significantly. A similar length requirement was reported for the binding of the Xenopus vigilin to nucleic acids (17). The large size of the vigilin binding sites strongly suggests that their 15 KH domains are actually involved in nucleic acid recognition.
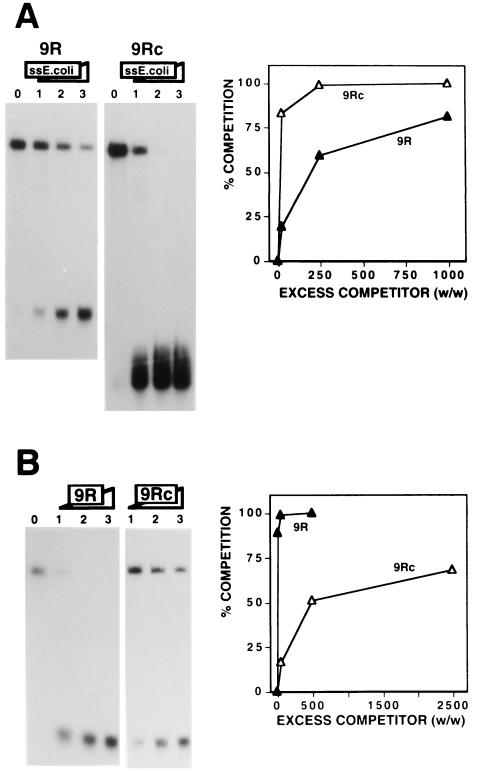
The binding of DDP1 to the dodeca-satellite C strand is highly sensitive to the extent of secondary structure of the DNA fragments. The affinity of DDP1 for oligo 9R is significantly higher than that for oligo 9Rc, as shown by the larger excess of single-stranded E. coli DNA required to compete binding to the former (Fig. 2A) as well as by the greater efficiency of oligo 9R as a competitor (Fig. 2B). Both DNA fragments are of similar lengths, containing nine repeats of the dodeca-satellite C strand that, in the case of oligo 9Rc, are perfect repetitions of the consensus sequence. The dodeca-satellite C-strand consensus sequence is slightly palindromic and, as a consequence, DNA fragments carrying perfect repetitions of this sequence form relatively stable fold-back structures under the experimental conditions used for DDP1 binding and EMSA. On the other hand, the repeats of oligo 9R are not perfect (Table 1) and, as a consequence, oligo 9R does not show any significant secondary structure. The faster electrophoretic mobility of oligo 9Rc than of oligo 9R (Fig. 2A) reflects the formation of fold-back structures on the former (27). Similar results were obtained when the affinity of DDP1 for oligo 4R, carrying four imperfect repeats, was compared to that for oligo 4Rc, carrying four perfect repeats of the C-strand consensus sequence (data not shown). These results indicate that the affinity of DDP1 is strongly influenced by the extent of secondary DNA structure of the substrate. Binding of Xenopus vigilin to the vitellogenin and dystrophin mRNAs was also found to be favored by mutations decreasing the degree of secondary structure of the substrate (17). Interestingly, natural dodeca-satellite sequences are rarely built by perfect repetitions of the consensus sequence (1, 24). As a consequence, the C strand of naturally occurring dodeca-satellite fragments shows in general a very low degree of secondary structure (13, 27) and is therefore a good substrate for DDP1 binding. Actually, DDP1 showed similar high affinities for several different naturally occurring C-strand fragments that, under these experimental conditions, were unstructured (data not shown).
FIG. 2.
Efficient binding of DDP1 to the dodeca-satellite C strand depends on the extent of secondary structure of the DNA substrate. (A) The binding of DDP1 to oligo 9R (panel 9R) and oligo 9Rc (panel 9Rc) is shown as a function of increasing excess quantities (weight to weight) of single-stranded (ss) E. coli DNA: 0 (lanes 0), 25 (lanes 1), 250 (lanes 2), and 1,000 (lanes 3). (B) The binding of DDP1 to oligo 9R is shown as a function of increasing amounts of oligo 9R (panel 9R) and oligo 9Rc (panel 9Rc). Excess quantities (weight to weight) of competitor used were as follows: panel 9R, 5 (lane 1), 50 (lane 2), and 500 (lane 3); and panel 9Rc, 50 (lane 1), 500 (lane 2), and 2,500 (lane 3). Lane 0 shows the binding obtained in the absence of any added competitor. Quantitative analysis of the results are shown to the right of each panel for oligo 9R and oligo 9Rc. See Table 1 for a description of the DNA fragments used in these experiments.
