Abstract
Background
Excessive lactate production, a hallmark of cancer, is largely formed by the reduction of pyruvate via lactate dehydrogenase (LDH) to l-lactate. Although d-lactate can also be produced from glucose via the methylglyoxal pathway in small amounts, less is known about the amount of d-lactate produced in cancer cells. Since the stereoisomers of lactate cannot be distinguished by conventional 1H NMR spectroscopy, a chiral NMR shift reagent was used to fully resolve the 1H NMR resonances of d- and l-lactate.
Methods
The production of l-lactate from glucose and d-lactate from methylglyoxal was first demonstrated in freshly isolated red blood cells using the chiral NMR shift reagent, YbDO3A-trisamide. Then, two different cell lines with high GLO1 expression (H1648 and H 1395) were selected from a panel of over 80 well-characterized human NSCLC cell lines, grown to confluence in standard tissue culture media, washed with phosphate-buffered saline, and exposed to glucose in a buffer for 4 h. After 4 h, a small volume of extracellular fluid was collected and mixed with YbDO3A-trisamide for analysis by 1H NMR spectroscopy.
Results
A suspension of freshly isolated red blood cells exposed to 5mM glucose produced l-lactate as expected but very little d-lactate. To evaluate the utility of the chiral NMR shift reagent, methylglyoxal was then added to red cells along with glucose to stimulate the production of d-lactate via the glyoxalate pathway. In this case, both d-lactate and l-lactate were produced and their NMR chemical shifts assigned. NSCLC cell lines with different expression levels of GLO1 produced both l- and d-lactate after incubation with glucose and glutamine alone. A GLO1-deleted parental cell line (3553T3) showed no production of d-lactate from glucose while re-expression of GLO1 resulted in higher production of d-lactate.
Conclusions
The shift-reagent-aided NMR technique demonstrates that d-lactate is produced from glucose in NSCLC cells via the methylglyoxal pathway. The biological role of d-lactate is uncertain but a convenient method for monitoring d-lactate production could provide new insights into the biological roles of d- versus l-lactate in cancer metabolism.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40170-021-00267-4.
Keywords: d- and l-lactate, Shift reagent-aided NMR, Cancer cells
Background
Glycolysis is the major pathway for the conversion of glucose to pyruvate in all mammalian cells. Under aerobic conditions, pyruvate is transported into mitochondria and oxidized in the TCA cycle whereas in tissues with limited oxygen availability, pyruvate is converted to lactate in the cytosol and exported from cells. Lactate production even in the setting of adequate oxygen (the Warburg effect) is observed in cultured cancer cells and in tumors. Lactate can also be imported and used as a respiratory fuel in some tumors [1]. In terms of whole-body metabolism, Hui, et al. [2] have determined in fed and fasted mice that the circulatory turnover of lactate is even higher than glucose turnover. This suggests that lactate may be a more important substrate than glucose for whole-body metabolism. In the brain, it is widely accepted that glucose is the predominant source of energy, although glucose oxidation occurs indirectly, with astrocytes taking up glucose from the blood, converting it to lactate via glycolysis, then exporting lactate for oxidation by neurons [3, 4]. This means that lactate must be converted back to pyruvate by neuronal lactate dehydrogenase (LDH) before being completely oxidized in the TCA cycle. These examples illustrate the concept of a lactate shuttle where lactate can freely be exchanged among cells, tissues, and organs under aerobic conditions [5]. One possible advantage of the astrocyte-neuron lactate shuttle in the brain is that it transfers an “extra” reducing equivalent from astrocytes to the more energy-demanding neurons. The amount of ATP generated from these extra reducing equivalents in neurons depends upon whether the lactate is converted to pyruvate via cytosolic LDH or, as has been reported in some tissues, via mitochondrial LDH [6–8].
Given that both d-lactate and l-lactate can be produced from glucose in some mammalian tissues, one must consider the potential role of d-lactate in cell metabolism. Methylglyoxal (MG), a highly reactive three-carbon glycating metabolite formed at the level of the triose phosphates [9], is rapidly converted to S-lactoylglutathione by glyoxalase-1 (GLO-1) and subsequently to d-lactate by glyoxalase-2 (GLO-2) to eliminate this reactive species in glycolytic cells. The amount of d-lactate measured in the plasma, liver, and skeletal muscle of rats is quite variable (nM to μM) depending upon nutritional state and presence of diabetes [10]. The amount of d-lactate in liver tissue is consistently about 15–17% that of l-lactate [10]. More recently, de Bari et al. [11] demonstrated that d-lactate is transported into isolated rat liver mitochondria and converted to pyruvate using a membrane-bound FAD/FMN flavoprotein on the matrix side of the inner mitochondrial membrane. The resulting pyruvate formed from d-lactate can be oxidized or converted to oxaloacetate and transported out of mitochondria via a d-lactate/OAA antiporter. Thus, both d-lactate and l-lactate can serve as three-carbon precursors of gluconeogenesis.
Why is d-lactate often ignored in modern biochemistry textbooks and by scientists investigating the role of the Warburg effect in cancer? One likely reason is that d- and l-lactate are difficult to distinguish using modern spectroscopic methods such as NMR or MRI. Kuchel et al. [12] demonstrated that one can use stretched gelatins in NMR tubes to resolve the NMR methyl resonances of d- and l-lactate based upon the differential dipolar splitting seen in their CH3 resonances. Although the method is quantitative, it is not easily adapted to routine measures of d- and l-lactate in biological samples. Hyperpolarized 13C-labeled MG has also been used to detect d-lactate as an end-product of the glyoxalase pathway in RBCs, in EL4 tumor-bearing mice, and in the liver and brain of live animals [13]. This exciting observation demonstrates that the glyoxalase pathway is active in these tissues, but it does not allow measurement of the relative amounts of d-lactate versus l-lactate produced in those tissues without the addition of exogenous MG in supraphysiological quantities. We introduce here a simple NMR method for resolving the resonances of d- and l-lactate in any biological sample by the addition of a chiral shift reagent (SR). The water-soluble, chiral agent, YbDO3A-(L-alanylamide) (Scheme 1) (referred to as Yb3 in a prior publication [14]) forms a 1:1 complex with d- and l-lactate with nearly equal affinity and the chemical shifts of the CH and CH3 proton resonances in those two complexes are well-resolved (the methyl resonances of d- and l-lactate differ by ~9.5 ppm depending upon temperature). In this study, this SR-aided 1H NMR method was used to demonstrate production of d-lactate in RBCs exposed to methylglyoxal and production of both d- and l-lactate in two different human cancer cell lines exposed to glucose alone.
Scheme 1.
