ABSTRACT
Hypoxia inhibits the tricarboxylic acid (TCA) cycle and leaves glycolysis as the primary metabolic pathway responsible for converting glucose into usable energy. However, the mechanisms that compensate for this loss in energy production due to TCA cycle inactivation remain poorly understood. Glycolysis enzymes are typically diffuse and soluble in the cytoplasm under normoxic conditions. In contrast, recent studies have revealed dynamic compartmentalization of glycolysis enzymes in response to hypoxic stress in yeast, C. elegans and mammalian cells. These messenger ribonucleoprotein (mRNP) structures, termed glycolytic (G) bodies in yeast, lack membrane enclosure and display properties of phase-separated biomolecular condensates. Disruption of condensate formation correlates with defects such as impaired synaptic function in C. elegans neurons and decreased glucose flux in yeast. Concentrating glycolysis enzymes into condensates may lead to their functioning as ‘metabolons’ that enhance rates of glucose utilization for increased energy production. Besides condensates, glycolysis enzymes functionally associate in other organisms and specific tissues through protein–protein interactions and membrane association. However, as discussed in this Review, the functional consequences of coalescing glycolytic machinery are only just beginning to be revealed. Through ongoing studies, we anticipate the physiological importance of metabolic regulation mediated by the compartmentalization of glycolysis enzymes will continue to emerge.
KEY WORDS: G body, Condensate, Glycolysis, Hypoxia
Summary: Glycolysis enzymes coalesce in non-membrane bound structures in response to energy stress in a wide variety of organisms. We review examples of this coalescence and evaluate what is known about the structure, function and formation of these structures.
Introduction
Glycolysis is a core energy-producing pathway in cells; it converts glucose to two net ATPs and pyruvates, which can then be utilized by the mitochondria to generate an additional 34 ATPs through oxidative phosphorylation in the tricarboxylic acid (TCA) cycle (Al Tameemi et al., 2019) (Fig. 1). Hypoxic stress precludes the function of the highly efficient oxidative phosphorylation pathway and limits energy production to glycolysis (Al Tameemi et al., 2019). Despite substantial work on cellular adaptations to hypoxia, how cells compensate for this decreased energy efficiency is not fully understood. Normally, the ten core glycolysis enzymes are diffusely localized throughout the cytoplasm (Huh et al., 2003) (Fig. 1). However, under hypoxic conditions, glycolysis enzymes become compartmentalized into cytoplasmic structures (Miura et al., 2013; Jang et al., 2016; Jin et al., 2017). For example, recent studies in yeast have revealed that glycolysis enzymes colocalize into glycolytic bodies (G bodies) in response to hypoxia, and analogous condensates form in Caenorhabditis elegans neurons (Miura et al., 2013; Jang et al., 2016; Jin et al., 2017) (Figs 2–4). Biophysical studies have demonstrated that these structures have properties of phase-separated condensates that are similar to those of stress granules and P bodies, thus joining a growing list of subcellular structures that are formed by phase separation (Hyman et al., 2014; Fuller et al., 2020; Jang et al., 2021). Mutants unable to form these structures have decreased viability or synaptic function in yeast and worms, respectively (Jang et al., 2016; Jin et al., 2017). The association of multiple glycolysis enzymes via phase separation may function to enhance the activity of the entire pathway and increase reaction rates critical for energy production, thereby forming a so-called ‘metabolon’ during hypoxic stress (Srere, 1987; Miura et al., 2013; Jang et al., 2016; Jin et al., 2017). Similar structures also form in cancer cell lines, raising the as-yet-unexplored possibility that analogous G body structures in humans may promote cancer cell survival (Jin et al., 2017). In this Review, we explore what is known about the structure, function and regulation of these glycolysis enzyme condensates, and highlight additional examples of compartmentalization of glycolysis enzymes.
Fig. 1.
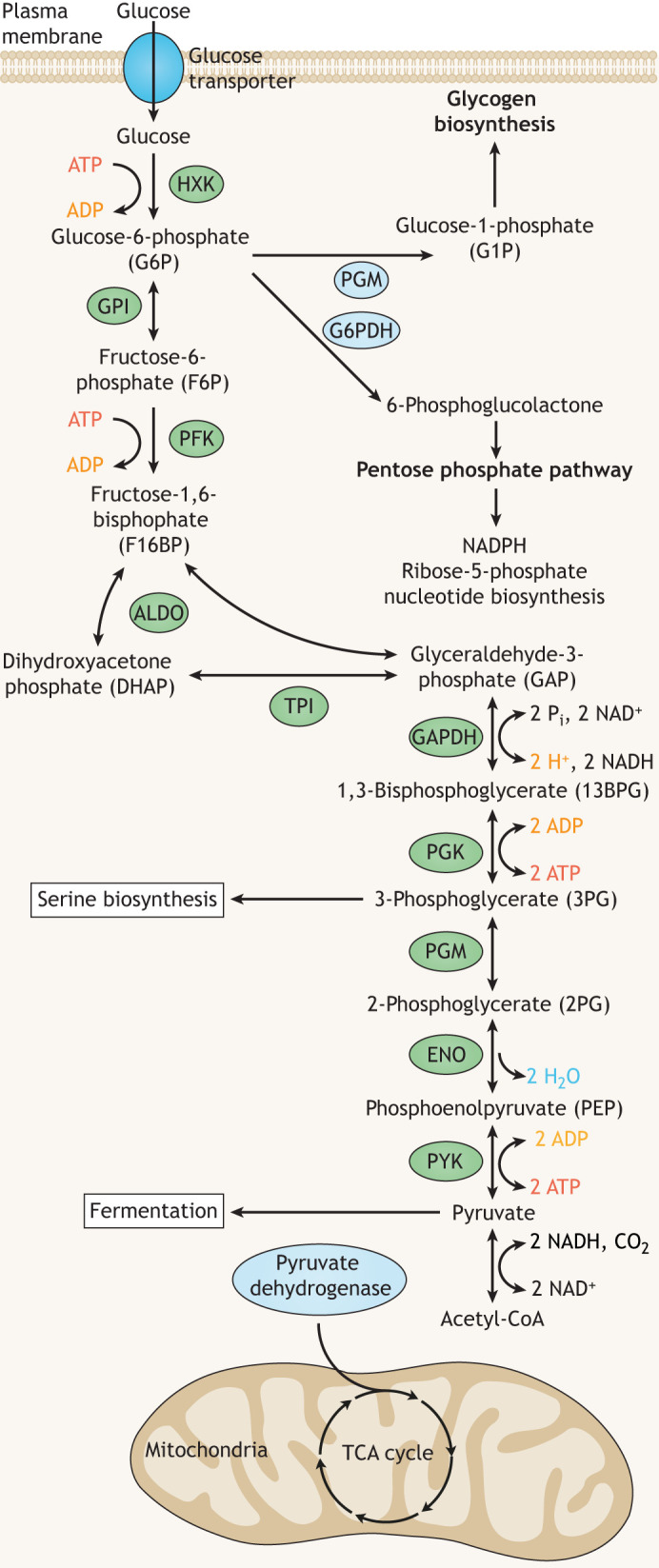
Glycolysis and related pathways. In the upstream glycolysis reactions, glucose is phosphorylated, isomerized to F6P, and phosphorylated once more, generating F16BP through the action of hexokinase (HXK), glucose phosphate isomerase (GPI) and phosphofructokinase (PFK), respectively. The reverse reaction of PFK is catalyzed by fructose-1,6-bisphosphatase (FBPase; not depicted). Next, F16BP is cleaved by aldolase (ALDO) into dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP). DHAP can interconvert with GAP through triose phosphate isomerase (TPI) activity, thus generating two equivalents of GAP per molecule of glucose. In an NAD+-dependent reaction, glyceraldehyde phosphate dehydrogenase (GAPDH) converts GAP into 1,3-bisphosphoglycerate (13BPG). Phosphoglycerate kinase (PGK) generates ATP when converting 13BPG into 3-phosphoglycerate (3PG), which is then converted into 2-phosphoglycerate (2PG) by phosphoglycerate mutase. Enolase (ENO) converts 2PG into phosphoenolpyruvate (PEP), the substrate of the final glycolytic enzyme, pyruvate kinase (PYK) which generates ATP and pyruvate. The reverse of this reaction is catalyzed by phosphoenolpyruvate carboxykinase (PEPCK; not depicted). At multiple steps, metabolites can enter other pathways beginning with glucose phosphate, which can be directed into glycogen biosynthesis or the pentose phosphate pathway by phosphoglucomutase (PGM) or glucose-6-phosphate dehydrogenase (G6PDH), respectively. Downstream, 3PG can be directed into serine biogenesis. Pyruvate from glycolysis is either converted into ethanol (yeast) or lactic acid (metazoans) by fermentation in the cytoplasm or enters the TCA cycle in the mitochondria generating additional ATP. Glycolysis enzymes are indicated in green ovals and enzymes of related pathways appear in blue ovals.
