Abstract
Background
Nowadays, both customers and producers prefer thin-tailed fat sheep. To effectively breed for this phenotype, it is important to identify candidate genes and uncover the genetic mechanism related to tail fat deposition in sheep. Accumulating evidence suggesting that post-transcriptional modification events of precursor-messenger RNA (pre-mRNA), including alternative splicing (AS) and alternative polyadenylation (APA), may regulate tail fat deposition in sheep. Differentially expressed transcripts (DETs) analysis is a way to identify candidate genes related to tail fat deposition. However, due to the technological limitation, post-transcriptional modification events in the tail fat of sheep and DETs between thin-tailed and fat-tailed sheep remains unclear.
Methods
In the present study, we applied pooled PacBio isoform sequencing (Iso-Seq) to generate transcriptomic data of tail fat tissue from six sheep (three thin-tailed sheep and three fat-tailed sheep). By comparing with reference genome, potential gene loci and novel transcripts were identified. Post-transcriptional modification events, including AS and APA, and lncRNA in sheep tail fat were uncovered using pooled Iso-Seq data. Combining Iso-Seq data with six RNA-sequencing (RNA-Seq) data, DETs between thin- and fat-tailed sheep were identified. Protein protein interaction (PPI) network, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were implemented to investigate the potential functions of DETs.
Results
In the present study, we revealed the transcriptomic complexity of the tail fat of sheep, result in 9,001 potential novel gene loci, 17,834 AS events, 5,791 APA events, and 3,764 lncRNAs. Combining Iso-Seq data with RNA-Seq data, we identified hundreds of DETs between thin- and fat-tailed sheep. Among them, 21 differentially expressed lncRNAs, such as ENSOART00020036299, ENSOART00020033641, ENSOART00020024562, ENSOART00020003848 and 9.53.1 may regulate tail fat deposition. Many novel transcripts were identified as DETs, including 15.527.13 (DGAT2), 13.624.23 (ACSS2), 11.689.28 (ACLY), 11.689.18 (ACLY), 11.689.14 (ACLY), 11.660.12 (ACLY), 22.289.6 (SCD), 22.289.3 (SCD) and 22.289.14 (SCD). Most of the identified DETs have been enriched in GO and KEGG pathways related to extracellular matrix (ECM). Our result revealed the transcriptome complexity and identified many candidate transcripts in tail fat, which could enhance the understanding of molecular mechanisms behind tail fat deposition.
Keywords: Sheep, Tail fat, Iso-seq, DET
Introduction
Post-transcriptional modification events of precursor-messenger RNA (pre-mRNA), including alternative splicing (AS) and alternative polyadenylation (APA), have attracted an increasing interest (Baralle & Giudice, 2017; Di Giammartino, Nishida & Manley, 2011; Gruber & Zavolan, 2019; Kornblihtt et al., 2013; Wang et al., 2015). In livestock, accumulated evidence suggests that AS (Fang et al., 2020; Leal-Gutierrez, Elzo & Mateescu, 2020; Xiang et al., 2018; Yuan et al., 2021) and APA (Deng et al., 2020; Jin et al., 2021) could contribute to the formation of important economic traits. For example, both AS (Motter, Corva & Soria, 2021) and APA (Nattrass et al., 2014) in calpastatin (CAST) were associated with meat tenderness in beef. RNA-sequencing (RNA-seq) is a way to accurately quantify the AS events, e.g., using the intron excision ratio (Li et al., 2018c). Unfortunately, it is unlikely to obtain the full-length transcripts and quantify the transcripts by RNA-seq because it is difficult to assemble full-length transcripts based on short reads and to exactly assign a short read to a certain transcript (Kovaka et al., 2019), which hamper the discovery of post-transcriptional events. The third-generation sequencing technology, such as PacBio isoform sequencing (Iso-Seq), which can directly produce full length transcripts (Rhoads & Au, 2015), providing an opportunity to discover novel genes and novel isoforms (Au et al., 2013; Beiki et al., 2019; Chen et al., 2017; Karlsson & Linnarsson, 2017) and to investigate the transcriptome complexity in mammals (Deng et al., 2020; Li et al., 2018b). In addition, the results from the previous study suggested that integrative analysis of Iso-Seq and RNA-seq data could accurately quantify the abundance of transcripts (Chao et al., 2019; Li et al., 2017). Thus, using Iso-Seq and RNA-seq could accurately identify the post-transcriptional modification events and differential expressed isoforms.
Hu sheep, a short fat-tailed sheep breed, is one of the most popular sheep breeds in China because of its high fecundity. In an intensive production system, Hu sheep is usually used as ewe crossing with elite ram breeds, e.g., Dorper, to produce meat. Backcross ((Dorper × Hu) × Hu, DHH) and grading up (Dorper × (Dorper × Hu), DDH) are two common hybridization methods in sheep meat production. The weight of the tail fat of their hybrid offspring is positively related to the composition of the Hu sheep genome. Nowadays, both customers and producers prefer a small amount of tail fat. To breed for this phenotype, it is important to identify the candidate genes linked with tail fat and understand the underlying mechanisms of fat deposition.
Many studies have attempted to identify the candidate gene associated with tail fat and understand the underlying mechanisms of fat deposition (Ahbara et al., 2018; Amane et al., 2020; Baazaoui et al., 2021; Bakhtiarizadeh & Alamouti, 2020; Dong et al., 2020; Han et al., 2021; Li et al., 2018a; Mastrangelo et al., 2019a; Mastrangelo et al., 2019b; Moioli, Pilla & Ciani, 2015; Moradi et al., 2012; Wei et al., 2015; Xu et al., 2017; Yuan et al., 2017; Zhang et al., 2019; Zhao et al., 2020; Zhi et al., 2018; Zhu et al., 2016). Previous studies suggested that microRNAs (Miao et al., 2015a; Pan et al., 2018), lncRNAs (Bakhtiarizadeh & Salami, 2019; He et al., 2020) and mRNAs (Bakhtiarizadeh et al., 2019; Kang et al., 2017a; Miao et al., 2015b; Wang et al., 2014) may regulate tail fat deposition in sheep. These results contribute to understanding the underlying mechanisms of fat deposition. However, due to the technological limitation, post-transcriptional modification events (e.g., AS and APA) in the tail fat of sheep remains unclear, which might play an important role in tail fat deposition. In the current study, Iso-Seq was used to uncover post-transcriptional modifications in sheep tail fat and combined with RNA-Seq to investigate differential expressed transcripts (DETs) in tail fat between DHH and DDH. Our results could reveal transcriptome complexity in sheep tail fat and characterize DETs between DHH and DDH sheep.
Materials & Methods
Animal tissues collection and RNA extraction
After wearing (2-month old), all animals were fed to 6-month old in house with complete formula granulated feed (DafengGe Feed Technology Co., Ltd.) at Suyang Sheep Industry Co., Ltd (Fengxian, Jiangsu, China). All animals were eating and drinking freely. In total, six unrelated 6-month old male sheep (DDH = 3, DHH = 3) with similar live weight (Table 1) were selected to slaughter for sampling. Animal experiments were approved by the Experimental Animal Ethical Committee of Yangzhou University (NO.202103294).
Table 1. Sample information.
| Cross type | Sample size | Live weight (kg) | Carcass weight (kg) | Tail fat (kg) |
|---|---|---|---|---|
| Backcross ((Dorper × Hu) × Hu sheep, DHH) | 3 | 41.53 ± 0.95 | 23.70 ± 0.64 | 0.78 ± 0.21 |
| Grading up (Dorper × (Dorper × Hu sheep), DDH) | 3 | 40.80 ± 4.57 | 22.00 ± 2.27 | 0.11 ± 0.03 |
| T-test t value | 0.27 | 1.27 | 5.44 | |
| T-test df | 2.17 | 2.32 | 2.10 | |
| T-test P value | 0.8094 | 0.3161 | 0.0288 |
After slaughter, tail fat tissues were sampled within 30 mins. Each fat sample was packed into a 1.5 ml cryotube. All the fat samples were quickly frozen in liquid nitrogen and stored at −80 °C. Total RNA was extracted from the tail fat using Trizol reagent Kit (TaKaRa, USA). The extraction protocol strictly follows the instructions of Kit. The quality of RNA samples was evaluated by Nanodrop 2,000 (Thermo Scientific™) and 2,100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). After quality control, all the RNA samples with RNA Integrity Number (RIN) greater than 7.0 and 28S/18S ratio greater than 1.0 were used for sequencing.
