Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a pandemic. Diagnostic testing for SARS-CoV-2 has continuously been challenged due to several variants with diverse spike (S) and nucleocapsid (N) protein mutations []. SARS-CoV-2 variant proliferation potentially affects N protein-targeted rapid antigen testing. In this study, rapid antigen and reverse transcription PCR (RT-PCR) tests were performed simultaneously in patients with suspected coronavirus disease 2019 (COVID-19). Direct whole genome sequencing was performed to determine the N protein variations, and the viral assemblies were uploaded to GISAID. The genomes were then compared with those of global virus strains from GISAID. These isolates belonged to the B.1.1.7 variant, exhibiting several amino acid substitutions, including D3L, R203K, G204R, and S235F N protein mutations. The T135I mutation was also identified in one variant case in which the rapid antigen test and RT-PCR test were discordantly negative and positive, respectively. These findings suggest that the variants undetected by the Panbio COVID-19 rapid antigen test may be due to the T135I mutation in the N protein, posing a potential diagnostic risk for commercially available antigen tests. Hence, we recommend concomitant paired rapid antigen tests and molecular diagnostic methods to detect SARS-CoV-2. False-negative results could be rapidly corrected using confirmatory RT-PCR results to prevent future COVID-19 outbreaks.
KEYWORDS: COVID-19, SARS-CoV-2, Rapid antigen test, B.1.1.7 variant, N protein
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), similar to other RNA viruses, continually mutates, and new variants appear and eventually become dominant. Several SARS-CoV-2 genes have a tendency to evolve, including those encoding the nucleocapsid (N) and spike (S) proteins (Dilucca et al., 2020) [Au?1]. By the end of December 2020, new SARS-CoV-2 variants with multiple accumulated mutations had emerged, and these variants of concern (VOCs) have reportedly been associated with increased transmissibility or decreased effectiveness of available diagnostic tools (Boehm et al., 2021) [Au?1].
Lateral flow antigen detection diagnostics have long been deployed for various infectious diseases because of their convenience and short turnaround times (<15 minutes) (Dinnes et al., 2021). Previous studies have demonstrated the advantages and clinical performance of these rapid antigen test devices (Jungnick et al., 2021), and their data have shown favorable performance in detecting SARS-CoV-2 VOCs using rapid antigen testing. High sensitivities have been observed in high viral load samples using commercial rapid antigen tests (Hayer et al., 2021). However, N protein mutations in SARS-CoV-2 VOCs may potentially lead to false-negative results with some rapid antigen tests, despite a high viral load (Boehm et al., 2021). Del Vecchio et al. reported that genetic variants of the N gene might impair the ability to utilize antigen tests for diagnosis and mass testing efforts aimed at controlling virus transmission and the emergence of SARS-CoV-2 genetic variants (Del Vecchio et al., 2021). Several samples failed to generate a positive result in the lower cycle threshold (Ct) values. Whole genome sequencing (WGS) analysis of these isolates showed that they belonged to the B.1.1.7 variant, and concordant or discordant reverse-transcription PCR (RT-PCR) and antigen assay results revealed several amino acid substitutions in the N protein.
The aim of this study was to identify novel N protein mutation sites that affect antigen test performance. The Panbio COVID-19 rapid antigen test was used to screen patients with suspected coronavirus disease 2019 (COVID-19). In addition, another pharyngeal specimen was subjected to nucleic acid amplification testing (NAAT).
2. Materials and methods
2.1. Study design
A total of 2496 samples were collected from patients admitted to Tri-Service General Hospital (Taipei City, Taiwan) between May and July 2021. Participants (symptomatic or non-symptomatic with a history of contact with a confirmed COVID-19 case) underwent both rapid antigen testing and RT-PCR molecular assay, using paired nasopharyngeal swabs (NPS). Both tests were performed and interpreted by on-site technicians on the same day.
2.2. SARS-CoV-2 rapid antigen testing
Two simultaneous NPS were collected from symptomatic and asymptomatic participants []. The first pharyngeal specimen was analyzed using the Panbio COVID-19 Ag Rapid Test Device (Abbott, IL, USA) as per the manufacturer's instructions.
2.3. Clinical specimens and SARS-CoV-2 RT-PCR testing
The second pharyngeal specimen was used for NAAT confirmation. SARS-CoV-2 infection was confirmed using RT-PCR, as described previously, with some modifications (Jian et al., 2021a). Briefly, automated sample-to-result SARS-CoV-2 RT-PCR testing was performed using the LabTurbo AIO 48 system (LabTurbo, New Taipei City, Taiwan), with the LabTurbo AIO COVID-19 RNA testing kit for SARS-CoV-2 multiplex real-time RT-PCR, simultaneously detecting the SARS-CoV-2 N1 and E genes.
