Abstract
Although immunoassays are the most widely used protein measurement method, aptamer-based methods such as the SomaScan platform can quantify up to 7,000 proteins per biosample, creating new opportunities for unbiased discovery. However, there is limited research comparing the consistency of biomarker-disease associations between immunoassay and aptamer-based platforms. In a substudy of the TRIBE-AKI cohort, preoperative and postoperative plasma samples from 294 patients with previous immunoassay measurements were analyzed using the SomaScan platform. Inter-platform Spearman correlations (rs) and biomarker-AKI associations were compared across 30 preoperative and 34 postoperative immunoassay-aptamer pairs. Possible factors contributing to inter-platform differences were examined including target protein characteristics, immunoassay and SomaScan coefficients of variation, other assay characteristics, and sample storage time. The median rs was 0.54 (IQR 0.34-0.83) in postoperative samples and 0.41 (IQR 0.21-0.69) in preoperative samples. We observed a trend of greater rs in biomarkers with greater concentrations; the Spearman correlation between the concentration of protein and the inter-platform correlation was 0.64 in preoperative pairs and 0.53 in postoperative pairs. Of proteins measured by immunoassays, we observed significant biomarker-AKI associations for 13 proteins preop and 24 postop; of all corresponding aptamers, 8 proteins preop and 12 postop. All proteins significantly associated with AKI as measured by SomaScan were also significantly associated with AKI as measured by immunoassay. All biomarker-AKI odds ratios were significantly different (P <0.05) between platforms in 14% of aptamer-immunoassay pairs, none of which had high (rs >0.50) inter-platform correlations. Although similar biomarker-disease associations were observed overall, biomarkers with high physiological concentrations tended to have the highest-confidence inter-platform operability in correlations and biomarker-disease associations. Aptamer assays provide excellent precision and an unprecedented coverage and promise for disease associations but interpretation of results should keep in mind a broad range of correlations with immunoassays.
Introduction
Immunoassays have been the standard method for measuring protein concentrations in biological samples since the 1960s 1. Although multiplexed immunoassays exist, cross-reactivity concerns limit multiplex assessments to groups of only 20-50 proteins 2. Given the >20,000 proteins produced by the human body, the limitation of measuring fewer than 100 proteins at a time necessitates hypothesis-based approaches, often involving preliminary animal models, to select a small fraction of putative proteins for assay-based quantification 3.
In recent years, interest has increased in aptamer-based technologies such as SomaScan assays by SomaLogic, which use slow off-rate modified aptamers (SOMAmers) that are currently able to simultaneously assess over 7,000 protein targets 4,5. Aptamers are short oligonucleotide segments that bind specific antigens, mimicking the function of antibodies. Compared with immunoassays, aptamer-based assays can detect much less abundant proteins 6. Although immunoassays provide absolute concentrations, aptamer-based platforms provide only relative quantifications.
Inter-platform comparisons are complicated by the difficulty of obtaining gold-standard references for protein quantification 7,8. Although immunoassays have been a standard method of protein quantification for several decades, approximately one-half of 9,300 protein-specific antibodies tested failed validation by consistency with experimental and/or bioinformatics data 9. Thus as we compare immunoassay and SomaScan measurements, we cannot comment on which platform most accurately quantifies these biomarkers without comparing them with a gold standard, such as immunoassays that have been fully validated for specificity, linearity, interference, and relevance for physiology or pathophysiology. Such assays are not available for the majority of the biomarkers in this study.
While the translatability of untargeted proteomic approaches to targeted validation is an important question to the biomarker discovery community, there is currently only limited research comparing immunoassay and aptamer-based assay platforms. Our group previously reported inter-platform Spearman correlations between 26 aptamer-immunoassay pairs in preoperative plasma samples (median 0.40, IQR 0.23-0.70) and 31 pairs in postoperative samples (median 0.73, IQR 0.40-0.91) in a cohort of 54 AKI patients 10. Another group also reported heterogeneous correlations across 9 clinically-validated immunoassays and their corresponding aptamers in plasma samples (median 0.85, IQI 0.56-0.89) 11. Recently, the Trans-Omics for Precision Medicine (TOPMed) consortium observed for 63 proteins measured by the SomaScan and Myriad RBM immunoassay platforms median correlations of 0.48 (IQR 0.32-0.71) in a cohort of 176 participants and 0.58 (IQR 0.37-0.76) in a cohort of 371 participants who were current and former smokers 8. These initial reports provide the rationale for additional studies that offer more comprehensive comparisons of different assay platforms using biosamples from individuals with a range of clinical conditions and extend these early correlative studies to comparisons of biomarker-disease associations. In addition to extending the study of inter-platform operability to disease association, we aim to identify patterns between disease association and correlation and with underlying factors including experimental, assay, and protein characteristics. These patterns will inform investigators considering or interpreting unbiased discovery approaches about their translatability to targeted immunoassays.
The SomaScan platform has demonstrated potential for identifying promising biomarkers for rheumatoid arthritis, Duchenne muscular dystrophy, and cardiovascular disease 12–14. However, comparisons of disease-biomarker associations as measured by both aptamer and immunoassay platforms have not yet been explored sufficiently, especially across a range of inter-platform correlations. Thus, our objective in this investigation was not only to evaluate correlations between immunoassay and SomaScan measurements preoperatively, postoperatively, and between time points, but also to compare disease-biomarker associations for a finite set of biomarkers assayed using both platforms in patients with acute kidney injury (AKI). We evaluated patients who were part of the multicenter Translational Research Investigating Biomarker Endpoints in Acute Kidney Injury (TRIBE-AKI) study, a prospective observational cohort study investigating kidney injury outcomes after cardiac surgery15. Approximately 2 million cardiac surgeries are performed around the world each year, and AKI is a frequent complication of cardiac surgery16. However, despite decades of research, no therapy has proven effective for human AKI. Large consensus conferences have called for a better clinical paradigm that detects AKI earlier and more reliably using new plasma and urine biomarkers17. Several plasma biomarkers measured by immunoassay platforms in TRIBE-AKI have been shown to be associated with kidney injury15,18–23. In the present investigation, we compared immunoassay and SomaScan measurements and their associations with AKI for a set of plasma biomarkers. Importantly, we tested factors that may impact inter-platform operability including experimental, target protein, immunoassay, and aptamer characteristics. These factors include aptamer cross-reactivity with homologous proteins and orthogonal validation of aptamers by mass spectrometry or inferred validation by cis-pQTLs, as evaluated by other groups 8,10,24.