The interaction of DDP1 with the dodeca-satellite C strand is of high affinity and specificity.
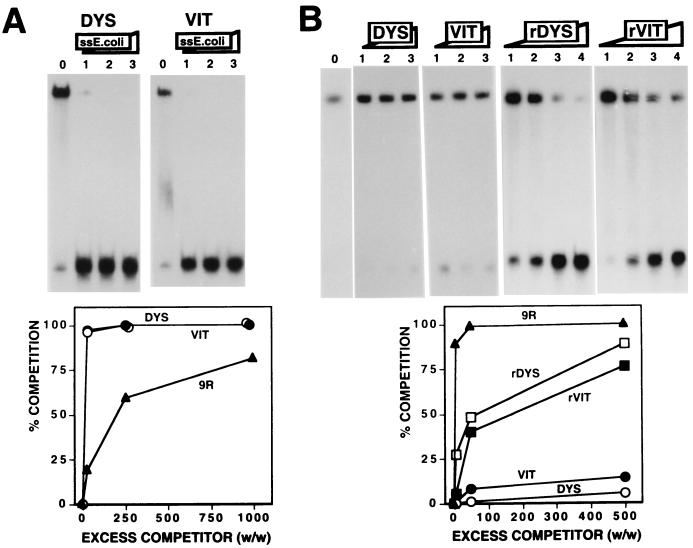
The results reported above indicate that, upon melting, the dodeca-satellite could provide sufficiently long unstructured DNA fragments for efficient DDP1 binding. Others have shown that vigilins can also specifically recognize various single-stranded DNA and RNA sequences (10, 12, 17, 37). In particular, the Xenopus vigilin was shown to specifically interact with sequences of the 3′ UTRs of the Xenopus vitellogenin and human dystrophin mRNAs (12, 17). The question then arises as to what extent the interaction of DDP1 with the dodeca-satellite is specific. Here, we have analyzed the relative affinity of DDP1 for the dodeca-satellite C strand and the vigilin binding sites of the 3′ UTRs of the vitellogenin and dystrophin mRNAs. Figure 3 shows the interaction of DDP1 with synthetic oligonucleotides spanning the vigilin binding sites of the vitellogenin (oligo VIT) and dystrophin (oligo DYS) mRNAs (17). To avoid any length dependence effects, these oligonucleotides were exactly the same length (104 nt) as oligo 9R (Table 1). As shown in Fig. 3A, single-stranded E. coli DNA competes the binding of DDP1 to oligo VIT and oligo DYS much more efficiently than to oligo 9R, indicating a significantly higher affinity for the latter. In good agreement with these results, binding to oligo 9R occurs at a protein concentration (2.5 μl) lower than that required for oligo DYS (6 μl) or oligo VIT (7 μl). Corroborating the lower affinity of DDP1 for the vigilin binding sites of the vitellogenin and dystrophin mRNAs, oligo VIT and oligo DYS compete the binding of DDP1 to oligo 9R very poorly, since no significant competition is observed even in the presence of a 500-fold excess of these competitors (Fig. 3B). Similar results were obtained when the RNA versions of oligo VIT and oligo DYS were used as competitors (Fig. 3B, rVIT and rDYS). Fragments rVIT and rDYS appear to compete DDP1 binding better than the corresponding DNA versions. However, significant binding is still observed in the presence of a 2,500-fold excess of the RNA competitors (Fig. 3B, rDYS and rVIT, lanes 4). On the other hand, the affinity of DDP1 for the RNA version of the dodeca-satellite C strand was shown not to be significantly different from that for its DNA form (8). These results show that DDP1 binds the dodeca-satellite C strand with a higher affinity than the vigilin binding sites of the Xenopus vitellogenin and human dystrophin mRNAs.
FIG. 3.