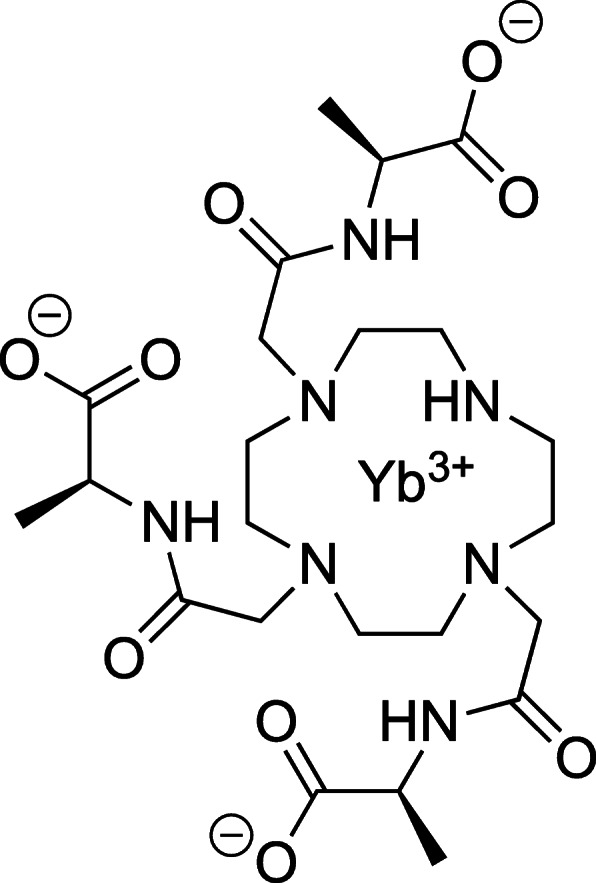
The chemical structure of the YbDO3A-(L-alanylamide) drawn as the (S)-isomer
Methods
General
All reagents and solvents were purchased from commercial sources and used as received without further purification. Preparative HPLC was performed on a Waters Delta Prep system with a Waters Atlantis Prep, T3, OBD, 5 μm, 30 mm X 250 mm column pump coupled to a Waters diode array UV detector. LC-MS experiments were carried out on a Waters Alliance LC system with a Atlantis T3 column (C18, 5 μ, 250 × 4.6 mm), connected to a Waters diode array UV detector, and an electrospray ionization mass spectrometer (ESI MS) using a Waters Q TOF-MS-XEVO ESI positive mode for detection. ICP-OES analysis of Yb3+ (Galbraith Laboratories, Inc., Knoxville, TN) was used to calibrate the concentration of Yb3 in the final stock solutions.
Preparation of shift reagent
Yb(III)-1,4,7,10-tetraazacyclododecane-1,4,7–tris(2-acetamido-L-alanine) (abbreviated Yb3) was prepared by mixing the ligand (prepared and purified as described previously[14]) with 0.95 equivalents of Yb(OH)3. The pH of the solution was adjusted to 5.5 and the reaction was stirred at room temperature overnight while the pH of the solution was maintained in the range of 5.5 – 6.0 by the addition of NaOH (or HCl). After filtration, the pH was adjusted to 7 with 1 N HCl and tested (xylenol orange) for free Yb3+ (none present). After filtering through a 2 μm membrane, the filtrate was lyophilized to give 0.60 g of Yb3 as a cream-colored solid. m/z (ESI-MS+): 731.02 (M+H)+ calculated for C23H39N7O9Yb 731.22.
NMR spectroscopy
1H, 13C, and CEST spectra were recorded on 9.4T Varian VNMRS direct-drive console spectrometer operating at 400 MHz (1H) and 100 MHz (13C), respectively. 1H NMR spectra were collected using a 90° pulse, 10,000 Hz sweep width, 19,979 data points, 2 s acquisition time, and a 1 s interpulse delay at 25°C averaged over 512 scans. Spectra were analyzed using ACD/SpecManager (Advanced Chemistry Development, Inc., Toronto, Canada). CEST spectra of Yb3 were collected by measuring the bulk water proton intensity after a series of 5 s frequency-selective pulses were applied prior to collection of the water signal (8 s delay between scans). The saturation frequency was arrayed in steps of 400 Hz. CEST Z-spectra are presented as a percent reduction of the intensity of the water signal (Mz/M0) versus saturation frequency.
Red blood cell experiments
The fresh blood, collected from a donor under an approved university protocol, was centrifuged at 2000 rpm for 10 min at 4°C. The plasma, including the buffy coat layer, was removed. The packed erythrocytes were washed in phosphate buffer (10 mM phosphate, 137 mM NaCl, 2.7 mM KCl, pH 7.4), resuspended in phosphate buffer (40% hematocrit), and exposed to either 5 mM glucose, 5 mM MG, or a mixture of 5 mM glucose plus 5 mM MG. After incubation for 15 min, 1 h, or 2 h, a 0.5-mL of buffer was collected and centrifuged, and Yb3 was added to a supernatant to a final concentration of 2.2 mM. For detection of d- and l-lactate directly in a suspension of red blood cells, RBCs were resuspended in phosphate buffer (10 mM phosphate, 137 mM NaCl, 2.7 mM KCl, pH 7.4) and incubated with 5 mM glucose, 5 mM MG, or no added substrate. After an incubation period of 2h, Yb3 was added to the 0.5 mL of packed RBCs at a final concentration of 2 mM.
Lung cancer cell lines
H1395 and H1648 cells were cultured in 100-mm plates in RPMI-1640 medium (Sigma, R8758) supplemented with 10% of dialyzed fetal bovine serum (FBS) (Sigma, F2442) and 20 units of penicillin-streptomycin (Sigma, P0781). The cells were starved for 12 h prior to the initiation of the experiment, then washed three times in phosphate-buffered saline (PBS) and incubated in a basal medium (Sigma, D5030) supplemented with 5 mM glucose or 5 mM MG for 4 h. The culture medium (10 mL) was then lyophilized and redissolved in 0.4 mL of buffer containing 5 mM Yb3 for NMR analysis. 3553T3 parental cells, a Glo1-deleted clone, and cells in which Glo1 was re-expressed were cultured in 100 mm culture plates with DMEM medium (Sigma, D5796) supplemented with 10% of FBS and 20 units of penicillin-streptomycin. When the cells reached confluence, the cells were washed with phosphate buffer and incubated for 4 h in basal medium (Sigma, D5030) supplemented with 5 mM MG and 5 mM glucose. As before, the culture media (7 mL) was then lyophilized and redissolved in 0.4 mL of buffer containing 10 mM Yb3 for NMR analysis. The number of cells used in each experiment (~3 x 106 for H1395 and H1648 cells and ~1.5 x 106 for 3553T3-derived cells) was determined using a TC20 Automated Cell Counter (Bio-Rad).