Methods
Cohort:
All patients were part of the TRIBE-AKI cohort, a prospective cohort study of adult patients undergoing cardiac surgery who are at high risk for post-operative AKI (at least AKIN stage 1)25. Patients were enrolled at 6 academic medical centers in North America from 2007 to 2012.
Sample collection:
All biosamples were collected preoperatively and postoperatively, soon after surgery when patients returned to the intensive care unit. Blood samples were initially collected via phlebotomy and stored in EDTA tubes. The tubes were subsequently centrifuged at room temperature for 15 minutes at 2000 rpm to separate plasma, which was then stored at −80 °C until all assays were performed. Samples were not subjected to repeated freeze/thaw cycles. All preoperative and postoperative samples were processed using the same protocol.
Biomarker measurement:
Immunoassay measurements were performed via Meso Scale Discovery (MSD), Beckman Coulter Access Array, Randox Biochip Array, or the Beckman Coulter Unicel assay between 2010 and 2015 for purposes of other TRIBE-AKI scientific studies 15,18–23. The mechanisms of the immunoassay platforms are: MSD, electro-chemiluminescence; Access, paramagnetic-chemiluminescence; Unicel, chemiluminescence; Biochip, multiplexed ELISA. Supplemental Table 1 lists the precise assays used for plasma biomarkers and sample storage time. All laboratory technicians were blinded to patients’ clinical information. Inter-assay coefficients of variation (%CVs) and intra-assay %CVs were evaluated from a subset of TRIBE plasma samples chosen at random. Inter-assay %CVs were determined by averaging the %CV of the same set of samples analyzed in duplicate on different plates on different days. Intra-assay %CVs were determined by averaging the %CV of the same set of samples analyzed in duplicate on the same plate.
In 2019, concentrations of protein analytes were quantified using a multiplexed modified DNA-based aptamer technology (SomaScan assay), which transforms individual protein concentrations into corresponding modified aptamer (SOMAmer reagent) concentrations that can be quantified by standard DNA microarrays in relative fluorescence units. The SomaScan V4 assay included 5,284 aptamers that mapped to 4,746 unique proteins or protein complexes in the Universal Protein Resource (UniProt) databases.
Protein analyte measurements underwent the SomaScan data standardization and normalization process 26. Hybridization control normalization was first applied to each sample based on a set of hybridization control sequences to correct for systematic biases during sample hybridization. Adaptive normalization by maximum likelihood was then applied to measures within a plate to remove sample or assay biases that may have occurred because of pipetting variation, variation in reagent concentrations, assay timing, and other sources of systematic variability within a single plate assay run. Finally, plate-to-plate variation was corrected using calibrator samples on each assay plate for each aptamer. Protein analytes with calibration factor greater or less than the median calibration factor (0.4) were excluded from all analyses.
Statistics:
Analyses were limited to patients with protein measurements on both platforms. Aptamers flagged by SomaLogic (epidermal growth factor, IL2, IL18, interferon gamma, and one of the 3 aptamers for KIM1) and immunoassay measurements with >50% of measurements outside of the limits of detection (preoperative TnI, NGAL, IL-1, and IL-4) were excluded. For immunoassay values at the lower or upper limits of detection, we used the value of the limit of quantification at its respective bound. SomaScan and immunoassay measurements were logarithmically-transformed (base 2), and values were truncated ±5 median absolute deviations from the median. We examined the change in protein values from preop to postop as the post:pre ratio (postop divided by preop) for patients with samples available at both time points. The majority of measurements demonstrated a normal distribution after logarithmic transformation.
Proteins were matched whenever possible by UniProt ID, considering additional annotation information whenever necessary. Spearman correlation coefficients were assessed between immunoassay measurements and SomaScan measurements. We classified the Spearman correlations in groups of low (<0.5), moderate (0.5-0.75), and high (>0.75) correlation 10,27,28.
We tested for statistically significant differences in inter-platform Spearman correlations from preoperative samples to postoperative samples using a two-tailed test of non-overlapping variables and dependent groups using Zou’s confidence interval, as implemented in the “cocor” library available for R29,30. For this analysis, we used the subset of patients for each biomarker whose plasma samples were quantified both preoperatively and postoperatively by both SomaScan and immunoassays.
We fit univariate logistic regression models to estimate the AKI odds ratio (OR) per doubling of biomarker concentration. ORs were compared between platforms using a two-sample z test with P <0.05. Target protein molecular weights and basal pIs were obtained from the PhosphoSitePlus database for post-translationally modified proteins31. Visual analysis and Spearman correlations were used to assess potential relationships of molecular weight, basal pI, inter-assay %CV, intra-assay %CV, assay type, and cross-reactivity with inter-platform measurement correlation and the ratio of aptamer-AKI OR and immunoassay-AKI OR. Information on nonspecific aptamer binding, mass spectrometry validation, and pQTLs was compiled and integrated from previous large SomaScan studies5,32. We evaluated storage time effects through linear regression between immunoassay measurements (log2-transformed) and storage time. All analyses were completed in R 3.6.2.
Study approval:
All patients provided written informed consent, and each institution’s review board approved the study protocol. The detailed methods used in the TRIBE-AKI cohort have been described previously 15,18,21
Results
Patient characteristics:
In our sample, 294 participants had both immunoassay biomarker measurements and SomaScan measurements for at least one biomarker at preoperative and postoperative time points. Details of individual biomarkers are reported in Supplemental Table 1. Of the 294 patients, 138 (41%) had AKI, as defined by at least Acute Kidney Injury Network (AKIN) stage 1 at any time during the index hospitalization; 84 (29%) patients had stage 1 AKI, 29 (10%) had stage 2 AKI, and 25 (9%) had stage 3 AKI. Other clinical characteristics are shown in Table 1.
Table 1: Participant characteristics by AKI status.
