Abstract
Introduction
Disease recurrence is a major concern in patients with localized prostate cancer (PCa) following treatment with radiotherapy (RT), and few studies have evaluated the clinical relevance of microRNAs (miRNAs) prior and post-RT.
Purpose
We aimed to investigate the significance of miRNAs in the outcomes of prostate cancer patients undergoing radiotherapy and to identify the related pathways through bioinformatics analysis.
Materials and Methods
The expression levels of miR-21, miR-106b, miR-141 and miR-375 involved in the response to radiotherapy were assessed by RT-qPCR in the serum of PCa patients (n=56) prior- and post-RT.
Results
Low expression levels of miR-106b prior-RT were associated with extracapsular extension and seminal vesicles invasion by the tumor (p=0.031 and 0.044, respectively). In the high-risk subgroup (n=47), post-RT expression levels of miR-21 were higher in patients with biochemical relapse (BR) compared to non-relapse (p=0.043). Also, in the salvage treatment subgroup (post-operative BR; n=20), post-RT expression levels of miR-21 and miR-106b were higher in patients with BR compared to non-relapse (p=0.043 and p=0.032, respectively). In the whole group of patients, high expression levels of miR-21 prior-RT and of miR-106b post-RT were associated with significantly shorter overall survival (OS; p=0.049 and p=0.050, respectively). No associations were observed among miR-141 and miR-375 expression levels with clinicopathological features or treatment outcome. Bioinformatics analysis revealed significant enrichment in DNA damage response pathways.
Conclusion
Circulating miRNAs prior or post-RT may hold prognostic implications in patients with PCa.
Keywords: prostate cancer, radiotherapy, salvage radiotherapy, circulating microRNAs, high risk, biochemical relapse
Introduction
Prostate cancer is the most commonly diagnosed cancer and a leading cause of cancer-related death in men worldwide.1 RT is a standard treatment option along with surgery, hormone therapy and chemotherapy.2 However, the efficacy of RT is impeded by inherent resistance of tumor cells.3
RT utilizes targeted ionizing radiation to induce DNA damage either directly or indirectly through reactive oxygen species (ROS).4 DNA damage response (DDR) pathways are activated to repair the damage or to induce arrest of cell-cycle progression or apoptotic cell death.5 Radioresistance mechanisms to RT-induced cell death or apoptosis include DNA repair,6 impaired apoptotic signaling after DNA damage,7 elevation of antioxidant enzymes,8 impaired intracellular signaling pathways such as Heat Shock Protein 90 (HSP90)9 and activation of epithelial to mesenchymal transition (EMT) pathways.10
Despite the progress in emerging biomarkers, such as PCA3 score and Prostate Health Index11 that improve the specificity of prostate-specific antigen (PSA) and provide useful information about disease aggressiveness, the results are otherwise inconclusive.12 In addition, there is a paucity of molecular biomarkers for the prediction of relapse in patients with localized prostate cancer receiving definitive RT as a single treatment modality or in the salvage setting.
MicroRNAs (miRNAs) are small non-coding RNAs (18–23nt) that regulate a plethora of physiological and pathological procedures.13 MiRNAs exert their function by regulating gene expression at post-transcriptional level.14 MiRNAs’ expression is deregulated in human malignancies and could either act as oncomirs or tumor suppressors, depending on the context.15 MiRNAs are significantly stable in tissues and in biological fluids such as serum and plasma.16 In the last few years, miRNAs have been proposed as promising biomarkers for cancer diagnosis, prognosis, and prediction of treatment outcome17 and have been recognized as liquid biopsy biomarkers.
Mounting evidence suggests that miRNAs expression is modulated after irradiation.18 Consequently, miRNAs regulate components involved in DDR machinery.18 MiR-21, miR-106b, miR-141 and miR-375, among others, have been shown to associate with the pathogenesis and prognosis of prostate cancer.19–22 Moreover, their expression is modulated post irradiation in prostate cancer cells.23 In addition, miR-141 and miR-106b have been proposed as potential prognostic biomarkers as they are differentially expressed in prostate cancer cell lines of different radiosensitivity.24 Mir-21 has also been demonstrated to mediate radioresistance25 as well as it was overexpressed in hypoxic conditions in prostate cancer cells,26 leading to more increased aggressiveness. However, the results from miRNA profiling studies investigating the prediction of biochemical recurrence after radical prostatectomy in patients with PCa, vary among studies.27 Furthermore, there are limited results regarding the role of miRNAs in the prediction of post-surgery RT outcomes27 or development of distant metastasis.28
In the present study, we investigated the role of circulating miRNAs as possible non-invasive biomarkers for monitoring patients undergoing RT. We assessed the serum miR-21, miR-106b, miR-141 and miR-375 expression levels prior- and post-RT in PCa patients undergoing RT and evaluated their significance in patients’ outcomes. We also performed a bioinformatic analysis to identify the key mRNA targets as well as the molecular pathways involved by the miRs of our study.
Materials and Methods
Patients and Sample Collection
In the current study, clinical and molecular data from 56 patients with histologically confirmed localized prostate adenocarcinoma who received definitive curative RT in the University Hospital of Heraklion, from January 2013 until July 2016, were prospectively collected. Patients were treated with 3-D conformal RT, which was the standard approach during that period in our department. RT was given as primary, adjuvant or salvage treatment. Radiation doses ranged from 64.8 Gy to 71 Gy, personalized per patient. Post radical prostatectomy (RP) biochemical relapse (BR) was defined as two consecutive rising PSA values ≥0.2 ng/mL.29 The BR post definitive RT was defined as a rise in PSA levels >2 ng/mL compared to its nadir value.30 In addition, patients were categorized based on their risk of relapse for localized prostate cancer according to ESMO guidelines31 and by D’Amico et al.32 More specifically, patients were categorized into three prognostic subgroups based on the following characteristics: 1) low risk: T1a-T2a, Gleason score=6, PSA<10 ng/mL; 2) intermediate risk: T2b-T2c, Gleason score=7, PSA=10–20 ng/mL; and 3) high risk: T≥T3a or Gleason score=8–10 or PSA>20 ng/mL.
Relapse-free survival (RFS) was defined as the time duration from the start of RT until BR or death. Overall survival (OS) was defined as the time duration from the start of RT until the time of death. Patients who had not progressed or they were alive at the time of data collection were censored for RFS or OS at the time of last follow-up, respectively.
Two peripheral whole blood samples of 3 mL each were collected from each patient by experienced nurses, with all precautions to avoid hemolysis. The first sample was taken just before the first and the second sample just after the last RT session, into tubes without anticoagulant. Coagulated samples were centrifuged at 3000 rpm for 10 min. Serum was carefully collected in 1.5 mL RNase-free tubes and immediately stored at −80°C until further process. Thawed frozen sera were centrifuged for 3 min at 11,000 g to remove residual cells and debris.
Approval Committee
This study was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the Human Ethics Committee of the University Hospital of Heraklion (ID: 16363). Written informed consent was obtained from all participants.