Comparison of the affinity of DDP1 for the vigilin binding sites of the Xenopus vitellogenin and human dystrophin mRNAs to its affinity for the dodeca-satellite C strand. (A) The binding of DDP1 to oligo DYS (panel DYS) and oligo VIT (panel VIT) is shown as a function of increasing excess quantities (weight to weight) of single-stranded (ss) E. coli DNA: 0 (lanes 0), 25 (lanes 1), 250 (lanes 2), and 1,000 (lanes 3). Quantitative analysis of the results is shown below for oligo DYS (○) and oligo VIT (●). The results obtained with oligo 9R (▴) (Fig. 2) are included for comparison. (B) The binding of DDP1 to oligo 9R is shown as a function of increasing excess quantities (weight to weight) of oligo DYS (panel DYS), oligo VIT (panel VIT), and the RNA transcripts corresponding to the DYS (panel rDYS) and VIT (panel rVIT) sequences: 5 (lanes 1), 50 (lanes 2), 500 (lanes 3), and 2,500 (lanes 4). Lane 0 shows the binding obtained in the absence of any added competitor. Quantitative analysis of the results is shown below for oligo DYS, oligo VIT, rDYS, and rVIT. The results obtained with oligo 9R (Fig. 2) are included for comparison. See Table 1 for a description of the DNA fragments used in these experiments.
As shown earlier, in Drosophila polytene chromosomes, DDP1 is found associated with the chromocenter spanning most of the pericentric heterochromatin (8), colocalizing with the dodeca-satellite-rich regions on chromosome 3, but also is present at the pericentric region of chromosome 2, where no dodeca-satellite sequences are detected; these findings suggest a possible association of DDP1 with other heterochromatin sequences. Consistent with this hypothesis, DDP1 also binds in vitro other centric satellite DNAs with significant affinity. Figure 4 shows the interaction of DDP1 with oligo CTCTT (Table 1), which corresponds to the pyrimidine-rich strand of the AAGAG satellite, the most abundant repetitive DNA of Drosophila, which is present at the pericentric region of chromosome 2 (23). As judged from the amount of nonspecific single-stranded E. coli DNA required to efficiently compete DDP1 binding (Fig. 4A), the affinity of DDP1 for oligo CTCTT is significantly higher than those for oligo 9Rc, oligo VIT, and oligo DYS, which all have lengths similar to oligo CTCTT. Similar results were obtained when the efficiency of oligo CTCTT to compete the binding of DDP1 to oligo 9R was analyzed (Fig. 4B). However, oligo CTCTT competes DDP1 binding to oligo 9R less efficiently than oligo 9R itself (Fig. 4B); a 5-fold excess of oligo 9R is sufficient to mostly abolish DDP1 binding, but significant binding is still detected in the presence of a 500-fold excess of oligo CTCTT. These results indicate that DDP1 shows a significant affinity for the unstructured pyrimidine strand of the Drosophila AAGAG satellite, albeit lower than that for the dodeca-satellite C strand.
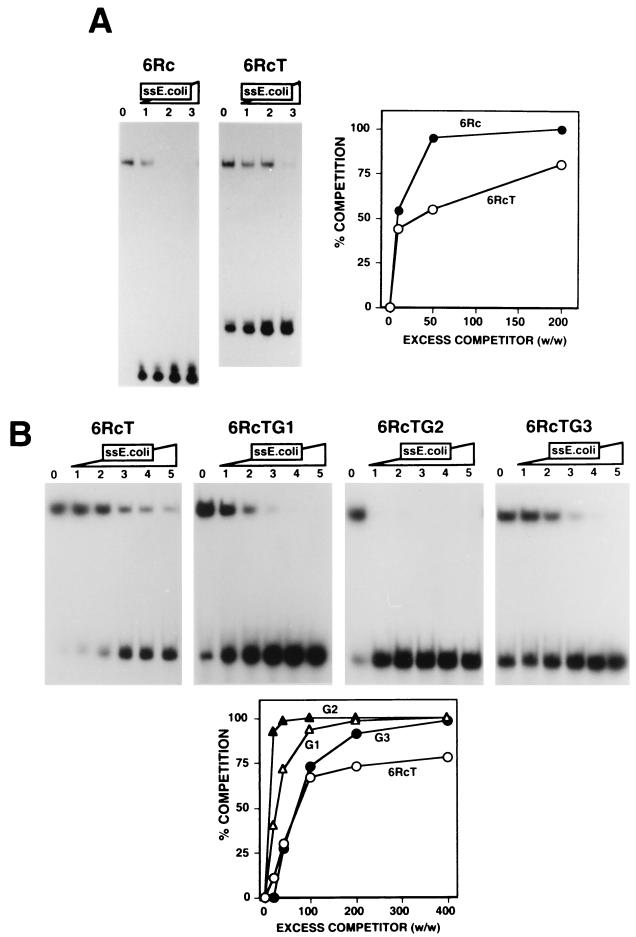
The high affinity of DDP1 for the dodeca-satellite C strand suggests that this interaction is sequence specific. Consistent with this interpretation, the substitution of part of the dodeca-satellite sequences of oligo 9R with unrelated DNA sequences results in a significant decrease in DDP1 binding (data not shown). To better understand the sequence determinants of the interaction of DDP1 with the dodeca-satellite C strand, the binding of DDP1 to oligo 6Rc, which carries six repeats of the C-strand consensus sequence, and to variants carrying specific base substitutions was analyzed (Table 1). The affinity of DDP1 for oligo 6RcT, in which the three central cytosine residues of each repeat were substituted with thymines (Table 1), is higher than that for oligo 6Rc (Fig. 5A). This increased affinity likely reflects the lower degree of secondary structure of oligo 6RcT. As mentioned above, oligo 6Rc forms relatively stable fold-back structures to which the central cytosines importantly contribute through the formation of very stable C · G pairs. Changing the three central cytosine residues to thymines prevents these base-pairing interactions and, therefore, the formation of the fold-back structures. The lower electrophoretic mobility of oligo 6RcT than of oligo 6Rc reflects the lack of secondary structure of the former (Fig. 5A).
