Abstract
Background
Tn5253, a composite Integrative Conjugative Element (ICE) of Streptococcus pneumoniae carrying tet(M) and cat resistance determinants, was found to (i) integrate at specific 83-bp integration site (attB), (ii) produce circular forms joined by a 84-bp sequence (attTn), and (iii) restore the chromosomal integration site. The purpose of this study is to functionally characterize the attB in S. pneumoniae strains with different genetic backgrounds and in other bacterial species, and to investigate the presence of Tn5253 attB site into bacterial genomes.
Results
Analysis of representative Tn5253-carryng transconjugants obtained in S. pneumoniae strains with different genetic backgrounds and in other bacterial species, namely Streptococcus agalactiae, Streptococcus gordonii, Streptococcus pyogenes, and Enterococcus faecalis showed that: (i) Tn5253 integrates in rbgA of S. pneumoniae and in orthologous rbgA genes of other bacterial species, (ii) integration occurs always downstream of a 11-bp sequence conserved among streptococcal and enterococcal hosts, (iii) length of the attB site corresponds to length of the duplication after Tn5253 integration, (iv) attB duplication restores rbgA CDS, (v) Tn5253 produced circular forms containing the attTn site at a concentration ranging between 2.0 × 10−5 to 1.2 × 10−2 copies per chromosome depending on bacterial species and strain, (vi) reconstitution of attB sites occurred at 3.7 × 10−5 to 1.7 × 10−2 copies per chromosome. A database search of complete microbial genomes using Tn5253 attB as a probe showed that (i) thirteen attB variants were present in the 85 complete pneumococcal genomes, (ii) in 75 pneumococcal genomes (88.3 %), the attB site was 83 or 84 nucleotides in length, while in 10 (11.7 %) it was 41 nucleotides, (iii) in other 19 bacterial species attB was located in orthologous rbgA genes and its size ranged between 17 and 84 nucleotides, (iv) the 11-bp sequence, which correspond to the last 11 nucleotides of attB sites, is conserved among the different bacterial species and can be considered the core of the Tn5253 integration site.
Conclusions
A functional characterization of the Tn5253 attB integration site combined with genome analysis contributed to elucidating the potential of Tn5253 horizontal gene transfer among different bacterial species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13100-021-00253-z.
Keywords: Mobile genetic elements (MGE), Integrative conjugative element (ICE), Conjugative transposon, Conjugation, Attachment site, Circular form, Tn5253, S. pneumoniae
Introduction
The acquisition of new genetic material by horizontal gene transfer (HGT) significantly drives bacterial genome evolution and is mediated by Mobile Genetic Elements (MGEs). The term “mobilome” is used to indicate the entire set of MGEs of the microbiome[1]. MGEs are responsible for the spread of resistance and virulence genes in the microbial communities [2–4]. Thus, to study the acquisition and dissemination of antibiotic determinants in a bacterial population, the characterization of mobilome is crucial [5]. Even though new metagenomic approaches, both whole and targeted [1, 6, 7] have been implemented, a functional study of MGEs is still required [8, 9]. Integrative and Conjugative Elements (ICEs) are MGEs commonly found in bacteria where they can constitute up to 25 % of the genome [5, 10–14]. One of the most studied ICE of gram-positive bacteria is Tn916, a conjugative transposon originally found in Enterococcus faecalis which carries the tet(M) tetracycline resistance gene and is considered the prototype of the Tn916-Tn1545 family of ICEs [15–19]. Conjugative transposons of the Tn916-Tn1545 family can insert at multiple integration sites in the chromosome [20], while other ICEs, like Tn5253, SXT, Tn5397, and ICESt1, integrate at a single specific site [21–26]. We previously characterized Tn5253, a 64,528-bp composite ICE of Streptococcus pneumoniae, containing the ICE Tn5251 of the Tn916-Tn1545 family and the Ωcat(pC194) element carrying tet(M) and cat resistance determinants, respectively [27–29]. Tn5253 was found integrated at 83-bp specific integration site (attB) located in the essential gene rbgA of the S. pneumoniae chromosome [26, 28, 30]. The ICE was shown to excise from the pneumococcal chromosome with production of (i) circular forms in which the ends of the element were joined by a 84-bp sequence (attTn) and (ii) a reconstituted chromosomal attB. Tn5253, once integrated into the chromosome, was flanked by the attL site, identical to attB, and the attR site, identical to attTn. Pneumococcal mobilome analysis showed the frequent presence of Tn5253-like elements in multidrug-resistant S. pneumoniae strains and the maintenance of the element in all derivative isolates [31–34]. In this work, in order to contribute to mobilome characterization, we first conducted a functional characterization of the Tn5253 integration site, by analyzing attB in Tn5253-carrying transconjugants obtained in S. pneumoniae strains with different genetic backgrounds and in strains belonging to other bacterial species. We then investigated the presence of the Tn5253 attB site into the complete microbial genomes available in public databases.