Values are N (%) or mean ± SD. CABG, coronary artery bypass graft; eGFR, estimated glomerular filtration rate.
| Characteristic | Overall | AKI | No AKI | |
|---|---|---|---|---|
| (N = 294) | (N = 138) | (N = 156) | ||
| Age at time of surgery, y | 71 ± 10 | 71 ± 10 | 71 ± 10 | |
| Men | 208 (70.7) | 100 (72.5) | 108 (69.2) | |
| White race | 279 (94.9) | 130 (94.2) | 149 (95.5) | |
| Diabetes | 117 (39.8) | 59 (42.8) | 58 (37.2) | |
| Hypertension | 230 (78.2) | 116 (84.1) | 114 (73.1) | |
| Congestive heart failure | 73 (24.8) | 44 (31.9) | 29 (18.6) | |
| Preoperative serum creatinine, mg/dL | 1.11 ± 0.394 | 1.17 ± 0.404 | 1.06 ± 0.380 | |
| Preoperative eGFR, mL/min/1.73 m2 | 67.3 ± 20.0 | 64.7 ± 20.7 | 69.6 ± 19.1 | |
| Type of surgery | Elective | 243 (82.7) | 103 (74.6) | 140 (89.7) |
| Urgent | 51 (17.3) | 35 (25.4) | 16 (10.3) | |
| CABG valve use | 232 (78.9) | 104 (75.4) | 128 (82.1) | |
Biomarker measurements:
Immunoassay measurements for 34 biomarkers were matched to 39 aptamers. Immunoassays were selected independently of aptamer results. All immunoassays measured in the TRIBE cohort targeting proteins matched by a SomaScan aptamer were included in this analysis.
Of 5,284 protein analytes quantified by SomaScan, 4,484 (85%) passed SomaLogic’s calibration filter quality control standard and were included in our analyses, including 35 of the 39 preliminary matched aptamers. After further excluding immunoassays with >50% of samples outside of assay detection limits, our analyses included 26 immunoassay biomarkers matched to 30 aptamers in preoperative samples and 30 immunoassay biomarkers matched to 34 aptamers in postoperative samples (Supplemental Table 2). After exclusion, all immunoassays in our analysis had ≤16% missing values in preop samples and ≤12% missing in postop samples. Some biomarkers were targeted by multiple aptamers, including vascular endothelial growth factor receptor 1 (VEGFR1), vascular endothelial growth factor d (VEGFd), kidney injury molecule 1 (KIM-1), and tumor necrosis factor receptor 2 (TNF-R2).
Immunoassay inter-assay %CVs were calculated from blind duplicates from the TRIBE study for 23 of the 30 immunoassays in this study, and had a median %CV of 9.4 (IQR 6.8-13.5) and a range 4.5-18.0. (Supplemental Table 1). SomaScan inter-assay %CVs for all 34 aptamers in our study were calculated from 197 pairs of blind duplicates from the Atherosclerosis Risk in Communities (ARIC) study, 5% of the cohort selected at random, and had a median %CV of 4.3 (IQR 3.9-5.4) and a range of 2.4-7.9 (Supplemental Table 2)33. While SomaScan inter-assay %CVs were significantly lower than immunoassay inter-assay %CVs, we were unable to measure enough duplicates in TRIBE using SomaScan to provide %CVs from the TRIBE cohort.
Correlation of immunoassay versus SomaScan measurements:
The median Spearman correlation coefficients for aptamer-immunoassay pairs were 0.54 (IQR 0.34-0.83) in postoperative samples and 0.41 (IQR 0.21-0.69) in preoperative samples (Supplemental Table 2). The median inter-platform correlation coefficient for the post:pre ratio (postoperative level divided by preoperative level) observed in samples was 0.43 (IQR 0.28-0.74). All aptamer-immunoassay pairs in our analyses were categorized into low (rs <0 .5), moderate (0.5≤ rs <0.75), or high (rs ≥0.75) correlation groups (Table 2). We observed overall greater inter-platform correlations in postoperative samples than in preoperative samples. For preoperative samples, 20% of pairs had high correlations, 27% had moderate correlations, and 53% had low correlations. For postoperative samples, 35% had high correlations, 24% had moderate correlations, and 41% had low correlations. Some of the biomarkers in clinical use and/or commonly used in research studies demonstrated high correlations (including tumor necrosis factor receptor 1 [TNF-R1], and TNF-R2, N-terminal pro-B-type natriuretic peptide [NT-proBNP], Cystatin C [CST3], neutrophil gelatinase-associated lipocalin [NGAL], interleukin 6 [IL-6], ST2, and troponin I [TnI]) and moderate correlations (creatine kinase-MB [CK-MB], galectin 3 [Gal-3], and high-sensitivity troponin T [hsTnT], KIM-1,). Of the 30 aptamer-immunoassay pairs in our analyses at both time points, 37% demonstrated significantly greater inter-platform correlations postoperatively by a median difference of 0.40 (range: 0.06-0.75), 47% did not significantly differ, and 16% were significantly lower by a median difference of 0.16 (range: 0.06-0.21).
Table 2: Correlation groups for 30 preoperative and 34 postoperative aptamer-immunoassay pairs.
Numbered biomarkers (e.g. VEGFd(1), VEGFd(2)) are used to identify unique immunoassay-aptamer pairs for biomarkers targeted by multiple aptamers. Details about each immunoassay-aptamer pair are described in Supplemental Table 2.
| Preoperative Correlation | Postoperative Correlation | ||||
|---|---|---|---|---|---|
|
| |||||
| Low: rs <0.5 |
Moderate: 0.5 ≤ rs <0.75 |
High: rs ≥0.75 |
Low: rs < 0.5 |
Moderate: 0.5 ≤ rs <0.75 |
High: rs ≥0.75 |
|
| |||||
| BNP-32 | hsTnT | NT-proBNP | BNP-32 | CK-MB | hsTnT |
| CK-MB | hFABP | CST3 | hFABP | IL-10 | NT-proBNP |
| MCP-1 | IL-6 | ST2 | IL-1 | VEGFR1(2) | TnI |
| IL-10 | IL-8 | TNF-R1 | IL-4 | VEGFd(1) | MCP-1 |
| IL-12 | VEGFd(1) | TNF-R2 (1) | IL-12 | KIM-1(1) | CST3 |
| IL-13 | VEGFd(2) | TNF-R2 (2) | IL-13 | YKL-40 | NGAL |
| TNFa | KIM-1(1) | TNFa | Gal3 | IL-6 | |
| bFGF | YKL-40 | bFGF | TNF-R1 | IL-8 | |
| PlGF | PlGF | VEGFR1(1) | |||
| VEGFR1(1) | VEGFd(2) | ST2 | |||
| VEGFR1(2) | VEGFc | TNF-R2(1) | |||
| VEGFc | VEGF | TNF-R2(2) | |||
| VEGF | Tie2 | ||||
| Tie2 | KIM-1(2) | ||||
| KIM-1(2) | |||||
| Gal3 | |||||
BNP-32, B-type natriuretic peptide 32; CST3, cystatin C; MCP-1, monocyte chemoattractant protein; PlGF, placenta growth factor; TNFα, tumor necrosis factor alpha; VEGF, vascular endothelial growth factor; VEGFc, vascular endothelial growth factor c.