RNA Isolation
Serum samples were subsequently spiked-in with known quantity of Caenorhabditis elegans microRNA, cel-mir-39 (miRNeasy Serum/Plasma spike-in control, Qiagen) as a control of extraction and amplification procedures. Specifically, serum samples of 300μL were spiked with 8.8×108 copies of cel-mir-39 microRNA and small RNAs were isolated by the NucleoSpin® miRNA Plasma Kit (Macherey-Nagel) according to manufacturer’s instructions.
Quantitative Real-Time PCR Analysis and miRNA Expression
TaqMan technology (Applied Biosystems) was used for reverse transcription and quantitative real-time polymerase chain reaction (RT-qPCR) reactions, according to manufacturer’s instructions. In brief, 2 μL of RNA input were used for cDNA synthesis in 10 μL reactions, including 2 μL of stem loop-specific primer for each miRNA, 0.1 μL dNTPs, 0.75μL reverse transcriptase, 1 μL 10x reaction buffer, 0.13 μL RNase inhibitor and 4.02 μL ddH2O. cDNAs were subsequently diluted threefold and 2 μL of diluted cDNA were used in 10 μL RT-qPCRusing TaqMan Universal Master mix II.no UNG including 0.5 μL primer, 5 μL 2× reaction mix and 2.5 μL ddH2O. All the assays were performed in triplicates. Appropriate negative controls were used in both cDNA synthesis and RT-qPCRs, where RNA input was replaced by H2O and no template control was used, respectively. TaqManTM Assay Ids of the examined microRNAs are depicted in Table S1. Relative changes in miRNA abundance in samples after RT were calculated using 2−ΔΔCt method using as reference cel-mir-39, using the following equation:
Relative change =2–[(Ct_miR#-Ct_cel-mir−39)before-(Ct_miR#-Ct_cel-mir−39)after]33
For the purposes of our analysis, patients were divided into two groups: 1) patients with relative change values of ≥1 are characterized by higher expression of each miRNA prior RT and 2) patients with relative change values of <1 are characterized by higher expression of each miRNA post RT.
Statistical Analysis
The statistical analysis was performed using the SPSS software package, version 22.0 (SPSS Inc. Chicago, IL). Descriptive statistics were applied to define and describe nominal and categorical values. Correlations of expression between the different miRNAs and were performed by Spearmans’ test. Spearmans’ correlation coefficiency was also applied to examine any potential correlation with miRNA levels expression levels before RT with PSA levels prior to RT. The chi-squared test was used to estimate potential associations between miRNA expression and clinicopathological characteristics. Mann–Whitney U-test was applied to examine the miRNA levels as continuous variables with the rates of biochemical recurrence post-RT and clinicopathological features. The associations between circulating miRNA expression levels and RFS or OS were assessed by Kaplan–Meier method, log rank test (Mantel–Cox) and Cox proportional hazard regression models. Patients were divided into high and low expression according to the median value for each miRNA expression. Patients with miRNA expression above or equal to the median values were characterized as having high, whereas patients with miRNA expression below the median as having low expression. Statistical significance was set at p≤0.05.
miRNA Gene Target and Pathway Enrichment Analysis
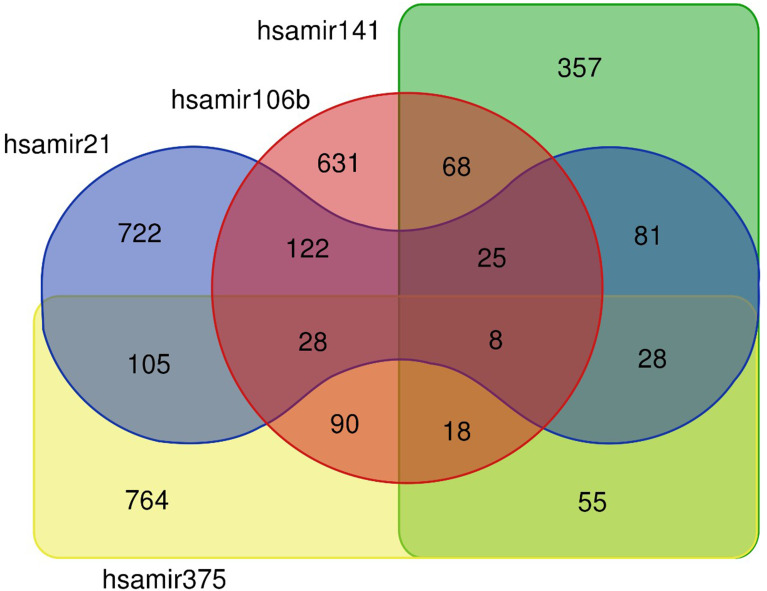
TarBase was used to retrieve the protein coding gene targets for each of the investigated miRNA.34 For each miRNA, we obtained the full set of human protein coding genes that were found associated in cancerous tissues. For all the four miRNAs, we were able to identify more than 1000 protein coding gene targets. Much smaller numbers of protein coding targets were found associated with prostate cancer tissues (181 targets for hsa-miR-106b-5p; 0 targets for hsa-miR-141-3p; 1 target for hsa-miR-21-3p and 1 target for hsa-miR-375) an effect probably associated with the lack of datasets from prostate cancer biopsies. Therefore, we decided to proceed with the larger number of genes associated with cancerous tissues, as mentioned above. Most of the targets were miRNA-specific; however, 628 out of 3048 targets (approximately 20%) were shared by at least two miRNAs (Figure 1). Gene and gene target overlaps were assessed with the use of the Jaccard similarity coefficient, which is given by the ratio of the two sets’ intersection over their union. We then analyzed the subset of 628 genes for particular functional enrichments. gProfiler35 was used to calculate the enrichment significance for Gene Ontology (GO) and for Biological Pathways (BP) as compiled by the Kyoto Encyclopedia of Genes and Genomes (KEGG). Protein–protein interaction (PPI) analysis was performed with the STRING database (STRING-DB; https://string-db.org/).36 Clustering was performed with Markov cluster (MCL) as implemented by STRING-DB.
Figure 1.
Venn diagram depicting common and specific protein-coding targets of the four analyzed miRNAs.
Results
Study Design and Patients’ Characteristics
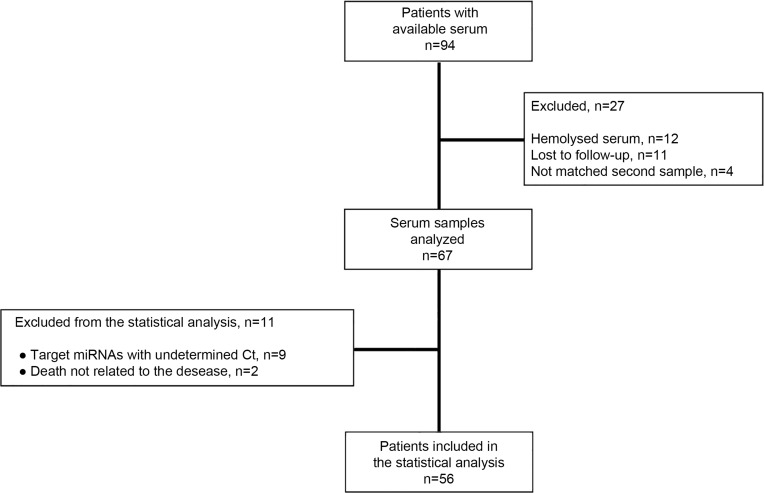
The flow chart of the study and patients’ characteristics are shown in Figure 2 and Table 1, respectively. A total of 56 patients were included in the study. The median age was 71 years (range 51–86 years), median PSA before RT was 0.5 (range 0–45.56), whereas 48.1% of the patients had a Gleason score 8–10 (Table 1). The median biochemical relapse-free survival (RFS) and overall survival (OS) have not yet been reached. After a median follow-up time of 4.8 years, 15 patients (26.8%) developed biochemical relapse (BR) and 9 (16.1%) patients died. PSA values distribution prior RT was significantly higher in patients who experienced biochemical relapse compared to those without relapse (Figure S1; Mann–Whitney U-test, p=0.011).