Western blots
Cells were lysed in RIPA buffer supplemented with the following additives: protease and phosphatase tablets (Roche), DTT (1 μg/ml), and benzamidine (1 μg/ml). Cleared lysates were resolved by 4–20% SDS-PAGE gels, transferred to nitrocellulose, and incubated with primary antibodies against Glyoxalase I (Novus Biologicals, NBP1-19015) and Beta Actin (Cell Signaling, 4970). Immunoreactive proteins were visualized by chemiluminescence (Pierce, 32106).
Results
Quantitative analysis of d- and l-lactate by NMR
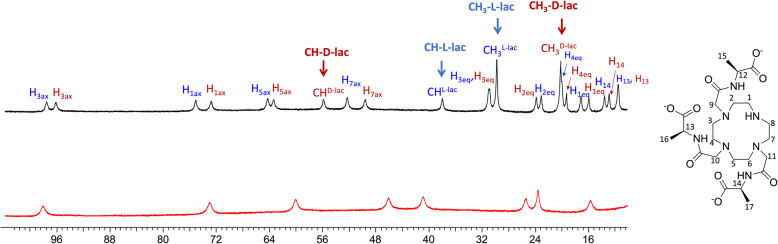
The fundamental basis of enantiomeric detection and quantification of d- and l-lactate by NMR was to add YbDO3A-(L-alanylamide)3 (abbreviated Yb3) as a paramagnetic shift reagent to biological samples [14]. Yb3 was selected over other amino acid amide options for several reasons: (1) ease of synthesis; (2) Yb3 exists in solution as a single stereoisomer (Λ(δδδδ) or Δ(λλλλ)) which, upon formation of a complex with lactate, yields single sharp CH and CH3 resonances for both d- and l-lactate; and (3) the paramagnetic properties of Yb3 result in large chemical shift differences between the bound resonances of d- and l-lactate [14]. The high-resolution 1H NMR spectrum of Yb3 used in this study is shown in Fig. 1. The downfield proton resonances in this complex, previously assigned using 2D EXSY NMR [14], reflect individual ligand protons in this highly asymmetric complex. Upon addition of either d- or l-lactate, two new proton resonances appear in the spectrum reflecting the methine and methyl protons of d-lactate·Yb3 or l-lactate·Yb3. The methyl resonance of d-lactate·Yb3 does overlap somewhat with the H4eq resonance of l-lactate·Yb3, so this needs to be taken into account in all quantitative measures. Fortunately, all of the highly shifted ethylenediamine protons split into two peaks, one reflecting d-lactate·Yb3 and the other reflecting l-lactate·Yb3, so the areas of these separated resonances provide an second analytical measure of the relative amounts of d- versus l-lactate in solution.
Fig. 1.
1H NMR spectra of an aqueous solution of Yb3 (6.3 mM) (bottom) and upon addition of d-lactate (15 mM) and l-lactate (15 mM) to the Yb3 sample (top), 9.4T; pH 7.2; 298K. The 1H NMR chemical shifts of all l-lactate ▪ Yb3(blue) and d-lactate▪ Yb3(red) protons were assigned as shown by the colored labels. The protons of the excess non-chelated lactate are far upfield in their normal diamagnetic positions (not shown here)
The upper panel of Fig. 1 shows the 1H spectrum of Yb3 after the addition of both d- and l-lactate (both in excess). The protons of unbound, excess lactate appear in their normal diamagnetic positions (Figure S2). This shows that the rate of lactate exchange between the lactate·Yb3 complexes and free lactate is slow in comparison to their frequency differences. The well-separated methyl protons (21.0 ppm and 30.4 ppm) and methine protons (56.7 ppm and 38.6 ppm) of d- and l-lactate, respectively, indicate that either pair of resonances or both pairs could be used to quantify the amount of d- versus l-lactate in biological samples. The near-equal intensities of the two methine or methyl resonances in the top spectrum of Fig. 1 suggest that the two enantiomers of lactate must have nearly equal binding affinities with Yb3.
To verify this, an additional 1H NMR titration experiment was performed. Upon the addition of increasing amounts of either d- or l-lactate (from 0 to 7 mM) to a fixed concentration of Yb3 aqueous solution (2 mM), the 1H-NMR signals of the bound lactate-methyl resonances gradually increased in intensity until a 1:1 complex was fully formed. At that point, further addition of lactate did not alter the intensity of the bound lactate methyl resonances further. These binding curves were fit to a simple 1:1 binding model (Equation S1 and Figure S1) [15] to yield dissociation constants (KD) of 914 ± 15 μM for d-lactate and 709 ±35 μM for l-lactate. Although the fitting results showed these binding constants did differ slightly as one would expect for molecules of differing chirality, this small difference translates to a correction factor of only 2–3% in the methine or methyl proton intensities of d- versus l-lactate in Fig. 1.
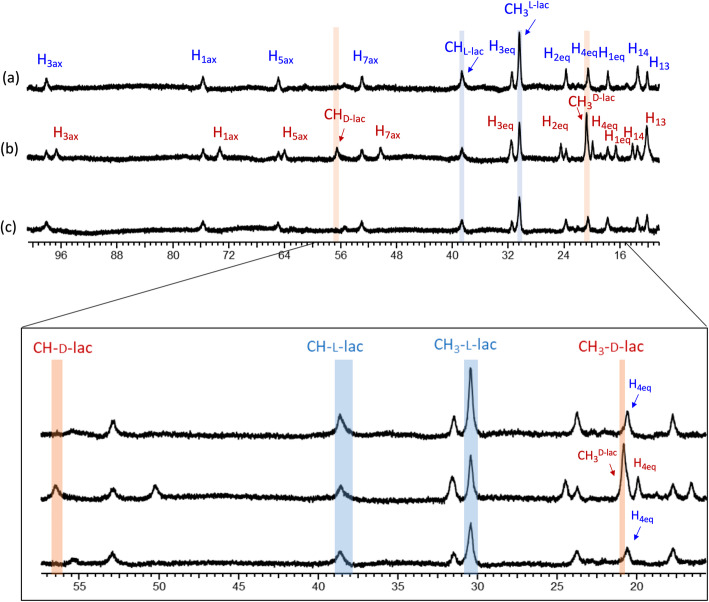
Production of d- and l-lactate in erythrocytes
Erythrocytes are known to have a glyoxalase pathway that converts methylglyoxal to d-lactate via glyoxalase 1 (GLO1) and glyoxalase 2 (GLO2) using glutathione as a co-factor [16]. To examine whether Yb3 detects both l-lactate and d-lactate production in human RBCs suspension, freshly isolated RBC suspensions in phosphate buffer (40% hematocrit) were incubated with either (a) 5 mM glucose, (b) 5 mM MG, or (C) no substrates. After incubation for 2 h at 37°C, a 2 mM Yb3 was added to the RBCs suspension before collection of their NMR spectra (Fig. 2). The spectrum in Fig. 2 a shows that 1.3 mM l-lactate was produced from glucose in RBCs; no d-lactate was evident in this spectrum. However, in RBCs incubated with MG alone, a significant amount of d-lactate (1.2 mM) was produced along with about 0.8 mM l-lactate (Fig. 2 b). Here, l-lactate must have been produced from intermediates remaining in the washed RBCs. In the RBC sample incubated without added substrates, nearly the same amount of l-lactate (1.1mM) was again produced from glycolytic intermediates (Fig. 2 c). Similar results were found in separate experiments where RBCs were incubated with these same substrates and the supernatants were separated from cells prior to the addition of the SR (Figure S3 and S4). This suggests that Yb3 remains extracellular in the experiments with cells present. We also collected CEST spectra of the supernatant samples and the areas of the CEST peaks for d- versus l-lactate⋅Yb3 complexes [14] gave quantitative values for D-Lac/L-Lac very similar to those measured by 1H NMR (Figure S5). The disadvantage of CEST spectroscopy for this measurement in comparison to 1H spectroscopy is that absolute concentrations are more difficult to obtain because the CEST intensities not only depend upon concentration but also the intensity of the applied B1 field and the amount of time the applied B1 is applied.