Immunoassay versus SomaScan biomarker-AKI associations:
Across both assay platforms, biomarker associations with AKI were stronger for postoperative measurements than for preoperative measurements. The median unadjusted ORs of AKI from postoperative samples measured by immunoassay was 1.46 per doubling (range: 0.72-6.72 and by SomaScan was 1.24 per doubling (range 0.39-5.97) (Figure 1, Supplemental Table 3). Of the 30 proteins measured by immunoassays, we observed significant biomarker-AKI associations for 13 proteins preoperatively and 24 proteins postoperatively, of which only postoperative PlGF was protective. Of all aptamers in our study, we observed significant associations for 8 proteins preoperatively and 12 proteins postoperatively, all of which had increased risk association with AKI. Interestingly, all proteins with significant biomarker-AKI associations as measured by SomaScan were also significantly associated with AKI as measured by immunoassay (Supplemental Table 4).
Figure 1. Biomarker-AKI ORs by platform for each inter-platform correlation group.
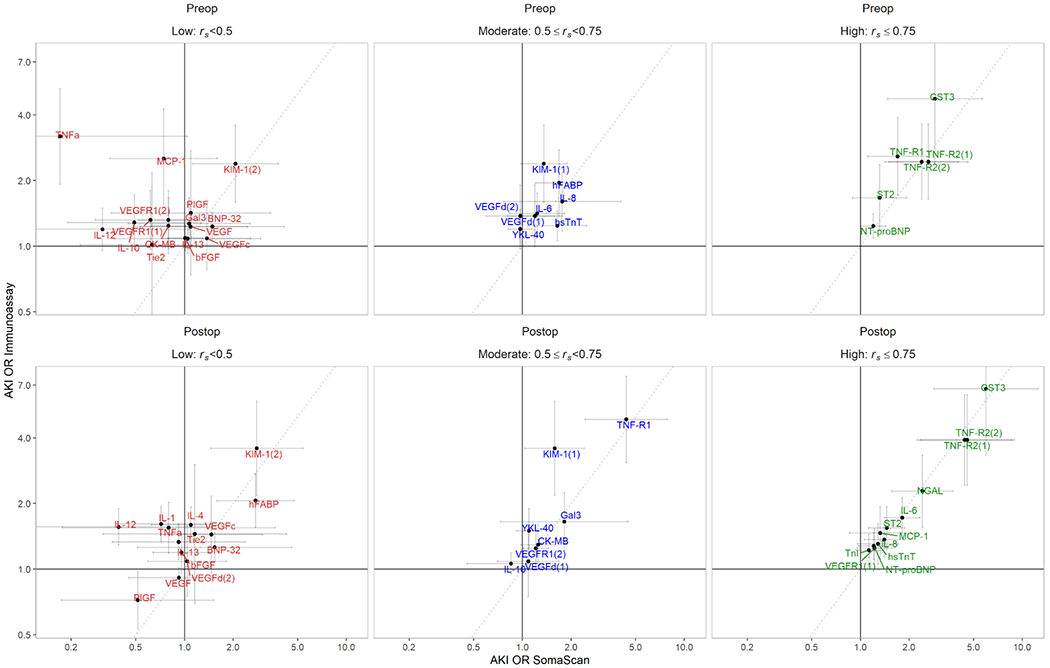
Biomarker-AKI associations were more similar in postoperative (Postop) plasma samples (bottom panels) than preoperative (Preop) plasma samples (top panels). Biomarkers with high inter-platform correlation (right panels, green) tended to have more consistent biomarker-AKI ORs per doubling between platforms than moderate- (blue, center) and low-correlation (left, red) biomarkers. Scatterplots of biomarker-AKI ORs per doubling, as measured by immunoassay (y-axis) and SomaScan (x-axis) platforms. Error bars represent 95% CIs for ORs. The dotted gray lines represent the identity (y = x) line. Preop/low-correlation TNFa SomaScan error bar and both Preop and Postop/high-correlation CST3 immunoassay error bars are truncated for visualization purposes.
We also observed that biomarkers with higher measurement correlations had more concordant ORs between the two assay platforms (Figure 1). In postoperative samples, significantly different (rs <0.05) ORs were observed for 14% of low-correlation biomarkers, 25% of moderate-correlation biomarkers, and none of the high-correlation biomarkers. In preoperative samples, 25% of low-correlation biomarkers, 13% of moderate-correlation biomarkers, and none of the high-correlation biomarkers demonstrated significantly different ORs. Overall, the platforms demonstrated similar disease associations, especially for biomarkers with high inter-platform correlations.
Among the five preoperative (CK-MB, MCP-1, IL-12, TNFa, KIM-1(1)) and four postoperative pairs (IL-1, IL-12, KIM-1(1), YKL-40) for which the biomarker-AKI ORs were significantly different between platforms, the conclusions of the studies based on a P <0.05 threshold would have changed for three preoperative (MCP-1, TNFa, KIM-1(1)) and three postoperative pairs (IL-1, IL-12, YKL-40). In all of these pairs, the immunoassay detected risk associations (P <0.05), whereas the corresponding SomaScan biomarker detected no significant associations.
Biomarker-AKI “protective” associations (OR <1, no P threshold) were more common in SomaScan measurements than in immunoassay measurements: postoperatively 24% versus 6% and preoperatively, 33% versus 0%, respectively. However, the only statistically significant protective association was by postoperative PlGF as measured by immunoassay; the rest of these putative protective associations were not statistically significant, and therefore, our conclusions related to “protective” proteins would not have changed based on which assay platform was used. Nonetheless, we report these observed trends because the significance of biomarker-disease associations vary with cohorts, diseases, sample sizes, and models.
Factors associated with inter-platform differences in biomarker correlations and biomarker-AKI associations:
We explored patterns in correlations and in the ratio of SomaScan-AKI ORs versus immunoassay-AKI ORs in relation to median biomarker concentrations, as measured by immunoassays, immunoassay type, immunoassay intra-assay %CV, immunoassay inter-assay %CV, target protein molecular weight, target protein basal isoelectric point, and biosample storage time. We also explored the role of aptamer cross-reactivity and possible orthogonal validation of aptamers by mass spectrometry and cis-protein quantitative trait loci (pQTLs), as tested in other studies, in inter-platform correlations and OR discrepancies 5,8,24. Factors tested in relation to the inter-platform correlations and OR discrepancies are summarized in Table 3.