Figure 2.
Flow chart of the study.
Table 1.
Patients’ Characteristics
| All Patients | ||
|---|---|---|
| Characteristics | N | % |
| Number of patients | 56 | |
| Age (years) | ||
| Median (range) | 71 (51–86) | |
| PSA before RT | ||
| Median (range) | 0.5 (0–45.56) | |
| Gleason score | ||
| 6 | 2 | 3.6 |
| 7 | 27 | 48.2 |
| 8–10 | 27 | 48.2 |
| RT dosage (Gy) | ||
| Mean (range) | 69 (63–72) | |
| Risk groups | ||
| Low | 0 | 0 |
| Intermediate | 9 | 16.1 |
| High | 47 | 83.9 |
| Radical prostatectomy | ||
| No | 17 | 30.4 |
| Yes | 39 | 69.6 |
| Resection | ||
| R0 | 18 | 46.1 |
| R1 | 21 | 53.8 |
| R2 | 0 | 0 |
| Biochemical relapse post-radical prostatectomy | ||
| No | 19 | 48.7 |
| Yes | 20 | 51.3 |
| Reason for RT administration | ||
| Adjuvant post R1 resections | 19 | 33.9 |
| Definitive treatment as single modality | 17 | 30.4 |
| Salvage treatment for rising PSA after surgery | 20 | 35.7 |
| Anti-androgen treatment during RT | ||
| Yes | 37 | 66.1 |
| No | 19 | 33.9 |
| Extra-prostatic extension | ||
| Yes | 31 | 55.3 |
| No | 13 | 23.2 |
| No data | 12 | 21.4 |
| Perineural invasion | ||
| Yes | 24 | 42.8 |
| No | 15 | 26.7 |
| No available data | 17 | 30.3 |
| Characteristics | N | % |
| Seminal vesicles invasion | ||
| Yes | 18 | 32.2 |
| No | 25 | 44.6 |
| No available data | 13 | 23.2 |
| Lymph node infiltration | ||
| Yes | 6 | 10.7 |
| No | 36 | 64.3 |
| No available data | 14 | 25.0 |
| Biochemical relapse post-RT | ||
| Yes | 15 | 26.8 |
| No | 41 | 73.2 |
| Death | ||
| Yes | 9 | 16.1 |
| No | 47 | 83.9 |
Abbreviations: Low risk, Τ1a-T2a, Gleason=6, PSA<10 ng/mL; intermediate risk, T2b-T2c, Gleason=7, PSA 10–20 ng/mL; high risk, Τ≥T3a or Gleason score=8–10 or PSA>20 ng/mL; Gy, Gray.
miRNA Expression and Statistical Correlations
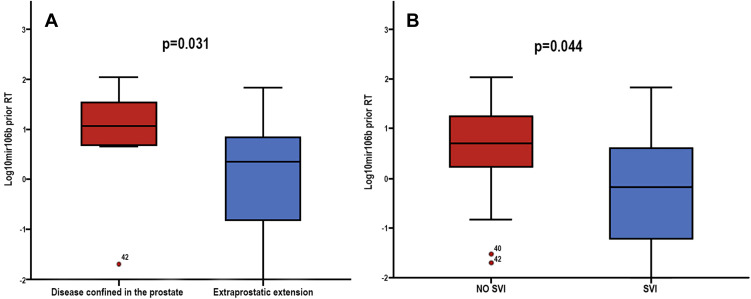
No correlations were observed among miRNAs expression and PSA levels prior RT administration (Figure S2). Low expression levels of miR-106b prior-RT were correlated with extracapsular extension and seminal vesicles invasion (Mann–Whitney U-test, p=0.031 and p=0.044, respectively; Table 2 and Figure 3). No other significant correlations were observed among miRNAs expression prior-RT and clinicopathological characteristics (Mann–Whitney U-test, p>0.05). Wilcoxon test revealed no significant differences among miRNAs expression prior and post-RT (Figure S3). However, significant correlations were observed between expression levels of the different miRNAs prior and post-RT (Table 3). A strong correlation was observed between miR-21 expression and miR-141 and miR-375 before RT (Spearman’s ρ, 0.764; p<0.001 and Spearman’s ρ, 0.699; p<0.001, respectively) and post-RT (Spearman’s ρ, 0.752; p<0.001 and Spearman’s ρ, 0.690; p<0.001, respectively). Furthermore, a strong correlation was observed between miR-141 and miR-375 before RT (Spearman’s ρ, 0.807; p<0.001) and post-RT (Spearman’s ρ, 0.844; p<0.001) (Table 3).
Table 2.
Correlation of miRNA Expression Levels with Clinical Parameters Prior-RT (p Values are Shown)
| Variable | miR-21 | miR-106b | miR-141 | miR-375 |
|---|---|---|---|---|
| Extraprostatic extension | 0.969 | 0.031* | 0.122 | 0.837 |
| PNI | 0.194 | 0.484 | 0.899 | 0.470 |
| SVI | 0.228 | 0.044* | 0.558 | 0.318 |
| LN infiltration | 0.746 | 0.175 | 0.983 | 0.834 |
| BR prior-RT | 0.182 | 0.848 | 0.960 | 0.279 |
| BR post-RT | 0.554 | 0.937 | 0.816 | 0.449 |
Notes: Mann–Whitney U-test, *p≤0.05.
Abbreviations: SVI, seminal vesicle invasion; PNI, perineural invasion; LN, lymph node; BR, biochemical relapse.
Figure 3.
Fold change of serum miR-106b expression levels prior-RT, according to (A) extraprostatic extension and (B) seminal vesicles invasion (SVI). Mann–Whitney U-test was used to determine statistically significant differences, and the results are displayed on box plots. Horizontal line depicts median, whereas the length of the boxes is the interquartile range that represents values between the 75th and 25th percentiles of individual fold change expression values. p Values are shown.
Table 3.