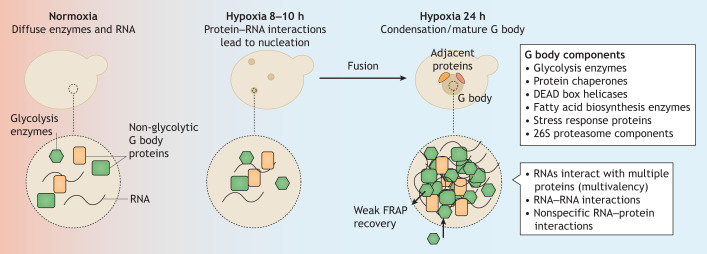
Fig. 2.
G body biogenesis, structure and composition. In normoxic conditions, glycolysis enzymes are diffuse throughout the cytoplasm, although they do associate with RNA (left). In hypoxia, RNA functions as a scaffold, promoting the formation of small complexes of glycolysis enzymes, RNA and additional non-glycolytic proteins. As cells remain in hypoxia, these small complexes fuse into large single granules that include additional proteins and are observable as bright foci with GFP-tagged reporters (right). G bodies are in close vicinity to other proteins despite lacking perfect overlap with their signals. Typical protein components are listed. G bodies have the physical properties of phase-separated gel-like granules as opposed to liquid assemblies, including stability in cell lysates, slow exchange with the cytosol, as measured in FRAP experiments, and slow fusion kinetics. Both the exchange of enzymes and fusion of single granules occurs on the timescale of minutes. RNA can modulate phase separation by several mechanisms (listed at bottom right).
Fig. 4.
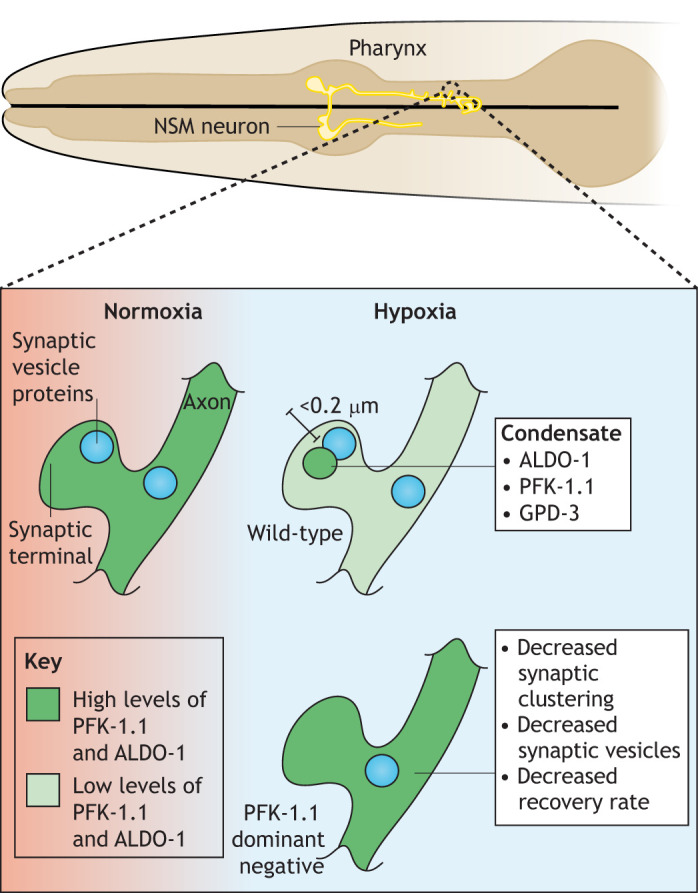
Pre-synaptic phase separation of glycolysis enzymes. Expanded view of an individual synaptic terminal in the neurosecretory motor neuron (NSM) of the pharynx of C. elegans. In normoxia, PFK-1.1 and ALDO-1 (green) are diffusely distributed, but within minutes of hypoxia occurring, they accumulate in condensates that are in close proximity with synaptic vesicles in a mutually dependent manner. GPD-3 (GAPDH) also localizes to these clusters. The condensates exhibit properties of liquid phase separation including the ability to fuse, fast dynamic formation and dissolution (within minutes of hypoxic treatment), fast exchange of proteins with the cytosol (measured by FRAP) and a propensity to become more solid as time in hypoxia increases. A dominant-negative allele of PFK-1.1 prevents this assembly, resulting in synaptic defects (bottom).
Protein composition of glycolytic granules
Of the glycolytic granules, yeast G bodies have the best-understood protein composition, based on proteomic studies that have been validated by in vivo colocalization of endogenous, fluorescently labeled candidate proteins residing in G bodies (Fig. 2). Of the glycolysis enzymes, Eno2 (enolase), Pfk1 and Pfk2, the α- and β-subunits of yeast phosphofructokinase (PFK), respectively, Fba1 (aldolase) and Cdc19 (pyruvate kinase, PYK) strongly colocalize into a large single G body under hypoxic stress in yeast (Miura et al., 2013; Jin et al., 2017). In C. elegans neurons, PFK, aldolase and glyceraldehyde-3-phosphate dehydrogenase (GPD-3; GAPDH) localize to glycolytic granules in hypoxia (Jang et al., 2016). PFK, which catalyzes the rate-limiting ATP-dependent conversion of fructose-6-phosphate into fructose-1,6-bisphosphate, has been found in all glycolytic granules identified to date (Miura et al., 2013; Jang et al., 2016; Kohnhorst et al., 2017; Jin et al., 2017) and serves as the canonical G body marker. Additional enzymes, including isoforms of GAPDH, PGK, phosphoglycerate mutase (PGM), Eno1 and Pyk2 localize to puncta resembling G bodies in a smaller subset of yeast cells (Jin et al., 2017). In addition to the glycolysis enzymes, many other proteins localize to yeast G bodies to varying degrees. These include 26S proteasome subunits, components of other metabolic pathways, such as fatty acid metabolism and trehalose biosynthesis, translation elongation factors, ribosomal proteins, signaling factors and protein chaperones (e.g. Hsp70, Hsp42, and Hsp26), which localize to G bodies in nearly all cells (Miura et al., 2013; Jin et al., 2017). Besides protein chaperones, which also localize to other condensates, including nucleoli during heat shock, and stress granules, where they are required for stress granule disassembly (Walters et al., 2015; Jain et al., 2016; Frottin et al., 2019), most of these proteins only sparingly colocalize with G bodies, and they often appear in puncta adjacent to Pfk2-labeled G body foci. This suggests that G bodies are either complex structures with multiple subcompartments or they associate with other condensates or filaments (Jin et al., 2017). Some of the proteins with weak colocalization, including several glycolysis enzymes, may only transiently localize to G bodies. Such dynamic recruitment of different sets of metabolic enzymes could rapidly reconfigure metabolism by altering the fate of glycolysis intermediates and the efficiency of individual steps of each reaction. Alternatively, the non-glycolytic proteins, such as proteasome subunits and translation-associated proteins, may be stably recruited to G bodies in a small fraction of cells. Finally, dynamic recruitment may indicate distinct stages of G body biogenesis. For instance, Eno2 foci precede the appearance of Cdc19 and PGM foci in hypoxic yeast, raising the possibility that Eno2 is required to form granules that subsequently recruit additional proteins (Yoshimura et al., 2021). Studies of the dynamics of various G body proteins will be required to differentiate between these possibilities.
In cancer cell lines, both glycolytic and gluconeogenic enzymes localize to puncta called glucosomes (Kohnhorst et al., 2017) (Fig. 5). Furthermore, many enzymes are shared between the opposing gluconeogenesis and glycolysis pathways, with allosteric regulation of human phosphofructokinase (PFKL, glycolysis) and fructose-1,6-bisphosphatase (FBPase, gluconeogenesis) largely governing which pathway predominates (Fig. 5). Both of these enzymes localize to glucosomes, in addition to phosphoenolpyruvate carboxykinase (PEPCK) and PYK (Kohnhorst et al., 2017); therefore, allosteric regulation of these enzymes within granules may influence flux through the opposing pathways. While PFK puncta increase in size in hypoxia in HepG2 cells, it is unknown whether gluconeogenesis enzymes also localize to these large granules (Jin et al., 2017). Proteomic analysis of glucosomes and hypoxic PFK puncta may help to define the composition of each granule. Are similar proteins recruited to these granules as to those in yeast G bodies? Do the larger hypoxic puncta comprise glucosomes that have fused or adjoined together, in which case there would be similar composition between the glucosome and hypoxic foci? Such studies may reveal principles of granule biogenesis and structure.
Fig. 5.