Library construction and RNA-sequencing
Library construction and sequencing of RNA-seq of six RNA samples were performed by a commercial sequencing service company (Frasergene Technology Co., Ltd, Wuhan, China). Briefly, the MGIEasy RNA Directional Library Prep Kit (MGI) and 1 μg of total RNA were used for library construction. Then, the library was sequenced on the MGISEQ-2000 platform. All sequencing data have been deposited in National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) database and can be accessed under the BioProject accession number PRJNA745517.
Iso-Seq library construction and sequencing
Six RNA samples (DDH = 3, DHH = 3) were pooled in equal quantities for Iso-Seq. Library preparation and Iso-Seq were performed by a commercial sequencing service company (Frasergene Technology Co., Ltd, Wuhan, China). Briefly, the SMARTer™ PCR cDNA synthesis kit (Takara Biotechnology, Dalian, China) was used to reverse-transcribed RNA into cDNA. Then, PCR Amplification was implemented by using 12–14 PCR cycles. PCR products were purified by the AMPure PB magnetic beads (Beckman Coulter, CA, USA). The BluePippin™ Size Selection System (Sage Science, MA, USA) was used for the size selection (>1 kb). Pacific Biosciences DNA Template Prep Kit 2.0 (Pacific Biosciences, CA, USA) was used for SMRTbell library construction. Agilent Bioanalyzer 2,100 system (Agilent Technologies, CA, USA) and Qubit fluorometer 2.0 (Life Technologies, CA, USA) were used for quality accession of the library. Then, the library was sequenced on the PacBio sequencing platform with 10 h sequencing movies.
Iso-Seq data processing
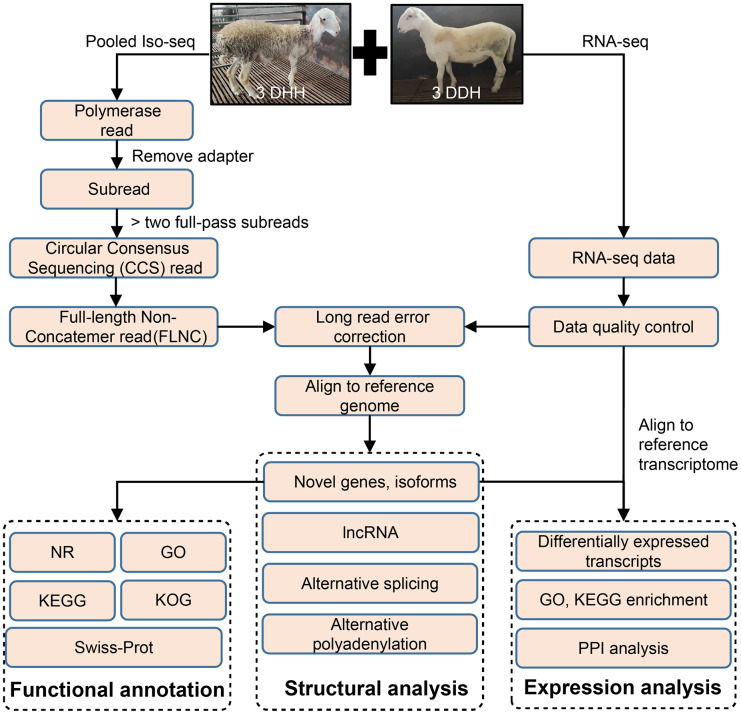
The pipeline of bioinformatics analysis was shown in Fig. 1. The raw reads of Iso-Seq were preprocessed using SMRT Link v8.0 (https://www.pacb.com/wp-content/uploads/SMRT-Link-Release-Notes-v8.0.pdf). The adapter of polymerase reads was remove to get Subreads. Circular Consensus sequencing (CCS) reads were obtained from the Subreads using the following parameters: minimum subread length = 50, maximum subread length = 15,000, minimum number of passes = 3, and minimum predicted accuracy = 0.99. Then, CCS were classified into full length reads by lima software. Full-length non-chimeric reads (FLNCs) were full-length CCS reads with 5′ and 3′ cDNA primers and polyA. The LoRDEC v0.9 software (Salmela & Rivals, 2014) was used for FLNC correction using the high-quality short reads with the default parameters. The quality of RNA-seq data was examined using SOAPnuke v2.1.0 (Chen et al., 2018). FLNCs were aligned to the sheep reference genome (http://asia.ensembl.org/Ovis_aries_rambouillet/Info/Index) using GMAP (Wu & Watanabe, 2005).
Figure 1. Overview of bioinformatics pipeline.
DDH, Dorper × (Dorper × Hu sheep); DHH, ((Dorper × Hu) × Hu sheep).
Novel genes and isoforms identification
Based on the results from alignments, FLNCs sharing the same splicing event were merged into one isoform. The isoforms with 5′ terminal region degraded were excluded for further analysis. Isoforms meet one of the following conditions were kept: (1) isoform was supported by at least two FLNCs; (2) isoform was supported by one FLNC whose Percentage Identity (PID) was greater than 99%; (3) all splicing sites in an isoform were fully supported by short reads; (4) all splicing junctions in an isoform were annotated by the reference genome. Isoforms overlapped over 20% of their length on the same strand were regarded as transcribing from the same gene locus (Wang et al., 2021). A gene locus was defined as a novel gene if it overlaps less than 20% of length with known genes. A novel isoform was defined as an isoform with a final splice site of 3′ ends changed or a new intron (exon) emerged.
Functional annotation of isoforms
To better understand the potential function of isoforms, they were aligned to Gene Ontology (GO) (Ashburner et al., 2000), Swiss-Prot (Gasteiger, Jung & Bairoch, 2001), NCBI non-redundant proteins (NR) and Cluster of Orthologous Groups of proteins (COG/KOG) (Tatusov et al., 2003) database using Diamond (Buchfink, Xie & Huson, 2015). Isoforms were also aligned to KEGG (https://www.genome.jp/kegg/) database using KOBAS (Xie et al., 2011).
lncRNA prediction
Novel isoforms with lengths greater than 200 nt were blast against the NR, KOG/KO and Swiss-Prot to remove transcripts with coding potential. A transcript with above annotated information was removed for further lncRNA prediction. Multiple software, including CPC 2.0 beta (default parameter) (Kang et al., 2017b), CNCI v2 (−m ve) (Sun et al., 2013), PLEK v1.2 (−minlength 200) (Li, Zhang & Zhou, 2014) and CPAT v1.2.4 (default parameter) (Wang et al., 2013), were used to predict lncRNAs from novel isoforms.
APA and AS analysis
The APA sites for each gene loci were detected using the TAPIS pipeline (Abdel-Ghany et al., 2016). AStalavista v3.2 software was used to detect AS events with default parameters (Foissac & Sammeth, 2007).
DETs detection
Reference genome annotation file and PacBio novel isoforms were merged to make a novel annotation file. The clean reads of each RNA-Seq library were aligned to the novel annotation file using the Bowtie2 software (-q --sensitive --dpad 0 --gbar 99999999 --mp 1,1 --np 1 --score-min L,0,-0.1 -I 1 -X 1000 --no-mixed --no-discordant -p 6) (Langmead & Salzberg, 2012). The RSEM software was used to quantify transcripts (Li & Dewey, 2011). DESeq2 R package (Love, Huber & Anders, 2014) was used to identify DETs between thin- and fat-tailed sheep samples. Transcripts with the false discovery rate (FDR) smaller than 0.05 and fold change greater than two were defined as significant DETs. The correlation between differentially expressed lncRNAs and DETs which were transcribed from tail fat related candidate genes was implemented using the ggcor R package (Huang et al., 2020).
Functional annotation of DETs
To extract the potential function of DETs, DETs were blast to GO database using Diamond (Buchfink, Xie & Huson, 2015) and KEGG database using KOBAS (Xie et al., 2011). FDR < 0.05 was regarded as the significant threshold. The GO enrichment results were visualized by the GOplot 1.0.2 R package (Walter, Sanchez-Cabo & Ricote, 2015). To explore the interaction between DETs and transcripts which were transcribed from the candidate genes linked with tail fat deposition, protein-protein interaction (PPI) networking analysis was implemented using the STRING database (Szklarczyk et al., 2017). The result of PPI was visualized by the Cytoscape v3.8.3 software (Shannon et al., 2003).