Correlation Coefficients Among the Four miRNAs
| Prior-RT | ||||
|---|---|---|---|---|
| miR-21 | miR-106 | miR-141 | miR-375 | |
| miR-21 | 1 | |||
| miR-106 | 0.414* | 1 | ||
| miR-141 | 0.764** | 0.518** | 1 | |
| miR-375 | 0.699** | 0.404* | 0.807** | 1 |
| Post-RT | ||||
| miR-21 | 1 | |||
| miR-106 | 0.373* | 1 | ||
| miR-141 | 0.752** | 0.414* | 1 | |
| miR-375 | 0.690** | 0.397* | 0.844** | 1 |
Notes: *p<0.01, **p<0.001.
miRNA Expression Levels and Clinical Outcome
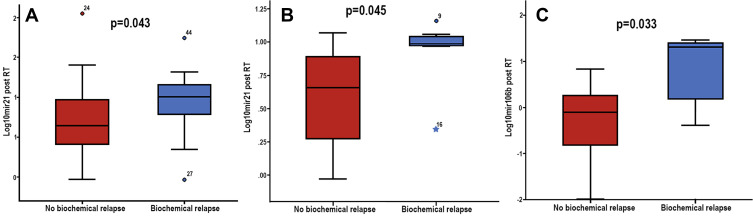
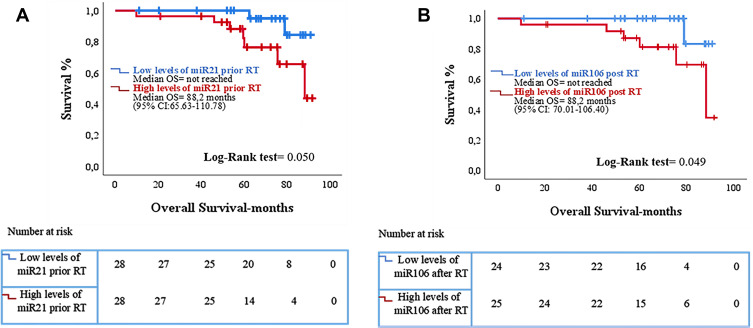
No significant differences were observed between expression levels of the examined miRNAs prior and post-RT and BR in the whole patients’ group. However, in the high-risk subgroup (n=47), patients who experienced BR after RT, had higher post-RT miR-21 expression levels compared to patients without relapse (p=0.043) (Table 4 and Figure 4A). Also, in the subgroup of patients who received RT as salvage treatment (n=20) higher post-RT miR-21 and miR-106 expression levels were observed in patients who experienced BR compared to those without relapse (p=0.043 and p=0.032, respectively) (Table 4 and Figure 4B and C). No associations were observed between miRNA expression and RFS. However, increased expression levels of miR-21 prior-RT and miR-106 post-RT were associated with significantly shorter OS (p=0.049 and p=0.050, respectively; Figure 5). No correlations were observed between the remaining two miRNAs and OS.
Table 4.
Association of miRNAs Expression Levels with Post-RT Biochemical Relapse in the Subgroups of High-Risk (n=47) and Salvage Treatment (n=20) Groups of Patients
| miR-21 | miR-106b | miR-141 | miR-375 | ||||
|---|---|---|---|---|---|---|---|
| Before RT | Post-RT | Before RT | Post-RT | Before RT | Post-RT | Before RT | Post-RT |
| High-risk group (n=47) | |||||||
| 0.334 | 0.043* | 0.841 | 0.444 | 0.678 | 0.119 | 0.279 | 0.147 |
| Salvage treatment group (n=20) | |||||||
| 0.866 | 0.045* | 0.526 | 0.033* | 0.928 | 0.190 | 0.499 | 0.128 |
Notes: Mann–Whitney U-test, *p<0.05.
Abbreviations: High risk, T≥T3a or Gleason score=8–10 or PSA>20 ng/mL; RT, radiotherapy.
Figure 4.
Mann–Whitney U-test demonstrating the differences in the distributions of (A) miR-21 fold change of expression post RT, in the high risk subgroup among those who experienced relapse and those without relapse, (B) mir21 and (C) miR-106b fold change of expression post RT, in the subgroup of salvage-treated among those who experienced biochemical relapse compared to those without relapse.
Figure 5.
Kaplan–Meier analysis for overall survival (OS) according to miRNA expression in the serum of prostate cancer patients (n = 56). Patients were classified into high and low expression groups according to the median value of each miRNA. OS in patients with high or low expression of (A) miR-21 prior-RT and (Β) miR-106 post-RT. Curves were compared using the log rank test. p Values are shown; RT, radiotherapy.
mRNA Target and Pathway Enrichment Analysis
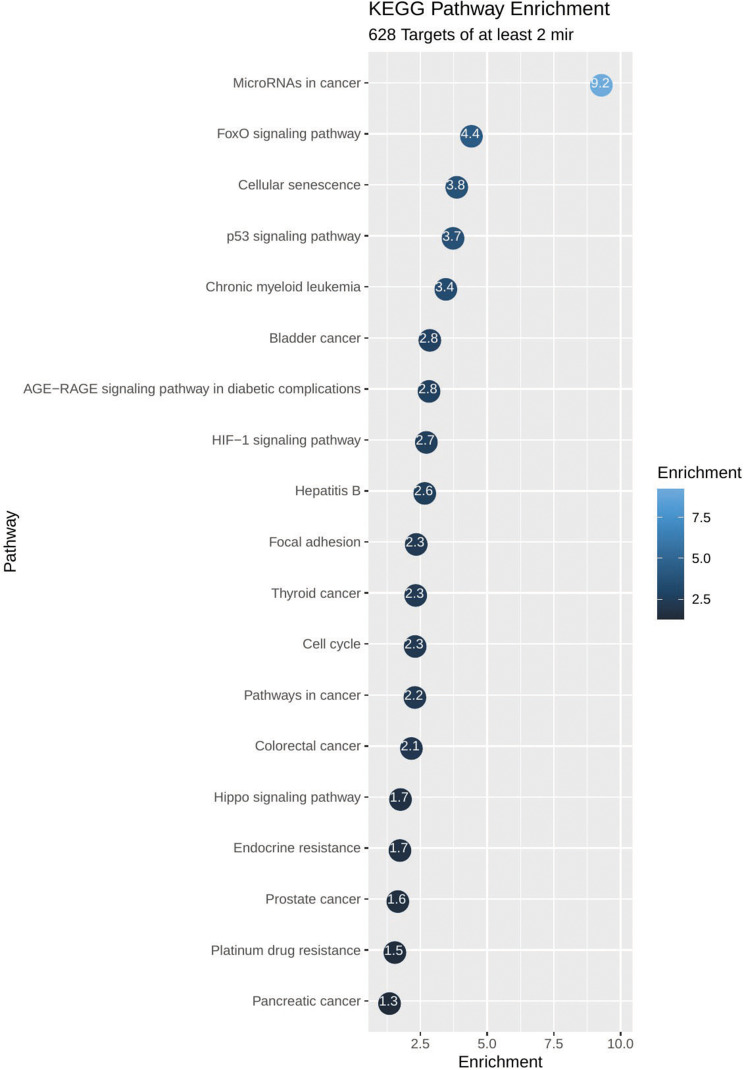
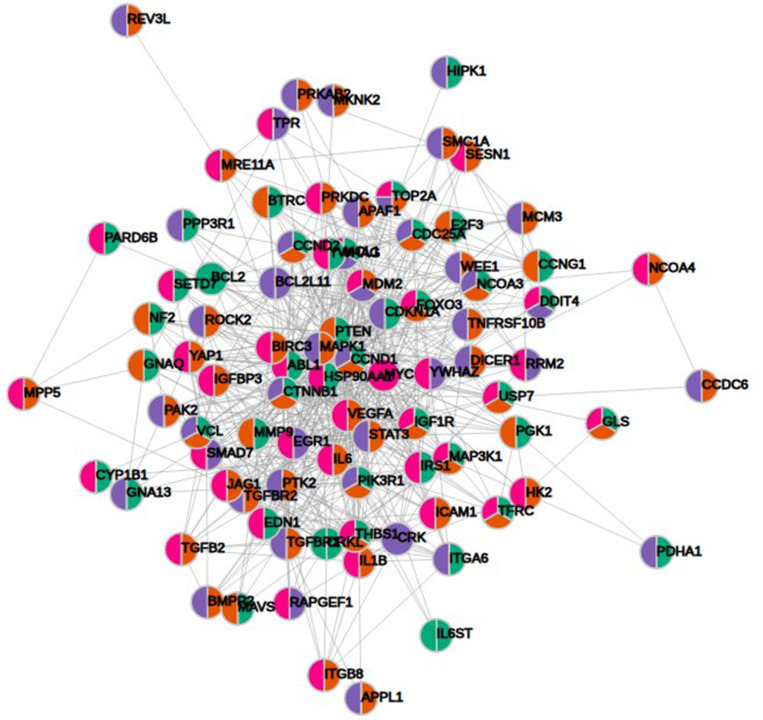