2.4. WGS of SARS-CoV-2
Of 2496 samples, 72 tested positive for SARS-CoV-2 by RT-PCR and 70 were scored as weakly positive or positive by the rapid test detecting the SARS-CoV-2 nucleocapsid protein. Here we focus on the SARS-CoV-2 variant (B.1.1.7) which might cause discordant result between paried antigen rapid test and RT-PCR. WGS of the five SARS-CoV-2 strains (denoted as TSGH42–46) was performed as described previously (Jian et al., 2021b; Jian et al., 2021c). All viral assemblies were uploaded to GISAID (https://www.gisaid.org/).
2.5. Amino acid variation mapping of SARS-CoV-2 genomes
CoV-GLUE (http://cov-glue.cvr.gla.ac.uk) was used to investigate the amino acid variations in these five strains and identify the possible novel N protein mutations in the SARS-CoV-2 lineage.
3. Results
3.1. Comparison of rapid antigen and RT-PCR test results
Data on the genome sequences of the five strains were deposited in the GISAID database, and these strains belonged to the B.1.1.7 variant (Table 1). The false-negative result obtained with the Panbio COVID-19 rapid antigen test was verified with three further tests with different lot numbers, and the same result was obtained.
Table 1.
SARS-CoV-2 variants identified by the Panbio COVID-19 Ag Rapid Test Device
| Reported case | Sex/age (years) | Accession ID | Lineage | N protein mutations | Panbio COVID-19 Ag Rapid Test | Ct valuea |
|---|---|---|---|---|---|---|
| Case 1 TSGH-42 | Male/48 | EPI_ISL_2693005 | B.1.1.7 | D3L, R203K, G204R, S235F | Positive | 17 |
| Case 2 TSGH-43 | Male/49 | EPI_ISL_2693006 | B.1.1.7 | D3L, R203K, G204R, S235F | Positive | 16 |
| Case 3 TSGH-44 | Female/2 | EPI_ISL_4096803 | B.1.1.7 | D3L, R203K, G204R, S235F | Positive | 20 |
| Case 4 TSGH-45 | Male/39 | EPI_ISL_4096805 | B.1.1.7 | D3L, R203K, G204R, S235F | Positive | 17 |
| Case 5 TSGH-46 | Male/44 | EPI_ISL_2693007 | B.1.1.7 | D3L, T135I, R203K, G204R, S235F | Negative | 16 |
Ct value: cycle threshold value for detecting the N1 gene.
3.2. Analysis of the N protein amino acid variants
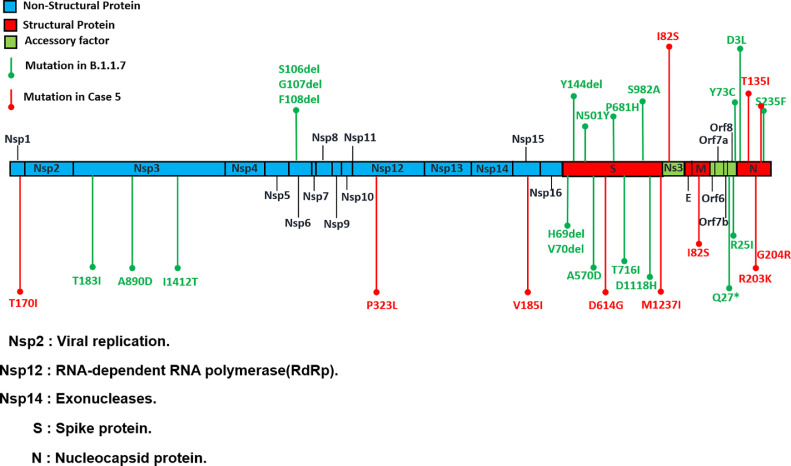
The analysis of the complete SARS-CoV-2 genomes of the five reported cases revealed several missense mutations. These mutations occurred in the N protein (Table 1) and included D3L, R203K, G204R, and S235F mutations in all five cases. A further investigation of the case with discordant results using the rapid antigen test and RT-PCR (case 5) was performed. Another novel amino acid substitution was observed in the T135I mutation in this case (Figure 1 ).
Figure 1.
Novel nucleotide substitutions found via whole genome sequencing analysis of the SARS-CoV-2 strain (case 5).
4. Discussion
The Abbott Panbio COVID-19 Ag Rapid Test Device, which has demonstrated moderate sensitivity and specificity, has been used as a screening tool (Fenollar et al., 2021). Through WGS analysis, this study revealed a pattern of amino acid substitution mapping around major epitopes of the N protein. The use of other rapid antigen testing methods that detect other SARS-CoV-2-specific genes or multi-target sites might be implemented in diagnostic laboratories as an alternative method. Inaccurate diagnostic test results will undermine efforts towards containing the COVID-19 pandemic. In the present study, case 5 with a high viral load was found to have a false-negative result by rapid antigen test [Au?1]. Such false-negative results could lead to more large-scale transmission [Au?1]. Sensitivity varies between rapid antigen tests and NAAT tests. Dinnes et al. reported an average sensitivity of 94.5% with Ct values <25 but 40.7% with Ct values >25 in 36 evaluations, while rapid molecular tests such as Xpert Xpress had an average sensitivity of 100% in two evaluations (Dinnes et al., 2021). As the inherent lower sensitivity may be offset by combination with NAAT methods, we propose implementing paired RT-PCR tests to counter the shortcomings of rapid antigen tests, especially their impaired ability to detect SARS-CoV-2 VOCs.