Results
PacBio Iso-Seq and bioinformatics analysis
In total, 487,822 polymerase reads were generated by Iso-Seq (Table 2). After quality control, 30,305,694 filtered subreads with a mean length of 1,474 bp and 360,901 CCS reads with an average depth of 71 passes were produced (Table 2). Finally, 271,718 FLNCs with an average length of 1,754 bp were used for further analysis (Table 2).
Table 2. Summarized information of Iso sequencing (Iso-Seq) data.
| Data type | Total number | Min length | Max length | Average length |
|---|---|---|---|---|
| Polymerase reads | 487,822 | 51 | 335,695 | 96,474 |
| Subreads | 30,305,694 | 51 | 250,276 | 1,474 |
| Circular consensus sequencing (CCS) reads | 360,901 | 62 | 15,123 | 2,107 |
| Full-length non-chimeric reads (FLNCs) | 271,718 | 50 | 8,211 | 1,754 |
Gene loci and isoforms detection
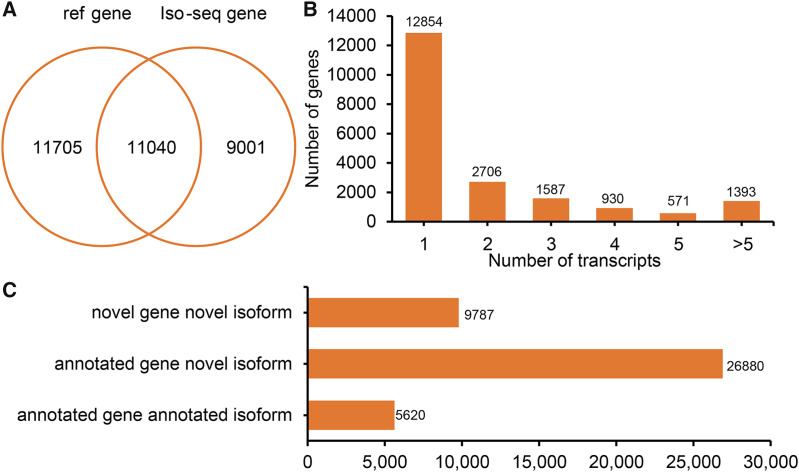
All FLNCs were corrected for sequencing errors and alignment position. According to the alignment position of each FLNC, gene loci and isoforms were identified. In total, 20,041 gene loci (11,040 known gene loci and 9,001 potential novel gene loci) were identified by Iso-Seq (Fig. 2A). A total of 7,187 (35.86%) detected genes generate two or more isoforms (Fig. 2B). Interestingly, the LIPE (lipase E, hormone-sensitive type) transcribed the greatest number of transcripts (113 novel transcripts and three known transcripts). In sheep tail fat, the majority of novel isoforms are transcribed from annotated genes (Fig. 2C). All novel gene loci and novel isoforms were added to the reference annotation file (Generic Feature Format, GFF, Table S1).
Figure 2. Genes and isoforms identified by Iso-seqencing (Iso-Seq) data.
(A) Venn plot of annotated genes and identified genes by Iso-Seq data. (B) Distribution of transcribed transcripts. (C) Hist plot of three type of transcripts.
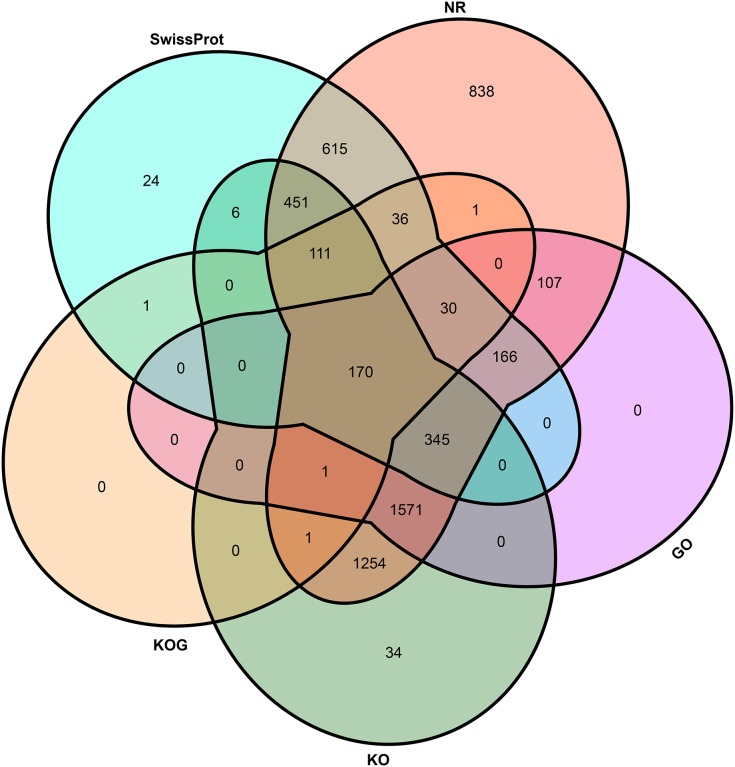
To elucidate the potential function of novel isoforms transcribed from novel genes, isoforms were annotated to public databases, including Gene Ontology (GO) (Ashburner et al., 2000), Swiss-Prot (Gasteiger, Jung & Bairoch, 2001), NCBI non-redundant proteins (NR), Cluster of Orthologous Groups of proteins (COG/KOG) (Tatusov et al., 2003) and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) database (Table S2). In total, above half novel transcripts (5,762, 58.87%) were annotated at least one public database, including 5,697 (58.21%) in NR, 2,390 (24.42%) in GO, 3,944 (40.30%) in KEGG, 351 (3.59%) in KOG and 1,955 (19.98%) in Swiss-Prot (Fig. 3).
Figure 3. Venn plot of annotated transcripts.
NR, NCBI non-redundant proteins; GO, gene ontology; KO, KEGG orthology; KOG, cluster of orthologous groups of proteins.
lncRNA prediction
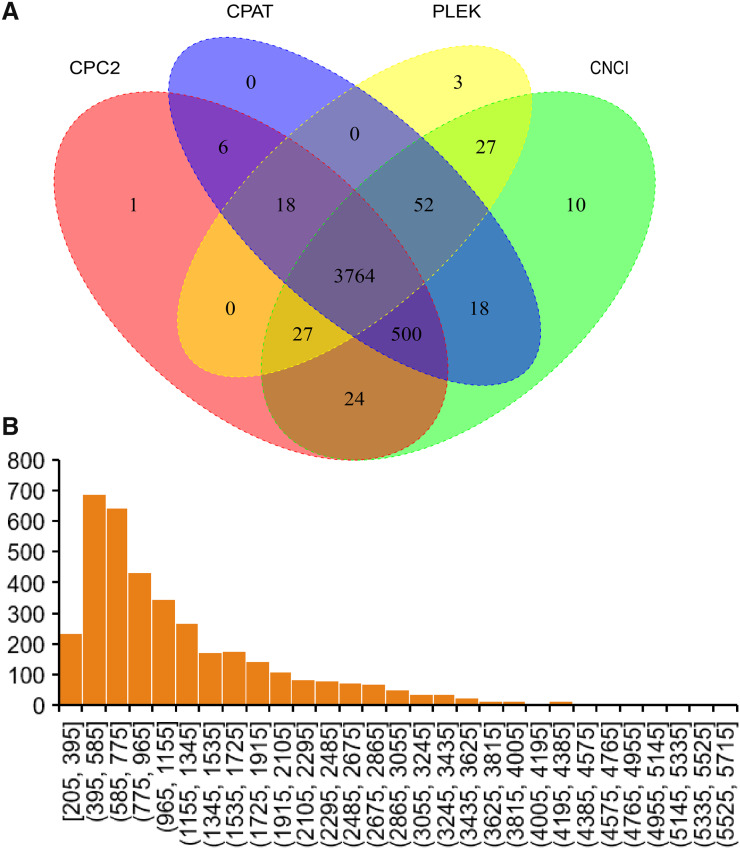
In the current study, four software were used to predict lncRNAs. Finally, 3,764 lncRNAs were predicted by all software (Fig. 4A). The sequence of predicted lncRNAs were documented in Table S3. The length of predicted lncRNAs ranged from 205 to 5,698 bp (Fig. 4B). The mean length of lncRNAs was 1,198 bp.
Figure 4. Identified lncRNAs.