Fig. 2.
Detection of d- and l-lactate formation in erythrocytes using Yb3 (2 mM) by 1H NMR e-buffered saline, pH 7.4 after incubation for 2 h at 37°C with a 5 mM glucose, b 5 mM MG, and c wo/substrate. The highlighted methyl and methine resonances are assigned to l-(blue) and d-(red) lactate
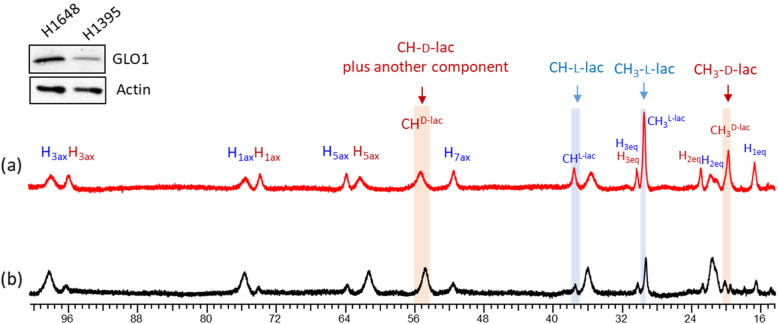
Production of d- and l-lactate in lung cancer cells
GLO1 has been reported to be overexpressed in several human cancer cell lines including leukemia, melanoma, prostate cancer, and breast cancer cells [17–20] and has been implicated in cancer progression and drug resistance [21, 22]. A recent study showed that expression of GLO1 is required for the growth of human-derived NSCLC xenografts in mice [23]. GLO2 also plays a role in androgen-dependent tumorigenesis in prostate cancer regulated by p53 [24]. To examine whether this SR-aided NMR-based method can be used to monitor the production of d- and l-lactate in tumor cells, two different cell lines with high GLO1 expression (H1648 and H 1395) were selected from a panel of over 80 well-characterized human NSCLC cell lines [25]. These cells were cultured in 5 mM glucose and glutamine added to RPMI media, grown to confluence, then washed and incubated with a modified buffer containing only 5 mM glucose and glutamine for sustained growth. After an additional 4 h, the supernatant was collected, freeze-dried, and dissolved in 0.4 mL of water containing 5 mM Yb3. The resulting 1H NMR spectra (Fig. 3) show that both cell lines make appreciable d-lactate in an apparent GLO1 expression-dependent manner (d-lac/L-lac = 0.63 in H1648 cells and 0.26 in H1395 cells).
Fig. 3.
11H NMR spectra of supernatant collected from a H1648 cells and b H1395 cells after incubation with glucose and glutamine for 4 h. After 4 h, 10 mL of media was collected, lyophilized, and redissolved into 0.4 mL water containing 5 mM Yb3 for analysis by 1H NMR (FA=45°, 9.4 T). The highlighted methyl and methine resonances are assigned to l-(blue) and d-(red) lactate. The Western blots (inset) show differences in GLO1 expression in H1648 and H1395 cells
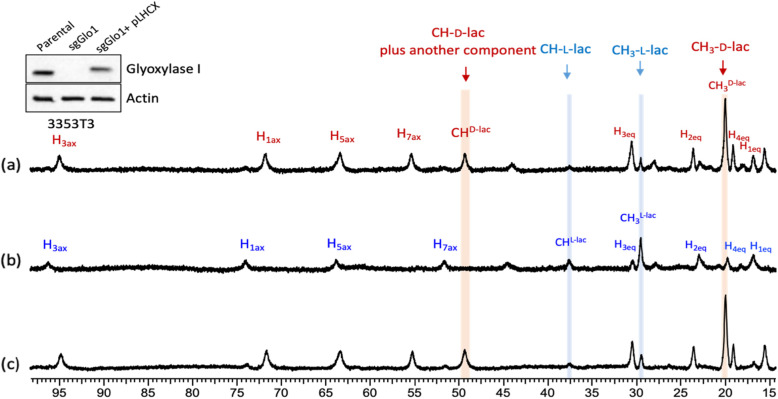
d-lactate production in GLO1-deleted and re-expressed clones
Additional experiments were designed to further test whether d-lactate production depends upon the expression of GLO1. Murine lung cancer cells (3553T3) expressing a guide RNA targeting Glo1 (sgGlo1) to knock out expression of this gene, and an isogenic line in which Glo1 was re-expressed (355T3 sgGlo1 pLHCX Glo1 )[23] were incubated in the presence of 5 mM MG for 4h. After this incubation period, 7 mL of supernatant was collected, freeze-dried, and redissolved in 0.4 mL of water containing 10 mM Yb3 for analysis by 1H NMR. As shown in Fig. 4, d-lactate was clearly evident in a Glo1 expression level-dependent manner with high levels of d-lactate produced from MG in the parental 3553T3 and 355T3 sgGlo1 pLHCX Glo1 cells, and no d-lactate detection in sgGlo1 cells. d-lactate was not detected after incubation with 5 mM glucose (Figure S6). The CEST spectra of these same samples (Fig. 5) reported identical d- and l-lactate ratios as those measured by 1H NMR.