FIG. 5.
Specific residues contribute importantly to the binding of DDP1 to the dodeca-satellite C strand. (A) The binding of DDP1 to oligo 6Rc (panel 6Rc) and oligo 6RcT (panel 6RcT) is shown as a function of increasing excess quantities (weight to weight) of single-stranded (ss) E. coli DNA: 0 (lanes 0), 10 (lanes 1), 50 (lanes 2), and 200 (lanes 3). Quantitative analysis of the results is shown on the right for oligo 6Rc and oligo 6RcT. (B) The binding of DDP1 to oligo 6RcT (panel 6RcT), oligo 6RcTG1 (panel 6RcTG1), oligo 6RcTG2 (panel 6RcTG2), and oligo 6RcTG3 (panel 6RcTG3) is shown as a function of increasing excess quantities (weight to weight) of single-stranded E. coli DNA: 0 (lanes 0), 20 (lanes 1), 40 (lanes 2), 100 (lanes 3), 200 (lanes 4), and 400 (lanes 5). Quantitative analysis of the results is shown below for oligo 6RcT, oligo 6RcTG1, oligo 6RcTG2, and oligo 6RcTG3. See Table 1 for a description of the DNA fragments used in these experiments.
Significant effects are observed when specific base substitutions are incorporated into the sequence of oligo 6RcT (Fig. 5B). A slight decrease in DDP1 affinity is observed when either the first or the third guanine residue of each repeat is changed to a cytosine, oligo 6RcTG1 or oligo 6RcTG3, respectively (Fig. 5B). However, when the second guanine is substituted by a cytosine, oligo 6RcTG2, the affinity of DDP1 decreases strongly (Fig. 5B). It must be noted that, according to their electrophoretic behavior, none of these three base substitutions has a significant effect on the extent of secondary structure of the corresponding DNA fragments. Interestingly, this second guanine residue is highly conserved in natural dodeca-satellite sequences (1, 24), being present in about 96% of all the repeats analyzed. In comparison, the first and third guanine residues mentioned above are less well conserved, being present in about 80 to 85% of the repeats. These results indicate that specific residues which are highly conserved in naturally occurring sequences contribute importantly to the interaction of DDP1 with the dodeca-satellite C strand.