(A) Veen plot of predicted lncRNAs by four software. (B) Length distribution of predicted lncRNAs.
Alternative polyadenylation events detection
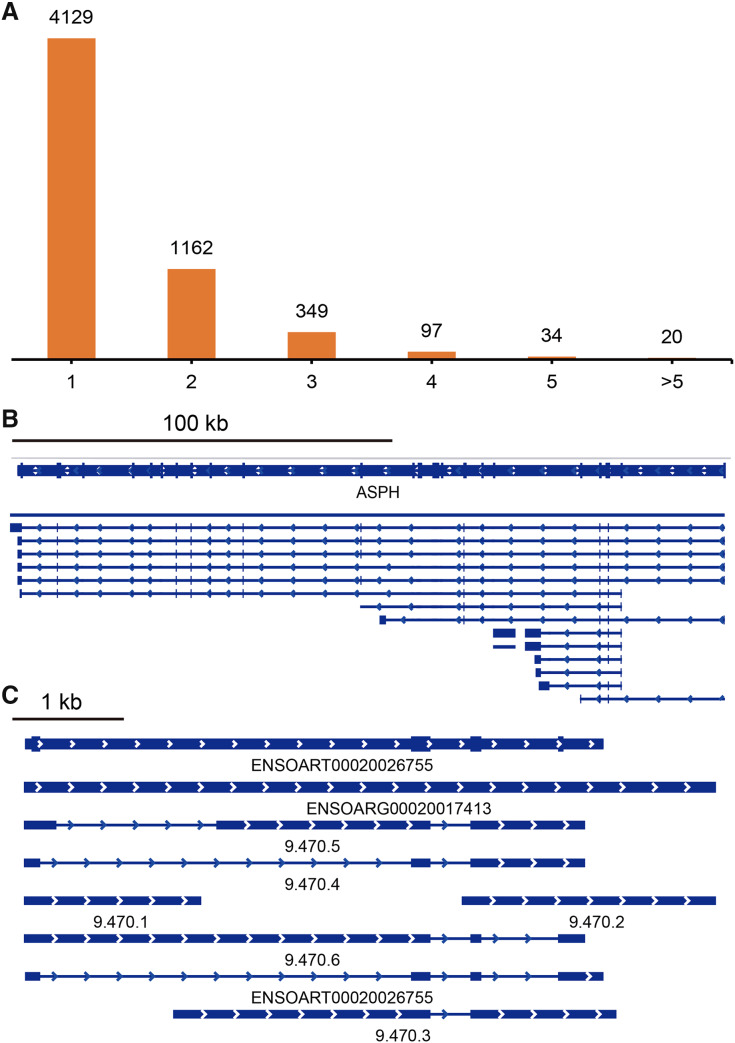
TAPIS pipeline was used to detect APA events in tail fat. In total, 5,791 detected genes had at least one poly(A) site (Fig. 5A, Table S4). Of them, 4,129 genes contained a single poly(A) site (Fig. 5A). The remaining 1,662 (28.70%) genes contained two or more poly(A) sites (Fig. 5A). The ENSOARG00020025751 (ASPH) and ENSOARG00020017413 (unannotated) contained the greatest number of distinct poly(A) sites, which were illustrated in Figs. 5B and 5C.
Figure 5. Identified alternative polyadenylation (APA) events.
(A) Distribution of the number of APA events. (B) APA events in ENSOARG00020025751 (ASPH). (C) APA events in ENSOARG00020017413 (unannotated).
Alternative splicing events detection
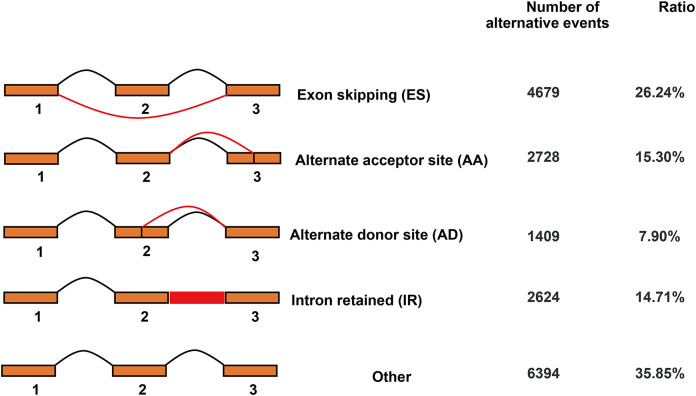
In total, 17,834 AS events were detected by Iso-Seq data. AS events were classified into four basic types and others (Fig. 6). Among four basic types, exon skipping (4,679, 26.24%) is the most prevalent AS event, followed by alternate acceptor site (2,728, 15.30%), intron retaining (2,624, 14.71%) and alternate donor site (1,409, 7.90%).
Figure 6. Classification of alternative splicing events.
Differentially expressed transcripts
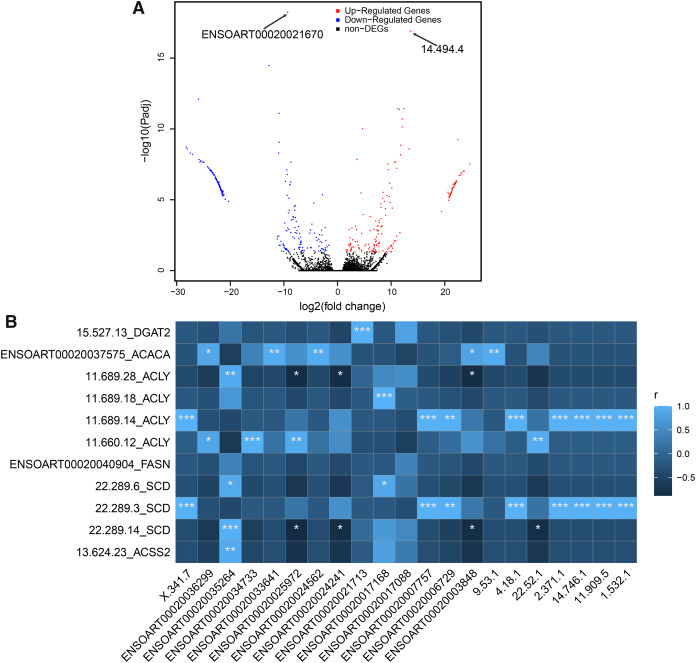
In our study, a total of 464 DETs between thin- and fat-tailed samples were identified (Fig. 7A, Table S5). The most significant differential transcript was ENSOART00020021670 (elongation factor 1-beta, FDR = 5.79e-19). Among these DETs, 21 DETs are differentially expressed lncRNAs, including 14 known lncRNAs and seven novel lncRNAs. The correlation between differentially expressed lncRNAs and DETs which transcribed from tail fat related candidate genes, including diacylglycerol O-acyltransferase 2 (DGAT2) (Bakhtiarizadeh & Alamouti, 2020), acetyl-CoA carboxylase alpha (ACACA) (Bakhtiarizadeh & Alamouti, 2020; Bakhtiarizadeh & Salami, 2019), ATP citrate lyase (ACLY) (Bakhtiarizadeh & Alamouti, 2020), fatty acid synthase (FASN) (Bakhtiarizadeh & Alamouti, 2020), stearoyl-CoA desaturase (SCD) (Kang et al., 2017a), and acyl-CoA synthetase short chain family member 2 (ACSS2) (Guangli et al., 2020), was investigated. The result suggests that 20 out of 21 lncRNAs (except lncRNA ENSOART00020017088) were significantly correlated with at least one transcripts (Fig. 7B).
Figure 7. Identified differentially expressed transcripts (DETs).
(A) Volcano plot of DETs. (B) Correlation of differentially expressed lncRNAs (xlab) and transcripts transcribed from tail fat related candidate genes (ylab). “*” denotes P < 0.05. “**” denotes P < 0.01. “***” denotes P < 0.001.