KEGG pathway analysis from 628 genes that are common targets for at least two miRNAs revealed strong functional association with a number of cancer types including prostate cancer, as well as for general miRNA deregulation in cancers. The top 19 most enriched, non-redundant biological processes ranked according to an adjusted p-value are shown in Table 5 and Figure 6. Interestingly, pathways related to DNA repair such as FoxO, p53 and Hippo signaling were significantly enriched (Table 5 and Figure 6). In an attempt to visualize the relationships between the protein coding genes targeted by the different miRNAs, we obtained the subset of genes that were enriched in at least one KEGG pathway (Table 5) and created their protein–protein interaction network with the use of STRING-DB. The network consisted of 90 genes is shown in Figure 7. Slight tendency for common occurrence of targets of hsa-miR-21 and hsa-miR-106b (blue-orange connecting edges, respectively; Figure 7) is visible in the network and also quantified in an increased Jaccard similarity for this particular combination (Table 6).
Table 5.
KEGG Enrichment Pathways for the Set of 628 Gene Targets Derived from the Combination of at Least Two miRNAs
| Pathway | Enrichment p-value | Genes |
|---|---|---|
| MicroRNAs in cancer | 5.75e-10 | TGFB2,ABL1,CRKL,MAPK1,DICER1,MMP9,CCND1,E2F3,VEGFA, CCNG1, GLS, CCND2, CDKN1A, MDM2, MYC, THBS1, CYP1B1, MCL1, BCL2L11, CDC25A, CRK, DDIT4, STAT3, IRS1, BCL2, PTEN, BMPR2 |
| FoxO signaling pathway | 4.24e-05 | TGFB2, MAPK1, TGFBR1, CCND1, FOXO3, CCND2, CDKN1A, PRKAB2, MDM2, IL6, IGF1R, SETD7, PIK3R1, BCL2L11, TGFBR2, STAT3, IRS1, PTEN, USP7 |
| Cellular senescence | 0.000151 | MRE11A, TGFB2, MAPK1, TGFBR1, CCND1, E2F3, FOXO3, CCND2, CDKN1A, MDM2, IL6, MYC, PIK3R1, IGFBP3, HIPK1, TGFBR2, CDC25A, BTRC, PTEN, PPP3R1 |
| p53 signaling pathway | 0.000208 | SESN1, CCND1, CCNG1, CCND2, APAF1, TNFRSF10B, CDKN1A, MDM2, THBS1, IGFBP3, BCL2, RRM2, PTEN |
| Chronic myeloid leukemia | 0.000393 | TGFB2, ABL1, CRKL, MAPK1, TGFBR1, CCND1, E2F3, CDKN1A, MDM2, MYC, PIK3R1, TGFBR2, CRK |
| AGE-RAGE signaling pathway in diabetic complications | 0.00166 | EDN1, ICAM1, TGFB2, MAPK1, TGFBR1, CCND1, VEGFA, EGR1, IL1B, IL6, PIK3R1, TGFBR2, STAT3, BCL2 |
| Bladder cancer | 0.00156 | MAPK1, MMP9, CCND1, E2F3, VEGFA, CDKN1A, MDM2, MYC, THBS1 |
| HIF-1 signaling pathway | 0.00209 | TFRC, EDN1, MKNK2, MAPK1, PGK1, VEGFA, CDKN1A, PDHA1, IL6, IGF1R, PIK3R1, HK2, STAT3, BCL2 |
| Hepatitis B | 0.0024 | MAVS, TGFB2, MAP3K1, MAPK1, MMP9, TGFBR1, CCND1, E2F3, APAF1, CDKN1A, IL6, MYC, PIK3R1, YWHAZ, STAT3, BCL2, PTEN |
| Focal adhesion | 0.00499 | BIRC3, VCL, ITGA6, CRKL, MAPK1, ITGB8, RAPGEF1, CCND1, VEGFA, CCND2, ROCK2, THBS1, IGF1R, PIK3R1, CRK, CTNNB1, PTK2, BCL2, PTEN, PAK2 |
| Thyroid cancer | 0.00527 | TPR, MAPK1, CCDC6, CCND1, CDKN1A, MYC, CTNNB1, NCOA4 |
| Cell cycle | 0.00548 | SMC1A, TGFB2, ABL1,CCND1, MCM3, E2F3, CCND2, CDKN1A, MDM2, MYC, CDC25A, YWHAZ, WEE1, YWHAG, PRKDC |
| Pathways in cancer | 0.00559 | BIRC3, TPR, HSP90AA1, ITGA6, TGFB2, ABL1,CRKL,MAPK1,MMP9, JAG1,TGFBR1, CCDC6, CCND1, E2F3, VEGFA, CCND2, GNA13, APAF1, NCOA3, CDKN1A, ROCK2, IL6ST, MDM2, IL6, MYC,I GF1R, PIK3R1, BCL2L11, GNAQ, APPL1, TGFBR2, CRK, CTNNB1, STAT3, PTK2, BCL2, PTEN, NCOA4 |
| Colorectal cancer | 0.00768 | TGFB2, MAPK1, TGFBR1, CCND1, CDKN1A, MYC, PIK3R1, BCL2L11, APPL1, TGFBR2, CTNNB1, BCL2 |
| Hippo signaling pathway | 0.0195 | MPP5, TGFB2, SMAD7, TGFBR1, CCND1, CCND2, PARD6B, MYC,YAP1, TGFBR2, YWHAZ, BTRC, CTNNB1, YWHAG, NF2, BMPR2 |
| Endocrine resistance | 0.0203 | MAPK1, MMP9, JAG1, CCND1, E2F3, NCOA3, CDKN1A, MDM2, IGF1R, PIK3R1, PTK2, BCL2 |
| Prostate cancer | 0.0248 | HSP90AA1, MAPK1, MMP9, CCND1, E2F3, CDKN1A, MDM2, IGF1R, PIK3R1, CTNNB1, BCL2, PTEN |
| Platinum drug resistance | 0.0318 | REV3L, BIRC3, MAPK1, APAF1, CDKN1A, TOP2A, MDM2, SLC31A1, PIK3R1, BCL2 |
| Pancreatic cancer | 0.0496 | TGFB2, MAPK1, TGFBR1, CCND1, E2F3, VEGFA, CDKN1A, PIK3R1, TGFBR2, STAT3 |
Figure 6.