Fig. 4.
1H NMR (400MHz) spectra of media collected from a sgGlo1 pLHCX cells (re-expressed GLO1), b sgGlo1 (GLO1 deleted cells), and c parental 3353T3 cells. Each cell line was incubated with 5 mM methylglyoxal (MG) in DMEM basal media for 4h, 37°C, and pH 7 and mixed with 10-mM Yb3 prior to collection of the NMR spectra. The highlighted methyl and methine resonances are assigned to l-(blue) and d-(red) lactate. Western blots showing GLO1 expression in 3353T3 cells
Fig. 5.
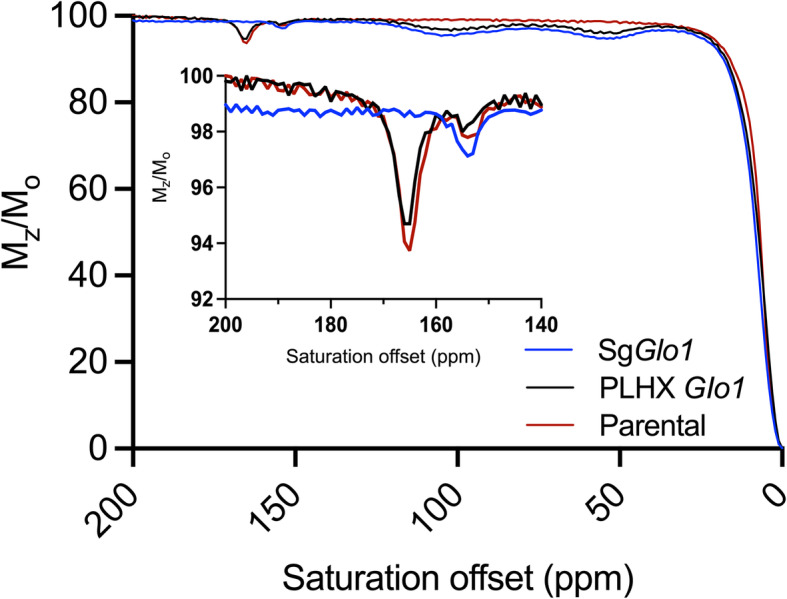
CEST spectra of the Yb3 complex (10 mM) containing 3353T3 cancer cell-cultured media which were obtained via incubation cells with 5 mM methylglyoxal (MG) in DMEM basal media (4 h at 37°C, pH ~ 7.0). The CEST peak at 165 ppm reflects d-lactate while the peak at 154 ppm reflects l-lactate in each cell line. Glo1 re-expressed cells, sgGlo1 pLHCX (black), Glo1 deleted cells, sgGlo1 (blue), and parental cells (red)
Discussion
Cancer promotes reprogramming of cell metabolism to support proliferation [26]. The highly glycolytic phenotype characteristic of most cancer cells has become an appealing therapeutic target [27, 28]. Elevated glucose metabolism inevitably produces more MG as a by-product [29, 30]. To counteract the toxic effects of MG, the glyoxalase pathway converts MG to d-lactate by sequential enzymatic reactions catalyzed by GLO1 and GLO2. Even though it has been reported that mammalian tissues are deficient in d-lactate dehydrogenase [31], Tubbs et al. showed that d-lactate is metabolized to pyruvate by the enzyme d-α-hydroxy acid dehydrogenase at a one-fifth rate of l-lactate [32]. A more recent study showed that prostate cancer cells metabolize d-lactate using d-lactate dehydrogenase, a mitochondrial membrane flavoprotein, the activity and protein level of which are higher in prostate cancer cells (PC-3) than in non-cancerous PNT1A cells [33]. Thus, detection of d-lactate in cancer in vivo could be important in that it represents a novel target for cancer diagnosis and prognosis of anti-cancer therapeutic strategies.
Potential applications of d-lactate detection are not limited to cancer metabolism. Although d-lactate is considered non-physiological in mammalian tissues, it does play a role in brain metabolism. The activity of the glyoxalase pathway is higher in astrocytes compared to neurons, consistent with higher glucose utilization and hence the greater requirement of cellular detoxification of MG in this cell type [34]. The link between MG cytotoxicity and advanced glycation end products (AGE) tightly correlates with the pathogenesis of the neurodegenerative disease [35]. A recent study showed that elevated levels of MG are present in the cerebrospinal fluid of patients with Alzheimer’s disease (AD) [36]. The GLO1 level in AD was found to correlate with the progression of the disease, increasing in early stages and then decreasing in middle to late stages of AD [37]. Other pathophysiological conditions also associated with increased serum and urinary d-lactate production are diabetic ketoacidosis and small bowel resection. Furthermore, significantly elevated serum d-lactate was also reported in infection, ischemia, and trauma [38]. In particular, d-lactate is also elevated in various pathogenic bacteria that can be a marker of infections [39]. Thus, a diagnostic method for the detection of d-lactate could provide new insights into the metabolic role of d-lactate in various disease processes. Current methods used to detect d-lactate rely upon either an enzymatic assay or liquid chromatography tandem mass spectrometry coupled with a chiral column, methods limited to analytical detection of d-lactate in tissue or cell extracts [38, 40].
In this study, we demonstrated the feasibility of using Yb3 for enantiomeric discrimination of d- and l-lactate produced in erythrocytes and cancer cells. Both d- and l-lactate bind to open coordination positions on Yb3 in a bidentate fashion and with similar binding affinities. Although not measured, the rate of exchange of lactate on and off the complex is slow on the NMR timescale (Δω >> kexch), where Δω is the difference in chemical shift of the bound versus free resonances measured in Hz and kexch is the rate of lactate exchange on and off the Yb3 complex. This results in distinct 1H NMR signals for unbound lactate versus Yb3-bound lactate. In the experiments described here, excess Yb3 was present in each NMR sample, so the relative intensities of the NMR signals of d- versus l-lactate directly reflect the quantitative levels of each enantiomer produced by cells during the incubation period. One of the nice features of this SR-aided technique is that one can quantify d- and l-lactate by integrating the areas of their well-separated methine or methyl resonances while taking advantage of the fact the ethylene signals of Yb3 also provide a second direct readout of the relative amounts of d-lactate:Yb3, l-lactate:Yb3, and unbound Yb3 present in each sample. A second nice feature is that the chemical shifts of the bound forms of lactate are independent of pH between 6 and 7.4 (Figure S7) so the method is applicable in cells that become acidic during the production of excess lactate.