Functional annotation of DETs
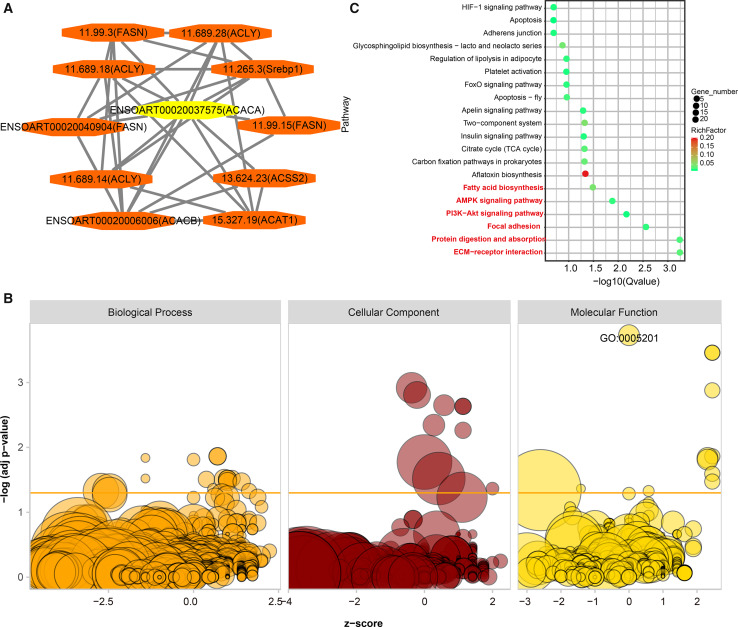
PPI analysis was implemented to identify functional similarities between the DETs. All of the networks were documented in Table S6. Here, we highlight an example. ACACA is a candidate gene related to tail fat deposition (Bakhtiarizadeh & Alamouti, 2020; Bakhtiarizadeh & Salami, 2019). In the current study, we found ten DETs (11.99.3, 11.689.28, 1.689.18, 11.265.3, ENSOART00020040904, 11.99.15, 11.689.14, 13.624.23, ENSOART00020006006, and 15.327.19) directly interactives with ENSOART00020037575 which was transcribed from ACACA (Fig. 8A). To further understand the function of DETs, GO and KEGG enrichment analyses were implemented. FDR < 0.05 was regarded as the significant threshold. All DETs were significantly enriched in 38 GO terms (Fig. 8B, Table S7). The most significant enriched GO term is extracellular matrix structural constituent (GO:0005201, FDR = 1.87e-4, Fig. 8B), including three annotated transcripts (ENSOART00020027615, ENSOART00020019556, and ENSOART00020033210) and nine novel transcripts (11.236.9, 10.185.4, 4.380.12, 4.380.22, 10.185.6, 24.240.21, 3.1134.2, 2.407.17, and 10.185.7). Detected DETs were significantly enriched in six KEGG pathways (Fig. 8C, Table S8). The most significant enriched pathway is Extracellular Matrix-receptor (ko04512), including three known transcripts (ENSOART00020039107, ENSOART00020027615, and ENSOART00020006511) and 11 novel transcripts (4.380.22, 11.236.16, 3.143.5, 10.185.4, 10.185.6, 10.185.7, 4.380.12, 12.528.5, 1.2008.6, 1.2007.14, and 11.236.9). The most significant GO term and KEGG pathway are extracellular matrix related indicating extracellular matrix play an important role in tail fat deposition.
Figure 8. Functional annotation of differentially expressed transcripts (DETs).
(A) DETs interative with ENSOART00020037575 (transcribed from ACACA). (B) Gene Ontology (GO) enrichment analysis of DETs. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DETs.
Discussion
In the current study, PacBio Iso-Seq was applied to uncovering the complexity of the transcriptome profile of the tail fat. Using Iso-Seq, 9,001 potential novel genes and 36,777 novel transcripts were detected (Fig. 2). The most notable gene was LIPE (lipase E, hormone-sensitive type) with 116 transcripts (113 novel transcripts and three known transcripts). The main function of hormone-sensitive lipase in adipose is to break triacylglycerol into fatty acids (Recazens, Mouisel & Langin, 2020). In humans, four exons (called exon B, A, T2 and T1) are alternatively used to produce different LIPE transcripts (Recazens, Mouisel & Langin, 2020). Similar AS patterns have also been found in rats, ewe, chickens, and fish (grass carp) (Bonnet et al., 1998; Cecilia et al., 1988; Sun et al., 2017). This suggests that Iso-Seq is promising in identifying novel gene loci and novel isoforms.
LncRNAs were predicted using Iso-Seq data to understand the transcriptome complexity of sheep tail fat. There are previous reports regarding lncRNA regulating tail fat deposition (Bakhtiarizadeh & Salami, 2019; Ma et al., 2018). These previous studies suggest that only a small portion of novel lncRNA identified in sheep tail fat were conserved (Bakhtiarizadeh & Salami, 2019). In the current study, we identified 3,764 lncRNA (Fig. 4, Table S3) and they could be useful for further investigating how lncRNA regulates tail fat deposition. Among them, 21 differentially lncRNAs were identified (Fig. 7B). It has been reported that lincRNA.3,473 interactive with ACACA may regulate fat deposition (Bakhtiarizadeh & Salami, 2019). In the current study, the positive significant correlation between several lncRNAs (ENSOART00020036299, ENSOART00020033641, ENSOART00020024562, ENSOART00020003848, and 9.53.1) and ACACA indicates that these lncRNAs may play a key role in tail fat deposition. The result that the majority of differential expressed lncRNAs (except lncRNA ENSOART00020017088) were significantly correlated with one or more transcripts transcribed from tail fat linked candidate genes could further validate their potential function in tail fat deposition. However, the genetic mechanism underlying lncRNAs regulate tail fat deposition still needs to be further investigated. In addition, APA was also investigated. In total, 1,662 genes contained at least two poly(A) sites were detected in sheep tail fat (Fig. 5). APA-mediated gene regulatory functions are tissue-specific (Di Giammartino, Nishida & Manley, 2011; Lianoglou et al., 2013; MacDonald, 2019; Tian & Manley, 2017). Generally, APA could affect mRNA stability, splicing, localization and translation by alter the sequence of regulatory elements, e.g., miRNA binding sites (Elkon, Ugalde & Agami, 2013; Tian & Manley, 2017). Although how the APA regulates tail fat deposition still needs further study, our results would be a useful resource for further analysis the function of APA in sheep tail fat.
The main function of AS is to increase the diversity of transcripts (Keren, Lev-Maor & Ast, 2010). As a result, transcripts expression abundance may change with different phenotypes. Using integrative analysis of Iso-Seq and RNA-seq data, hundreds of DETs were identified between thin- and fat-tailed sheep (Fig. 7A). One of DETs 15.527.13 was transcribed from DGAT2. DGAT2 has been reported to affect tail fat deposition in sheep (Bakhtiarizadeh & Alamouti, 2020) and back fat deposition in pig (Yin et al., 2012). A novel transcript 13.624.23, also a DET, was transcribed from ACSS2. Four DETs, 11.689.28, 11.689.18, 11.689.14, and 11.660.12, were transcribed from ACLY. ACSS2 and ACLY have been identified as candidate genes related to tail fat deposition in sheep (Bakhtiarizadeh & Alamouti, 2020; Guangli et al., 2020). Cytosolic acetyl CoA (Ac-CoA), the central precursor for lipid biosynthesis in mammals, is produced by ACLY from mitochondria-derived citrate or by ACSS2 from acetate (Vysochan et al., 2017). ENSOART00020040904 was transcribed from FASN. Three DETs (22.289.6, 22.289.3 and 22.289.14) were transcribed from SCD. ENSOART00020037575 was transcribed from ACACA. FASN, ACACA (Bakhtiarizadeh & Alamouti, 2020) and SCD (Kang et al., 2017a) are candidate genes related to tail fat deposition. These three gene, FASN (Leonard et al., 2004), SCD (Garcia-Fernandez et al., 2010; Kang et al., 2017a) and ACACA affect fatty acid composition (Cronan & Waldrop, 2002). These suggest that many novel transcripts were transcribed from well-studied candidate genes linked to lipid synthesis and may regulate tail fat deposition, highlighting alternative splicing is a complicated process in regulating tailed fat deposition.
