Abstract
The role of human herpesvirus 6 (HHV-6) in disease beyond primary infection remains unclear. We have developed and validated a new reverse transcription-PCR (RT-PCR) assay for HHV-6 that can determine the presence of HHV-6 in clinical specimens and differentiate between latent and replicating virus. Peripheral blood mononuclear cells from 109 children were evaluated for HHV-6 by RT-PCR, DNA PCR, and viral culture. Of these samples, 106 were suitable for analysis. A total of 20 samples were positive for HHV-6 by culture and DNA PCR, of which 19 were positive by RT-PCR (sensitivity, 95%). All 28 samples from children that were negative by viral culture, but positive by DNA PCR, were negative for viral transcripts by our RT-PCR assay. One positive RT-PCR result was observed in 56 samples that were negative by tissue culture and DNA PCR. This indicates a low rate of false-positive results (1.2%) and a specificity of 98.8%. This RT-PCR assay can reliably differentiate between latent and actively replicating HHV-6 and should allow insight into the pathogenesis of this ubiquitous virus.
Human herpesvirus 6 (HHV-6) is a ubiquitous T-lymphotropic virus that infects essentially all children by 36 months of age. Primary infection causes an undifferentiated febrile illness, with a subset of children exhibiting the classic manifestations of roseola infantum (8). Similar to all human herpesviruses, HHV-6 is hypothesized to persist lifelong in a latent form following initial infection. This concept is supported by the detection of HHV-6 DNA in the peripheral blood mononuclear cells (PBMCs) and central nervous systems of normal adults and children (4, 8, 10).
Several diseases of both normal and immunocompromised adults have been associated with HHV-6, including meningoencephalitis, fever following solid-organ transplantation, and Hodgkin's disease (3). In bone marrow transplant patients, HHV-6 is thought to cause interstitial pneumonitis and bone marrow suppression following transplantation. HHV-6 has also been associated with multiple sclerosis (3).
The pathogenic role of HHV-6 beyond primary infection, in diseases postulated to be associated with this virus, remains unclear. Detection of HHV-6 by DNA PCR does not indicate whether HHV-6 persists in a latent or a reactivated state, which would be necessary to determine whether HHV-6 possibly causes disease or is only coincidental. Our aim, therefore, was to develop and validate a sensitive and specific reverse transcription-PCR (RT-PCR) assay for HHV-6 which could differentiate between latent and actively replicating virus, for use in clinical and research settings.
(This work was presented in part at the annual meeting of the American Society for Virology, Vancouver, British Columbia, Canada, July 1998 [abstract W51-9].)
MATERIALS AND METHODS
Subjects.
Whole-blood specimens collected in EDTA (0.125 to 4 ml) were used for this study. Blood samples were obtained from 109 children, ≤3 years of age, who had blood available following an evaluation for fever or seizures at the Strong Memorial Hospital Emergency Department, Rochester, N.Y., between 14 March and 20 December 1997. Patients were evaluated as clinically indicated by members of the medical staff not involved in this study. The group contained 60 boys (55%) and 49 girls (45%), with a mean age of 12.2 months (range, 1 to 36 months).
The research protocol was approved by the human subjects review board of the University of Rochester Medical Center, Rochester, N.Y.
Serology.
Patient samples which had been stored for 72 h or less at 4°C were chosen for the study. Following centrifugation, plasma was removed and serologic testing was performed by an indirect fluorescent antibody assay for immunoglobulin G, as previously described (8). Briefly, patient sera were tested by serial twofold dilutions on HSB2 cells infected with a previously described clinical isolate of HHV-6 that contains a mixture of HHV-6 variants A and B (R-1) (8). A positive result was defined as a titer greater than 20, with the end point identified as the highest dilution positive by specific immunofluorescence (8).
Viral isolation.