Which other variables must be considered to ensure this method would provide quantitative measures of d- and l-lactate production in rodent tumor models? If one assumes that sufficient Yb3 can be injected into an animal to achieve an extracellular concentration of 1 mM, similar to that after a typical injected dose of a Gd-based contrast agent, and if the extracellular concentration of d- and l-lactate produced by a tumor at steady-state is 100 and 600 μM, respectively, then the 1H NMR signals would provide a direct readout of the absolute concentrations of d- and l-lactate. If, however, the local extracellular concentrations of d- and l-lactate are much higher, for example, 1 and 6 mM, then there would not be sufficient Yb3 present to fully form complexes with all lactate. However, even in this circumstance, the relative intensities of the methyl resonances of the d- and l-lactate complexes with Yb3 would reflect the relative amounts of the two enantiomers (after an appropriate correction factor) since the binding constants of the two complexes are nearly equal. Ultimately, the lower detection limits for detection of d- and l-lactate will be determined by the sensitivity of the imaging coil and field strength of the scanner used for 1H detection. The biocompatibility and toxicity of Yb3 are yet to be evaluated, but other lanthanide DOTA-amide-type complexes have proven safe for injection into animals at relatively high doses [41].
Conclusions
The chiral shift reagent method presented here allows for rapid, simultaneous monitoring of d- and l-lactate production in cancer cells by either high-resolution 1H NMR or CEST NMR without additional chiral separation methods. Both isomers of lactate form bidentate complexes with Yb3 with nearly equal affinities and the paramagnetic SR properties of Yb3 reagent shift the proton resonances of d- and l-lactate well-downfield and well-separated from each other. Integration of the respective methyl resonances or the -OH CEST signals provides a quantitative measure of d- versus l-lactate. The method was used to measure the production of d- and l-lactate in two different human NSCLC cell lines over a 4-h period where it was shown that both cell lines produce an appreciable d-lactate from glucose in an apparent GLO1 expression dependent manner. The biological significance of d-lactate production in these cells will require further studies, but the importance of the SR-aided NMR method reported here is that it offers other investigators a simple method for monitoring d- versus l-lactate production in living cells.
Supplementary Information
Additional file 1: Figure S1. Titration curves for d- and l-lactate. Figure S2. Full 1H NMR spectrum of d-lactate (15 mM) and l-lactate (15 mM) addition to Yb3 (6.3 mM) showing the presence of excess free lactate CH3 and CH resonances in their normal diamagnetic positions (inserted). Figure S3. Detection of d- and l-lactate formation in erythrocytes using Yb3 (2.2 mM) by 1H NMR spectra of the supernatant of erythrocytes (40 % hematocrit) in phosphate buffered saline, pH 7.4 after incubation with (a) 5 mM glucose for 2 h (b) 5 mM methyl glyoxal for 2 h (c) 5 mM glucose for 30 min followed by addition of 5 mM MG for additional 1.5 h and (d) 5 mM methyl glyoxal for 2 h (d) wo/substrate for 2 h at 37°C. Figure S4. Detection of d- and l-lactate formation in erythrocytes at each time point during 2 h incubation using Yb3 (2.2 mM) by 1H NMR spectra of the supernatant of erythrocytes (40 % hematocrit) in phosphate buffered saline, pH 7.4 (a) 5 mM glucose for 2 h (b) 5 mM glucose for 30 min followed by addition of 5 mM MG for additional 1.5 h and (C) 5 mM methyl glyoxal for 2 h (d) wo/substrate for 2 h at 37°C. The data was normalized with no substrate incubation data as background correction. Figure S5. CEST spectra of the Yb3 complex (5mM) in RBC cultured media. Presaturation pulse of 5s with B1 of 15uT was applied at 298K using 9.4 NMR spectrometer: RBCs (40% hematocrit) was incubated in Phosphate-buffered saline (pH ~7.0, 2 h at 37 °C) with 5mM glucose; Glc 5mM, 5mM glucose 30min incubated then add 5mM methyl glyoxal; Glc + MG, and 5mM methyl glyoxal; MG 5mM. The CEST peaks are assignable at 168ppm to d-lactate and at 157ppm are l- lactate. Figure S6. 1H NMR (400MHz) spectra of media collected from (a) sgGlo1 pLHCX cells (re-expressed GLO1) (b) sgGlo1 (GLO1 deleted cells) and (c) parental 3353T3 cells. Each cell line was incubated with 5mM glucose in DMEM basal media for 4 h, 37 °C, pH 7. Subsequently, a 0.5 mL volume of media was collected and mixed with 10 mM Yb3 prior to collection of the NMR spectra. The highlighted methyl resonances are assigned to l-(blue) and d-(red) lactate. Figure S7. 1H NMR spectra of an aqueous solution of Yb3 (6 mM) in the presence of d-lactate (15 mM) and l-lactate (15 mM) at pH 6.0 and 7.4. The chemical shifts of CH3-L-lac and CH3-D-lac were insensitive to pH over this range. The signals of excess unbound d- and l-lactate appeared in their normal diamagnetic positions (not shown here). Table S1. 1H NMR shift (ppm) of l-lactate·Yb3 and d-lactate·Yb3 of RBCs supernatant sample.
Acknowledgements
We thank Dr. Matthew G. Vander Heiden and Alba Luengo, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, for providing murine lung cancer (3553T3) cell lines.
Abbreviations
- LDH
Lactate dehydrogenase
- GLO-1
Glyoxalase-1
- GLO-2
Glyoxalase-2
- NMR
Nuclear magnetic resonance
- SR
Shift reagent
- EXSY
Exchange spectroscopy
- CEST
Chemical exchange saturation transfer
- MG
Methylglyoxal
- Yb3
Yb(III)-1,4,7,10-tetraazacyclododecane-1,4,7–tris(2-acetamido-l-alanine)
- AGE
Advanced glycation end products
Authors’ contributions
Conceptualization and supervision: ADS and CFGCG. Study design: ADS, CFGCG, RJD, and EHS. Investigation and methodology: ADS, CFGCG, and EHS. Data curation and formal analysis: ADS, CFGCG, and EHS. Data acquisition: EHS, SC, BF, and RA. Writing the original draft: ADS, EHS, and CFGCG. The authors read and approved the final manuscript.
Funding
ADS acknowledges the partial financial support of this work from the National Institutes of Health (CA115531 and EB02584), the Robert A. Welch Foundation (AT-584), and the STEP-UP program (R25 DK113659) for support of Raul Ayala during a summer research rotation. CFGCG acknowledges the Coimbra Chemistry Centre (UID/QUI/00313/2019 and POCI-01-0145-FEDER-027996) of the University of Coimbra, and a Fulbright visiting scholarship. R.J.D. is supported by the Howard Hughes Medical Institute and R35 CA22044901 from the National Institutes of Health.