Table 3:
Factors influencing inter-platform correlation and biomarker-AKI association discrepancies
| Factors | Inter-platform Correlation | Biomarker-AKI Association Discrepancy | Results |
|---|---|---|---|
| Biomarker concentration (among samples within a biomarker, and across biomarkers) | Strong | Unclear beyond rs:OR relationship | Figures 2 and 3, Supplemental Figure 1 |
| Immunoassay type | Weak; more research needed | None; more research needed | Supplemental Figure 2 |
| SomaScan inter-assay %CV | None-to-weak | None | Supplemental Figure 3 |
| Immunoassay inter-assay %CV | None | None | Supplemental Figure 3 |
| Immunoassay intra-assay %CV | None | None | Supplemental Figure 4 |
| Target molecular weight | None | None | Supplemental Figure 5 |
| Target basal pI | None | None | Supplemental Figure 6 |
| Storage time | None | - | Supplemental Figure 7 |
| Aptamer cross-reactivity with homologous proteinsA | None; more research needed | Low-correlation cross-reactive were more concordant; more research needed | Supplemental Figure 8 |
| Aptamer orthogonal validation by mass spectrometryB | Some; more research needed (sparse) | - | cystatin C, CK-MB, Gal3, hFABP, bFGF |
| Aptamer orthogonal validation by cis-pQTLB | Weak; more research needed | None | BNP-32, NT-proBNP, CK-MKB, CST3, PlGF, VEGFc, VEGF, Tie2, Kim-1, YKL-40, Gal3, ST2, and TNF-R2 |
Among the biomarker pairs included in our analysis at each timepoint, we observed a strong correlation between biomarker median immunoassay concentrations and inter-platform measurement correlations (Figure 2). The Spearman correlation between the molar concentration of protein and the inter-platform correlation was 0.64 in preoperative pairs and 0.53 in postoperative pairs.
Figure 2: Across biomarkers, elevated median immunoassay concentrations are associated with greater inter-platform correlations.
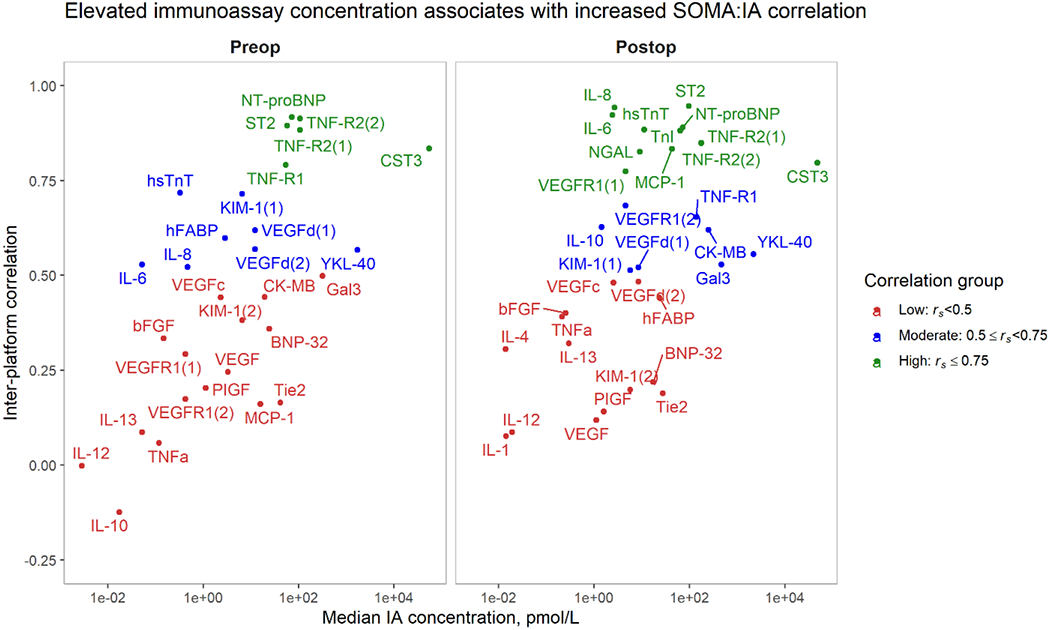
Biomarkers with greater median immunoassay concentration tend to have greater inter-platform Spearman correlations (rs). This trend is observed in both preoperative samples (left) and postoperative samples (right). Median immunoassay (IA) concentrations (x-axis) versus inter-platform Spearman correlations. Correlation groups are indicated by color.
We observed that higher-concentration biomarkers also tended to have more consistent biomarker-AKI ORs between platforms (Supplemental Figure 1) and observed the opposite with lower-concentration biomarkers. However, we are unable to distinguish a distinct relationship between biomarker concentrations and biomarker-AKI association discrepancies, beyond their relationship with inter-platform correlations and, subsequently, the relationship between inter-platform correlation and inter-platform biomarker-AKI association concordance.
We also explored how changes in biomarker concentrations from preop to postop affected inter-platform correlations of biomarkers included in our study at both timepoints. We hypothesized that the greater inter-platform correlations observed in postop samples could be attributed in part to elevated postop biomarker concentrations in plasma (Figure 3). This is particularly relevant for immunoassays, especially in preop samples where we may encounter issues of linearity or variation in the range near the lower limit of detection. We observed that many biomarkers with highly elevated postop immunoassay concentrations (compared with preoperative concentrations) demonstrated significantly higher inter-platform correlations in postop samples than in preop samples. In contrast, biomarkers without substantial postop increases in concentration had relatively smaller inter-platform differences between the two time points. We did not observe decreases postoperatively of an order of magnitude in any proteins quantified in this study. This suggests that among samples of a given biomarker, those with higher concentrations tend to have more consistent measurements between the immunoassay and SomaScan platforms.
Figure 3. Biomarkers with elevated postoperative concentrations have higher inter-platform correlations than preoperatively.