Functional enrichment plot of the set of 628 genes targeted by at least two miRNAs. Pathways related to prostate cancer, cellular functions, miRNAs deregulation and to DNA repair are shown to be significantly enriched. On the plot, each bubble represents a term and includes the significance of the enrichment as -log10 (p-value).
Figure 7.
Protein–protein interaction network of 90 protein coding gene targets of at least two miRNAs with significant KEGG enrichments. Color-coding depends on the associated miRNA (blue, hsa-miR-21; orange, hsa-miR-106b; green, hsa-miR-141 and magenta, hsa-miR-375).
Table 6.
Common Protein Gene Targets with Pathway Enrichment Between miRΝΑ Calculated as Jaccard Index (2*intersection/Union)
| miR (Total Genes) | hsa-miR-141 (45) | hsa-miR-21 (62) | hsa-miR-106b (47) | hsa-miR-375 (43) |
|---|---|---|---|---|
| hsa-miR-141 | 0.633 | 0.479 | 0.529 | |
| hsa-miR-21 | 0.717 | 0.633 | ||
| hsa-miR-106b | 0.286 |
Discussion
In the present study, we assessed the expression levels of four miRNAs in serum prior- and post-RT and evaluated their prognostic significance in PCa patients. We found that low expression levels of miR-106b prior-RT were correlated with extraprostatic extension and seminal vesicles invasion. In addition, patients in the high-risk subgroup who experienced BR had significantly higher distributions of post-RT levels of miR-21. Furthermore, in the salvage RT-treated subgroup, patients who experienced BR had significantly higher post RT mir21 and mir106b distribution levels compared to those without recurrent disease. Most importantly, high expression levels of miR-21 prior and of miR-106b post-RT were associated with shorter OS. Finally, bioinformatics analysis of mRNA targets revealed interesting associations related to DNA repair pathways regulated by the examined miRNAs.
MiR-21 is a well-known oncomir with regulatory roles in cell proliferation, apoptosis, epithelial-mesenchymal transition (EMT) and chemoresistance.37 MiR-21 mediates androgen-induced prostate cancer cell proliferation and is sufficient for androgen-dependent tumors to overcome castration-mediated growth arrest.38,39 Also, miR-21 promotes prostate cancer cell proliferation by targeting PTEN40 whereas, by regulating MARCKS it promotes apoptosis resistance and invasion in prostate cancer cells.41
MiR-21 is commonly up-regulated in cancer tissue in several types of malignancies, including PCa,42 thus it has been proposed as a potential biomarker in the prognosis of PCa patients.37 Specifically, deregulated circulating miR-21 has been shown to distinguish PCa patients from healthy adults and local from metastatic disease.43 It has also been proposed as predictor of chemotherapy efficacy in metastatic hormone refractory disease.44 In addition, up-regulated expression of tissue miR-21 is an independent predictor of 5-yr biochemical recurrence.45 This is in accordance with our results that higher circulating levels of miR-21 prior-RT were associated with increased rates of BR in the subgroup of high-risk patients and those receiving salvage RT. Furthermore, we showed for the first time in this study that high expression levels of circulating miR-21 prior-RT are associated with shorter OS.
MiR-106b is a member of the miR-106b-25 cluster, which is frequently deregulated in several types of cancer, including PCa and has been associated with cancer progression.46 It has been found to play dual role in cancer development, acting either as oncomir or as tumor suppressor.47 MiR-106b has been shown to regulate apoptosis and focal adhesion-related pathways.48 Also, miR-106b promotes cell proliferation and invasion49 and in another study was shown that its hypoxia-induced over-expression results in a more aggressive phenotype.46 It has also been shown that miR-106b is highly expressed in metastatic tissues compared to normal and is correlated with bone metastasis.21 In contrast, we herein found that lower expression levels of miR-106b before RT were correlated with locally advanced disease.
In irradiated prostate cancer cells, miR-106b increases radioresistance by overriding p21-activated cell cycle arrest.50 In addition, similar results were obtained for colorectal51 and cervical cancer cells.52 Over-expression of miR-106b was also correlated with increased risk of early recurrence.21 In accordance with the above results, we observed that higher post-RT levels of miR-106b were associated with increased rates of BR in the subgroup of patients receiving salvage RT. Moreover, we found that higher post-RT levels are associated with shorter OS. As it is noted, limited data exist regarding miR-106b role in clinical samples, and therefore its prognostic significance shown for the first time in our study, merits further evaluation in PCa.
MiR-141 also plays a major role in PCa, targeting genes that regulate apoptosis, cell proliferation and EMT transition.53 It was one of the first circulating miRNAs associated with PCa pathogenesis and poor prognosis.54 MiR-375 is involved in cell proliferation and EMT transition in PCa.54 It also stimulates cell growth and invasion and impairs apoptosis.54 Higher expression levels of miR-375 and miR-141 are associated with worse prognosis, high Gleason scores, positive lymph nodes, castration resistance and metastatic disease.55 Although their expression prior and post-RT was strongly correlated with that of miR-21, probably due to underlying common mechanisms of regulation, we did not find significant associations with pathological features or with patients’ outcome.
KEGG pathway enrichment analysis revealed 19 significantly enriched pathways mostly related to cancer. Among these, p53, FoxO and Hippo signaling are widely reported to be correlated with the DDR.56–58 Interestingly, major regulatory components of the above pathways such as MDM2, FOXO3 and YAP1 were included in the 90 genes that were enriched in at least one KEGG pathway. Furthermore, the increased Jaccard similarity for common occurrence of targets of miR-21 and miR-106b suggest a functional link between these two miRNAs related to DDR.
Most of our knowledge about the role of miRNAs in radiation response comes from in vitro studies. Among others, mir-21 was upregulated during RT in breast cancer stem cell cultures and was associated with negative prognostic factors such as ki-67 and triple-negative phenotype in breast cancer patients.59 In non-small cell lung cancer (NSCLC) cells, mir-21 increased radioresistance either by upregulating hypoxia-inducible factor 1a60 or by downregulating PTEN, SNX1, and SGPP1 expression and increasing Akt phosphorylation61 or even by inhibiting programmed cell death.62 Inhibition of mir-21 decreased radioresistance in esophageal cancer cells through activation of PTEN63 and in malignant glioma cells through inhibition of PI3k/AKT pathway64 or increased expression of PDCD4 and hMSH2.65 It is also linked to radioresistance due to DNA double-strand breaks repair.25,66 Over-expression of mir-106b in cervical cancer cells resulted in decreased radiosensitivity by negatively regulating Immediate Early Response 3 (IER3) gene.52 In the same study, Growth Arrest-Specific 5 (GAS5) noncoding RNA “sponged” mir-106b, resulting in increased radiosensitivity.