PPI analysis is an effective way to search functional similarities between the DETs. For example, ENSOART00020037575 (transcribed from ACACA) directly interactives with 11.265.3 (transcribed from sterol regulatory element binding transcription factor 1, Srebp1) implying that Srebp1 may be linked with tailed fat deposition (Fig. 8A). Srebp1 has been reported to affect tail traits and lipid metabolism in sheep (Liang et al., 2020), which could validate our predicted result. In addition to PPI analysis, the functionality of DETs were also been investigated by GO and KEGG enrichment analyses. Interestingly, the most significant GO term (GO:0005201, extracellular matrix structural constituent, Fig. 8B, Table S7) and KEGG pathway (ko04512, ECM-receptor interaction, Fig. 8C, Table S8) is extracellular matrix related. Previous studies suggest that differentially expressed genes (DEGs) between fat-tail tissue of fat-tailed sheep and thin-tailed sheep were significantly enriched in the ECM-receptor interaction pathway (Bakhtiarizadeh et al., 2019; Li et al., 2018a). The studies in cattle and human adipose tissue also suggest that ECM plays an important role in adipogenesis (Casado-Diaz et al., 2017; Lee et al., 2013). Consistent with these results, in the present study, many novel DETs were significantly enriched in ECM-related GO terms and pathways, which suggests that Iso-Seq combined with the RNA-seq is a useful way to identification DETs.
Conclusions
In the present study, PacBio Iso-Seq was used for producing comprehensive transcriptomic data of tail fat of sheep, result in 9,001 potential novel gene loci, 17,834 AS events, 5,791 APA events and 3,764 lncRNAs. Combined with Iso-Seq and RNA-Seq data, hundreds of DETs between thin- and fat-tailed sheep were identified. Among them, 21 differentially expressed lncRNAs, such as ENSOART00020036299, ENSOART00020033641, ENSOART00020024562, ENSOART00020003848 and 9.53.1 may regulate tail fat deposition. Many novel transcripts were identified as DETs, including 15.527.13 (DGAT2), 13.624.23 (ACSS2), 11.689.28 (ACLY), 11.689.18 (ACLY), 11.689.14 (ACLY), 11.660.12 (ACLY), 22.289.6 (SCD), 22.289.3(SCD) and 22.289.14 (SCD), highlighting AS is a complicated process in regulating tailed fat deposition. Identified DETs were most significantly enriched in ECM-related GO terms and KEGG pathways suggesting their important roles in lipid metabolism. Our result revealed the transcriptome complexity and identified many candidate transcripts in tail fat, which could enhance the understanding of the mechanisms of tail fat deposition.
Supplemental Information
Funding Statement
This research was funded by the National Natural Science Foundation of China (31872333, 32172689), National Natural Science Foundation of China-CGIAR (32061143036), Major New Varieties of Agricultural Projects in Jiangsu Province (PZCZ201739), The Projects of Domesticated Animals Platform of the Ministry of Science, Key Research and Development Plan (modern agriculture) in Jiangsu Province (BE2018354), Jiangsu Agricultural Science and Technology Innovation Fund (CX(18)2003), and Natural Science Foundation of Jiangsu Province (BK20210811). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Zehu Yuan conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Ling Ge performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Jingyi Sun performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Weibo Zhang performed the experiments, prepared figures and/or tables, and approved the final draft.
Shanhe Wang performed the experiments, prepared figures and/or tables, and approved the final draft.
Xiukai Cao performed the experiments, prepared figures and/or tables, and approved the final draft.
Wei Sun conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
Animal experiment were approved by the Experimental Animal Ethical Committee of Yangzhou University (NO.202103294).
Data Availability
The following information was supplied regarding data availability:
The data is available at NCBI SRA: PRJNA745517.
References
- Abdel-Ghany et al. (2016).Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, Ben-Hur A, Reddy AS. A survey of the sorghum transcriptome using single-molecule long reads. Nature Communications. 2016;7(1):11706. doi: 10.1038/ncomms11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahbara et al. (2018).Ahbara A, Bahbahani H, Almathen F, Al Abri M, Agoub MO, Abeba A, Kebede A, Musa HH, Mastrangelo S, Pilla F, Ciani E, Hanotte O, Mwacharo JM. Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Frontiers in Genetics. 2018;9:699. doi: 10.3389/fgene.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amane et al. (2020).Amane A, Belay G, Nasser Y, Kyalo M, Dessie T, Kebede A, Getachew T, Entfellner JD, Edea Z, Hanotte O, Tarekegn GM. Genome-wide insights of Ethiopian indigenous sheep populations reveal the population structure related to tail morphology and phylogeography. Genes & Genomics. 2020;42(10):1169–1178. doi: 10.1007/s13258-020-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner et al. (2000).Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au et al. (2013).Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, Wong WH. Characterization of the human ESC transcriptome by hybrid sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(50):E4821–E4830. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baazaoui et al. (2021).Baazaoui I, Bedhiaf-Romdhani S, Mastrangelo S, Ciani E. Genome-wide analyses reveal population structure and identify candidate genes associated with tail fatness in local sheep from a semi-arid area. Animal. 2021;15(4):100193. doi: 10.1016/j.animal.2021.100193. [DOI] [PubMed] [Google Scholar]
- Bakhtiarizadeh & Alamouti (2020).Bakhtiarizadeh MR, Alamouti AA. RNA-seq based genetic variant discovery provides new insights into controlling fat deposition in the tail of sheep. Scientific Reports. 2020;10(1):13525. doi: 10.1038/s41598-020-70527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtiarizadeh & Salami (2019).Bakhtiarizadeh MR, Salami SA. Identification and expression analysis of long noncoding RNAs in fat-tail of sheep breeds. G3: Genes, Genomes, Genetics. 2019;9(4):1263–1276. doi: 10.1534/g3.118.201014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtiarizadeh et al. (2019).Bakhtiarizadeh MR, Salehi A, Alamouti AA, Abdollahi-Arpanahi R, Salami SA. Deep transcriptome analysis using RNA-seq suggests novel insights into molecular aspects of fat-tail metabolism in sheep. Scientific Reports. 2019;9(1):9203. doi: 10.1038/s41598-019-45665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle & Giudice (2017).Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nature Reviews Molecular Cell Biology. 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiki et al. (2019).Beiki H, Liu H, Huang J, Manchanda N, Nonneman D, Smith TPL, Reecy JM, Tuggle CK. Improved annotation of the domestic pig genome through integration of Iso-Seq and RNA-seq data. BMC Genomics. 2019;20(1):344. doi: 10.1186/s12864-019-5709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet et al. (1998).Bonnet M, Faulconnier Y, Flechet J, Hocquette JF, Leroux C, Langin D, Martin P, Chilliard Y. Messenger RNAs encoding lipoprotein lipase, fatty acid synthase and hormone-sensitive lipase in the adipose tissue of underfed-refed ewes and cows. Reproduction Nutrition Development. 1998;38(3):297–307. doi: 10.1051/rnd:19980310. [DOI] [PubMed] [Google Scholar]
- Buchfink, Xie & Huson (2015).Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Casado-Diaz et al. (2017).Casado-Diaz A, Anter J, Muller S, Winter P, Quesada-Gomez JM, Dorado G. Transcriptomic analyses of adipocyte differentiation from human mesenchymal stromal-cells (MSC) Journal of Cellular Physiology. 2017;232(4):771–784. doi: 10.1002/jcp.25472. [DOI] [PubMed] [Google Scholar]
- Cecilia et al. (1988).Cecilia H, Todd GK, Karen LS, Gudrun F, Staffan N, Chad GM, John ES, Camilla H, Robert SS, Thuluvancheri M, Aldons JL, Per B, Michael CS. Hormone-sensitive lipase: sequence, expression, and chromosomal localization to 19 cent-q13.3. Science. 1988;241(4872):1503–1506. doi: 10.1126/science.3420405. [DOI] [PubMed] [Google Scholar]