Altogether, these results show that, although DDP1 binds in vitro a variety of different substrates, its interaction with the dodeca-satellite C strand is of the highest affinity. Sequence-specific interactions importantly contribute to the efficient binding of DDP1 to the dodeca-satellite C strand. It is known that nucleic acid recognition by the KH fold is, to some extent, sequence specific and that homologous KH domains of slightly different amino acid sequences show different nucleic acid binding preferences in vitro (3, 4, 10, 17, 19, 31, 32, 37). It is therefore likely that the lower affinity of DDP1 for the rest of the single-stranded nucleic acids tested here reflects less favorable sequence-specific interactions. However, sequence-specific interactions are not the only factor governing the efficient binding of DDP1 or vigilins in general to single-stranded nucleic acids. As shown here and elsewhere (17), both the length and the degree of secondary structure of the substrate also have a strong influence on the binding of vigilins. Actually, the affinity of DDP1 for oligo 9Rc, which contains nine repeats of the C-strand consensus sequence and is structured, is not significantly different from that for oligo VIT or oligo DYS. This finding suggests that DNA sequences other than the dodeca-satellite C strand could also be recognized by vigilins, provided that they are sufficiently long and unstructured. The localization of DDP1 at the pericentric heterochromatin on chromosome 2 of Drosophila, together with its significant affinity for the pyrimidine strand of the AAGAG satellite, suggests that this DNA could also be a substrate for DDP1 binding in vivo. Our results do not exclude at all the possibility that vigilins could also bind in vivo specific RNAs. Increasing evidence suggests that RNA binding proteins, such as hnRNP K, also play a role in DNA metabolism (26, 28, 33–36). The mechanisms regulating in vivo the binding of these proteins to their different possible target nucleic acids are still largely unknown.
A model for the interaction of DDP1 with the dodeca-satellite C strand.
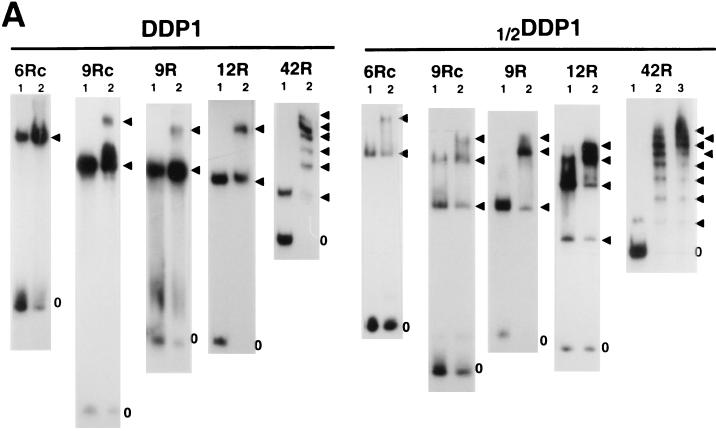
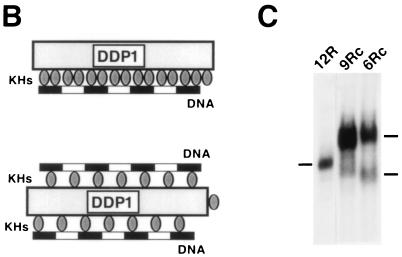
Determination of the stoichiometry of the interaction of DDP1 with the dodeca-satellite C strand suggests that DDP1 contains two independent nucleic acid binding surfaces. When DDP1 is bound to DNA fragments of the dodeca-satellite C strand in the presence of increasing protein concentrations, the formation of complexes accommodating more than one protein molecule is observed. Figure 6A shows the determination of the stoichiometry of the complexes formed with various C-strand fragments. For these experiments, the binding of 1/2DDP1, a shorter protein construct that carries only the first seven KH domains of DDP1, was also analyzed. The maximum number of protein molecules that can be incorporated into the complex depends on the length of the DNA fragment and the size of the protein construct (Table 2). For instance, the shortest, oligo 6Rc, accommodates only one DDP1 but two 1/2DDP1 molecules, while fragment 12R can accommodate up to two DDP1 or four 1/2DDP1 molecules (Fig. 6A). From these data, the number of protein KH domains per dodeca-satellite repeat involved in the interaction can be estimated (Table 2). In all cases, the stoichiometry of the complexes closely corresponds to two KH domains per dodeca-satellite repeat. This estimate is less precise for complexes formed with oligo 9R and oligo 9Rc. These DNA fragments can accommodate up to two DDP1 molecules; however, even at very high protein concentrations, only a small percentage of complexes contains two DDP1 molecules (Fig. 6A), indicating that the second DDP1 molecule enters the complex with great difficulty. Therefore, in these cases, the actual stoichiometry of the complexes will be somewhere between those of the complexes containing one and two DDP1 molecules, 1.7 and 3.3 KH domains per DNA repeat, respectively. Similarly, fragment 42R accommodates at least eight 1/2DDP1 molecules. However, in this case, complexes of higher stoichiometry are also formed, but they could not be resolved electrophoretically (Fig. 6A). These results indicate that the interaction of DDP1 with the dodeca-satellite C strand occurs at a defined stoichiometry of approximately two KH domains per DNA repeat.
FIG. 6.