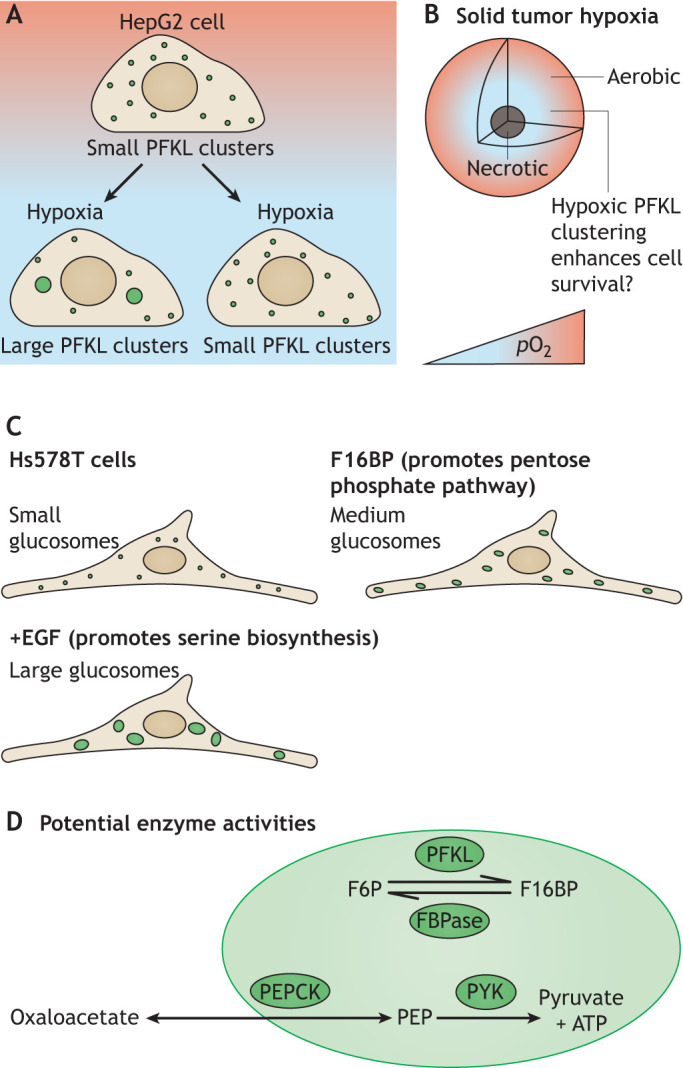
Compartmentalization in mammalian cancer cells. (A) In HepG2 cells, a hepatocarcinoma-derived cell line, PFKL is localized in small clusters in normoxic conditions. Hypoxia generates large clusters of PFKL in an RNA-dependent manner as treatment with RNase prevents their formation. (B) Hypoxic conditions in the interior of solid tumors may induce an increase in the size of PFKL clusters to enhance glycolytic activity and promote survival. (C) Glucosomes, constitutive assemblies of glycolysis enzymes, are present in Hs578T cells and other cancer cell lines even in the absence of hypoxia. Treatment with metabolites that promote alternative glucose utilization pathways (serine biosynthesis or the pentose phosphate pathway) leads to changes in granule size which may correlate with flux of metabolites into these alternative pathways. Glucosomes may be identical with the small PFKL clusters in A. (D) Glucosomes contain both glycolytic and gluconeogenic enzymes, allowing potential control of glycolytic and gluconeogenic flux through PFKL (glycolysis) or FBPase (gluconeogenesis) activity via conversion of F6P and F16BP. Downstream reactions can promote gluconeogenesis by conversion of oxaloacetate from the TCA cycle to PEP via PEPCK or complete glycolysis by conversion of PEP into pyruvate and ATP catalyzed by PYK.
Protein–protein interactions are likely important for localization to glycolytic granules. In C. elegans glycolytic condensates, aldolase (ALDO-1) and PFK (PFK-1.1) recruitment are mutually dependent (Fig. 4) (Jang et al., 2021). Moreover, many glycolysis enzymes function in quaternary structures; for example, PFK is an octamer (Sträter et al., 2011; Schöneberg et al., 2013). In yeast, this octamer is comprised of four α (Pfk1) and four β (Pfk2) subunits, stabilized by interactions between each set of subunits (Banaszak et al., 2011; Sträter et al., 2011). The multiple interactions between domains of human PFKL monomers facilitate filamentation, which is morphologically distinct from spherical condensates typical of G bodies and C. elegans glycolytic condensates (Jang et al., 2016; Webb et al., 2017; Jin et al., 2017). In yeast, several glycolysis enzymes form filaments, including Cdc19, Pfk1 and Pfk2, and glucokinase (Glk1), although the relationship between filamentation and spherical granule formation remains unclear (Shen et al., 2016; Noree et al., 2019; Stoddard et al., 2020). Filaments could act as scaffolds that recruit additional enzymes. Glycolysis enzymes, including hexokinase (HXK), triose phosphate isomerase (TPI), glucose phosphate isomerase (GPI), aldolase, GAPDH and enolase associate with F-actin, which acts as a scaffold, in yeast, promoting interactions between glycolysis enzymes (Waingeh et al., 2006; Araiza-Olivera et al., 2013). Importantly, the actin-associated enzymes only partially overlap with G body proteins and thus likely represent a distinct complex. Moreover, the strongest interaction between actin and a glycolysis enzyme is with GAPDH (Waingeh et al., 2006). In contrast, GAPDH only rarely localizes to G bodies, highlighting the different composition of each complex. Finally, the biophysical properties of actin-associated glycolysis enzyme complexes in yeast have yet to be explored and they may not constitute condensates. Yeast Glk1 filamentation resembles folding of actin (Stoddard et al., 2020). By mimicking the shape of F-actin, glucokinase may provide similar binding sites to function as a scaffold for additional glycolysis enzymes, although this possibility has not yet been explored.
Characterization of the proteomes of glycolytic granules from different species will be important for understanding the overall composition and potential function of these granules. Additionally, studies comparing granule composition under various environmental conditions will elucidate dynamic protein recruitment to glycolytic granules. Finally, evaluating granule composition in mutants, in which different granule components are deleted, will help identify the genetic requirements for granule assembly, structure, maintenance and regulation.
Phase separation of glycolysis enzymes
Recently, phase separation has emerged as an organizing process that can rapidly concentrate specific proteins and RNAs in structures termed ‘condensates’ in response to a variety of environmental cues (Hyman et al., 2014). Condensates form with a range of physical properties, from those resembling liquid droplets, such as nucleoli and yeast P bodies, to solid gels, such as yeast stress granules, while other condensates possess both gel- and liquid-like components (Brangwynne et al., 2011; Kroschwald et al., 2015; Jain et al., 2016; Putnam et al., 2019). For example, in C. elegans P granules, MEG-3 adopts a gel phase to serve as a solid scaffold for PGL-3, which has more liquid-like properties (Putnam et al., 2019).
Several lines of evidence suggest that G bodies in yeast and glycolysis enzyme structures in C. elegans neurons are condensates formed via phase separation but possess distinct physical properties (Fig. 2). First, G bodies lack membranes, as revealed by electron microscopy (Jin et al., 2017). Second, recruitment of Pfk2 to G bodies relies on multivalent interactions through both its N- and C-terminal regions, which are connected by a disordered linker (Sträter et al., 2011; Fuller et al., 2020). Third, G bodies in a and α yeast cells can fuse during mating (Fuller et al., 2020). Consistent with the slow (10–20 min) recovery kinetics measured by fluorescence recovery after photobleaching (FRAP), G bodies within mating cells fuse slowly, often taking many minutes for two smaller granules to form into a single large G body (Fuller et al., 2020). In contrast, liquid-like P bodies fuse within seconds of contact and have components that display near full FRAP recovery within seconds (Kroschwald et al., 2015). In C. elegans, hypoxia-induced assemblies of glycolysis enzymes have more liquid-like properties than their yeast counterparts, fusing within two seconds of contact (Jang et al., 2021). Similar fusion timescales have been observed for PFKL in cell culture overexpression experiments (Webb et al., 2017). Unlike in yeast, C. elegans hypoxic condensates display strong FRAP recovery, within tens of seconds, although there is less recovery with prolonged exposure to hypoxic conditions, indicating a transition to a more gel-like state (Jang et al., 2021) (Fig. 4). A fourth line of evidence that G bodies form via phase separation is that their size decreases following treatment with the aliphatic alcohol 1,6-hexanediol, although G bodies do not completely dissolve (Fuller et al., 2020). It is tempting to speculate that partial dissolution may reflect a dynamic shell around G bodies, akin to what is present on stress granules (Jain et al., 2016). A dynamic shell could explain the variation in colocalization observed for different G body components (Jin et al., 2017). Fifth, the initial formation of granules in C. elegans occurs in regions of the axon with high local concentrations of PFK-1.1, indicating that it has a propensity to phase separate in a concentration-dependent manner (Jang et al., 2021). Finally, G bodies appear stable and can be biochemically purified from a cell lysate, rather than dissolving in dilute conditions (Miura et al., 2013; Jin et al., 2017; Fuller et al., 2020). Thus, in yeast, G bodies appear to form gels that maintain their structure, even when moved to a dilute medium, exchange proteins with the cytoplasm and fuse slowly, while C. elegans granules are more fluid and dynamic. However, even in yeast, multiple smaller foci are initially observed prior to the appearance of a single G body per cell upon extended periods in hypoxia, suggesting that these smaller puncta fuse to form the larger mature G body (Jin et al., 2017). Therefore, liquid states could precede the gel-like state of G bodies, as has been observed in disease states for stress granules and in C. elegans hypoxic granules (Patel et al., 2015; Murakami et al., 2015; Bolognesi et al., 2016; Mateju et al., 2017; Fuller et al., 2020; Jang et al., 2021).