Results and discussion
Tn5253 integration sites and circularization in different pneumococcal transconjugants
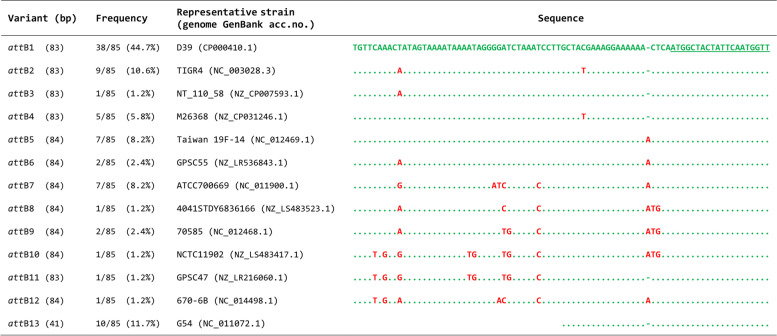
Representative Tn5253-carrying transconjugants were obtained in S. pneumoniae with different genetic backgrounds, namely TIGR4, A66 and SP18-BS74 [28] (Table 1). DNA sequence analysis of Tn5253-chromosome junction fragments showed that: (i) Tn5253 integration occurred at a specific integration site (attB) located in rbgA gene of the pneumococcal chromosome [26], (ii) attL was identical to attB and (iii) attR was identical to attTn, as already described for D39 and its derivative strains [26], and that (iv) attB sites among these pneumococcal strains were not identical, with their size varying from 41 nucleotides (variant attB13 in SP18-BS74) to 83 nucleotides (variant attB2, in TIGR4 and A66) (Fig. 1). We also analysed the nucleotide sequence of Tn5253 junction fragments in the original Tn5253-carrying clinical strain BM6001 and DP1322, in which Tn5253 was transferred by transformation of a crude lysate from BM6001 [35]. attL sequences of BM6001 and DP1322 were identical and belonged to a 84 bp-long variant (attB5, Fig. 1), since Tn5253 integration occurred via homologous recombination between DNA sequences beyond Tn5253 att sites. Tn5253 was found to excise from pneumococcal chromosome with consequent production of circular forms, containing the attTn site, and reconstitution of attB site [26]. To investigate if different pneumococcal genetic backgrounds influence the excision and circularization of Tn5253, quantitative PCR on cell lysates was used to quantify the excision of Tn5253 and attB reconstitution in liquid pneumococcal cultures (Table 2). Interestingly, the transconjugant FR56, derived from SP18-BS74, produced Tn5253 circular forms and reconstituted attB site at very high frequency (1.2 × 10−2 and 1.9 × 10−3 copies per chromosome, respectively). However, these results did not correlate with the conjugation frequency, which was 6.1 × 10−6, indicating that the frequency of circularization is not the only limiting factor of the conjugation process. Neither circular forms nor reconstituted attB of Tn5253 could be detected in the TIGR4 background (<3.6 × 10−5 and to <3.5 × 10−4, respectively), correlating with the absence of conjugal transfer (<9.9 × 10−8). Analysis of Tn5253 integration, in pneumococci with different genetic backgrounds, revealed that the element always integrates downstream of nucleotide position 20 of rbgA coding sequence (CDS) (Fig. 1). rbgA is an essential gene encoding the ribosomal biogenesis GTPase protein involved in the 50S ribosome subunit assembly [36]. Integration of Tn5253 leads to the duplication of the integration site restoring the CDS and preserving cell viability. Site specific integration of MGEs often occurs at the 5’ or 3’ end of genes, such as those coding for tRNAs or ribosomal proteins, which are essential and conserved among different bacterial species. This characteristic allows to overpass the single species border and favors the spread of MGEs within bacterial communities.
Table 1.
Bacterial strains and relevant properties
| Strain | Relevant propertiesa | Genome Genbank acc. no., [Reference] |
|---|---|---|
| Streptococcus pneumoniae | ||
| A66 | Avery’s strain, clinical isolate, serotype 3 | LN847353.1, draft genome, [41, 42] |
| HB565 | A66 derivative, carrying str-1, SmR | [39, 43] |
| FR39 | HB565 transconjugant derivative, carrying Tn5253, SmR, TcR, CmR | This study |
| TIGR4 | Clinical isolate, serotype 4 | NC_003028.3, [44] |
| FP47 | TIGR4 derivative, carrying nov-1, NovR | [29] |
| FR54 | FP47 transconjugant derivative, carrying Tn5253, NovR, TcR, CmR | [29] |
| SP18-BS74 | Clinical isolate, serotype 18 C | NZ_ABAE01000001.1, draft genome, [45] |
| FR55 | SP18-BS74 derivative, carrying str-1, SmR | [28] |
| FR56 | FR55 transconjugant derivative, carrying Tn5253, SmR, TcR, CmR | [28] |
| Other streptococci | ||
| H36B | S. agalactiae, clinical isolate, serotype Ib | NZ_LN847353.1, [46] |
| FR67 | H36B transconjugant derivative, carrying Tn5253, TcR, CmR | [29] |
| SF370 | S. pyogenes, clinical isolate, serotype M1 | AE004092.2, [47] |
| FR68 | H36B transconjugant derivative, carrying Tn5253, TcR, CmR | [29] |
| V288 | S. gordonii Challis, clinical isolate | NC_009785.1, [48, 49] |
| GP204 | V288 derivative, carrying str-204; SmR | [50] |
| FR43 | GP204 transconjugant derivative, carrying Tn5253, SmR, TcR, CmR | [29] |
| Enterococcus faecalis | ||
| OG1 | Clinical isolate, formely named 2SaR | [51] |
| OG1RF | OG1 derivative, FusR, RifR | NC_017316.1, [52, 53] |
| OG1SS | OG1 derivative, SpeR, SmR | [15, 54] |
| FR49 | OG1SS transconjugant derivative, carrying Tn5253, SpeR, SmR, TcR, CmR | [28] |
| JH2 | Clinical isolate | [55] |
| JH2-2 | JH2 derivative, FusR, RifR | NZ_KI518257.1, draft genome, [55] |
| FR50 | JH2-2 transconjugant derivative, carrying Tn5253, FusR, RifR, TcR, CmR | [28] |
a str-41 and str-204 indicate point mutations conferring resistance to streptomycin, while nov-1 to novobiocin. Sm, streptomycin; Tc, tetracycline,
Cm, chloramphenicol; Nov, novobiocin; Fus, fusidic acid; Rif, rifampicin; Spe, spectinomycin;
Fig. 1.