Determination of the stoichiometry of the DDP1–C-strand complexes. (A) The binding of DDP1 and 1/2DDP1 to oligo 6Rc (panels 6Rc), oligo 9Rc (panels 9Rc), oligo 9R (panels 9R), fragment 12R (panels 12R), and fragment 42R (panels 42R) is presented as a function of increasing protein concentrations: a threefold-higher protein concentration was used in lanes 2 than in lanes 1. The arrowheads indicate protein-DNA complexes of increasing stoichiometry. The position of the free DNA probe is indicated by zero. (B) Possible models for the interaction of DDP1 with the dodeca-satellite C strand (see the text for details). (C) High-resolution EMSA of the complexes containing one DDP1 molecule formed with fragment 12R (lane 12R), oligo 9Rc (lane 9Rc), and oligo 6Rc (lane 6Rc).
TABLE 2.
Stoichiometry of the complexes of DDP1 with C-strand fragments of different lengths
| DNA fragment | No. of:
|
||
|---|---|---|---|
| Dodeca repeats | DDP1 moleculesa | 1/2DDP1 moleculesa | |
| Oligo 6Rc | 6 | 1 (2.5) | 2 (2.3) |
| Oligo 9R | 9 | 1 (1.7)–2 (3.3) | 3 (2.3) |
| Oligo 9Rc | 9 | 1 (1.7)–2 (3.3) | 3 (3.3) |
| Fragment 12R | 12 | 2 (2.5) | 4 (2.3) |
| Fragment 42R | 42 | 6 (2.1) | >8 (>1.3) |
The number of KH domains per dodeca-satellite repeat is shown in parentheses. DDP1 and 1/2DDP1 contain 15 and 7 KH domains, respectively.
Two possible models can account for these results. Either each DNA repeat is recognized by two KH domains (Fig. 6B, top) or, alternatively, each KH domain binds one DNA repeat, but the organization of the KH domains is such that there are two nucleic acid binding surfaces in the protein (Fig. 6B, bottom). This second possibility appears more likely. First, although it is not precisely known how many bases are directly involved in the interaction with a single KH domain, it appears unlikely that a dodeca-satellite C-strand repeat which is only 11 to 12 nt long would be sufficiently long to accommodate two relatively large KH domains. In this respect, it is interesting to note that the shortest sequence known to be bound by a single KH domain is 15 nt long (4). Second, the binding of two KH domains to a single dodeca-satellite C-strand repeat would necessarily imply that each of the two KH domains recognizes a different nucleotide sequence; therefore, equivalent protein-DNA interactions would take place only at every other KH domain of DDP1. However, given the repetitive character of both substrates, it appears more likely that each KH domain would maintain equivalent molecular interactions with the dodeca-satellite C strand. Furthermore, if DDP1 contained two nucleic acid binding surfaces, a second DNA fragment could be accommodated in the complexes formed with short DNA fragments but not in those formed with large fragments. Consistent with this hypothesis, the complexes formed with oligo 6Rc and oligo 9Rc which, under routine EMSA conditions, behave as a single molecular species containing only one DDP1 molecule, are resolved into two closely migrating species when subjected to a longer electrophoretic run (Fig. 6C, 9Rc and 6Rc). On the other hand, under these high-resolution EMSA conditions, only a single complex is observed with fragment 12R (Fig. 6C, 12R). All these considerations strongly suggest that DDP1 contains two nucleic acid binding surfaces. Interestingly, in crystals of the KH3 domain of Nova-1 or Nova-2, the lattice is composed of symmetric tetramers of independent KH domains that define two opposite nucleic acid binding surfaces and two different protein-protein interfaces (22).