- Chao et al. (2019).Chao Q, Gao ZF, Zhang D, Zhao BG, Dong FQ, Fu CX, Liu LJ, Wang BC. The developmental dynamics of the Populus stem transcriptome. Plant Biotechnology Journal. 2019;17(1):206–219. doi: 10.1111/pbi.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2017).Chen SY, Deng F, Jia X, Li C, Lai SJ. A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Scientific Reports. 2017;7(1):7648. doi: 10.1038/s41598-017-08138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2018).Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, Zhang X, Wang J, Yang H, Fang L, Chen Q. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Giga Science. 2018;7(1):1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan & Waldrop (2002).Cronan JE, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Progress in Lipid Research. 2002;41(5):407–435. doi: 10.1016/S0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Deng et al. (2020).Deng L, Li L, Zou C, Fang C, Li C. Characterization and functional analysis of polyadenylation sites in fast and slow muscles. BioMed Research International. 2020;2020(1):2626584. doi: 10.1155/2020/2626584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino, Nishida & Manley (2011).Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Molecular Cell. 2011;43(6):853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong et al. (2020).Dong K, Yang M, Han J, Ma Q, Han J, Song Z, Luosang C, Gorkhali NA, Yang B, He X, Ma Y, Jiang L. Genomic analysis of worldwide sheep breeds reveals PDGFD as a major target of fat-tail selection in sheep. BMC Genomics. 2020;21(1):800. doi: 10.1186/s12864-020-07210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon, Ugalde & Agami (2013).Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature Reviews Genetics. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Fang et al. (2020).Fang L, Cai W, Liu S, Canela-Xandri O, Gao Y, Jiang J, Rawlik K, Li B, Schroeder SG, Rosen BD, Li CJ, Sonstegard TS, Alexander LJ, Van Tassell CP, VanRaden PM, Cole JB, Yu Y, Zhang S, Tenesa A, Ma L, Liu GE. Comprehensive analyses of 723 transcriptomes enhance genetic and biological interpretations for complex traits in cattle. Genome Research. 2020;30(5):790–801. doi: 10.1101/gr.250704.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac & Sammeth (2007).Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Research. 2007;35(Web Server):W297–W299. doi: 10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez et al. (2010).Garcia-Fernandez M, Gutierrez-Gil B, Garcia-Gamez E, Sanchez JP, Arranz JJ. Detection of quantitative trait loci affecting the milk fatty acid profile on sheep chromosome 22: role of the stearoyl-CoA desaturase gene in Spanish Churra sheep. Journal of Dairy Science. 2010;93(1):348–357. doi: 10.3168/jds.2009-2490. [DOI] [PubMed] [Google Scholar]
- Gasteiger, Jung & Bairoch (2001).Gasteiger E, Jung E, Bairoch A. SWISS-PROT: connecting biomolecular knowledge via a protein database. Current Issues in Molecular Biology. 2001;3:47–55. doi: 10.21775/cimb.003.047. [DOI] [PubMed] [Google Scholar]
- Gruber & Zavolan (2019).Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nature Reviews Genetics. 2019;20(10):599–614. doi: 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- Guangli et al. (2020).Guangli Y, Huan Z, Shuhong Z, Zhiqiang L, Fengyi G, Guan W, Qiankun W, Xiangyu L, Yongfeng Y, Ming L. Identification of the genetic basis for the large-tailed phenotypic trait in Han sheep through integrated mRNA and miRNA analysis of tail fat tissue samples. Research Square. 2020 doi: 10.21203/rs.3.rs-107294/v1. [DOI] [Google Scholar]
- Han et al. (2021).Han J, Guo T, Yue Y, Lu Z, Liu J, Yuan C, Niu C, Yang M, Yang B. Quantitative proteomic analysis identified differentially expressed proteins with tail/rump fat deposition in Chinese thin-and fat-tailed lambs. PLOS ONE. 2021;16(2):e0246279. doi: 10.1371/journal.pone.0246279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2020).He X, Wu R, Yun Y, Qin X, Chen L, Han Y, Wu J, Sha L, Borjigin G. Transcriptome analysis of messenger RNA and long noncoding RNA related to different developmental stages of tail adipose tissues of sunite sheep. 2020 doi: 10.21203/rs.3.rs-131480/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2020).Huang H, Zhou L, Chen J, Wei T. Ggcor: extended tools for correlation analysis and visualization. R package version 0.9.8. 2020. https://gitee.com/dr_yingli/ggcor. [26 October 2021]. https://gitee.com/dr_yingli/ggcor
- Jin et al. (2021).Jin W, Zhu Q, Yang Y, Yang W, Wang D, Yang J, Niu X, Yu D, Gong J. Animal-APAdb: a comprehensive animal alternative polyadenylation database. Nucleic Acids Research. 2021;49(D1):D47–D54. doi: 10.1093/nar/gkaa778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang et al. (2017a).Kang D, Zhou G, Zhou S, Zeng J, Wang X, Jiang Y, Yang Y, Chen Y. Comparative transcriptome analysis reveals potentially novel roles of Homeobox genes in adipose deposition in fat-tailed sheep. Scientific Reports. 2017a;7(1):14491. doi: 10.1038/s41598-017-14967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang et al. (2017b).Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Research. 2017b;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson & Linnarsson (2017).Karlsson K, Linnarsson S. Single-cell mRNA isoform diversity in the mouse brain. BMC Genomics. 2017;18(1):126. doi: 10.1186/s12864-017-3528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren, Lev-Maor & Ast (2010).Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nature Reviews Genetics. 2010;11(5):345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- Kornblihtt et al. (2013).Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nature Reviews Molecular Cell Biology. 2013;14(3):153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Kovaka et al. (2019).Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biology. 2019;20(1):278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead & Salzberg (2012).Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal-Gutierrez, Elzo & Mateescu (2020).Leal-Gutierrez JD, Elzo MA, Mateescu RG. Identification of eQTLs and sQTLs associated with meat quality in beef. BMC Genomics. 2020;21(1):104. doi: 10.1186/s12864-020-6520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2013).Lee HJ, Jang M, Kim H, Kwak W, Park W, Hwang JY, Lee CK, Jang GW, Park MN, Kim HC, Jeong JY, Seo KS, Kim H, Cho S, Lee BY. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction is involved in the depot-specific adipogenesis in cattle. PLOS ONE. 2013;8(6):e66267. doi: 10.1371/journal.pone.0066267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard et al. (2004).Leonard AE, Pereira SL, Sprecher H, Huang Y-S. Elongation of long-chain fatty acids. Progress in Lipid Research. 2004;43(1):36–54. doi: 10.1016/S0163-7827(03)00040-7. [DOI] [PubMed] [Google Scholar]
- Li, Zhang & Zhou (2014).Li A, Zhang J, Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics. 2014;15(1):311. doi: 10.1186/1471-2105-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Dewey (2011).Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2018a).Li B, Qiao L, An L, Wang W, Liu J, Ren Y, Pan Y, Jing J, Liu W. Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition. BMC Genomics. 2018a;19(1):338. doi: 10.1186/s12864-018-4747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2017).Li Y, Dai C, Hu C, Liu Z, Kang C. Global identification of alternative splicing via comparative analysis of SMRT-and Illumina-based RNA-seq in strawberry. Plant Journal. 2017;90(1):164–176. doi: 10.1111/tpj.13462. [DOI] [PubMed] [Google Scholar]
- Li et al. (2018b).Li Y, Fang C, Fu Y, Hu A, Li C, Zou C, Li X, Zhao S, Zhang C, Li C. A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing. DNA Research. 2018b;25(4):421–437. doi: 10.1093/dnares/dsy014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2018c).Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, Pritchard JK. Annotation-free quantification of RNA splicing using LeafCutter. Nature Genetics. 2018c;50(1):151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang et al. (2020).Liang C, Qiao L, Han Y, Liu J, Zhang J, Liu W. Regulatory roles of SREBF1 and SREBF2 in lipid metabolism and deposition in two Chinese representative fat-tailed sheep breeds. Animals (Basel) 2020;10(8):1317. doi: 10.3390/ani10081317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou et al. (2013).Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes & Development. 2013;27(21):2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, Huber & Anders (2014).Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2018).Ma L, Zhang M, Jin Y, Erdenee S, Hu L, Chen H, Cai Y, Lan X. Comparative transcriptome profiling of mRNA and lncRNA related to tail adipose tissues of sheep. Frontiers in Genetics. 2018;9:365. doi: 10.3389/fgene.2018.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald (2019).MacDonald CC. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond (2018 update) Wiley Interdisciplinary Reviews: RNA. 2019;10(4):e1526. doi: 10.1002/wrna.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo et al. (2019a).Mastrangelo S, Bahbahani H, Moioli B, Ahbara A, Al Abri M, Almathen F, da Silva A, Belabdi I, Portolano B, Mwacharo JM, Hanotte O, Pilla F, Ciani E. Novel and known signals of selection for fat deposition in domestic sheep breeds from Africa and Eurasia. PLOS ONE. 2019a;14(6):e0209632. doi: 10.1371/journal.pone.0209632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo et al. (2019b).Mastrangelo S, Moioli B, Ahbara A, Latairish S, Portolano B, Pilla F, Ciani E. Genome-wide scan of fat-tail sheep identifies signals of selection for fat deposition and adaptation. Animal Production Science. 2019b;59(5):835. doi: 10.1071/an17753. [DOI] [Google Scholar]