The differences in physical properties between yeast G bodies and C. elegans PFK-1.1 condensates mirror the differing material properties between yeast stress granules, which resemble solid aggregates, and mammalian stress granules, which resemble a viscous liquid (Kroschwald et al., 2015). One possible explanation for the discrepancy in material properties is that yeast G bodies require many hours to form, whereas C. elegans PFK-1.1 condensates form quickly within 10 min under hypoxia. Therefore, the prolonged period required to form yeast G bodies may provide the time for these condensates to mature from a liquid to gel-like state (Jang et al., 2016; Jin et al., 2017; Jang et al., 2021). Moreover, extended periods in hypoxia lead to a decreased amplitude, but not half time, of PFK-1.1 FRAP recovery in granules (Jang et al., 2021). Thus, C. elegans granules and yeast G bodies may form via a similar mechanism, but the latter represents a much later stage of maturation in the liquid-to-solid transition.
Interestingly, the yeast Cdc19 PYK accumulates in stress granules during glucose starvation, likely through phase separation that is dependent on an intrinsically disordered region in the enzyme (Saad et al., 2017; Grignaschi et al., 2018). However, neither G bodies nor PFK-1.1 condensates in C. elegans neurons seem to contain the stress granule markers Pab1 and TIAR, respectively (Jin et al., 2017; Jang et al., 2021). Thus, these hypoxia-induced granules are distinct from stress granules, and Cdc19 recruitment under hypoxic conditions may have a different role rather than just its sequestration.
The role of RNA in G body formation and structure
Like other condensates, RNA localizes to G bodies. Most known G body proteins can bind to RNA, despite lacking canonical RNA-binding domains (Fuller et al., 2020). Sequencing of mRNAs crosslinked to purified messenger ribonucleoprotein (mRNP) complexes has revealed that most glycolysis enzymes bind RNA in normoxic conditions (Castello et al., 2012; Beckmann et al., 2015; Matia-González et al., 2015; Fuller et al., 2020). RNA binding by glycolysis enzymes is highly conserved in bacteria, plants and metazoans (Sysoev et al., 2016; Despic et al., 2017; Albihlal and Gerber, 2018; Huang et al., 2018; Shchepachev et al., 2019). Included among the hundreds of distinct transcripts bound by glycolysis enzymes are those of the glycolysis enzymes themselves, which have a large number of experimentally validated protein-binding sites (Freeberg et al., 2013; Shchepachev et al., 2019; Fuller et al., 2020). Targeting RNases to sites of G body formation prevents G bodies from forming, indicating that RNA is required for their nucleation. Moreover, targeting RNases to existing G bodies causes them to fracture into multiple smaller granules, suggesting that RNA stabilizes mature condensates (Fuller et al., 2020).
RNA could contribute to G body formation by concentrating and sequestering protein components through multiple binding sites (i.e. multivalency), thereby acting as a scaffold (Garcia-Jove Navarro et al., 2019; Dutagaci et al., 2021) (Fig. 2). Specific mRNA–protein interactions can be important for condensate formation. For example, in vitro, the yeast Whi3 protein partitions into either gel or liquid droplets depending on which of its mRNA substrates is available through interactions largely driven by secondary structure (Zhang et al., 2015; Langdon et al., 2018). Moreover, RNA alone can phase separate, and specific homotypic RNA–RNA interactions are emerging as an organizing principle in germ granule composition in Drosophila melanogaster (Trcek et al., 2015, 2020; Jain and Vale, 2017; Van Treeck et al., 2018). In contrast, nonspecific interactions between RNA and intrinsically disordered regions can also drive phase separation (Protter et al., 2018).
Whether specific protein–mRNA interactions underlie RNA-mediated G body formation is unclear. Sequencing of G body RNAs and localization by single-molecule fluorescence in situ hybridization (smFISH) demonstrate that many different RNAs reside in G bodies, although only a small fraction (<2%) of a given mRNA species localizes to G bodies (Fuller et al., 2020). Despite the fact that small quantities of particular mRNA species are concentrated, there is a wide variability in mRNA enrichment in G bodies. G body RNAs show a substantial, but incomplete, overlap with RNAs bound by glycolysis enzymes in normoxic conditions, suggesting that glycolysis enzymes are bound by RNA in the cytoplasm and then recruited to G bodies together (Fuller et al., 2020). The lack of complete overlap could be due to the presence of other RNA binding proteins in G bodies or altered RNA binding in hypoxic conditions. In yeast, altered environmental conditions, such as glucose or nitrogen starvation, lead to substantial changes in protein occupancy on mRNAs, including those of G body-associated mRNAs and glycolysis enzyme mRNAs themselves (Freeberg et al., 2013). Additional studies will be necessary to determine whether specific mRNA–protein and RNA–RNA interactions are required for G body structure and formation.
While P bodies and stress granules incorporate a large fraction of the transcriptome, they recruit certain mRNAs to a far larger extent than others, indicating some degree of specificity in recruitment (Hubstenberger et al., 2017; Khong et al., 2017). For example, unlike in G bodies, which recruit at most 2% of a single mRNA species, up to 95% of certain mRNA species, such as AHNAK and DYNC1H1, localize to stress granules (Khong et al., 2017). However, most stress granule mRNAs are enriched in stress granules relative to the cytoplasm to a lesser extent. mRNAs in stress granules and P bodies are poorly translated (Khong et al., 2017). However, given that the bulk of each of the mRNAs identified in G bodies localizes elsewhere, it seems unlikely that the minor fraction sequestered into G bodies significantly impacts the overall translational output of the mRNA for a specific protein. Nevertheless, additional studies will be required to determine the fate of mRNAs that localize to G bodies.
Regulation of G body assembly
Stress induces the formation of many condensates within the cell, such as stress granules, P bodies and inclusion bodies. As previously mentioned, hypoxic stress, in particular, induces the self-association of glycolysis enzymes into large granules in yeast, C. elegans and human cancer cells (Figs 2, 4 and 5) (Miura et al., 2013; Jang et al., 2016; Jin et al., 2017). Several genetic determinants of G body assembly have been identified and implicate energy-sensing homeostasis pathways in G body regulation (Fig. 3). In particular, rapamycin treatment, which inhibits TORC1, the complex in yeast homologous to the mammalian target of rapamycin complex (mTORC1), prevents the formation of Eno2–GFP puncta in yeast, demonstrating that TORC1 activity is required for G body formation (Miura et al., 2013). Additionally, Ira2, a negative regulator of the pro-growth yeast RAS/cAMP pathway, which activates the downstream protein kinase A (PKA), localizes to G bodies in less than 2% of cells and is required for their formation, although it is unknown whether this requirement is due to signaling or direct physical interactions (Jin et al., 2017). Finally, the AMP-activated protein kinase Snf1, which is activated in response to high AMP-to-ATP ratios and promotes glycolytic activity, is required for normal G body formation in yeast (Miura et al., 2013; Jin et al., 2017; Yoshimura et al., 2021). By controlling protein synthesis, TORC1 and PKA may modify the proteomic response to hypoxia, thereby affecting G body formation. Alternatively, these factors may directly phosphorylate G body proteins or regulators and control the association of G body enzymes. However, while Pfk2 is progressively phosphorylated in a Snf1-dependent manner in hypoxia, Pfk2 localization to G bodies appears to be independent of its phosphorylation (Jin et al., 2017).
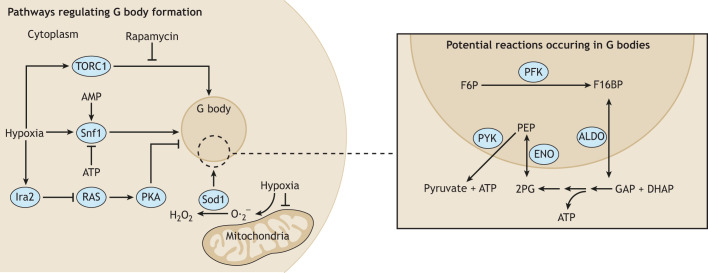
Fig. 3.
Genetic regulation of G body formation. Hypoxia triggers activation of signaling pathways that are involved in energy homeostasis and required for G body formation (shown on the left). Inhibition of the TORC1 complex by rapamycin treatment prohibits G body formation. In addition to TORC1, Snf1, the yeast ortholog of the AMP-activated protein kinase (AMPK), and Ira2, a negative regulator of the pro-growth pathway (RAS to PKA), is required for G body formation. Hypoxia also triggers the production of superoxide radicals through mitochondrial dysfunction, and the expression of Sod1, the cytoplasmic enzyme that degrades superoxide radicals, is involved in G body formation. Many of the glycolysis enzymes strongly localize to G bodies, including the sequential set of enzymes, PFK and aldolase (see diagram on the right). However, TPI and GAPDH do not strongly localize to G bodies, suggesting the reactions they catalyze occur in the cytoplasm. Downstream of GAPDH, enolase and PYK, catalyzing the final two steps of glycolysis, localize to G bodies, suggesting that both the upstream and far downstream reactions occur in G bodies.