To the best of our knowledge, the present study is among the first to correlate circulating miRNAs with outcomes of PCa patients undergoing RT. Limitations of the current study include the small cohort of patients with limited statistical power and the lack of validation in an independent patient group. Also, due to the small number of events post-RT, the associations of high miR-21 and miR-106b expression levels with shorter OS were not validated as independent predictors in a multivariate analysis. However, significant advantages strengthen the power of our results such as 1) pre-analytical and analytical parameters were thoroughly examined to avoid bias in miRNAs detection and quantification,67 2) blood samples and clinical information were prospectively collected and 3) follow-up time was adequate for our patient cohort. Nevertheless, further studies in an independent cohort of PCa patients are needed to confirm our findings.
In summary, we herein show that serum miR-106b expression levels assessed prior-RT were correlated with locally advanced disease. Also, higher prior- or post-RT serum miR-21 and miR-106b expression levels were correlated with BR in the high risk and salvage RT-treated subgroups and with OS in the whole group of patients. Furthermore, using a combination of functional enrichment and protein–protein interaction networks, we showed that these miRNAs are involved in cancer-related biological processes and particularly in DNA repair pathways. The above findings further confirm preclinical evidence for their potential role in mechanisms of radioresistance in PCa. Therefore, miR-21 and miR-106b assessment in the serum of PCa patients during the disease time course merits further evaluation to address their potential role as non-invasive biomarkers for monitoring patients who could benefit from RT.
Conclusion
We herein demonstrate that the expression levels of miRNAs evaluated in the serum of patients undergoing radiotherapy may hold significant prognostic implications in PCa. Bioinformatic analysis suggests that the unfavorable prognostic role of miR-21 and miR-106b could be related to their functional involvement in pathways related to DDR. Further studies are needed to investigate the potential functional role of these miRNAs in PCa.
Acknowledgments
We would like to thank Sevasti Dara for her help in graphic design. We are also grateful to all patients who signed the consent form and participated in this study.
Disclosure
The authors declare that they have no conflicts of interest regarding this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Halperin EC. Perez & Brady’s Principles and Practice of Radiation Oncology. Philadelphia: Wolters Kluwer; 2019. [Google Scholar]
- 3.Huang RX, Zhou PK. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduction Targeted Ther. 2020;5(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. doi: 10.1016/j.redox.2018.101084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim Biophys Acta. 2016;1863(12):2977–2992. doi: 10.1016/j.bbamcr.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee P, Choudhary GS, Sharma A, et al. PARP inhibition sensitizes to low dose-rate radiation TMPRSS2-ERG fusion gene-expressing and PTEN-deficient prostate cancer cells. PLoS One. 2013;8(4):e60408. doi: 10.1371/journal.pone.0060408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biau J, Chautard E, Verrelle P, Dutreix M. Altering DNA repair to improve radiation therapy: specific and multiple pathway targeting. Front Oncol. 2019;9:1009. doi: 10.3389/fonc.2019.01009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan M, Ahmed KM, Coleman MC, Spitz DR, Li JJ. Nuclear factor-kappaB and manganese superoxide dismutase mediate adaptive radioresistance in low-dose irradiated mouse skin epithelial cells. Cancer Res. 2007;67(7):3220–3228. doi: 10.1158/0008-5472.CAN-06-2728 [DOI] [PubMed] [Google Scholar]
- 9.Skvortsova I, Skvortsov S, Stasyk T, et al. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics. 2008;8(21):4521–4533. doi: 10.1002/pmic.200800113 [DOI] [PubMed] [Google Scholar]
- 10.Lee SY, Jeong EK, Ju MK, et al. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer. 2017;16(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopes Vendrami C, McCarthy RJ, Chatterjee A, et al. The utility of prostate specific antigen density, prostate health index, and prostate health index density in predicting positive prostate biopsy outcome is dependent on the prostate biopsy methods. Urology. 2019;129:153–159. doi: 10.1016/j.urology.2019.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw GL, Thomas BC, Dawson SN, et al. Identification of pathologically insignificant prostate cancer is not accurate in unscreened men. Br J Cancer. 2014;110(10):2405–2411. doi: 10.1038/bjc.2014.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 15.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzimagiorgis G, Michailidou EZ, Kritis A, Markopoulos AK, Kouidou S. Recovering circulating extracellular or cell-free RNA from bodily fluids. Cancer Epidemiol. 2011;35(6):580–589. doi: 10.1016/j.canep.2011.02.016 [DOI] [PubMed] [Google Scholar]
- 17.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. doi: 10.7150/thno.11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbé M, Hoey C, Ray J, et al. microRNAs identified in prostate cancer: correlative studies on response to ionizing radiation. Mol Cancer. 2020;19(1):63. doi: 10.1186/s12943-020-01186-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Lieberman R, Pan J, et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol Cancer. 2016;15(1):70. doi: 10.1186/s12943-016-0556-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melbø-Jørgensen C, Ness N, Andersen S, et al. Stromal expression of MiR-21 predicts biochemical failure in prostate cancer patients with Gleason score 6. PLoS One. 2014;9(11):e113039. doi: 10.1371/journal.pone.0113039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson RS, Yi M, Esposito D, et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene. 2013;32(35):4139–4147. doi: 10.1038/onc.2012.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzales JC, Fink LM, Goodman OB Jr, Symanowski JT, Vogelzang NJ, Ward DC. Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin Genitourin Cancer. 2011;9(1):39–45. doi: 10.1016/j.clgc.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 23.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46(2):298–311. doi: 10.1016/j.ejca.2009.10.027 [DOI] [PubMed] [Google Scholar]
- 24.McDermott N, Meunier A, Wong S, Buchete V, Marignol L. Profiling of a panel of radioresistant prostate cancer cells identifies deregulation of key miRNAs. Clin Transl Radiat Oncol. 2017;2:63–68. doi: 10.1016/j.ctro.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B, Wang X, Hu S, et al. miR-21-mediated radioresistance occurs via promoting repair of DNA double strand breaks. J Biol Chem. 2017;292(8):3531–3540. doi: 10.1074/jbc.M116.772392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Bao B, Ahmad A, Kong D, et al. Hypoxia induced aggressiveness of prostate cancer cells is linked with deregulated expression of VEGF, IL-6 and miRNAs that are attenuated by CDF. PLoS One. 2012;7(8):e43726. doi: 10.1371/journal.pone.0043726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pashaei E, Pashaei E, Ahmady M, Ozen M, Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS One. 2017;12(6):e0179543. doi: 10.1371/journal.pone.0179543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen PL, Haddad Z, Ross AE, et al. Ability of a genomic classifier to predict metastasis and prostate cancer-specific mortality after radiation or surgery based on needle biopsy specimens. Eur Urol. 2017;72(5):845–852. doi: 10.1016/j.eururo.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 29.Cookson MS, Aus G, Burnett AL, et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association prostate guidelines for localized prostate cancer update panel report and recommendations for a standard in the reporting of surgical outcomes. J Urol. 2007;177(2):540–545. [DOI] [PubMed] [Google Scholar]