Following density gradient centrifugation (Histopaque 1077; Sigma Diagnostics, St. Louis, Mo.), PBMCs were resuspended in 1.8 ml of medium and divided for DNA PCR, RT-PCR, and viral culture in aliquots of 0.2, 1.0, and 0.6 ml, respectively. Viral isolation was performed as previously described (7). Positive cultures were confirmed by indirect immunofluorescent staining with monoclonal antibodies directed against HHV-6 variant A (2D6; gift of B. Chandran), HHV-6 variant B (MAB8535; Chemicon, Temecula, Calif.), and HHV-7 (KR-4; gift of K. Yaminishi).
DNA PCR.
DNA PCR for HHV-6 and HHV-7 was performed on proteinase K-digested samples as previously described (1, 7, 8).
RT-PCR.
PBMCs for the RT-PCR were pelleted, placed in Tri-Reagent LS (Molecular Research Center, Inc., Cincinnati, Ohio), and stored at −70°C.
The range of PBMCs in the RT-PCR aliquot was 1.0 × 105 to 2.78 × 107 cells/ml, with a mean of 3.21 × 106 cells/ml. Total RNA was extracted from PBMCs frozen in Tri-Reagent LS via the manufacturer's protocol. The total RNA was then resuspended in 20 μl of diethyl pyrocarbonate-treated water with 40 U of RNase inhibitor (Perkin-Elmer, Norwalk, Conn.).
RT-PCR was performed on specimens with the Access RT-PCR system (Promega, Madison, Wis.), with primers derived from the published HHV-6 sequence for the U100 region (12). All RT-PCRs and DNA PCRs were performed without knowledge of the tissue culture results. The external primers, which amplify both HHV-6 variant A and HHV-6 variant B, are 5′ CTAAATTTTCTACCTCCGAAATGT 3′ and 5′ GAGTCCATGAGTTAGAAGATT 3′. These primers span a well-defined splice junction so that amplified cDNA can be distinguished from viral genomic DNA. Each RT-PCR mixture contained 10 μl of total RNA from each patient sample in 40 μl of mixture which contained 5 U of avian myeloblastosis virus (AMV) reverse transcriptase, 5 U of Thermus flavus DNA polymerase, 10 μl of AMV/T. flavus 5× reaction buffer, 50 mM MgSO4, and 10 mM each deoxynucleoside triphosphate. The thermocycling profile was constructed as follows: 48°C for 45 min; 94°C for 5 min; 35 cycles of 57°C for 45 s, 72°C for 45 s, and 94°C for 1 min; and 1 cycle each of 57°C for 2 min and 72°C for 8 min.
The nested amplification reaction was performed with 1 μl of the first-round RT-PCR product in a final volume of 20 μl. The mixture contained 1.5 mM MgCl, 0.2 mM each deoxynucleoside triphosphate, 5 U of Taq, and 0.75 mM internal primers 5′ ACTACTACCTTAGAAGATATAG 3′ and 5′ AAGCGCGTGCAGGTTTCCCAA 3′. Results were obtained by electrophoresis with 2.5% agarose gels followed by ethidium bromide staining and confirmed by Southern blot hybridization as previously described (8). The probe used for the Southern blot hybridization, 5′ GAGCTGATCGTCAGTATCGAG 3′, anneals to the region internal to the nested primers.
Controls.
Ten microliters of each total RNA sample was used in a separate, equivalent reaction mixture, during first-round amplification, to detect the histidyl-tRNA synthetase gene as a control for adequate cellular RNA and to exclude the presence of inhibitors in each sample (14). Cord blood mononuclear cells (CBMCs), infected with a previously described clinical isolate (R-1), and culture-negative cord blood RNA were used as the positive and negative controls for both rounds of amplification (8).
Cloning.
Cloning of RT-PCR products was performed with the Original TA cloning kit (Invitrogen, Carlsbad, Calif.) with the pCR 2.1 vector via the manufacturer's protocol. The positive-control sample, R-1, was sequenced by protocols for the Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.).