The results reported here and elsewhere (8) indicate that DDP1 is a single-stranded DNA binding protein whose affinity for double-stranded DNA is extremely low. Therefore, the association of DDP1 with heterochromatin suggests that this specialized chromosomal structure contains regions of single-stranded DNA that could be recognized by DDP1. The formation of single-stranded DNA at heterochromatin blocks could originate from their characteristic enrichment on highly repetitive satellite DNA sequences, which is likely to promote strand slippage events during DNA replication. In this respect, the dodeca-satellite, like general satellites showing Pu-Py strand asymmetry, appear especially suited for the binding of DDP1 or vigilins in general, since its two strands show drastically different structural properties (6, 7, 13, 14, 27). The pyrimidine-rich strand could remain unstructured, providing long single-stranded DNA stretches, as required for efficient DDP1 binding. On the other hand, the high tendency of the purine-rich strand to form intramolecular fold-back structures could help prevent reannealing of the two complementary strands. Mechanisms involving specific protein-DNA interactions could stabilize, propagate, or even induce the formation of single-stranded DNA at heterochromatin. DDP1 could play such a role(s). DDP1 could also contribute to some of the characteristic properties of heterochromatin. Heterochromatin appears to be involved in homologous as well as ectopic chromosome pairing both at the centromeric regions and throughout the chromosome (2, 9, 11, 16, 18). In Drosophila, the frequent association of the bwD locus with the centric heterochromatin of chromosome 2 or the clustering of different heterochromatin regions to form the chromocenter likely involves heterochromatin-mediated pairing events (9, 11). The possibility that DDP1 contains two nucleic acid binding surfaces suggests a potential contribution to heterochromatin pairing. A single DDP1 molecule could bind noncontiguous single-stranded DNA stretches, linking together heterochromatin regions of the same chromosome or of sister chromosomes and thereby contributing to chromosome pairing and/or cohesion.
ACKNOWLEDGMENTS
This work was financed by grants from the Spanish DGES (PB96-812) and the CIRIT of the Generalitat de Catalunya (SGR97-55). A.C. was a recipient of a doctoral fellowship from the CIRIT. This work was carried out within the context of the Centre de Referència en Biotecnologia of the Generalitat de Catalunya.
REFERENCES
- 1.Abad J P, Carmena M, Baars S, Saunders R D C, Glover D M, Ludeña P, Sentis C, Tyler-Smith C, Villasante A. Dodecasatellite: a conserved C+G-rich satellite from centromeric heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci USA. 1992;89:4663–4667. doi: 10.1073/pnas.89.10.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Cranston G. Mutations derepressing silent chromatin domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 3.Ashley C T, Jr, Wilkinson K, Reines D, Warren S T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 4.Buckanovich R J, Darnell R B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmena M, Abad J P, Villasante A, González C. The Drosophila melanogaster dodecasatellite is closely linked to the centromere and can form connections between sister chromatids during mitosis. J Cell Sci. 1993;105:41–50. doi: 10.1242/jcs.105.1.41. [DOI] [PubMed] [Google Scholar]
- 6.Catasti P, Gupta G, Garcia A E, Ratliff R, Hong L, Yau P, Moyzis R K, Bradbury E M. Unusual structures of the tandem repetitive DNA sequences located at human centromeres. Biochemistry. 1994;33:3819–3830. doi: 10.1021/bi00179a005. [DOI] [PubMed] [Google Scholar]
- 7.Chou S-H, Zhu L, Reid B R. The unusual structure of the human centromere (GGA)2 motif. J Mol Biol. 1994;244:259–268. doi: 10.1006/jmbi.1994.1727. [DOI] [PubMed] [Google Scholar]
- 8.Cortés A, Huertas D, Fanti L, Pimpinelli S, Marsellach F X, Piña B, Azorín F. DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J. 1999;18:3820–3833. doi: 10.1093/emboj/18.13.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csink A K, Henikoff S. Genetic modification of heterochromatin association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- 10.Dejgaard K, Leffers H. Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur J Biochem. 1996;241:425–431. doi: 10.1111/j.1432-1033.1996.00425.x. [DOI] [PubMed] [Google Scholar]