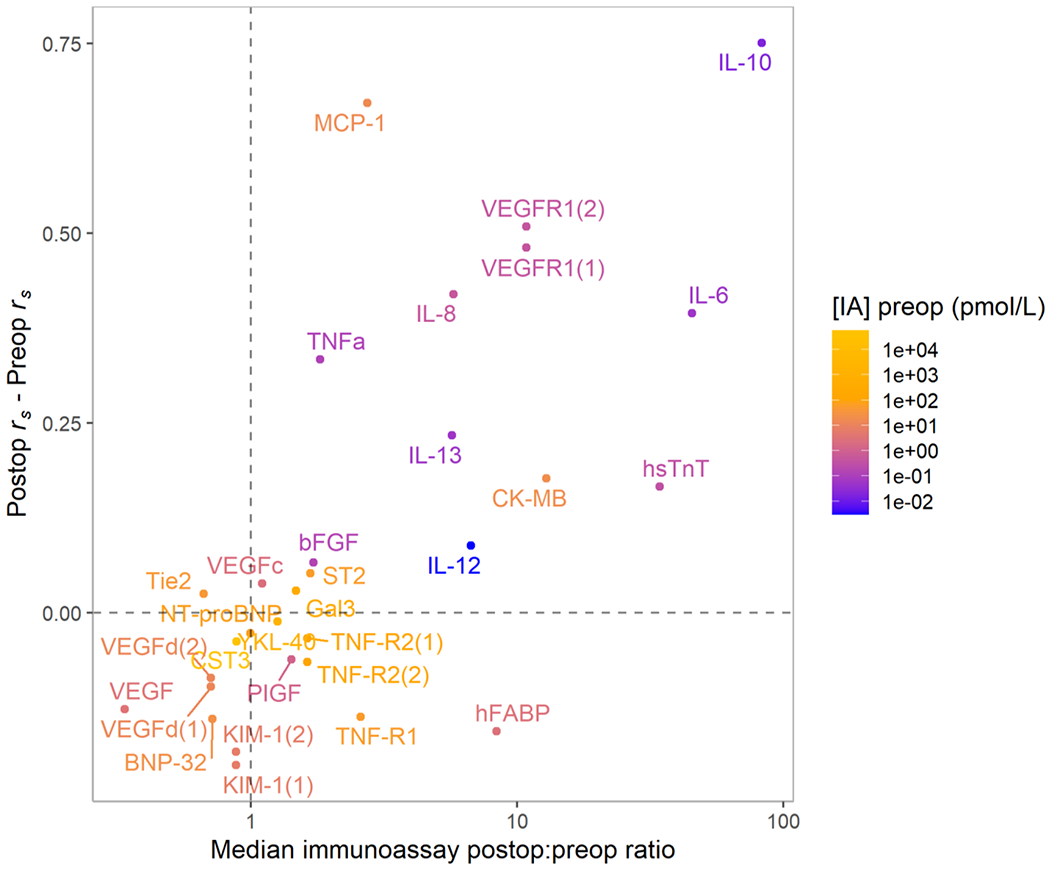
Scatterplot of difference between inter-platform Spearman correlation (rs) observed in postoperative samples versus preoperative samples (y-axis) versus the median increase observed in participants’ postoperative versus preoperative immunoassay protein concentrations ([IA] = concentration immunoassay). Color indicates the preoperative immunoassay protein concentration to contextualize the change in protein levels from the preoperative to postoperative time point. We observed large increases in inter-platform correlations in biomarkers that were greatly elevated postoperatively, such as IL-10. The proteins for which these increases in inter-platform correlation were observed also tended to be present at low concentrations in preoperative samples. Biomarkers that did not experience large median changes had smaller differences in inter-platform correlations between time points, as seen in the cluster around the origin.
Similarly, we observed inter-platform correlations of protein post:pre ratio tended to be greater and more similar to postoperative correlations in biomarkers substantially elevated postoperatively, including hsTnT, CK-MB, IL-6, IL-8, IL-10, and VEGFR1. (Supplemental Table 2). Conversely, we observed lower post:pre ratio correlations for biomarkers without substantial increases from preoperative to postoperative samples, including BNP-32, NT-proBNP, VEGF, VEGFd, CST3, and Tie2.
Thus, we observed the association between biomarker concentrations and inter-platform correlations not only among the 30 different biomarkers in our analysis (Figure 2), but also for a single biomarker measured at different timepoints with different concentration distributions (Figure 3).
In our investigation, 24 (70%) of 34 immunoassays were measured using the MSD platform, with the remainder from Coulter Access (4, 12%), UniCel (3, 9%), and Biochip (3, 9%). We compared only MSD immunoassays vs SomaScan to partially address the heterogeneity of immunoassay mechanisms in our comparison, and found overall similar results. Between MSD and SomaScan, we observed median rs postop of 0.52 (IQR 0.32-0.77) and preop of 0.44 (IQR 0.17-0.59). We continued to observe the strong association between median biomarker concentration and inter-platform operability, with Spearman correlation 0.54 postop and 0.69 preop. No relationship was found across all immunoassay type and either inter-platform correlations or inter-platform association discrepancies (Supplemental Figure 2).
While we hypothesized that we might observe a correlation between inter-platform correlation and %CVs of either platform, overall %CVs on either platform were similar across low, moderate, and high-correlation proteins (Supplemental Figure 3). We observed only with a weak trend between lower SomaScan %CV and greater inter-platform correlation. Additionally, no strong relationships between immunoassay intra-assay %CVs and inter-platform correlations or biomarker-AKI discrepancies were observed (Supplemental Figure 4).
Target protein molecular weights ranged from 3.4 kDa to 134 kDa, and target protein basal pIs ranged from 4.54 to 11.18 (Supplemental Table 1). We did not observe a relationship between either target protein molecular weight and target protein basal pI versus either inter-platform correlations or inter-platform OR discrepancies (Supplemental Figures 5 and 6).
Biosamples were collected in various years, and immunoassay measurements for different biomarkers were performed in various years. Among biomarkers in this study, samples had a median storage time ranging from 1.2 to 6.9 years before measurement by immunoassay (Supplemental Table 1). All SomaScan measurements were completed at the same time, with a median storage time of 10.8 years. No storage time effects were observed in either immunoassay or SomaScan measurements (Supplemental Figure 7).
The idea that the cross-reactivity of binding reagents for structurally-related proteins in either aptamer-based assays or immunoassays could play a role in inter-platform discrepancies has been previously suggested 8,10. Using the results of cross-reactivity tests conducted by Williams et al., we explored relationships between aptamer cross-reactivity and both inter-platform correlations and biomarker-AKI OR discrepancies 24. Cross-reactivity testing results were available for 31 of 34 aptamers in our study, 8 of which demonstrated cross-reactivity and 23 of which did not. We found similar rates of moderate-to-high inter-platform correlation between aptamers that did not demonstrate cross-reactive binding (61% in postoperative samples, 42% in preoperative samples) and those that did (50% in postoperative samples, 50% in preoperative samples) (Supplemental Figure 8). We observe more significantly different low-correlation pairs in the group where aptamers did not demonstrate cross-reactivity, and concordant ORs in both groups between moderate-correlation and high-correlation pairs.