G body formation has also been linked to mitochondrial function, given that mitochondrial inhibitors, such as antimycin or CCCP, disrupt Eno2–GFP localization to G bodies (Miura et al., 2013). This inhibition could be related to the generation of mitochondrial reactive oxygen species (ROS) that occurs upon a shift to hypoxia (Görlach et al., 2015). N-acetyl cysteine, an antioxidant, also inhibits the formation of Eno2–GFP foci (Miura et al., 2013). Moreover, Sod1, the primary superoxide dismutase in yeast, localizes to G bodies at a low frequency and is required for G body formation (Fig. 3) (Jin et al., 2017). Further studies will be required to determine the molecular mechanisms by which these pathways regulate G body formation, and whether they also impact the formation of G body-like structures in C. elegans and mammalian cells.
Finally, granule formation may be sensitive to the metabolic needs of a cell. Glucosome size in mammalian cancer cell lines in normoxic conditions varies depending on addition of metabolites that differentially promote glucose flux, the pentose phosphate pathway or serine synthesis (Fig. 5) (Kohnhorst et al., 2017). Similarly, G body formation in yeast requires glucose (Jin et al., 2017). Therefore, the composition of glycolysis enzyme granules may vary depending on available nutrients. G bodies can recruit a number of different metabolic enzymes, including the enzymes of fatty acid synthesis (Jin et al., 2017). Some of these enzymes and G body RNAs are recruited to the periphery of the G body rather than perfectly overlapping with G body markers, possibly indicating transient association with these granules (Jin et al., 2017; Fuller et al., 2020). It is interesting to speculate that compositional changes of these assemblies, in response to altered nutrient availability, could promote activity of pathways besides glycolysis by supplying ATP or substrates.
Physiological function of glycolysis enzyme granules
A variety of functions have been proposed for glycolytic granules. Mutant C. elegans strains lacking the ability to form condensates have decreased synaptic function (Jang et al., 2016). Loss of PFK-1.1, the primary PFK isoform in C. elegans neurons, leads to disruption of presynaptic vesicle clustering specifically during hypoxia at the same location where the condensates form (Jang et al., 2016). Overexpression of a dominant-negative allele that impairs PFK-1.1 localization also results in similar defects in vesicle clustering and loss of pfk-1.1 causes decreased recovery of synapses (Fig. 4) (Jang et al., 2016). Thus, condensate formation in C. elegans neurons may occur to enhance glycolytic rates during energy stress to maintain synaptic transmission between neurons.
In yeast, mutants unable to form G bodies display decreased glucose flux and accumulate upstream glycolysis metabolites (Miura et al., 2013; Jin et al., 2017). Normoxic cells with diffuse glycolysis enzymes consume glucose at a slower rate than cells taken from hypoxia that have G bodies (Jin et al., 2017). Finally, cells lacking G bodies are at a competitive growth disadvantage compared to cells that are able to form G bodies (Fuller et al., 2020). The phenotypes of cells lacking G bodies suggest that glycolysis enzymes phase separate to enhance energy production and promote cell survival (Fig. 3). These physiological findings make it unlikely that G bodies are aggregates of misfolded and inactive proteins, as is the case for other compartments, such as insoluble protein deposits (IPODS), juxta-nuclear quality control compartments (JUNQs), or aggresomes (Rothe et al., 2018). Direct measurements of enzyme activity in purified G bodies are needed to confirm that this is the case.
If G body-mediated enhancement of glucose flux is conserved in human cancer-derived cells, glycolytic granules could promote hypoxic cell survival and proliferation in the hypoxic environment of solid tumors (Petrova et al., 2018; Al Tameemi et al., 2019). Clustering of glycolysis enzymes in glucosomes is constitutive in several cancer cell lines, but is modified under conditions affecting energy production (Kohnhorst et al., 2017; Jin et al., 2017). In particular, hypoxia, which creates energy stress by depriving cells of respiration, enhances clustering of PFKL in the HepG2 hepatocarcinoma-derived cell line (Jin et al., 2017). Thus, clustering may function as a method to enhance pathway activity when energy production is limited. Indeed, hypoxic exposure has been shown to enhance cancer cell proliferation (Das et al., 2008). Moreover, glucosomes may have a role in normoxic conditions to enhance cancer cell viability (Kohnhorst et al., 2017). One possibility is that condensates of glycolysis enzymes promote Warburg metabolism, or aerobic glycolysis, in which cancer cells primarily utilize glycolysis and fermentation rather than respiration through the TCA cycle to generate ATP (Potter et al., 2016). Hypoxic exposure could promote the formation of granules that subsequently promote proliferation and cancer metabolism in normoxic conditions.
Enzyme activity in glycolytic granules
Enzyme activity may be modulated in assemblies of enzymes in condensates. For example, yeast aminoacylation enzymes aggregate in heat shock but maintain their activity (Wallace et al., 2015). Clustered glycolysis enzymes may function to enhance glycolytic rates through several possible mechanisms, including concentrating metabolites, altering enzyme reaction rates or directly supporting substrate channeling, whereby sequential enzymes associate with one another so that they can directly transfer the product of the first reaction to the second enzyme as its substrate. Enzyme reaction rates can vary substantially for glycolysis enzymes, especially PFK, which is subject to both activating and inhibitory allosteric regulation (Locasale, 2018). Formation of large complexes can affect metabolic enzyme activity. For instance, acetyl-CoA carboxylase can increase its reaction rates when forming filaments, whereas yeast glucokinase is strongly inhibited and held in an inactive conformation in filaments (Kleinschmidt et al., 1969; Stoddard et al., 2020; Park and Horton, 2019). Similarly, enzymes in condensates may adopt conformations that either favor or disfavor their activity. Alternatively, substrates may partition differentially in the cytosol and condensate phase. In vitro, dextranase partitioned in a multi-phase droplet system has increased activity due to the concentration of substrate in another phase, thereby relieving substrate inhibition (Kojima and Takayama, 2018). Moreover, pyrenoids, liquid condensates of RuBisCo involved in carbon fixation in green algae, enhance reaction rates and oppose a futile reverse reaction by concentrating the substrate, CO2, in the condensate (Freeman Rosenzweig et al., 2017; Wunder et al., 2018). Some of the many glycolysis metabolites may become concentrated in granules, or inhibitors such as ATP may be excluded, achieving similar results. One final possibility is that glycolysis enzymes may form a metabolon that supports substrate channeling (Srere, 1987). This limits diffusion and potential utilization by other cellular processes, and can enhance reaction rates for enzymes with extremely high turnover rates or low concentrations of substrate (Sweetlove and Fernie, 2018). Indeed, the low substrate availability after the extended periods (>12 h) of hypoxia that are required for G body formation in yeast may necessitate substrate channeling (Jin et al., 2017). Another condensate, the purinosome, which forms in response to purine depletion in mammalian cells and is comprised of purine biosynthesis enzymes, may function through substrate channeling (An et al., 2008). Through a combination of isotope labeling experiments and a novel in situ mass spectrometry approach, purinosomes have recently been shown to concentrate purine biosynthesis intermediates in vivo and support substrate channeling (Pareek et al., 2020). Similar methods may be applicable to studying condensates of glycolysis enzymes and reveal functional mechanisms.
Other forms of compartmentalization of glycolysis enzymes
In metazoans, several glycolysis enzymes colocalize in muscle tissue (Clarke and Masters, 1975; Menard et al., 2014). A striking example of the importance of this association has been observed in D. melanogaster flight muscle (Fig. 6A). GAPDH, aldolase, PGK, PGM, TPI and glycerol phosphate dehydrogenase (GPDH) all localize to muscle fibers in flight muscle tissue (Wojtas et al., 1997; Sullivan et al., 2003). Mutants lacking the muscle-specific isoform of GPDH fail to recruit the other enzymes to sarcomeres (Wojtas et al., 1997; Sullivan et al., 2003). Expression of a separate GPDH isoform fails to rescue sarcomere recruitment of glycolysis enzymes in the absence of the muscle isoform and results in a loss of the ability to fly, indicating that recruitment of these enzymes to sarcomeres promotes muscle function (Wojtas et al., 1997). In contrast, disruption of most of the individual glycolysis enzymes by P-element excision leads to only a small or no change in wing beat frequency, a measure of the function of flight muscle, while GPDH disruption decreases wing beat frequency (Eanes et al., 2006; Merritt et al., 2006). The association of these enzymes with actin may aid their recruitment, but is unlikely to scaffold these complexes due to the lack of actin in M-lines and Z-lines (Waingeh et al., 2006; Lange et al., 2020). Alternatively, GPDH may directly recruit additional enzymes and itself interact with proteins at M- and Z-lines, although this possibility has not yet been reported. Several pairs of sequential pathway enzymes, including TPI and GAPDH, and PGK and PGM, localize to M-lines and Z-lines of sarcomeres (Fig. 6A), although there is no direct evidence of substrate channeling, which would require in vitro measurements of activity of Z-line and M-line-associated glycolysis enzymes (Sullivan et al., 2003). Thus, concentration of these enzymes may power flight muscle by converting a local high-concentration pool of metabolites into usable energy or by directly interacting with flight muscle at a low copy number. Alternatively, flight muscle glycolysis enzymes may facilitate mitochondrial activity. Analysis of additional enzyme localization, studies of mutants with impaired localization but not activity, and reconstitution of enzyme activity in this muscle tissue system would clarify function and test whether M-line association of glycolysis enzymes supports substrate channeling and constitutes a metabolon.