Availability of data and materials
All data and materials are fully described in the manuscript. A copy of all data analyzed is available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
All procedures involving human sample collection were carried out under a protocol approved by the institutional review board of the University of Texas Southwestern Medical Center.
Consent for publication
This manuscript is not concurrently submitted elsewhere, and all authors are aware of and fully agree with its contents.
Competing interests
R.J.D. is the advisor for Agios Pharmaceuticals and Vida Ventures. The other authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate metabolism in human lung tumors. Cell. 2017;171(2):358–371. e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bélanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14(6):724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Pellerin L, Magistretti PJ. Sweet sixteen for ANLS. J Cereb Blood Flow Metab. 2012;32(7):1152–1166. doi: 10.1038/jcbfm.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks GA. The science and translation of lactate shuttle theory. Cell Metab. 2018;27(4):757–785. doi: 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Brooks GA, Dubouchaud H, Brown M, Sicurello JP, Butz CE. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc Natl Acad Sci U S A. 1999;96(3):1129–1134. doi: 10.1073/pnas.96.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto T, Hussien R, Brooks GA. Colocalization of MCT1, CD147, and LDH in mitochondrial inner membrane of L6 muscle cells: evidence of a mitochondrial lactate oxidation complex. Am J Physiol Endocrinol Metab. 2006;290(6):E1237–E1244. doi: 10.1152/ajpendo.00594.2005. [DOI] [PubMed] [Google Scholar]
- 8.Passarella S, de Bari L, Valenti D, Pizzuto R, Paventi G, Atlante A. Mitochondria and l-lactate metabolism. FEBS Lett. 2008;582(25-26):3569–3576. doi: 10.1016/j.febslet.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 9.Cooper RA, Anderson A. The formation and catabolism of methylglyoxal during glycolysis in Escherichia coli. FEBS Lett. 1970;11(4):273–276. doi: 10.1016/0014-5793(70)80546-4. [DOI] [PubMed] [Google Scholar]
- 10.Kondoh Y, Kawase M, Kawakami Y, Ohmori S. Concentrations of d-lactate and its related metabolic intermediates in liver, blood, and muscle of diabetic and starved rats. Res Exp Med (Berl) 1992;192(6):407–414. doi: 10.1007/BF02576298. [DOI] [PubMed] [Google Scholar]
- 11.de Bari L, Atlante A, Guaragnella N, Principato G, Passarella S. d-lactate transport and metabolism in rat liver mitochondria. Biochem J. 2002;365(2):391–403. doi: 10.1042/bj20020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuchel PW, Pagès G, Naumann C. Chiral compartmentation’ in metabolism: enzyme stereo-specificity yielding evolutionary options. FEBS Lett. 2013;587(17):2790–2797. doi: 10.1016/j.febslet.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 13.Shishmarev D, Kuchel PW, Pagès G, Wright GJ, Hesketh RL, Kreis F, et al. Glyoxalase activity in human erythrocytes and mouse lymphoma, liver and brain probed with hyperpolarized 13C-methylglyoxal. Commun Biol. 2018;1(1):232. doi: 10.1038/s42003-018-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Martins AF, Zhao P, Tieu M, Esteban-Gómez D, McCandlesset GT, et al. Enantiomeric recognition of d- and l-lactate by CEST with the aid of a paramagnetic shift reagent. J Am Chem Soc. 2017;139(48):17431–17437. doi: 10.1021/jacs.7b08292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Martins AF, Mai Y, Zhao P, Funk AM, Clavijo Jordan MV, et al. Imaging extracellular lactate in vitro and in vivo using CEST MRI and a paramagnetic shift reagent. Chem Eur J. 2017;23(8):1752–1756. doi: 10.1002/chem.201604558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornalley PJ. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269(1):1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto H, Mashima T, Kizaki A, Dan S, Hashimoto Y, Naito M, et al. Glyoxalase I is involved in resistance of human leukemia cells to antitumor agent-induced apoptosis. Blood. 2000;95(10):3214–3218. doi: 10.1182/blood.V95.10.3214. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DH, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ESC. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4(11):1686–1696. doi: 10.1074/mcp.M400221-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Antognelli C, Buono CD, Ludovini V, Gori S, Talesa VN, Crinò L, et al. CYP17, GSTP1, PON1 and GLO1gene polymorphisms as risk factors for breast cancer: an Italian case-control study. BMC Cancer. 2009;9(1):115. doi: 10.1186/1471-2407-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanuik TL, Ueda T, Le N, Haile S, Yong TMK, Thomson T, et al. Novel biomarkers for prostate cancer including noncoding transcripts. Am J Pathol. 2009;175(6):2264–2276. doi: 10.2353/ajpath.2009.080868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santarius T, Bignell GR, Greenman CD, Widaa S, Chen L, Mahoney CL, et al. GLO1-a novel amplified gene in human cancer. Genes Chromosom Cancer. 2010;49(8):711–725. doi: 10.1002/gcc.20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornalley PJ, Rabbani N. Glyoxalase in tumourigenesis and multidrug resistance. Semin Cell Dev Biol. 2011;22(3):318–325. doi: 10.1016/j.semcdb.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Luengo A, Abbott KL, Davidson SM, Hosios AM, Faubert B, Chan SH, et al. Reactive metabolite production is a targetable liability of glycolytic metabolism in lung cancer. Nat Commun. 2019;10(1):5604. doi: 10.1038/s41467-019-13419-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antognelli C, Ferri I, Bellezza G, Siccu P, Love HD, Talesa VN, et al. Glyoxalase 2 drives tumorigenesis in human prostate cells in a mechanism involving androgen receptor and p53-p21 axis. Mol Carcinog. 2017;56(9):2112–2126. doi: 10.1002/mc.22668. [DOI] [PubMed] [Google Scholar]