- 30.Roach M 3rd, Hanks G, Thames H Jr, et al. Defining biochemical failure following radiotherapy with or without hormonal therapy in men with clinically localized prostate cancer: recommendations of the RTOG-ASTRO phoenix consensus conference. Int J Radiat Oncol Biol Phys. 2006;65(4):965–974. doi: 10.1016/j.ijrobp.2006.04.029 [DOI] [PubMed] [Google Scholar]
- 31.Parker C, Castro E, Fizazi K, et al. Prostate cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31(9):1119–1134. doi: 10.1016/j.annonc.2020.06.011 [DOI] [PubMed] [Google Scholar]
- 32.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280(11):969–974. doi: 10.1001/jama.280.11.969 [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 34.Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–159. doi: 10.1093/nar/gku1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–815. doi: 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bautista-Sánchez D, Arriaga-Canon C, Pedroza-Torres A, et al. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol Ther Nucleic Acids. 2020;20:409–420. doi: 10.1016/j.omtn.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9(5):923–929. doi: 10.4161/cc.9.5.10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribas J, Ni X, Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69(18):7165–7169. doi: 10.1158/0008-5472.CAN-09-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Guo JX, Shao ZQ. miR-21 targets and inhibits tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion: an experimental study. Asian Pac J Trop Med. 2017;10(1):87–91. doi: 10.1016/j.apjtm.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 41.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383(3):280–285. doi: 10.1016/j.bbrc.2009.03.077 [DOI] [PubMed] [Google Scholar]
- 42.Ghorbanmehr N, Gharbi S, Korsching E, Tavallaei M, Einollahi B, Mowla SJ. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate. 2019;79(1):88–95. doi: 10.1002/pros.23714 [DOI] [PubMed] [Google Scholar]
- 43.Yaman Agaoglu F, Kovancilar M, Dizdar Y, et al. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32(3):583–588. doi: 10.1007/s13277-011-0154-9 [DOI] [PubMed] [Google Scholar]
- 44.Zhang HL, Yang LF, Zhu Y, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–331. doi: 10.1002/pros.21246 [DOI] [PubMed] [Google Scholar]
- 45.Li T, Li RS, Li YH, et al. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol. 2012;187(4):1466–1472. doi: 10.1016/j.juro.2011.11.082 [DOI] [PubMed] [Google Scholar]
- 46.Liang H, Studach L, Hullinger RL, Xie J, Andrisani OM. Down-regulation of RE-1 silencing transcription factor (REST) in advanced prostate cancer by hypoxia-induced miR-106b~25. Exp Cell Res. 2014;320(2):188–199. doi: 10.1016/j.yexcr.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang C, Dou R, Yin T, Ding J. MiRNA-106b-5p in human cancers: diverse functions and promising biomarker. Biomed Pharmacother. 2020;127:110211. doi: 10.1016/j.biopha.2020.110211 [DOI] [PubMed] [Google Scholar]
- 48.Yin W, Chen J, Wang G, Zhang D. MicroRNA106b functions as an oncogene and regulates tumor viability and metastasis by targeting LARP4B in prostate cancer. Mol Med Rep. 2019;20(2):951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi N, Park J, Lee JS, et al. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015;6(27):23533–23547. doi: 10.18632/oncotarget.4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Shi XB, Nori D, et al. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate. 2011;71(6):567–574. doi: 10.1002/pros.21272 [DOI] [PubMed] [Google Scholar]
- 51.Zheng L, Zhang Y, Liu Y, et al. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Liu L, Li G, et al. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 2019;126:994–1001. doi: 10.1016/j.ijbiomac.2018.12.176 [DOI] [PubMed] [Google Scholar]
- 53.Filella X, Foj L. miRNAs as novel biomarkers in the management of prostate cancer. Clin Chem Lab Med. 2017;55(5):715–736. doi: 10.1515/cclm-2015-1073 [DOI] [PubMed] [Google Scholar]
- 54.Kanwal R, Plaga AR, Liu X, Shukla GC, Gupta S. MicroRNAs in prostate cancer: functional role as biomarkers. Cancer Lett. 2017;407:9–20. doi: 10.1016/j.canlet.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 55.Brase JC, Johannes M, Schlomm T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–616. doi: 10.1002/ijc.25376 [DOI] [PubMed] [Google Scholar]
- 56.Furth N, Aylon Y, Oren M. p53 shades of Hippo. Cell Death Differ. 2018;25(1):81–92. doi: 10.1038/cdd.2017.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. TIG. 2012;28(3):128–136. doi: 10.1016/j.tig.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pefani DE, O’Neill E. Hippo pathway and protection of genome stability in response to DNA damage. FEBS J. 2016;283(8):1392–1403. doi: 10.1111/febs.13604 [DOI] [PubMed] [Google Scholar]
- 59.Griñán-Lisón C, Olivares-Urbano MA, Jiménez G, et al. miRNAs as radio-response biomarkers for breast cancer stem cells. Mol Oncol. 2020;14(3):556–570. doi: 10.1002/1878-0261.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang S, Wang R, Yan H, Jin L, Dou X, Chen D. MicroRNA-21 modulates radiation resistance through upregulation of hypoxia-inducible factor-1α-promoted glycolysis in non-small cell lung cancer cells. Mol Med Rep. 2016;13(5):4101–4107. doi: 10.3892/mmr.2016.5010 [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Zhang C, Hu L, et al. Abnormal expression of miR-21 and miR-95 in cancer stem-like cells is associated with radioresistance of lung cancer. Cancer Invest. 2015;33(5):165–171. doi: 10.3109/07357907.2015.1019676 [DOI] [PubMed] [Google Scholar]
- 62.Jiang L-P, He C-Y, Zhu Z-T. Role of microRNA-21 in radiosensitivity in non-small cell lung cancer cells by targeting PDCD4 gene. Oncotarget. 2017;8(14):23675–23689. doi: 10.18632/oncotarget.15644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus. 2013;26(8):823–831. doi: 10.1111/j.1442-2050.2012.01389.x [DOI] [PubMed] [Google Scholar]
- 64.Gwak HS, Kim TH, Jo GH, et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7(10):e47449. doi: 10.1371/journal.pone.0047449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chao TF, Xiong HH, Liu W, Chen Y, Zhang JX. MiR-21 mediates the radiation resistance of glioblastoma cells by regulating PDCD4 and hMSH2. J Huazhong Univ Sci Technol Med Sci. 2013;33(4):525–529. doi: 10.1007/s11596-013-1153-4 [DOI] [PubMed] [Google Scholar]
- 66.Tang S, Liu B, Liu M, et al. Ionizing radiation-induced growth in soft agar is associated with miR-21 upregulation in wild-type and DNA double strand break repair deficient cells. DNA Repair (Amst). 2019;78:37–44. doi: 10.1016/j.dnarep.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57(6):833–840. doi: 10.1373/clinchem.2010.157198 [DOI] [PubMed] [Google Scholar]