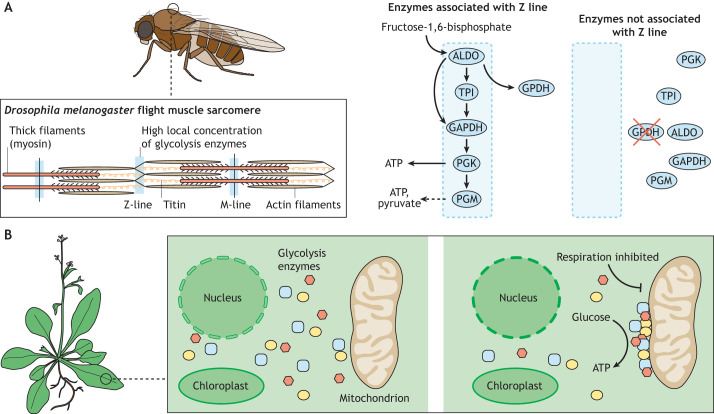
Fig. 6.
Non-condensate compartmentalization in plants and insects. (A) Colocalization of glycolysis enzymes in D. melanogaster flight muscle fibers. M-lines and Z-lines associate with glycolysis enzymes (indicated with blue). Glycolysis enzymes associated with Z-lines are shown on the right. In the presence of GPDH, multiple sequential glycolysis enzymes associate with the Z-line, allowing them to convert fructose-1,6-bisphosphate to ATP and pyruvate. Deletion of muscle GPDH disrupts localization of the other glycolysis enzymes, resulting in flight defects. (B) Substrate channeling on the mitochondrial membrane in plants. In Arabidopsis thaliana, glycolysis enzymes (yellow, blue and purple shapes) are diffusely localized in the cytosol. Inhibition of respiration triggers the accumulation of each glycolysis enzyme on mitochondrial membranes. These complexes promote substrate channeling through the glycolytic pathway, efficiently converting glucose into usable ATP.
In addition, glycolysis enzymes have been found to directly interact with mitochondria in a number of circumstances. Mitochondria in yeast co-purify with glycolysis enzymes, including HXK (Morgenstern et al., 2017). Moreover, in plants such as Arabidopsis thaliana, functional glycolysis enzymes both associate with the mitochondria and reside in the cytosol (Fig. 6B) (Giegé et al., 2003). Mitochondrial association of glycolysis enzymes has been linked to mitochondria–chloroplast interactions (Zhang et al., 2020). Activity of all glycolysis enzymes is detected in mitochondrial fractions. The amount of mitochondria-associated glycolytic activity increases when respiration is inhibited – especially for downstream enzymes – indicating that this localization may function to enhance energy production when respiration is limited (Graham et al., 2007). In support of this hypothesis, isotope labeling experiments measuring activity of glycolysis enzymes associated with mitochondria indicate that the glycolysis enzymes on mitochondria can support direct substrate channeling (Fig. 6B) (Graham et al., 2007; Zhang et al., 2020). Dilution with unlabeled glycolysis intermediates led to either no decrease or less than expected decreases in the rate of accumulation of 13C-labeled glycolysis intermediates derived from [13C]glucose (or [13C]F16BP), indicating that metabolites were efficiently channeled through sequential reactions and not outcompeted by freely diffusing unlabeled intermediates (Graham et al., 2007). The downstream reactions converting 2-phosphoglycerate (2PG) into phosphoenolpyruvate (PEP) then pyruvate also appear to be channeled using the same approach (Zhang et al., 2020). These sequential downstream glycolysis enzymes – PGM, enolase and PYK – form a specific complex on mitochondria and achieve a 20-fold increase in catalytic efficiency (Zhang et al., 2020). Thus, glycolysis enzymes can support substrate channeling when concentrated on mitochondrial membranes.
The glycolysis enzymes are also able to associate with the plasma membrane in erythrocytes, a cell type solely reliant on glycolysis for energy production, in both mouse and humans (Campenella et al., 2005, 2008; Puchulu-Campenella et al., 2013). PFK, aldolase, GAPDH, PYK and lactate dehydrogenase all associate on the membrane only in normoxic conditions, unlike in G bodies and C. elegans neuronal condensates (Campenella et al., 2005). These enzymes are recruited through interactions with the membrane protein, band 3 (Campenella et al., 2005; Puchulu-Campenella et al., 2013).
Finally, glycolysis enzymes are organized in membrane-bound compartments related to peroxisomes, called glycosomes, in a variety of organisms, including Trypanosomes (Quiñones et al., 2020). These structures have been extensively reviewed (Michels et al., 2005, 2021; Haanstra et al., 2016) and require transport in membrane-bound compartments rather than relying on protein–protein or RNA–protein interactions to form and are beyond the scope of this Review.
Outlook and future directions
Evidence is mounting that glycolysis enzymes associate with one another in certain contexts. Compartmentalization is often induced by environmental stimuli requiring increased energy output, such as hypoxia or in the presence of respiratory chain inhibitors (Graham et al., 2007; Miura et al., 2013; Jang et al., 2016; Jin et al., 2017; Fuller et al., 2020; Yoshimura et al., 2021). In other cases, compartmentalization is constitutive in tissues with high energy demands, although it may also relate to disease states in cancer cell lines (Wojtas et al., 1997; Kohnhorst et al., 2017). However, our understanding of the function and pervasiveness of this phenomenon is hampered by a dearth of examples, all from widely different systems, and a reliance on correlative data. The mechanisms governing formation of these structures may widely vary. While condensate formation by phase separation appears to be responsible for yeast G bodies and C. elegans neuronal granules, phase separation has not been tested as an organizing process for other glycolysis enzyme structures. Formation of soluble, multienzyme complexes could occur without relying on phase separation. Even membrane-bound compartments of glycolysis enzymes can form (i.e. the glycosome), demonstrating a variety of strategies to concentrate glycolysis enzymes.
Even in yeast, we are only beginning to understand the pathways regulating glycolytic compartmentalization. Future studies in yeast and other organisms will be required to define the genetics of G body formation and function. Furthermore, there is evidence that composition within granules changes in various environments, although the underlying genetics and functional significance of these changes is unclear (Kohnhorst et al., 2017; Jin et al., 2017). Furthermore, our understanding of the function of these condensates is minimal. With a few notable exceptions (Graham et al., 2007), direct substrate channeling has not been observed and understanding the function and mechanism of the condensates will require biochemical reconstitution of glycolytic activity in glycolytic granules or adaptation of novel methods to observe metabolites and glycolysis enzyme activity in vivo (Pareek et al., 2020). Finally, the observation that glycolysis enzymes form granules in cancer cells raises the possibility that compartmentalization of glycolysis enzymes is related to cancer progression and promotes Warburg metabolism. Exploration of this intriguing hypothesis is important and will be aided by a better understanding of genetic regulation in other systems to facilitate physiological studies in vitro and in vivo.
Acknowledgements
The authors thank Amelia Alessi, Mindy Clark and Margaret Starostik of the Kim laboratory for helpful comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our work is supported by a grant from the National Institutes of Health (NIH) R01 GM129301. Deposited in PMC for release after 12 months.