To investigate the effect of aptamer-binding specificity on inter-platform correlations and inter-platform biomarker-disease association discrepancies, we compared the results from Emilsson et al. 5, who confirmed mass spectrometry binding in multiple media as a means of orthogonal validation for aptamer specificity. Only five of 34 analyzed aptamers were tested by mass spectrometry, one of which was confirmed in plasma (cystatin C, high preoperative/high postoperative correlation), 3 of which were confirmed in serum but not plasma (CK-MB, Gal3, human heart-type fatty acid binding protein [hFABP]), and one of which, basic fibroblast growth factor (bFGF), was confirmed only in cell lysates. Interestingly, although cystatin C demonstrated high inter-platform correlations at both time points, biomarkers that were confirmed only in serum or in cell lysates and failed to be confirmed in plasma had low to moderate correlations. However, there is little overlap between the biomarkers in our study and those in the Emilsson study, limiting our ability to draw further conclusions regarding orthogonal validation by mass spectrometry.
“Inferred validation” through cis-pQTLs identified by Emilsson et al.5 in the AGES-Reykjavik cohort was available for 15 of the 34 aptamers in our study (Table 3). Inferred validation by cis-pQTLs has been suggested by Raffield et al.8 to support inter-platform correlations. However, we did not observe significant enrichment in moderate- to high-correlations in the 15 biomarkers with cis-pQTL inferred validations; 33% high-, 20% moderate-, and 47% low-correlation in postoperative samples; and 33% high-, 13% moderate-, and 54% low- correlation in preoperative samples. No relationships with biomarker-disease association discrepancies were found beyond those noted above for correlations.
Discussion
We observed a broad range of correlations between immunoassay and SomaScan measurements for 34 immunoassay-aptamer pairs from preoperative and postoperative plasma samples from subjects undergoing cardiac surgery. Biomarkers with greater plasma concentrations, as measured by immunoassays, demonstrated both stronger inter-platform correlations and more concordant biomarker-AKI associations. More biomarkers demonstrated moderate- to high-correlations postoperatively (59%) than preoperatively (47%), and we observed significantly greater inter-platform correlations in postoperative samples than in preoperative samples in 37% of the immunoassay-aptamer pairs. Although high-correlation aptamer-immunoassay pairs had no statistically significant inter-platform discrepancies in AKI ORs, significant inter-platform AKI OR discrepancies were observed when inter-platform measurement correlations were low to moderate. Within these biomarkers, more significant (P<0.05) AKI associations were found by immunoassay than by SomaScan, and all significant biomarkers as measured by SomaScan were significant by immunoassay. We did not find strong relationships between immunoassay type, inter-assay and intra-assay CVs, molecular weight, basal pI, or storage time and either inter-platform correlations or biomarker-AKI associations. We also did not find a strong relationship between either aptamer cross-reactivity or inferred validation by cis-pQTLs versus inter-platform concordance using cross-reactivity and orthogonal validation data from previous studies.
A novel part of our investigation is the comparison of disease-biomarker associations for measurements obtained from the two assay platforms for 34 postoperative and 30 preoperative aptamer-immunoassay pairs. Demonstrating similar biomarker-disease associations across many biomarkers, especially high-correlation biomarkers, gives investigators confidence in translating findings from high-throughput proteomics platforms to those based on standard immunoassays, as one might in a nontargeted proteomics approach. Immunoassays identified more biomarkers as significantly associated with AKI, and interestingly, all proteins as measured by SomaScan with significant biomarker-AKI associations were also significantly associated as measured by immunoassay. Despite greater postoperative correlations and similar fraction discrepant odds ratios, agreement in significance of biomarker-AKI associations was lower in postop samples than in preop samples. Possible systemic biases that may contribute to this pattern of discrepancy include firstly, that biomarkers are traditionally discovered and validated through immunoassays, and that even when using different immunoassay platforms, the antigen binding behaviors of immunoassays are more similar than the oligonucleotide binding of an aptamer. Secondly, part of the SomaScan normalization pipeline includes scaling protein dilution bins to match their medians to those measured in an external healthy reference population. In individuals and populations with broad disruptions from a healthy plasma proteome, such as cardiac surgery patients, this normalization could potentially attenuate the strength of associations by reducing more extreme values if they are associated with outcome.
Our inter-platform correlation findings are similar to those reported previously by our group and by other groups8,10,34. Our SomaScan-immunoassay correlations are similar to correlations reported between two different immunoassays for a single protein target; researchers who compared measurements of a range of 20 biomarkers in two different commercially available Luminex xMap-based immunoassays from different manufacturers observed inter-immunoassay correlations >0.50 for 60% of biomarkers34. Further examples of inter-immunoassay platform comparison studies include one reporting 39% of proteins with correlation >0.50, and another reporting 35% of proteins with correlation >0.5035,36. Altogether, SomaScan-immunoassay correlations were moderately correlated on average, with improved correlation in high-concentration biomarkers. We observed this trend both across multiple measurements of single biomarkers at two time points, and across different biomarkers found at different physiological levels. Biomarker-disease associations corresponded well between platforms, with few significantly different ORs per biomarker doubling, even among low-correlation aptamers (rs<0.5). High-correlation aptamers demonstrated especially similar ORs.
Another novel part of our investigation is the characterization of the correlation between measured differences between preoperative and postoperative time points. We observe that these correlations have a comparable distribution to correlations of preoperative samples, lending confidence to inter-platform operability of high-correlation aptamer-immunoassay pairs across diverse physiological conditions. Greater biosample protein abundance for specific biomarkers can give investigators further confidence in inter-platform operability. We found that both inter-platform correlations and inter-platform AKI OR concordance improved in samples with higher biomarker concentrations. We observed this increase in inter-platform concordance not only in postoperative samples with significant increases in protein levels versus preoperative samples, but also when comparing the 30 different biomarkers in the study. Biomarkers with greater endogenous concentrations (e.g., cystatin C, as opposed to IL-10) as measured by immunoassays tended to have greater inter-platform correlations and more concordant AKI ORs. This observation was unexpected, as one of our preconceptions was that so long as our measurements were within the limits of detection and linearity range of our assays, we should see comparable accuracy among immunoassays. This observation could be in part due to reduced variability in either assay platform at greater protein concentrations as measurements enter a range of the immunoassay and/or aptamer-based platform with improved assay accuracy and linearity.