- Miao et al. (2015a).Miao X, Luo Q, Qin X, Guo Y. Genome-wide analysis of microRNAs identifies the lipid metabolism pathway to be a defining factor in adipose tissue from different sheep. Scientific Reports. 2015a;5(1):18470. doi: 10.1038/srep18470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao et al. (2015b).Miao X, Luo Q, Qin X, Guo Y, Zhao H. Genome-wide mRNA-seq profiling reveals predominant down-regulation of lipid metabolic processes in adipose tissues of small tail Han than Dorset sheep. Biochemical and Biophysical Research Communications. 2015b;467(2):413–420. doi: 10.1016/j.bbrc.2015.09.129. [DOI] [PubMed] [Google Scholar]
- Moioli, Pilla & Ciani (2015).Moioli B, Pilla F, Ciani E. Signatures of selection identify loci associated with fat tail in sheep. Journal of Animal Science. 2015;93(10):4660–4669. doi: 10.2527/jas.2015-9389. [DOI] [PubMed] [Google Scholar]
- Moradi et al. (2012).Moradi MH, Nejati-Javaremi A, Moradi-Shahrbabak M, Dodds KG, McEwan JC. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genetics. 2012;13(1):10. doi: 10.1186/1471-2156-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motter, Corva & Soria (2021).Motter MM, Corva PM, Soria LA. Expression of calpastatin isoforms in three skeletal muscles of Angus steers and their association with fiber type composition and proteolytic potential. Meat Science. 2021;171(3):108267. doi: 10.1016/j.meatsci.2020.108267. [DOI] [PubMed] [Google Scholar]
- Nattrass et al. (2014).Nattrass GS, Cafe LM, McIntyre BL, Gardner GE, McGilchrist P, Robinson DL, Wang YH, Pethick DW, Greenwood PL. A post-transcriptional mechanism regulates calpastatin expression in bovine skeletal muscle. Journal of Animal Science. 2014;92(2):443–455. doi: 10.2527/jas.2013-6978. [DOI] [PubMed] [Google Scholar]
- Pan et al. (2018).Pan Y, Jing J, Zhao J, Jia X, Qiao L, An L, Li B, Ma Y, Zhang Y, Liu W. MicroRNA expression patterns in tail fat of different breeds of sheep. Livestock Science. 2018;207(Suppl. 1):7–14. doi: 10.1016/j.livsci.2017.11.007. [DOI] [Google Scholar]
- Recazens, Mouisel & Langin (2020).Recazens E, Mouisel E, Langin D. Hormone-sensitive lipase: sixty years later. Progress in Lipid Research. 2020;82(Supplement):101084. doi: 10.1016/j.plipres.2020.101084. [DOI] [PubMed] [Google Scholar]
- Rhoads & Au (2015).Rhoads A, Au KF. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela & Rivals (2014).Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30(24):3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon et al. (2003).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun et al. (2017).Sun J, Yang Z, Xiao P, Liu Y, Ji H, Du Z, Chen L. Two isoforms of hormone-sensitive lipase b are generated by alternative exons usage and transcriptional regulation by insulin in grass carp (Ctenopharyngodon idella) Fish Physiology and Biochemistry. 2017;43(2):539–547. doi: 10.1007/s10695-016-0308-1. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2013).Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research. 2013;41(17):e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk et al. (2017).Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov et al. (2003).Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4(1):41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian & Manley (2017).Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nature Reviews Molecular Cell Biology. 2017;18(1):18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vysochan et al. (2017).Vysochan A, Sengupta A, Weljie AM, Alwine JC, Yu Y. ACSS2-mediated acetyl-CoA synthesis from acetate is necessary for human cytomegalovirus infection. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(8):E1528–E1535. doi: 10.1073/pnas.1614268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, Sanchez-Cabo & Ricote (2015).Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2013).Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Research. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2021).Wang W, Wang L, Wang L, Tan M, Ogutu CO, Yin Z, Zhou J, Wang J, Wang L, Yan X. Transcriptome analysis and molecular mechanism of linseed (Linum usitatissimum L.) drought tolerance under repeated drought using single-molecule long-read sequencing. BMC Genomics. 2021;22(1):109. doi: 10.1186/s12864-021-07416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2014).Wang X, Zhou G, Xu X, Geng R, Zhou J, Yang Y, Yang Z, Chen Y. Transcriptome profile analysis of adipose tissues from fat and short-tailed sheep. Gene. 2014;549(2):252–257. doi: 10.1016/j.gene.2014.07.072. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang Y, Liu J, Huang BO, Xu YM, Li J, Huang LF, Lin J, Zhang J, Min QH, Yang WM, Wang XZ. Mechanism of alternative splicing and its regulation. Biomedical Reports. 2015;3(2):152–158. doi: 10.3892/br.2014.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2015).Wei C, Wang H, Liu G, Wu M, Cao J, Liu Z, Liu R, Zhao F, Zhang L, Lu J, Liu C, Du L. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics. 2015;16(1):194. doi: 10.1186/s12864-015-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu & Watanabe (2005).Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Xiang et al. (2018).Xiang R, Hayes BJ, Vander Jagt CJ, MacLeod IM, Khansefid M, Bowman PJ, Yuan Z, Prowse-Wilkins CP, Reich CM, Mason BA, Garner JB, Marett LC, Chen Y, Bolormaa S, Daetwyler HD, Chamberlain AJ, Goddard ME. Genome variants associated with RNA splicing variations in bovine are extensively shared between tissues. BMC Genomics. 2018;19(1):521. doi: 10.1186/s12864-018-4902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie et al. (2011).Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research. 2011;39(suppl_2):W316–322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2017).Xu SS, Ren X, Yang GL, Xie XL, Zhao YX, Zhang M, Shen ZQ, Ren YL, Gao L, Shen M, Kantanen J, Li MH. Genome-wide association analysis identifies the genetic basis of fat deposition in the tails of sheep (Ovis aries) Animal Genetics. 2017;48(5):560–569. doi: 10.1111/age.12572. [DOI] [PubMed] [Google Scholar]
- Yin et al. (2012).Yin Q, Yang H, Han X, Fan B, Liu B. Isolation, mapping, SNP detection and association with backfat traits of the porcine CTNNBL1 and DGAT2 genes. Molecular Biology Reports. 2012;39(4):4485–4490. doi: 10.1007/s11033-011-1238-8. [DOI] [PubMed] [Google Scholar]
- Yuan et al. (2017).Yuan Z, Liu E, Liu Z, Kijas JW, Zhu C, Hu S, Ma X, Zhang L, Du L, Wang H, Wei C. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Animal Genetics. 2017;48(1):55–66. doi: 10.1111/age.12477. [DOI] [PubMed] [Google Scholar]
- Yuan et al. (2021).Yuan Z, Sunduimijid B, Xiang R, Behrendt R, Knight MI, Mason BA, Reich CM, Prowse-Wilkins C, Vander Jagt CJ, Chamberlain AJ, MacLeod IM, Li F, Yue X, Daetwyler HD. Expression quantitative trait loci in sheep liver and muscle contribute to variations in meat traits. Genetics Selection Evolution. 2021;53(1):8. doi: 10.1186/s12711-021-00602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2019).Zhang T, Gao H, Sahana G, Zan Y, Fan H, Liu J, Shi L, Wang H, Du L, Wang L, Zhao F. Genome-wide association studies revealed candidate genes for tail fat deposition and body size in the Hulun Buir sheep. Journal of Animal Breeding and Genetics. 2019;136(5):362–370. doi: 10.1111/jbg.12402. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2020).Zhao F, Deng T, Shi L, Wang W, Zhang Q, Du L, Wang L. Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types. Animals (Basel) 2020;10(5):773. doi: 10.3390/ani10050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi et al. (2018).Zhi D, Da L, Liu M, Cheng C, Zhang Y, Wang X, Li X, Tian Z, Yang Y, He T, Long X, Wei W, Cao G. Whole genome sequencing of hulunbuir short-tailed sheep for identifying candidate genes related to the short-tail phenotype. G3: Genes, Genomes, Genetics. 2018;8:377–383. doi: 10.1534/g3.117.300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2016).Zhu C, Fan H, Yuan Z, Hu S, Ma X, Xuan J, Wang H, Zhang L, Wei C, Zhang Q, Zhao F, Du L. Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Scientific Reports. 2016;6(1):27822. doi: 10.1038/srep27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The data is available at NCBI SRA: PRJNA745517.