References
- Al Tameemi, W., Dale, T. P., Al-Jumaily, R. M. K. and Forsyth, N. R. (2019). Hypoxia-modified cancer cell metabolism. Front. Cell Dev. Biol. 7, 4. 10.3389/fcell.2019.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albihlal, W. S. and Gerber, A. P. (2018). Unconventional RNA–binding proteins: an uncharted zone in RNA biology. FEBS Lett. 592, 2917-2931. 10.1002/1873-3468.13161 [DOI] [PubMed] [Google Scholar]
- An, S., Kumar, R., Sheets, E. D. and Benkovic, S. J. (2008). Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103-106. 10.1126/science.1152241 [DOI] [PubMed] [Google Scholar]
- Araiza-Olivera, D., Chiquete-Felix, N., Uribe-Carvajal, S., Sampedro, J. G., Peña, A., Mujica, A. and Uribe-Carvajal, S. (2013). In Saccharomyces cerevisiae a glycolytic metabolon is stabilized by F-Actin. FEBS J. 280, 3887-3905. 10.1111/febs.12387 [DOI] [PubMed] [Google Scholar]
- Banaszak, K., Mechin, I., Obmolova, G., Oldham, M., Chang, S. H., Ruiz, T., Radermacher, M., Kopperschläger, G. and Rypniewski, W. (2011). The crystal structures of eukaryotic phosphofructokinases from baker's yeast and rabbit skeletal muscle. J. Mol. Biol. 407, 284-297. 10.1016/j.jmb.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Beckmann, B. M., Horos, R., Fischer, B., Castello, A., Eichelbaum, K., Alleaume, A.-M., Schwarzl, T., Curk, T., Foehr, S., Huber, W.et al. (2015). The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 6, 10127. 10.1038/ncomms10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi, B., Lorenzo Gotor, N., Dhar, R., Cirillo, D., Baldrighi, M., Tartaglia, G. G. and Lehner, B. (2016). A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Reports 16, 222-231. 10.1016/j.celrep.2016.05.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne, C. P., Mitchison, T. J. and Hyman, A. A. (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334-4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenella, M. E., Chu, H. and Low, P. (2005). Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl Acad. Sci. USA 102, 2402-2407. 10.1073/pnas.0409741102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenella, M. E., Chu, H., Wandersee, N. J., Peters, L. L., Mohandas, N., Gilligan, D. M. and Low, P. S. (2008). Characterization of glycolytic enzyme interactions with murine erythrocyte membranes in wild-type and membrane protein knockout mice. Blood 112, 3900-3906. 10.1182/blood-2008-03-146159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., Davey, N. E., Humphreys, D. T., Preiss, T., Steinmetz, L. M.et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393-1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Clarke, F. M. and Masters, C. J. (1975). On the association of glycolytic enzymes with structural proteins of skeletal muscle. Biochim. Biophys. Acta 381, 37-46. 10.1016/0304-4165(75)90187-7 [DOI] [PubMed] [Google Scholar]
- Das, S., Srikanth, M. and Kessler, J. A. (2008). Cancer stem cells and glioma. Nat. Clin. Pract. Neurol. 4, 427-435. 10.1038/ncpneuro0862 [DOI] [PubMed] [Google Scholar]
- Despic, V., Dejung, M., Gu, M., Krishnan, J., Zhang, J., Herzel, L., Straube, K., Gerstein, M. B., Butter, F. and Neugebauer, K. M. (2017). Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res. 27, 1184-1194. 10.1101/gr.215954.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutagaci, B., Nawrocki, G., Goodluck, J., Ashkarran, A. A., Hoogstraten, C. G., Lapidus, L. J. and Feig, M. (2021). Charge-driven condensation of RNA and proteins suggests broad role of phase separation in cytoplasmic environments. eLife 10, e64004. 10.7554/eLife.64004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes, W. F., Merritt, T. J. S., Flowers, J. M., Kumagai, S., Sezgin, E. and Zhu, C.-T. (2006). Flux control and excess capacity in the enzymes of glycolysis and their relationship to flight. Proc. Natl Acad. Sci. USA 103, 19413-19418. 10.1073/pnas.0607095104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeberg, M. A., Han, T., Moresco, J. J., Kong, A., Yang, Y.-C., Lu, Z., Yates, J. R. and Kim, J. K. (2013). Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 14, R13. 10.1186/gb-2013-14-2-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Rosenzweig, E. S., Xu, B., Kuhn Cuellar, L., Martinez-Sanchez, A., Schaffer, M., Strauss, M., Cartwright, H. N., Ronceray, P., Plitzko, J. M., Förster, F.et al. (2017). The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148-162.e19. 10.1016/j.cell.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin, F., Schueder, F., Tiwary, S., Gupta, R., Körner, R., Schlichthaerle, T., Cox, J., Jungmann, R., Hartl, F. U. and Hipp, M. S. (2019). The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342-347. 10.1126/science.aaw9157 [DOI] [PubMed] [Google Scholar]
- Fuller, G. G., Han, T., Freeberg, M. A., Moresco, J. J., Ghanbari Niaki, A., Roach, N. P., Yates, J. R., Myong, S. and Kim, J. K. (2020). RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. eLife 9, e48480. 10.7554/eLife.48480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Jove Navarro, M.,, Kashida, S., Chouaib, R., Souquere, S., Pierron, G., Weil, D. and Gueroui, Z. (2019). RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 10, 3230. 10.1038/s41467-019-11241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, P., Heazlewood, J. L., Roessner-Tunali, U., Millar, A. H., Fernie, A. R., Leaver, C. J. and Sweetlove, L. J. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140-2151. 10.1105/tpc.012500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach, A.,, Dimova, E. Y., Petry, A., Martínez-Ruiz, A., Hernansanz-Agustín, P., Rolo, A. P., Palmeira, C. M. and Kietzmann, T. (2015). Reactive oxygen species, nutrition, hypoxia and diseases: Problems solved? Redox Biol. 6, 372-385. 10.1016/j.redox.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, J. W. A., Williams, T. C. R., Morgan, M., Fernie, A. R., Ratcliffe, R. G. and Sweetlove, L. J. (2007). Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19, 3723-3738. 10.1105/tpc.107.053371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignaschi, E., Cereghetti, G., Grigolato, F., Kopp, M. R. G., Caimi, S., Faltova, L., Saad, S., Peter, M. and Arosio, P. (2018). A hydrophobic low-complexity region regulates aggregation of the yeast pyruvate kinase Cdc19 into amyloid-like aggregates in vitro. J. Biol. Chem. 293, 11424-11432. 10.1074/jbc.RA117.001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanstra, J. R., González-Marcano, E. B., Gualdrón-López, M. and Michels, P. A. M. (2016). Biogenesis, maintenance and dynamics of glycosomes in trypanosomatid parasites. Biochimica Biophysica. Acta 1863, 1038-1048. 10.1016/j.bbamcr.2015.09.015 [DOI] [PubMed] [Google Scholar]
- Huang, R., Han, M., Meng, L. and Chen, X. (2018). Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl Acad. Sci. USA 115, E3879-E3887. 10.1073/pnas.1718406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger, A., Courel, M., Bénard, M., Souquere, S., Ernoult-Lange, M., Chouaib, R., Yi, Z., Morlot, J.-B., Munier, A., Fradet, M.et al. (2017). P-Body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144-157. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Huh, W.-K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. and O'shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686-691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Hyman, A. A., Weber, C. A. and Jülicher, F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39-58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Jain, A. and Vale, R. D. (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243-247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A. and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487-498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S., Nelson, J. C., Bend, E. G., Rodríguez-Laureano, L., Tueros, F. G., Cartagenova, L., Underwood, K., Jorgensen, E. M. and Colón-Ramos, D. A. (2016). Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron 90, 278-291. 10.1016/j.neuron.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S., Xuan, Z., Lagoy, R. C., Jawerth, L. M., Gonzalez, I. J., Singh, M., Prashad, S., Kim, H. S., Patel, A., Albrecht, D. R.et al. (2021). Phosphofructokinase relocalizes into subcellular compartments with liquid-like properties in vivo. Biophys. J. 120, 1170-1186. 10.1016/j.bpj.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., Fuller, G. G., Han, T., Yao, Y., Alessi, A. F., Freeberg, M. A., Roach, N. P., Moresco, J. J., Karnovsky, A., Baba, M.et al. (2017). Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Reports 20, 895-908. 10.1016/j.celrep.2017.06.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R. and Parker, R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808-820.e5. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt, A. K., Moss, J. and Lane, D. M. (1969). Acetyl coenzyme A carboxylase: filamentous nature of the animal enzymes. Science 166, 1276-1278. 10.1126/science.166.3910.1276 [DOI] [PubMed] [Google Scholar]
- Kohnhorst, C. L., Kyoung, M., Jeon, M., Schmitt, D. L., Kennedy, E. L., Ramirez, J., Bracey, S. M., Luu, B. T., Russell, S. J. and An, S. (2017). Identification of a multienzyme complex for glucose metabolism in living cells. J. Biol. Chem. 292, 9191-9203. 10.1074/jbc.M117.783050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, T. and Takayama, S. (2018). Membraneless compartmentalization facilitates enzymatic cascade reactions and reduces substrate inhibition. ACS Appl. Mater. Interfaces 10, 32782-32791. 10.1021/acsami.8b07573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald, S., Maharana, S., Mateju, D., Malinovska, L., Nüske, E., Poser, I., Richter, D. and Alberti, S. (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807. 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon, E. M., Qiu, Y., Ghanbari Niaki, A., Mclaughlin, G. A., Weidmann, C. A., Gerbich, T. M., Smith, J. A., Crutchley, J. M., Termini, C. M., Weeks, K. M.et al. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922-927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, S., Pinotsis, N., Agarkova, I. and Ehler, E. (2020). The M-band: the underestimated part of the sarcomere. Biochim. Biophys. Acta 1867, 118440. 10.1016/j.bbamcr.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale, J. W. (2018). New concepts in feedback regulation of glucose metabolism. Curr. Opin. Syst. Biol. 8, 32-38. 10.1016/j.coisb.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju, D., Franzmann, T. M., Patel, A., Kopach, A., Boczek, E. E., Maharana, S., Lee, H. O., Carra, S., Hyman, A. A. and Alberti, S. (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669-1687. 10.15252/embj.201695957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matia-González, A. M., Laing, E. E. and Gerber, A. P. (2015). Conserved mRNA-binding proteomes in eukaryotic organisms. Nat. Struct. Mol. Biol. 22, 1027-1033. 10.1038/nsmb.3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard, L., Maughan, D. and Vigoreaux, J. (2014). The structural and functional coordination of glycolytic enzymes in muscle: Evidence of a metabolon? Biology 3, 623-644. 10.3390/biology3030623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, T., Sezgin, E., Zhu, C.-T. and Eanes, W. F. (2006). Triglyceride pools, flight and activity variation at the Gpdh locus in Drosophila melanogaster. Genetics 172, 293-304. 10.1534/genetics.105.047035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels, P. A. M., Moyersoen, J., Krazy, H., Galland, N., Herman, M. and Hannaert, V. (2005). Peroxisomes, glyoxysomes and glycosomes (Review). Mol. Membr. Biol. 22, 133-145. 10.1080/09687860400024186 [DOI] [PubMed] [Google Scholar]