- 25.Chen PH, Cai L, Huffman K, Yang C, Kim J, Faubert B, et al. Metabolic diversity in human non-small cell lung cancer cells. Mol Cell. 2019;76(5):838–851.e5. doi: 10.1016/j.molcel.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh J, Suh EH, Sharma G, Khemtong C, Sherry AD, Kovacs Z. Probing carbohydrate metabolism using hyperpolarized 13C-labeled molecules. NMR Biomed. 2019;32(10):e4018. doi: 10.1002/nbm.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabbani N, Xue M, Weickert MO, Thornalley PJ. Multiple roles of glyoxalase 1-mediated suppression of methylglyoxal glycation in cancer biology-involvement in tumour suppression, tumour growth, multidrug resistance and target for chemotherapy. Semin Cancer Biol. 2018;49:83–93. doi: 10.1016/j.semcancer.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Bellahcène A, Nokin MJ, Castronovo V, Schalkwijk C. Methylglyoxal-derived stress: an emerging biological factor involved in the onset and progression of cancer. Semin Cancer Biol. 2018;49:64–74. doi: 10.1016/j.semcancer.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Uribarri J, Oh MS, Carroll HJ. D-lactic acidosis. A review of clinical presentation, biochemical features, and pathophysiologic mechanisms. Medicine (Baltimore) 1998;77(2):73–82. doi: 10.1097/00005792-199803000-00001. [DOI] [PubMed] [Google Scholar]
- 32.Tubbs PK. The metabolism of D-alpha-hydroxy acids in animal tissues. Ann N Y Acad Sci. 1965;119(3):920–926. doi: 10.1111/j.1749-6632.1965.tb47452.x. [DOI] [PubMed] [Google Scholar]
- 33.de Bari L, Moro L, Passarella S. Prostate cancer cells metabolize d-lactate inside mitochondria via a d-lactate dehydrogenase which is more active and highly expressed than in normal cells. FEBS Lett. 2013;587(5):467–473. doi: 10.1016/j.febslet.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Bélanger M, Yang J, Petit JM, Laroche T, Magistretti PJ, Allaman I. Role of the glyoxalase system in astrocyte-mediated neuroprotection. J Neurosci. 2011;31(50):18338–18352. doi: 10.1523/JNEUROSCI.1249-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen F, de Souza DF, Silveira DL, Hoefel AL, Fontoura JB, Tramontina AC, et al. Methylglyoxal alters glucose metabolism and increases AGEs content in C6 glioma cells. Metab Brain Dis. 2012;27(4):531–539. doi: 10.1007/s11011-012-9329-3. [DOI] [PubMed] [Google Scholar]
- 36.Kuhla B, Lüth HJ, Haferburg D, Boeck K, Arendt T, Münch G. Methylglyoxal, glyoxal, and their detoxification in Alzheimer’s disease. Ann N Y Acad Sci. 2005;1043(1):211–216. doi: 10.1196/annals.1333.026. [DOI] [PubMed] [Google Scholar]
- 37.Kuhla B, Boeck K, Lüth HJ, Schmidt A, Weigle B, Schmitz M, et al. Age-dependent changes of glyoxalase I expression in human brain. Neurobiol Aging. 2006;27(6):815–822. doi: 10.1016/j.neurobiolaging.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Ewaschuk JB, Naylor JM, Zello GA. d-lactate in human and ruminant metabolism. J Nutr. 2005;135(7):1619–1625. doi: 10.1093/jn/135.7.1619. [DOI] [PubMed] [Google Scholar]
- 39.Smith SM, Eng RH, Campos JM, Chmel H. D-lactic acid measurements in the diagnosis of bacterial infections. J Clin Microbiol. 1989;27(3):385–388. doi: 10.1128/jcm.27.3.385-388.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheijen JL, Hanssen NM, van de Waarenburg MP, Jonkers DM, Stehouwer CD, Schalkwijk CG. l(+) and d(-) lactate are increased in plasma and urine samples of type 2 diabetes as measured by a simultaneous quantification of l(+) and d(-) lactate by reversed-phase liquid chromatography tandem mass spectrometry. Exp Diabetes Res. 2012;2012:234812. doi: 10.1155/2012/234812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Zhang S, Soesbe TC, Yu J, Vinogradov E, Lenkinski RE. pH imaging of mouse kidneys in vivo using a frequency-dependent paraCEST agent. Magn Reson Med. 2016;75(6):2432–2441. doi: 10.1002/mrm.25844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Titration curves for d- and l-lactate. Figure S2. Full 1H NMR spectrum of d-lactate (15 mM) and l-lactate (15 mM) addition to Yb3 (6.3 mM) showing the presence of excess free lactate CH3 and CH resonances in their normal diamagnetic positions (inserted). Figure S3. Detection of d- and l-lactate formation in erythrocytes using Yb3 (2.2 mM) by 1H NMR spectra of the supernatant of erythrocytes (40 % hematocrit) in phosphate buffered saline, pH 7.4 after incubation with (a) 5 mM glucose for 2 h (b) 5 mM methyl glyoxal for 2 h (c) 5 mM glucose for 30 min followed by addition of 5 mM MG for additional 1.5 h and (d) 5 mM methyl glyoxal for 2 h (d) wo/substrate for 2 h at 37°C. Figure S4. Detection of d- and l-lactate formation in erythrocytes at each time point during 2 h incubation using Yb3 (2.2 mM) by 1H NMR spectra of the supernatant of erythrocytes (40 % hematocrit) in phosphate buffered saline, pH 7.4 (a) 5 mM glucose for 2 h (b) 5 mM glucose for 30 min followed by addition of 5 mM MG for additional 1.5 h and (C) 5 mM methyl glyoxal for 2 h (d) wo/substrate for 2 h at 37°C. The data was normalized with no substrate incubation data as background correction. Figure S5. CEST spectra of the Yb3 complex (5mM) in RBC cultured media. Presaturation pulse of 5s with B1 of 15uT was applied at 298K using 9.4 NMR spectrometer: RBCs (40% hematocrit) was incubated in Phosphate-buffered saline (pH ~7.0, 2 h at 37 °C) with 5mM glucose; Glc 5mM, 5mM glucose 30min incubated then add 5mM methyl glyoxal; Glc + MG, and 5mM methyl glyoxal; MG 5mM. The CEST peaks are assignable at 168ppm to d-lactate and at 157ppm are l- lactate. Figure S6. 1H NMR (400MHz) spectra of media collected from (a) sgGlo1 pLHCX cells (re-expressed GLO1) (b) sgGlo1 (GLO1 deleted cells) and (c) parental 3353T3 cells. Each cell line was incubated with 5mM glucose in DMEM basal media for 4 h, 37 °C, pH 7. Subsequently, a 0.5 mL volume of media was collected and mixed with 10 mM Yb3 prior to collection of the NMR spectra. The highlighted methyl resonances are assigned to l-(blue) and d-(red) lactate. Figure S7. 1H NMR spectra of an aqueous solution of Yb3 (6 mM) in the presence of d-lactate (15 mM) and l-lactate (15 mM) at pH 6.0 and 7.4. The chemical shifts of CH3-L-lac and CH3-D-lac were insensitive to pH over this range. The signals of excess unbound d- and l-lactate appeared in their normal diamagnetic positions (not shown here). Table S1. 1H NMR shift (ppm) of l-lactate·Yb3 and d-lactate·Yb3 of RBCs supernatant sample.
Data Availability Statement
All data and materials are fully described in the manuscript. A copy of all data analyzed is available from the corresponding author upon reasonable request.