Allelic variants of the Tn5253 attB integration site in S. pneumoniae. Tn5253 attB is located in the essential pneumococcal rbgA gene. In the 85 complete S. pneumoniae genomes, 13 allelic variants of attB were found. The 83-bp variant 1 (attB1) is the most frequent, carried by 38 genome strains (44.7 % of the analyzed genomes), including D39 strain, its rough derivative R6, and A66 strain, and was used as reference for the alignment. Variant 2 is carried by TIGR4 and other 8 genome strain and contains 2 nucleotide substitutions. BM6001, the original Tn5253-carrying clinical strain, and DP1322, in which Tn5253 was transferred by transformation from BM6001, harbour variant 5. SP18-BS74, whose draft genome is available, harbours the 41-bp variant 13. Within the sequences, identical nucleotides are indicated by periods, substitutions in red. For better alignment, dashes are inserted. The 20 nucleotides belonging to the rbgA coding sequence are underlined
Table 2.
PCR quantification of Tn5253 circular forms and reconstituted attB integration sites in Tn5253-carrying transconjugants
| Strain | Genetic background | Circular Forms | Reconstituted attB Site | Conjugation Frequency Mean (Range)a |
|---|---|---|---|---|
| FR39 | S. pneumoniae A66 | 2.0 × 10−5 ± 1.9 × 10−5 | <6.9 × 10−6 ± 7.1 × 10−8 | 4.4 × 10−7 (3.2 × 10−7 - 5.8 × 10−7) |
| FR54 | S. pneumoniae TIGR4 | <3.6 × 10−5 ± 2.6 × 10−6 | <3.5 × 10−4 ± 2.6 × 10−5 | <9.9 × 10−8 (<9.6 × 10−8 - <1.3 × 10−7) |
| FR56 | S. pneumoniae SP18-BS74 | 1.2 × 10−2 ± 7.5 × 10−5 | 1.9 × 10−3 ± 1.0 × 10−4 | 6.1 × 10−6 (3.5 × 10−6 - 9.5 × 10−6) |
| FR67 | S. agalactiae H36B | 2.9 × 10−5 ± 8.7 × 10−6 | 3.0 × 10−4 ± 1.0 × 10−4 | 1.1 × 10−6 (3.2 × 10−7 - 2.1 × 10−6) |
| FR68 | S. pyogenes SF370 | 3.0 × 10−5 ± 1.0 × 10−5 | 3.7 × 10−5 ± 6.2 × 10−6 | 9.5 × 10−4 (3.3 × 10−5 - 2.6 × 10−3) |
| FR43 | S. gordonii V288 | 6.5 × 10−5 ± 4.4 × 10−5 | 7.9 × 10−5 ± 1.2 × 10−5 | 8.3 × 10−7 (1.2 × 10−7 - 2.0 × 10−6) |
| FR49 | E. faecalis OG1SS | 1.4 × 10−4 ± 9.2 × 10−5 | 6.8 × 10−3 ± 1.0 × 10−4 | <1.8 × 10−8 (<1.0 × 10−9 - <3.9 × 10−8) |
| FR50 | E. faecalis JH2-2 | <2.7 × 10−7 ± 7.5 × 10−8 | 1.7 × 10−2 ± 1.3 × 10−3 | <2.7 × 10−8 (<9.9 × 10−9 - <5.0 × 10−8) |
aFrequency refers to mating experiments where S. pneumoniae FP10 or FP11was the conjugation recipient [29]; conjugation frequency is expressed as
CFU of transconjugants per CFU of donors; each result is the mean of at least three mating experiments
Tn5253 integration sites and circularization in Streptococcus and Enterococcus.