In vitro transcription.
HHV-6 variant A cDNA containing the U100 region (gift of B. Chandran) was linearized with the BamHI restriction enzyme at the end of the insertion (Life Technologies, Inc., Gaithersburg, Md.) and then purified by dialysis. The in vitro transcription reaction was performed with T7 RNA polymerase (MBI Fermentas, Amherst, N.Y.). Briefly, the reaction mixture contained 50 U of T7 RNA polymerase, 10 μl of 5× transcription buffer, 1 μg of linearized DNA, 50 U of RNase inhibitor (Perkin-Elmer), and 2.0 mM each ribosomal nucleoside triphosphate; it was incubated at 37°C for 120 min.
RESULTS
During assay development, the specificity of the HHV-6 primers was evaluated. Initially, the HHV-6 RT-PCR primers were compared to all viral sequence information in the National Center for Biotechnology Information. No matches were identified. RT-PCR for HHV-6 was then performed on RNA purified from single clinical isolates of HHV-7, human cytomegalovirus (HCMV), herpes simplex virus 2, and varicella-zoster virus (results not shown). All reactions were negative for the HHV-6 product by gel electrophoresis with ethidium bromide staining and Southern blot hybridization. Additionally, the RT-PCR product of the positive-control sample, R-1, was cloned and sequenced and verified as being of HHV-6 origin with approximately 98% sequence identity.
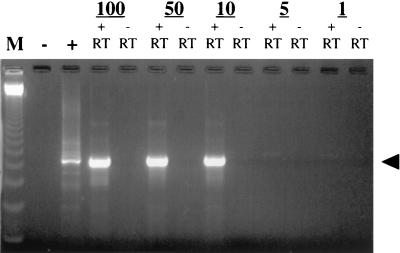
The sensitivity of this assay was also determined. HHV-6 variant A RNA was produced by in vitro transcription. This method was chosen in order to generate large quantities of pure gp105 RNA transcripts which could be reliably measured. Known copy numbers were tested by RT-PCR. Ten copies or 4.3 × 10−3 fg of RNA in a background of 0.21 μg of negative cord blood RNA was reproducibly detected by gel electrophoresis with ethidium bromide (Fig. 1). These results were confirmed by Southern blot hybridization.
FIG. 1.
Sensitivity assay for HHV-6 RT-PCR, showing template RNA copies (numbered at the top). M, marker with 123-bp ladder; −, negative control; +, positive control; + RT, with reverse transcriptase; − RT, without reverse transcriptase; arrowhead, 500 bp.
Following these initial validation studies, a total of 109 patient specimens were tested for HHV-6 by viral isolation, DNA PCR, and RT-PCR. Three specimens were excluded from further analysis because of a lack of adequate cellular material, as shown by a negative RT-PCR for histidyl-tRNA synthetase mRNA. Figure 2 shows representative results of the remaining 106 RT-PCR analyses.
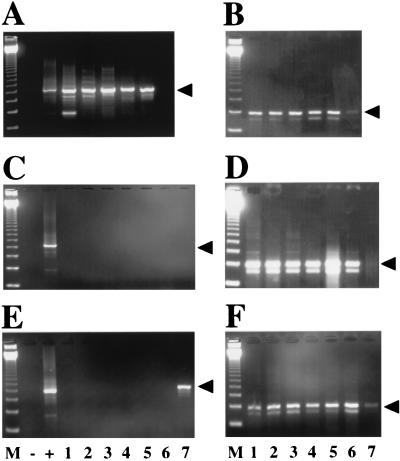
FIG. 2.