- Michels, P. A. M., Villafraz, O., Pineda, E., Alencar, M. B., Cáceres, A. J., Silber, A. M. and Bringaud, F. (2021). Carbohydrate metabolism in trypanosomatids: new insights revealing novel complexity, diversity and species-unique features. Exp. Parasitol. 224, 108102. 10.1016/j.exppara.2021.108102 [DOI] [PubMed] [Google Scholar]
- Miura, N., Shinohara, M., Tatsukami, Y., Sato, Y., Morisaka, H., Kuroda, K. and Ueda, M. (2013). Spatial reorganization of Saccharomyces cerevisiae enolase to alter carbon metabolism under hypoxia. Eukaryot. Cell 12, 1106-1119. 10.1128/EC.00093-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, M., Stiller, S. B., Lübbert, P., Peikert, C. D., Dannenmaier, S., Drepper, F., Weill, U., Höß, P., Feuerstein, R., Gebert, M.et al. (2017). Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Reports 19, 2836-2852. 10.1016/j.celrep.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, T., Qamar, S., Lin, J. Q., Schierle, G. S. K., Rees, E., Miyashita, A., Costa, A. R., Dodd, R. B., Chan, F. T. S., Michel, C. H.et al. (2015). ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678-690. 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree, C., Begovich, K., Samilo, D., Broyer, R., Monfort, E. and Blanchoin, L. (2019). A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol. Biol. Cell 30, 2721-2736. 10.1091/mbc.E19-04-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek, V., Tian, H., Winograd, N. and Benkovic, S. J. (2020). Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 368, 283-290. 10.1126/science.aaz6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C. K. and Horton, N. C. (2019). Structures, functions, and mechanisms of filament forming enzymes: a renaissance of enzyme filamentation. Biophys. Rev. 11, 927-994. 10.1007/s12551-019-00602-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., Stoynov, S., Mahamid, J., Saha, S., Franzmann, T. M.et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066-1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Petrova, V., Annicchiarico-Petruzzelli, M., Melino, G. and Amelio, I. (2018). The hypoxic tumour microenvironment. Oncogenesis 7, 10. 10.1038/s41389-017-0011-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, M., Newport, E. and Morton, K. J. (2016). The Warburg effect: 80 years on. Biochem. Soc. Trans. 44, 1499-1505. 10.1042/BST20160094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter, D. S. W., Rao, B. S., Van Treeck, B., Lin, Y., Mizoue, L., Rosen, M. K. and Parker, R. (2018). Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Reports 22, 1401-1412. 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchulu-Campenella, E., Chu, H., Anstee, D. J., Galan, J. A., Tao, W. A. and Low, P. S. (2013). Identification of the components of a glycolytic enzyme metabolon on the human red blood cell membrane. J. Biol. Chem. 288, 848-858. 10.1074/jbc.M112.428573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam, A., Cassani, M., Smith, J. and Seydoux, G. (2019). A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 26, 220-226. 10.1038/s41594-019-0193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones, W., Acosta, H., Gonçalves, C. S., Motta, M. C. M., Gualdrón-López, M. and Michels, P. A. M. (2020). Structure, properties, and function of glycosomes in Trypanosoma cruzi. Front. Cell Infect. Microbiol. 10, 25. 10.3389/fcimb.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe, S., Prakash, A. and Tyedmers, J. (2018). The insoluble protein deposit (IPOD) in yeast. Front. Mol. Neurosci. 11, 237. 10.3389/fnmol.2018.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad, S., Cereghetti, G., Feng, Y., Picotti, P., Peter, M. and Dechant, R. (2017). Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat. Cell Biol. 19, 1202-1213. 10.1038/ncb3600 [DOI] [PubMed] [Google Scholar]
- Schöneberg, T., Kloos, M., Brüser, A., Kirchberger, J. and Sträter, N. (2013). N. Structure and allosteric regulation of eukaryotic 6-phosphofructokinases. Biol. Chem. 394, 977-993. 10.1515/hsz-2013-0130 [DOI] [PubMed] [Google Scholar]
- Shchepachev, V., Bresson, S., Spanos, C., Petfalski, E., Fischer, L., Rappsilber, J. and Tollervey, D. (2019). Defining the RNA interactome by total RNA-associated protein purification. Mol. Syst. Biol. 15, e8689. 10.15252/msb.20188689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.-J., Kassim, H., Huang, Y., Li, H., Zhang, J., Li, G., Wang, P.-Y., Yan, J., Ye, F. and Liu, J.-L. (2016). Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J. Genet. Genomics 43, 393-404. 10.1016/j.jgg.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere, P. A. (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89-124. 10.1146/annurev.bi.56.070187.000513 [DOI] [PubMed] [Google Scholar]
- Stoddard, P. R., Lynch, E. M., Farrell, D. P., Dosey, A. M., Dimaio, F., Williams, T. A., Kollman, J. M., Murray, A. W. and Garner, E. C. (2020). Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science 367, 1039-1042. 10.1126/science.aay5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträter, N., Marek, S., Kuettner, E. B., Kloos, M., Keim, A., Brüser, A., Kirchberger, J. and Schöneberg, T. (2011). Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases. FASEB J. 25, 89-98. 10.1096/fj.10-163865 [DOI] [PubMed] [Google Scholar]
- Sullivan, D. T., Macintyre, R., Fuda, N., Fiori, J., Barrilla, J. and Ramizel, L. (2003). Analysis of glycolytic enzyme co-localization in Drosophila flight muscle. J. Exp. Biol. 206, 2031-2038. 10.1242/jeb.00367 [DOI] [PubMed] [Google Scholar]
- Sweetlove, L. J. and Fernie, A. R. (2018). The role of dynamic enzyme assemblies and substrate channeling in metabolic regulation. Nat. Commun. 9, 2136. 10.1038/s41467-018-04543-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoev, V. O., Fischer, B., Frese, C. K., Gupta, I., Krijgsveld, J., Hentze, M. W., Castello, A. and Ephrussi, A. (2016). Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat. Commun. 7, 12128. 10.1038/ncomms12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek, T., Grosch, M., York, A., Shroff, H., Lionnet, T. and Lehmann, R. (2015). Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 6, 7962. 10.1038/ncomms8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek, T., Douglas, T. E., Grosch, M., Yin, Y., Eagle, W. V. I., Gavis, E. R., Shroff, H., Rothenberg, E. and Lehmann, R. (2020). Sequence-independent self-assembly of germ granule mRNAs into homotypic clusters. Mol. Cell 78, 941-950. 10.1016/j.molcel.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck, B., Protter, D. S. W., Matheny, T., Khong, A., Link, C. D. and Parker, R. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 115, 2734-2739. 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waingeh, V. F., Gustafson, C. D., Kozliak, E. I., Lowe, S. L., Knull, H. R. and Thomasson, K. A. (2006). Glycolytic enzyme interactions with yeast and skeletal muscle F-Actin. Biophys. J. 90, 1371-1384. 10.1529/biophysj.105.070052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, E. W. J., Kear-Scott, J. L., Pilipenko, E. V., Schwartz, M. H., Laskowski, P. R., Rojek, A. E., Katanski, C. D., Riback, J. A., Dion, M. F., Franks, A. M.et al. (2015). Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286-1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, R. W., Muhlrad, D., Garcia, J. and Parker, R. (2015). Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 21, 1660-1671. 10.1261/rna.053116.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, B. A., Dosey, A. M., Wittmann, T., Kollman, J. M. and Barber, D. L. (2017). The glycolytic enzyme phosphofructokinase-1 assembles into filaments. J. Cell Biol. 216, 2305-2313. 10.1083/jcb.201701084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas, K., Slepecky, N., Von Kalm, L. and Sullivan, D. (1997). Flight muscle function in Drosophila requires colocalization of glycolytic enzymes. Mol. Biol. Cell 8, 1665-1675. 10.1091/mbc.8.9.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder, T., Cheng, S. L. H., Lai, S.-K., Li, H.-Y. and Mueller-Cajar, O. (2018). The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 9, 5076. 10.1038/s41467-018-07624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, Y., Hirayama, R., Miura, N., Utsumi, R., Kuroda, K., Ueda, M. and Kataoka, M. (2021). Small-scale hypoxic cultures for monitoring the spatial reorganization of glycolytic enzymes in Saccharomyces cerevisiae. Cell Biol. Int. 45, 1776-1783. 10.1002/cbin.11617 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Elbaum-Garfinkle, S., Langdon, E. M., Taylor, N., Occhipinti, P., Bridges, A. A., Brangwynne, C. P. and Gladfelter, A. S. (2015). RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220-230. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Sampathkumar, A., Kerber, S. M.-L., Swart, C., Hille, C., Seerangan, K., Graf, A., Sweetlove, L. and Fernie, A. R. (2020). A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat. Commun. 11, 4509. 10.1038/s41467-020-18234-w [DOI] [PMC free article] [PubMed] [Google Scholar]