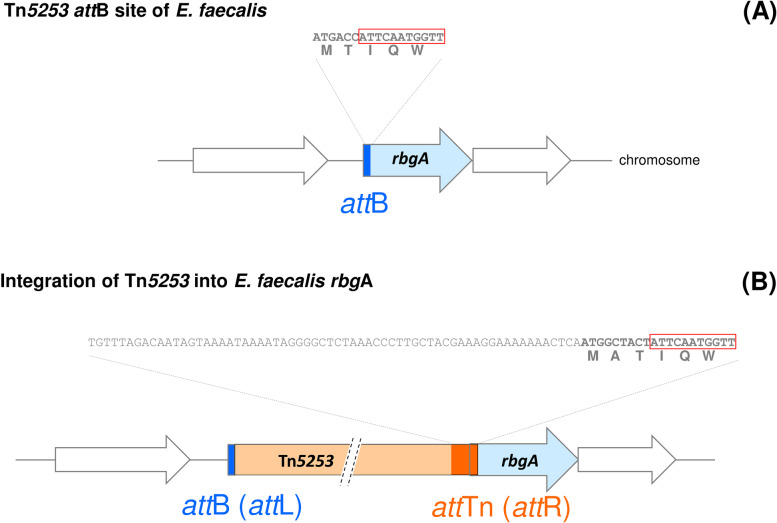
We then extended Tn5253 functional analysis to streptococci and enterococci characterizing Tn5253 circular forms and integration sites in the transconjugants obtained in S. agalactiae H36B, S. pyogenes SF370, S. gordonii V288, E. faecalis OG1SS and JH2-2 backgrounds (Table 1). In all bacterial hosts, Tn5253 integration occurred in the orthologous rbgA genes (Fig. 2). As found in S. pneumoniae, in all the bacterial hosts tested: (i) attL was identical to attB regardless of the bacterial strain harbouring the element and attR was identical to attTn suggesting a polarization of Tn5253 integration process, (ii) integration always occurs downstream of a 11-bp conserved sequence, namely the last 11 nucleotides of attB sites, (iii) length of the attB site corresponds to length of the duplication after Tn5253 integration, (iv) attB site duplication restores rbgA CDS. It is worth to note that in E. faecalis, attB duplication modifies the rbgA predicted gene product (Fig. 3). Tn5253 produced circular forms at a similar frequency in S. agalactiae (2.9 × 10−5 copies per chromosome) and S. pyogenes (3.0 × 10−5 copies per chromosome), while no circular forms were detected in E. faecalis JH2-2 genetic background (<2.7 × 10−7 copies per chromosome) (Table 2). Reconstituted attB sites were found in all streptococci tested at a frequency ranging between 3.7 × 10−5 (in S. pyogenes) to 1.7 × 10−2 (in E. faecalis JH2-2 background) copies per chromosome. In E. faecalis, Tn5253 excision and circularization are strain dependent: a representative transconjugant obtained in OG1SS background produced circular forms and reconstituted attB site at 1.4 × 10−4 and 6.8 × 10−3 copies per chromosome, respectively; transconjugant FR50, obtained in JH2-2 background, produced reconstituted attB site at a frequency of 1.7 × 10−2 copies per chromosome but did not produce circular forms (<2.7 × 10−7). Conjugation frequency was lower than circularization frequency in all the tested strains except in S. pyogenes FR68. Many other factors are likely to be important in the conjugation process such as the expression of a capsular polysaccharide [37], the cell wall thickness, the surface charges, and the ability of the conjugation pore to establish a stable contact between cells from different species.
Fig. 2.
Tn5253 attB integration sites in the orthologous rbgA genes of other bacterial species. Genome sequence analysis identified Tn5253 attB sites in the orthologous rbgA genes of 18 other bacterial species with a size ranging between 33 nucleotides of S. gordonii to 84 nucleotides of Streptococcus mitis. The 17-bp E. faecalis attB was at first experimentally found by PCR and sequencing the Tn5253-chromosomal junction fragments of our E. faecalis transconjugants. Then the 17 nucleotides were used as probes for database interrogation. Inside the same bacterial species, different strains can harbour different allelic variants (up to 7 in S. equi). The sequence of the most represented allelic variant was used for the sequence alignment and its frequency is reported. The S. pneumoniae D39 attB variant 1 was used as reference. Tn5253 chromosomal integration, in the original S. pneumoniae host, as in the other functionally characterized streptococcal and enterococcal hosts (shaded), occurs always downstream of a 11-bp conserved sequence, namely the last 11 nucleotides of attB sites. These 11 nucleotides (boxed in blue) are conserved also among the attB sites of other bacterial species. Within the sequences, identical nucleotides are indicated by periods, substitutions are in red. For better alignment, dashes are inserted. The 20 nucleotides belonging to the rbgA coding sequence are underlined
Fig. 3.