HHV-6 RT-PCR results. Results of ethidium bromide staining of agarose gel electrophoresis of specimens obtained from representative patient samples are shown. (A) Lanes 1 to 6, RT-PCR of specimens from children with acute HHV-6 infection (defined as isolation of HHV-6 in culture, a positive DNA PCR, and a negative antibody titer). Lane 6 demonstrates the negative RT-PCR result from a specimen from one of these children. Arrowhead, 500 bp. (B) Corresponding histidyl-tRNA synthetase RT-PCR of samples in panel A. Arrowhead, 237 bp. (C) Lanes 1 to 7, RT-PCR of samples from children with prior HHV-6 infection (negative HHV-6 culture, positive DNA PCR, and negative RT-PCR). (D) Corresponding histidyl-tRNA synthetase RT-PCR of samples in panel C. (E) Lanes 1 to 7, RT-PCR of samples from children with no evidence of prior infection with HHV-6 (negative HHV-6 culture and negative DNA PCR). Lane 7 demonstrates the positive RT-PCR result from a sample from one child. (F) Corresponding histidyl-tRNA synthetase RT-PCR of samples in panel E. M, marker with 123-bp ladder; +, positive control; −, negative control.
A total of 20 children had acute HHV-6 infection with active viral replication, as demonstrated by the isolation of HHV-6 in culture, a positive DNA PCR, and a negative antibody titer (8 children were <6 months of age and had persistent maternal antibodies). Of these samples, 19 were positive by RT-PCR (sensitivity, 95%).
All 28 samples from children with evidence of previous HHV-6 infection (as reflected by a negative viral culture but a positive DNA PCR and a positive titer to HHV-6) were found to be negative for viral transcripts by our RT-PCR assay. This suggests that HHV-6 persists in a latent form in human PBMCs following primary infection. One positive RT-PCR result was observed in 56 samples that were negative by tissue culture and DNA PCR (i.e., HHV-6 negative). Of these 56 samples, 15 had a negative antibody titer, including the sample that was RT-PCR positive. The remaining samples had a geometric mean titer of 320, with a mean patient age of 4 months, suggesting the persistence of maternal antibodies. This indicates a low rate of false-positive results (1.2%, 1 of 84 samples). The specificity, therefore, was 98.8% (84 of 85) with a positive predictive value of 95% (19 of 20) and a negative predictive value of 98.8% (84 of 85 samples) (Table 1).
TABLE 1.
DNA PCR, viral culture, and RT-PCR results from samples from 106 patients tested for HHV-6
| Result of viral culture (DNA PCR result) | RT-PCR resulta
|
Predictive value (%)
|
Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|---|---|
| + | − | Positive | Negative | |||
| Positive (positive) | 19 | 3b | 95 | 95 | ||
| Negative (negative) | 1 | 55 | 98.8c | 98.8c | ||
| Negative (positive) | 0 | 28 | ||||
+, positive; −, negative.
Samples from two patients were positive for HHV-7 and HHV-6 by viral culture and DNA PCR but negative for HHV-6 by RT-PCR; these are excluded from analysis (see text).
Calculated by using both the Negative (negative) and Negative (positive) values.
Two patients had evidence of acute infection with HHV-7. Both HHV-7 and HHV-6 were isolated in culture, and DNA PCR results were positive for both viruses. The children had elevated antibody titers to HHV-6, consistent with prior infection with HHV-6. The RT-PCR assay was also negative in both patient specimens, suggesting prior latent infection with HHV-6. It is noteworthy that HHV-6 was not identified in culture from these two specimens until days 24 and 25 (routine positive cultures for HHV-6 are identified by days 10 to 14). This suggests that exogenous, latent HHV-6 was reactivated in vitro by the patient's primary HHV-7 infection, a phenomenon which has been previously reported (9). A cord blood sample used for refeeding one patient's culture was found to be positive for latent HHV-6 DNA on subsequent PCR testing and may have been the source of the late-appearing HHV-6 isolate in this case.
DISCUSSION
We have developed a novel RT-PCR assay for HHV-6. The primers chosen amplify mRNA transcripts corresponding to a major structural antigen of the virus, gp105. This antigen is expressed as a late γ gene and, therefore, should be indicative of productive viral replication (11). Furthermore, the primers were designed to amplify across well-defined splicing boundaries within gp105 mRNA. Thus, the assay allows one to readily distinguish between amplification of mRNA and amplification of residual DNA contamination.