- 11.Dernburg A F, Broman K W, Fung J C, Marshall W F, Philips J, Agard D A, Sedat J W. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 12.Dodson R E, Shapiro D J. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J Biol Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 13.Ferrer N, Azorín F, Villasante A, Gutiérrez C, Abad J P. Centromeric dodeca-satellite DNA sequences form fold-back structures. J Mol Biol. 1995;245:8–21. doi: 10.1016/s0022-2836(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 14.Grady D L, Ratliff R L, Robinson D L, McCanlies E C, Meyne J, Moyzis R K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci USA. 1992;89:1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray D M, Hung S H, Johnson K H. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 17.Kanamori H, Dodson R E, Shapiro D J. In vitro genetic analysis of the RNA binding site of vigilin, a multi-KH-domain protein. Mol Cell Biol. 1998;18:3991–4003. doi: 10.1128/mcb.18.7.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karpen G H, Le M H, Le H. Centric heterochromatin and the efficiency of achiasmate dysjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 19.Krüse C, Grünweller A, Notbohm H, Kügler S, Purschke W G, Müller P K. Evidence for a novel cytoplasmic tRNA-complex containing the KH-multidomain protein vigilin. Biochem J. 1996;329:247–252. doi: 10.1042/bj3200247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüse C, Grünweller A, Willkomm D K, Pfeiffer T, Hartmann R K, Müller P K. tRNA is entrapped in similar, but distinct, nuclear and cytoplasmic ribonucleoprotein complexes, both of which contain vigilin and elongation factor 1α. Biochem J. 1998;329:615–621. doi: 10.1042/bj3290615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kügler S, Grünweller A, Probst C, Klinger M, Müller P K, Krüse C. Vigilin contains a functional nuclear localisation sequence and is present in both the cytoplasm and the nucleus. FEBS Lett. 1996;382:330–334. doi: 10.1016/0014-5793(96)00204-9. [DOI] [PubMed] [Google Scholar]
- 22.Lewis H A, Chen H, Edo C, Buckanovich R J, Yang Y Y L, Musunuru K, Zhong R, Darnell R B, Burley S K. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 23.Lohe A R, Hilliker A J, Roberts P A. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losada A, Abad J P, Villasante A. Organization of DNA sequences near the centromere of Drosophila melanogaster Y chromosome. Chromosoma. 1997;106:503–512. doi: 10.1007/s004120050272. [DOI] [PubMed] [Google Scholar]
- 25.McKnight G L, Reasoner J, Gilbert T, Sundquist K O, Hokland B, McKernan P A, Champagne J, Johnson C J, Bailey M C, Holly R, O'Hara P J, Oram J F. Cloning and expression of a cellular high density lipoprotein-binding protein that is up-regulated by cholesterol loading of cells. J Biol Chem. 1992;267:12131–12141. [PubMed] [Google Scholar]
- 26.Michelotti G A, Michelotti E F, Pullner A, Duncan R C, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Lombardía M, Cortés A, Huertas D, Eritja R, Azorín F. Tandem 5′-GA:GA-3′ mismatches account for the high stability of the fold-back structures formed by the centromeric Drosophila dodeca-satellite. J Mol Biol. 1998;277:757–762. doi: 10.1006/jmbi.1998.1646. [DOI] [PubMed] [Google Scholar]
- 28.Ostrowski J, Van Seuningen I, Seger R, Rauch C T, Sleath P R, McMullen B A, Bomsztyk K. Purification, cloning, and expression of a murine phosphoprotein that binds the kB motif in vitro identify it as the homolog of the human heterogeneous nuclear ribonucleoprotein K protein. J Biol Chem. 1994;269:17626–17634. [PubMed] [Google Scholar]
- 29.Plenz G, Kügler S, Schnittger S, Rieder H, Fonatsch C, Müller P K. The human vigilin gene: identification, chromosomal localisation and expression pattern. Hum Genet. 1994;93:575–582. doi: 10.1007/BF00202827. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt C, Henkel B, Pöschl E, Zorbas H, Purschke W G, Gloe T R, Müller P K. Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionarily conserved domains. Eur J Biochem. 1992;206:625–634. doi: 10.1111/j.1432-1033.1992.tb16967.x. [DOI] [PubMed] [Google Scholar]
- 31.Siebel C W, Admon A, Rio D C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 32.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interaction among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, Dreyfuss G, Levens D. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J Biol Chem. 1993;268:18249–18258. [PubMed] [Google Scholar]
- 34.Tomonaga T, Levens D. Activating transcription from single-stranded DNA. Proc Natl Acad Sci USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomonaga T, Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 36.Tomonaga T, Michelotti G A, Libutti D, Uy A, Sauer B, Levens D. Unrestraining genetic processes with a protein-DNA hinge. Mol Cell. 1998;1:759–764. doi: 10.1016/s1097-2765(00)80075-1. [DOI] [PubMed] [Google Scholar]
- 37.Weber V, Wernitzing A, Hager G, Harata M, Frank P, Wintersberger U. Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur J Biochem. 1997;249:309–317. doi: 10.1111/j.1432-1033.1997.00309.x. [DOI] [PubMed] [Google Scholar]
- 38.Wintersberger U, Kühne C, Karwan A. Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast. 1995;11:929–944. doi: 10.1002/yea.320111004. [DOI] [PubMed] [Google Scholar]