Tn5253 attB site and integration in E. faecalis chromosome. (A) Tn5253 attB (represented as a blue box) in E. faecalis is composed by 17 nucleotides which correspond to the first 17 nucleotides of the rbgA CDS (light blue arrow). In streptococci and enterococci, Tn5253 always integrates downstream of a conserved 11-nucleotides sequence (boxed in red). The nucleotide sequence of attB and deduced amino acid sequence are reported. (B) Site specific integration of Tn5253 into rbgA causes integration site duplication, restoring an intact CDS. The integration of Tn5253 into bacterial chromosome seems to be polarized, since attTn (orange box) always flanks the element (light orange box) at the right end. In E. faecalis integration site duplication results in the acquisition of an additional codon (GCT ◊ Alanine) in the rbgA CDS. attB and attTn sites are not scaled. The 84 nucleotides sequence of attTn and the deduced amino acid sequence of RbgA N-terminal end are reported. Amino acids single-letter code is used
Genome sequence analysis of Tn5253 attB site in S. pneumoniae
To integrate biological data, a genome-wide investigation of Tn5253 attB among pneumococci was carried out. The database of 85 complete S. pneumoniae genomes (accessed in August 2021) was interrogated by using as a query the 83-bp attB. Sequence homology analysis identified thirteen allelic variants of attB. (Fig. 1, Table S1). In 75 genomes (88.3 %), the attB site was 83 or 84 nucleotides in length, while in 10 (11.7 %) it was 41 nucleotides. The 83-bp attB variant 1 is the most frequent variant, carried by 38 genome strains (44.7 % of the analyzed genomes), including D39 strain, its rough derivative R6, and the classical type 3 Avery’s strain A66. Variant 2 is carried by TIGR4 and other 8 genome strains (10.6 %) and contains two nucleotide substitutions. Variant 13 is harboured by G54 and other 9 genome strains (11.7 %) and contains only the last 41 nucleotides of variant 1. In addition, SP18-BS74, whose draft genome is available, also harbours variant 13. Variants 5 and 7 are carried by 7 strains (8.2 %), variant 4 by 5 strains (5.8 %), variants 6 and 9 by 2 strains (2.4 %). The remaining 5 variants (3, 8, 10, 11 and 12) were found in only in one strain. In thirteen pneumococcal genomes, carrying the attB variants 1, 2, 7, 11 and 12, Tn5253-like elements were integrated into the pneumococcal chromosome, resulting in the duplication of the attB site.
Genome sequence analysis of Tn5253 attB site in other bacterial species
Genome analysis was extended to the 58,138 complete microbial genomes (accessed in August 2021). Homology search identified the Tn5253 attB site in 18 other bacterial species, including the functionally characterized S. agalactiae, S. pyogenes,S. gordonii, hosts (Fig. 2, Table S2). The 17-bp E. faecalis attB was at first experimentally found by sequencing the Tn5253-chromosomal junction fragments obtained by inverse PCR from our E. faecalis transconjugants. Then the 17 nucleotides were used as a probe for database interrogation. Tn5253 attB was located in orthologous rbgA genes and its size ranged between 17 nucleotides of E. faecalis to 84 nucleotides of Streptococcus mitis. Alignment of the attB sequences obtained from the different bacterial species confirms the presence of the 11-bp conserved sequence. Theoretically, all of these attB sites allow Tn5253 integration, however only in one genome, namely Streptococcus mitis SVGS_061, a Tn5253-like element was found integrated, producing attB duplication.
Conclusions
In the present paper we conducted a functional characterization of Tn5253 attB site in S. pneumoniae and other streptococcal and enterococcal species and found that: (i) during conjugal transfer, Tn5253 integrated in S. pneumoniae rbgA gene or in the orthologous rbgA genes of the other bacterial hosts, (ii) Tn5253 produced circular forms containing the attTn site and the frequency was species- and strain-dependent, (iii) reconstitution of attB site was species- and strain-dependent. Through a DNA homology search conducted in the complete microbial genome database, we also found that: (i) thirteen allelic variants of the Tn5253 attB site were present in the complete S. pneumoniae genomes and their size ranged from 41 to 84 nucleotides, (ii) in other bacterial species, Tn5253 attB is located in orthologous rbgA genes with a size ranging between 17 and 84 nucleotides. Tn5253 integration, in the original S. pneumoniae host, as in the other streptococcal and enterococcal hosts, occurs always downstream of a 11-bp conserved sequence located in the rbgA CDS. Genome analysis revealed that the 11 nucleotides, corresponding to the last 11 nucleotides of the attB sites, are conserved also among the attB sites of other bacteria and can be considered the core of the integration site. In conclusion, even if a huge number of bacterial genomes is available, an in-silico analysis and a functional characterization of the mobilome is reported only in few cases. In this work, a functional characterization of the Tn5253 attB integration site, combined with genome sequence analysis, contributed to elucidating the potential of Tn5253 horizontal gene transfer among different bacterial species.
Materials and methods
Bacterial strains, growth, and mating conditions
Bacterial strains and their relevant properties are reported in Table 1. Both streptococcal and enterococcal strains were grown in tryptic soy broth or tryptic soy agar (Difco) supplemented, where appropriate, with antibiotics. Plate mating conjugation experiments were performed as previously described [38]. Briefly, donor and recipient cells were grown until the end of exponential phase and mixed at a 1:10 ratio, then were collected by centrifugation, plated and incubated for 4 h. Cells were harvested by scraping the plates and recombinant strains were selected by a multilayer plating procedure in presence of the appropriate antibiotics. Transconjugant FR39 was obtained from a mating experiment where FP58 [29] was the donor of Tn5253 and HB565, a streptomycin resistance derivative of type 3 Avery strain A66 [14, 39, 40], was the recipient.