HHV-6 RT-PCR used on more than 100 clinical specimens was found to be highly sensitive, detecting 95% of primary HHV-6 infections in children, with a 98.8% specificity and negative predictive value. In comparison, a plasma DNA PCR assay developed by Secchiero et al. to diagnose acute HHV-6 infection has a lower sensitivity (13). In initial studies, plasma DNA PCR for HHV-6 detected primary HHV-6 infection in 6 of 7 infants (13); subsequent studies demonstrated a sensitivity of only approximately 66% (5).
The highly sensitive nature of our assay could explain the single discordant result that we obtained. One positive specimen was RT-PCR positive but DNA PCR negative and tissue culture negative. This could be due to a false-positive RT-PCR. Alternatively, the RT-PCR assay may be sufficiently more sensitive to detect earlier infection than are standard tissue culture and DNA PCR techniques. Our DNA PCR assay can reliably detect 10 copies of the HHV-6 genome (7). However, total specimen volume, processing, and amplification techniques are distinctly different between the two PCR assays, making comparisons difficult. Additionally, neither PCR is designed to be quantitative.
Our new RT-PCR assay may help in delineating the pathogenesis of HHV-6 by differentiating acute or reactivated infection from past or latent infection. Currently, viral isolation, serology, and DNA PCR are used most frequently to detect HHV-6 infection. Although viral isolation indicates active infection and, combined with serology, can determine primary infection, viral isolation is difficult, costly, and time-consuming. More commonly, DNA PCR and serology are used for diagnosis. These assays, however, cannot differentiate between latent and reactivated infections.
RT-PCR assays have been demonstrated to reliably differentiate latency from reactivation in patients with other herpesvirus infections. The major immediate-early and late mRNA transcripts of HCMV detected by RT-PCR have been linked to viral replication (2, 6). The assay for HCMV was also compared to viral isolation and HCMV DNA PCR techniques (6). The RT-PCR had a lower sensitivity but a much higher specificity, suggesting that the RT-PCR assay was a more useful clinical tool (6).
Our RT-PCR assay proved useful in determining the pathogenesis of dual HHV-6 and HHV-7 infections in two patients. Although both HHV-6 and HHV-7 were isolated in culture and detected by DNA PCR, the RT-PCR was negative for HHV-6, and serology indicated past infection with HHV-6. Without this conflicting information from the RT-PCR, the recognition of subsequent contamination of exogenous, latent HHV-6 DNA from a CBMC suspension used to refeed the cultures would not have been suspected or detected. Latent HHV-6 has been shown to be reactivated by HHV-7 in vitro (9). Also, the presence of HHV-6 DNA in CBMCs has been described (4). Alternatively, reactivation of the patient's own HHV-6 in culture is also a possibility.
In conclusion, this RT-PCR assay appears to reliably differentiate between latent and actively replicating HHV-6. This is a potentially powerful tool for both clinical and basic science investigations and also for greater insight into the pathogenesis of this ubiquitous virus.
ACKNOWLEDGMENTS
Studies were supported in part by a General Clinical Research Center grant (5 MO1 RR00044) from the National Center for Research Resources, National Institutes of Health, and also by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (RO1 AI33020-06, KO4 AI01240, and RO1 AI34232).
We thank Maria Gouveia for her technical support in performing all the serologic assays on the specimens.