Rahman et al. observed that a high-frequency co-occurring mutation ratio (R203K and G204R) destabilized and decreased overall N protein structural flexibility (Rahman et al., 2021). The replacement of amino acid T by I might have affected coated antibodies that recognize the SARS-CoV-2 N protein epitope.
In conclusion, this study highlights the importance of the possibility of the N protein mutation in the B.1.1.7 VOC escaping detection by antigen tests in COVID-19 cases. Given the widespread proliferation of SARS-CoV-2 VOCs, additional sequencing data from new strains are required to test this hypothesis.
Author contributions
Conceptualization: Jung-Chung Lin, Kuo-Ming Ye, Chien-Wen Chen, De-Yu Lin, and Feng-Yee Chang. Data curation: Ming-Jr Jian, Hsing-Yi Chung, and Kuo-Sheng Hung. Formal analysis: Hung-Sheng Shang and Ming-Jr Jian. Investigation: Hsing-Yi Chung, Chih-Kai Chang, and Cherng-Lih Perng. Methodology: Chih-Kai Chang and Cherng-Lih Perng. Resources: Hung-Sheng Shang and Kuo-Sheng Hung. Software: Kuo-Sheng Hung. Supervision: Hung-Sheng Shang, Jung-Chung Lin, Kuo-Ming Ye, Chien-Wen Chen, Feng-Yee Chang, and Cherng-Lih Perng. Writing – original draft and revised draft: Ming-Jr Jian. Writing – review and editing: Hung-Sheng Shang.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Funding
This study was supported by Tri-Service General Hospital, Taipei, Taiwan, ROC (grant number TSGH-D-110100). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Ethics statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Tri-Service General Hospital (TSGHIRB No. C202005041), registered on February 8, 2021.
Patient consent
Written informed consent was obtained from the participants for the publication.
References
- Boehm E, Kronig I, Neher RA, Eckerle I, Vetter P, Kaiser L, et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021;27:1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M, Forcelloni S, Georgakilas AG, Giansanti A, Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12:498. doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vecchio C, Brancaccio G, Brazzale AR, Lavezzo E, Onelia F, Franchin E, et al. Emergence of N antigen SARS-CoV-2 genetic variants escaping detection of antigenic tests. medRxiv 2021. https://doi.org/10.1101/2021.03.25.21253802.
- Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2021;3 doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, et al. Evaluation of the Panbio COVID-19 rapid antigen detection test device for the screening of patients with COVID-19. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.02589-20. e02589–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer J, Kasapic D, Zemmrich C. Real-world clinical performance of commercial SARS-CoV-2 rapid antigen tests in suspected COVID-19: A systematic meta-analysis of available data as of November 20, 2020. Int J Infect Dis. 2021;108:592–602. doi: 10.1016/j.ijid.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian MJ, Chung HY, Chang CK, Lin JC, Yeh KM, Chiu SK, et al. Novel automated sample-to-result SARS-CoV-2 laboratory-developed RT-PCR assay for high-throughput testing using LabTurbo AIO 48 system. Clin Chim Acta. 2021;514:54–58. doi: 10.1016/j.cca.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian MJ, Chung HY, Chang CK, Lin JC, Yeh KM, Chen CW, et al. Clinical comparison of three sample-to-answer systems for detecting SARS-CoV-2 in B.1.1.7 lineage emergence. Infect Drug Resist. 2021;14:3255–3261. doi: 10.2147/IDR.S328327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian MJ, Chung HY, Chang CK, Hsieh SS, Lin JC, Yeh KM, et al. Investigation of one familial cluster of COVID-19 in Taiwan: Differentiation of genetic variation among isolates and implications for epidemiological investigation and surveillance by genomic assay. Infect Drug Resist. 2021;14:971–977. doi: 10.2147/IDR.S298451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnick S, Hobmaier B, Mautner L, Hoyos M, Haase M, Baiker A, et al. Detection of the new SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in five SARS-CoV-2 rapid antigen tests (RATs), Germany, March 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.16.2100413. https://doi.org/2100413.10.2807/1560-7917.ES.2021.26.16.2100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Islam MR, Alam A, Islam I, Hoque MN, Akter S, et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J Med Virol. 2021;93:2177–2195. doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]