Bacterial lysate preparation
Bacterial cultures (1 ml) were harvested in exponential phase (OD590 about 0.2, roughly corresponding to 5 × 108 CFU/ml) and centrifuged at 11,000 x g for 2 min. Pneumococcal lysates were obtained by using lysis solution (0.1 % DOC, 0.008 % SDS) as already reported [26]. Streptococcal and enterococcal cell pellets were resuspended in 90 µl protoplasting buffer (25 % sucrose, 100 mM Tris pH 7.2, 5 mM EDTA), then lysozyme (for E. faecalis) or mutanolysin (for S. agalactiae, S. gordonii and S. pyogenes) was added at a final concentration of 1 mg/ml or 20 µg/ml respectively and mixtures were incubated at 37 °C for 1 h. Protoplasts were centrifuged at 3,000 x g for 15 min, resuspended in 100 µl of dH2O, heated at 85 °C for 5 min and kept on ice until use.
PCR, inverse PCR, sequencing
PCR experiments and direct DNA sequencing of PCR amplicons were carried out essentially as already described [28, 29]. Briefly, PCR reactions were carried out in a 25-µl reaction mixture containing DreamTaq buffer 1X, 100 µM dNTPs, 1.5 mM MgCl2, 10 pmol of each primer, 0.2 U of DreamTaq enzyme, 1 µl bacterial culture. Inverse PCR, for amplifying the Tn5253-chromosome junctions, was performed with pairs of divergent primers targeting the Tn5253 ends as described [29]. 100 ng of each unpurified PCR fragment were used as template in sequencing reactions carried out with the BigDye Terminator v3.1 Cycle Sequencing Kit.
Quantitative Real time PCR
A LightCycler 1.5 apparatus (Roche) and the KAPA SYBR FAST qPCR kit Master Mix Universal (2X) (Kapa Biosystems) were used for Real Time PCR experiments according to the protocol extensively described [26]. Quantification of Tn5253 circular intermediates and reconstituted pneumococcal attB was obtained with the primer pairs IF327/IF328 and IF496/IF356, respectively [26]. Reconstituted attB site was quantified in S. agalactiae with the primer pair IF560/IF561 which amplified a 353 bp fragment, in S. gordonii with IF544/IF545 which amplified a 396 bp fragment, in S. pyogenes with IF509/IF510 which amplified a 249 bp fragment, in E. faecalis with IF525/IF532 which amplified a 480 bp fragment (Table S3). A standard curve for the gyrB gene was used to standardize results and melting curve analysis was performed to differentiate the amplified products from primer dimers as reported [26].
Microbial database interrogation and sequence analysis
Homology searches of the databases available at the National Center for Biotechnology Information were conducted using the Microbial Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC.
=MicrobialGenomes), selecting the complete genomes database. Default parameters were used and only alignments with significant e-values were considered. We built a stand-alone database containing only genomes of interest to be searched with BLAST software to confirm the results. Sequence analysis was carried out with BioEdit 7.2.5 (http://bioedit.software.informer.
com/). Multiple DNA sequence alignments were performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Supplementary Information
Additional file 1: Table S1. Blast searches output of the S. pneumoniae complete genomes. For each variant, name, length, sequence, strain host, GenBank accession number, are reported.
Additional file 2: Table S2. Blast searches output of the complete microbial genomes. For each attB site, name, length, sequence, bacterial species host, GenBank accession number, are reported.
Additional file 3: Table S3. Oligonucleotide primers.
Additional file 4: Table S4. Quantitative PCR data. The threshold cycles (Cts) relative to circular forms (CI), reconstitution of attB site, and chromosomal reference gene (gyrB) quantification are reported. Standard curves, slope and intercept values are reported.
Acknowledgements
Not applicable.
Authors’ contributions
FS, GP, FI conceived and designed the study, FS, VF, AR, EL carried out the experiments, FS, VF, GP, FI performed data analysis, FS, FI drafted the first version of the manuscript, FS, VF, GP, FI drafted subsequent versions of the manuscript, GP received funds for the study. All authors read and approved the final manuscript.