REFERENCES
- 1.Berneman Z N, Ablashi D V, Li G, Eger-Fletcher M, Reitz M S, Jr, Hung C, Brus I, Komaroff A L, Gallo R C. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc Natl Acad Sci USA. 1992;89:10552–10556. doi: 10.1073/pnas.89.21.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitsch A, Kirchner H, Dupke R, Bein G. Cytomegalovirus transcripts in peripheral blood leukocytes of actively infected transplant patients detected by reverse transcription-polymerase chain reaction. J Infect Dis. 1993;167:740–743. doi: 10.1093/infdis/167.3.740. [DOI] [PubMed] [Google Scholar]
- 3.Braun D K, Dominguez G, Pellett P E. Human herpesvirus 6. Clin Microbiol Rev. 1997;10:521–567. doi: 10.1128/cmr.10.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caserta M T, Hall C B, Schnabel K C, McIntyre K, Long C, Costanza M, Dewhurst S, Insel R, Epstein L G. Neuroinvasion and persistence of human herpesvirus 6 in children. J Infect Dis. 1994;170:1586–1589. doi: 10.1093/infdis/170.6.1586. [DOI] [PubMed] [Google Scholar]
- 5.Clark D A, Kidd I M, Collingham K E, Tarlow M, Ayeni T, Riordan A, Griffiths P C, Emery V C, Pillay D. Diagnosis of primary human herpesvirus 6 and 7 infections in febrile infants by polymerase chain reaction. Arch Dis Child. 1997;77:42–45. doi: 10.1136/adc.77.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gozlan J, Salord J, Chouaid C, Duvivier C, Picard O, Meyohas M, Petit J. Human cytomegalovirus (HCMV) late mRNA detection in peripheral blood of AIDS patients: diagnostic value for HCMV disease compared with those of viral culture and HCMV DNA detection. J Clin Microbiol. 1993;31:1943–1945. doi: 10.1128/jcm.31.7.1943-1945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall C B, Caserta M T, Schnabel K C, Long C, Epstein L G, Insel R A, Dewhurst S. Persistence of human herpesvirus 6 according to site and variant: possible greater neurotropism of variant A. Clin Infect Dis. 1998;26:132–137. doi: 10.1086/516280. [DOI] [PubMed] [Google Scholar]
- 8.Hall C B, Long C E, Schnabel K C, Schnabel K C, Caserta M T, McIntyre K M, Costanza M, Knott A, Dewhurst S, Insel R A, Epstein L G. Human herpesvirus-6 infection in children. N Engl J Med. 1994;331:432–438. doi: 10.1056/NEJM199408183310703. [DOI] [PubMed] [Google Scholar]
- 9.Katsafanas G, Schirmer E, Wyatt L, Frenkel N. In vitro activation of human herpesviruses 6 and 7 from latency. Proc Natl Acad Sci USA. 1996;93:9788–9792. doi: 10.1073/pnas.93.18.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luppi M, Barozzi P, Maiorana A, Marasca R, Torelli G. Human herpesvirus 6 infection in normal human brain tissue. J Infect Dis. 1994;169:943–944. doi: 10.1093/infdis/169.4.943. [DOI] [PubMed] [Google Scholar]
- 11.Mirandola P, Menegazzi P, Merighi S, Ravaioli T, Cassai E, Di Luca D. Temporal mapping of transcripts in herpesvirus 6 variants. J Virol. 1998;72:3837–3844. doi: 10.1128/jvi.72.5.3837-3844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeiffer B, Thomson B, Chandran B. Identification and characterization of a cDNA derived from multiple splicing that encodes envelope glycoprotein gp105 of human herpesvirus 6. J Virol. 1995;69:3490–3500. doi: 10.1128/jvi.69.6.3490-3500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Secchiero P, Carrigan D R, Asano Y, Benedetti L, Crowley R W, Komaroff A L, Gallo R C, Lusso P. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J Infect Dis. 1995;171:273–280. doi: 10.1093/infdis/171.2.273. [DOI] [PubMed] [Google Scholar]
- 14.Tsui F W, Siminovitch L. Isolation, structure and expression of mammalian genes for histidyl-tRNA synthetase. Nucleic Acids Res. 1987;15:3349–3367. doi: 10.1093/nar/15.8.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]