Funding
This work was supported by the Italian Ministry of Education, University and Research (MIUR-Italy) under grant number 20177J5Y3P (call “Progetti di Ricerca di Rilevante Interesse Nazionale – Bando 2017”).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Carr VR, Shkoporov A, Hill C, Mullany P, Moyes DL. Probing the Mobilome: Discoveries in the Dynamic Microbiome. Trends Microbiol. 2021;29(2):158–70. doi: 10.1016/j.tim.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Ghaly TM, Gillings MR. Mobile DNAs as Ecologically and Evolutionarily Independent Units of Life. Trends Microbiol. 2018;26(11):904–12. doi: 10.1016/j.tim.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev. 2018;31(4). [cited 2021 Aug 26] Available from: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed]
- 4.Gyles C, Boerlin P. Horizontally Transferred Genetic Elements and Their Role in Pathogenesis of Bacterial Disease. Vet Pathol. 2014;51(1):328–40. doi: 10.1177/0300985813511131. [DOI] [PubMed] [Google Scholar]
- 5.Botelho J, Schulenburg H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2021;29(1):8–18. doi: 10.1016/j.tim.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Jørgensen TS, Kiil AS, Hansen MA, Sørensen SJ, Hansen LH. Current strategies for mobilome research. Front Microbiol. 2015;5:750. doi: 10.3389/fmicb.2014.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K, Kim D-W, Cha C-J. Overview of bioinformatic methods for analysis of antibiotic resistome from genome and metagenome data. J Microbiol. 2021;59(3):270–80. doi: 10.1007/s12275-021-0652-4. [DOI] [PubMed] [Google Scholar]
- 8.Saak CC, Dinh CB, Dutton RJ. Experimental approaches to tracking mobile genetic elements in microbial communities. FEMS Microbiol Rev. 2020;44(1):606–30. doi: 10.1093/femsre/fuaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 2014;38(4):720–60. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 10.Paulsen IT. Role of Mobile DNA in the Evolution of Vancomycin-Resistant Enterococcus faecalis. Science. 2003;28(5615):2071–4. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 11.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155(5):376–86. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8(8):552–63. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 13.Johnson CM, Grossman AD. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoro F, Iannelli F, Pozzi G. Genomics and Genetics of Streptococcus pneumoniae. Microbiol Spectr. 2019;7(3). [DOI] [PMC free article] [PubMed]
- 15.Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of ‘conjugal’ transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice LB. Tn916 Family Conjugative Transposons and Dissemination of Antimicrobial Resistance Determinants. Antimicrob Agents Chemother. 1998;42(8):1871–7. doi: 10.1128/aac.42.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17(6):251–8. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Mullany P, Williams R, Langridge GC, Turner DJ, Whalan R, Clayton C, Lawley T, Hussain H, McCurrie K, Morden N, Allan E, Roberts AP. Behavior and Target Site Selection of Conjugative Transposon Tn916 in Two Different Strains of Toxigenic Clostridium difficile. Appl Environ Microbiol. 2012;78(7):2147–53. doi: 10.1128/AEM.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santoro F, Vianna ME, Roberts AP. Variation on a theme; an overview of the Tn916/Tn1545 family of mobile genetic elements in the oral and nasopharyngeal streptococci. Front Microbiol. 2014;5:535. doi: 10.3389/fmicb.2014.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gawron-Burke C, Clewell DB. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982;300(5889):281–4. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- 21.Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32(1):99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 22.Burrus V, Roussel Y, Decaris B, Guédon G. Characterization of a Novel Integrative Element, ICESt1, in the Lactic Acid Bacterium Streptococcus thermophilus. Appl Environ Microbiol. 2000;66(4):1749–53. doi: 10.1128/aem.66.4.1749-1753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burrus V, Pavlovic G, Decaris B, Guédon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48(2):77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
- 24.Burrus V, Waldor MK. Control of SXT Integration and Excision. J Bacteriol. 2003;185(17):5045–54. doi: 10.1128/JB.185.17.5045-5054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Smith MCM, Mullany P. The Conjugative Transposon Tn5397 Has a Strong Preference for Integration into Its Clostridium difficile Target Site. J Bacteriol. 2006;188(13):4871–8. doi: 10.1128/JB.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santoro F, Romeo A, Pozzi G, Iannelli F. Excision and Circularization of Integrative Conjugative Element Tn5253 of Streptococcus pneumoniae. Front Microbiol. 2018;31:1779. doi: 10.3389/fmicb.2018.01779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provvedi R, Manganelli R, Pozzi G. Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett. 1996;135(2–3):231–6. doi: 10.1111/j.1574-6968.1996.tb07994.x. [DOI] [PubMed] [Google Scholar]
- 28.Iannelli F, Santoro F, Oggioni MR, Pozzi G. Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2013/12/02 ed. 2014;58(2):1235–9. [DOI] [PMC free article] [PubMed]
- 29.Santoro F, Oggioni MR, Pozzi G, Iannelli F. Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett. 2010;308(2):150–8. doi: 10.1111/j.1574-6968.2010.02002.x. [DOI] [PubMed] [Google Scholar]
- 30.Vijayakumar A. Nucleotide sequence analysis of the termini and chromosomal locus involved in site-specific integration of the streptococcal conjugative transposon Tn5252. J Bacteriol. 1993;175(9). [DOI] [PMC free article] [PubMed]
- 31.Croucher NJ, Walker D, Romero P, Lennard N, Paterson GK, Bason NC, Mitchell AM, Quail MA, Andrew PW, Parkhill J, Bentley SD, Mitchell TJ. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23FST81. J Bacteriol. 2009;191(5):1480–9. doi: 10.1128/JB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson-Begg SK, Roberts AP, Hall LMC. Diversity of putative Tn5253-like elements in Streptococcus pneumoniae. Int J Antimicrob Agents. 2009;33(4):364–7. doi: 10.1016/j.ijantimicag.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Mingoia M, Tili E, Manso E, Varaldo PE, Montanari MP. Heterogeneity of Tn5253-like composite elements in clinical Streptococcus pneumoniae isolates. Antimicrob Agents Chemother. 2011;55(4):1453–9. doi: 10.1128/AAC.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;28(6016):430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith MD, Hazum S, Guild WR. Homology among tet determinants in conjugative elements of streptococci. J Bacteriol. 1981;148(1):232–40. doi: 10.1128/jb.148.1.232-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uicker WC, Schaefer L, Britton RA. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol Microbiol. 2006;59(2):528–40. doi: 10.1111/j.1365-2958.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 37.Dahmane N, Robert E, Deschamps J, Meylheuc T, Delorme C, Briandet R, Leblond-Bourget N, Guédon E, Payot S. Impact of Cell Surface Molecules on Conjugative Transfer of the Integrative and Conjugative Element ICESt3 of Streptococcus thermophilus. Kivisaar M, editor. Appl Environ Microbiol. 2018;84(5). [DOI] [PMC free article] [PubMed]
- 38.Iannelli F, Santoro F, Fox V, Pozzi G. A Mating Procedure for Genetic Transfer of Integrative and Conjugative Elements (ICEs) of Streptococci and Enterococci. Methods Protoc. 2021;4(3):59. doi: 10.3390/mps4030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iannelli F, Chiavolini D, Ricci S, Oggioni MR, Pozzi G. Pneumococcal Surface Protein C Contributes to Sepsis Caused by Streptococcus pneumoniae in Mice. Infect Immun. 2004;72(5):3077–80. doi: 10.1128/IAI.72.5.3077-3080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearce BJ, Iannelli F, Pozzi G. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res Microbiol. 2002;5. [DOI] [PubMed]
- 41.Hahn C, Harrison EM, Parkhill J, Holmes MA, Paterson GK. Draft Genome Sequence of the Streptococcus pneumoniae Avery Strain A66. Genome Announc. 2015;3(3). [DOI] [PMC free article] [PubMed]
- 42.RAVIN AW Reciprocal capsular transformations of pneumococci. J Bacteriol. 1959;77(3):296–309. doi: 10.1128/jb.77.3.296-309.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernheimer HP, Wermundsen IE. Unstable Binary Capsulated Transformants in Pneumococcus. J Bacteriol. 1969;98(3):1073–9. doi: 10.1128/jb.98.3.1073-1079.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete Genome Sequence of a Virulent Isolate of Streptococcus pneumoniae. Science. 2001;293(5529):498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 45.Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. Comparative Genomic Analyses of Seventeen Streptococcus pneumoniae Strains: Insights into the Pneumococcal Supragenome. J Bacteriol. 2007;15(22):8186–95. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, DeBoy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJB, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial ‘pan-genome’. Proc Natl Acad Sci. 2005;27(39):13950–5. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci. 2001;10(8):4658–63. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vickerman MM, Iobst S, Jesionowski AM, Gill SR. Genome-Wide Transcriptional Changes in Streptococcus gordonii in Response to Competence Signaling Peptide. J Bacteriol. 2007;189(21):7799–807. doi: 10.1128/JB.01023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macrina FL, Jones KR, Wood PH. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis) J Bacteriol. 1980;143(3):1425–35. doi: 10.1128/jb.143.3.1425-1435.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pozzi G, Musmanno RA, Renzoni EA, Oggioni MR, Cusi MG. Host-vector system for integration of recombinant DNA into chromosomes of transformable and nontransformable streptococci. J Bacteriol. 1988;170(4):1969–72. doi: 10.1128/jb.170.4.1969-1972.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gold OG, Jordan HV, van Houte J. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch Oral Biol. 1975;20(7):473-IN15. doi: 10.1016/0003-9969(75)90236-8. [DOI] [PubMed] [Google Scholar]
- 52.Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci. 1978;75(7):3479–83. [DOI] [PMC free article] [PubMed]
- 53.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9(7):R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunny GM, Craig RA, Carron RL, Clewell DB. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid. 1979;2(3):454–65. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 55.Jacob AE, Hobbs SJ. Conjugal Transfer of Plasmid-Borne Multiple Antibiotic Resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117(2):360–72. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Blast searches output of the S. pneumoniae complete genomes. For each variant, name, length, sequence, strain host, GenBank accession number, are reported.
Additional file 2: Table S2. Blast searches output of the complete microbial genomes. For each attB site, name, length, sequence, bacterial species host, GenBank accession number, are reported.
Additional file 3: Table S3. Oligonucleotide primers.
Additional file 4: Table S4. Quantitative PCR data. The threshold cycles (Cts) relative to circular forms (CI), reconstitution of attB site, and chromosomal reference gene (gyrB) quantification are reported. Standard curves, slope and intercept values are reported.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.