In preoperative samples, several biomarker immunoassays were restricted by their limits of quantification, including biomarkers excluded from the analyses because >50% of samples were outside the limits of immunoassay quantification and biomarkers included in the analysis with <50% of samples outside the limits of quantification. Though samples were within the immunoassay manufacturer’s stated limits of quantification of acceptable levels of precision and accuracy, lower precision and accuracy near the limits of quantification may challenge inter-platform operability. However, we are unable to comment on which platform more accurately quantifies biomarkers levels without gold-standard references for protein quantification in this matrix. The lack of a gold-standard for protein quantification is a challenge that the field of proteomics has yet to fully address; while mass-spectrometry has been viewed by some as a gold-standard, it faces limitations in quantifying low-abundance proteins and proteins with both mature and immature forms in circulation. Low inter-platform correlations for biomarkers found at picomolar to nanomolar concentrations raise questions of accuracy, precision, and linearity in either platform. In future studies, inter-platform comparisons of immunoassays and aptamers could be supported by reference measurements from a fully validated clinical immunoassay or ratiometric comparisons of liquid chromatography-mass spectrometry measurements to a heavy-labeled standard with proven linearity or a standard curve 7.
Cross-reactivity and negative cooperative binding may also contribute to inter-platform discrepancies. The idea that cross-reactivity could result in a lack of inter-platform concordance has been described previously 8,10. We did not find a relationship between higher inter-platform correlation and cross-reactivity in the 8 cross-reactive and 23 non-cross-reactive aptamer-immunoassay pairs we studied. One limitation in exploring the role of cross-reactivity in inter-platform correlations is our inability to assess immunoassay cross-reactivity using these data due to differences between aptamer and antibody binding domains.
Contrary to our initial hypothesis, we did not observe greater inter-platform correlation between biomarkers with lower %CVs on both platforms. This suggests that inter-platform discrepancies are primarily attributable to systematic differences between platforms rather than experimental imprecision. Such reasons for discrepancies in aptamer-immunoassay correlations may include unique binding reagent-target epitope interactions and/or differences in epitope-binding availabilities in the unique binding matrices of each assay. Both immunoassay and aptamer-based platforms quantify proteins by measuring binding-reagent binding with protein epitopes. In a complex matrix like blood, an epitope may not be available for binding for a variety of reasons (e.g., it is hidden by another molecule that binds at or near the epitope site). Thus, some discrepancies between binding reagents, whether antibodies or aptamers, are expected.
SomaScan is a promising aptamer-based platform with an unprecedented scope and throughput for screening of proteins in various clinical settings. Investigators should have confidence in the inter-platform operability of quantification and disease association of biomarkers found in greater physiological abundance in circulation. Without a true gold standard for protein quantification, consistent evidence across multiple studies is needed to establish the translatability of results and interoperability of quantification between different kinds of assay platforms. In this study we have qualified the inter-platform operability of 30 plasma proteins associated with AKI across preoperative and postoperative samples from 294 cardiac surgery patients. We found all proteins significantly associated with AKI as measured by SomaScan were also significantly associated with AKI as measured by immunoassay. Our study contributes to the evidence that associations established in the unbiased SomaScan platform translate to immunoassay associations. Additionally, there is emerging evidence for classifying some aptamer-immunoassay pairs as highly correlated and others as poorly correlated, where highly correlated assay pairs all showed similar clinical associations with AKI. Orthogonal validation through other methods, such as mass spectrometry and cis-pQTLs, has potential for further validation of aptamer technology for biomarker discovery and validation. Future research in broad settings with reasonable sample sizes will strengthen confidence in the use of aptamer-based technologies compared with immunoassays
Background.
Aptamer-based proteomic technologies hold a promise for the identification of novel biomarkers in biofluids that enable unbiased discovery with an unprecedented scope of over 7000+ protein targets. While biomarker discovery is a research focus in acute kidney injury (AKI), unbiased proteomics in biofluid proteins is of interest throughout translational research.
Translational Significance.
This study examines consistency in inter-platform correlation and biomarker-AKI odds ratios between SomaScan, an unbiased aptamer-based platform, and immunoassays, the standard method in clinical settings, for 34 biomarker-pairs at two clinical timepoints. Among the ten target protein, immunoassay, and aptamer characteristics tested for relationships with inter-platform operability, we found a strong association between improved inter-platform operability and higher biomarker concentrations.
Acknowledgements
This work was supported by the NIH (R01HL085757 to CRP, TRIBE-AKI Consortium). CRP was also supported by NIH grants U01DK082185 and the P30DK079310 O’Brien Kidney Center Grant and is a member of the NIH-sponsored CKD BIOCon (U01DK106962) and KPMP Consortium (UH3DK114866). JVB was supported by grants from the National Institute of Health/NIDDK U01DK106962 and R01DK072381. VSR is supported in part by Contracts NO1-HC-25195, HHSN268201500001I and 75N92019D00031 from the National Heart, Lung and Blood Institute, grants RF1AG063507 and R01HL132320, the Evans Medical Foundation, and the Jay and Louis Coffman Endowment from the Department of Medicine, Boston University School of Medicine. All authors have read the journal’s authorship agreement and have reviewed and approved the manuscript.
List of abbreviations
- AKI
acute kidney injury
- OR
odds ratio
- CV
coefficient of variation
- rs
Spearman correlation
- SD
standard deviation
- TRIBE-AKI
Translational Research Investigating Biomarker Endpoints in Acute Kidney Injury
- SOMA
Slow Off-rate Modified Aptamers
- TOPMed
Trans-Omics for Precision Medicine
- Cis-pQTL
cis-protein quantitative trait locus
- AKIN
Acute Kidney Injury Network
- MSD
Meso Scale Discovery
- UniProt
Universal Protein Resource
- hFABP
human heart-type fatty acid-binding protein
- VEGF
vascular endothelial growth factor
- hsTnT
high-sensitivity troponin T
- NT-proBNP
N-terminal-pro hormone BNP
- CK-MB
creatine kinase myocardial band
- TnI
troponin I
- bFGF
basic fibroblast growth factor
- PlGF
placental growth factor
- VEGFR1
vascular endothelial growth factor receptor 1
- VEGFd
vascular endothelial growth factor d
- VEGFc
vascular endothelial growth factor c
- Tie2
angiopoietin-1 receptor
- MCP-1
monocyte chemoattractant protein-1
- KIM-1
kidney injury molecule 1
- YKL-40
chitinase 3-like protein 1
- Gal3
Galectin 3
- ST2
soluble interleukin 1 receptor-like 1
- TNF-R1
tumor necrosis factor receptor 1
- TNF-R2
tumor necrosis factor receptor 2
- IL-1
interleukin 1
- IL-4
interleukin 4
- IL-6
interleukin 6
- IL-8
interleukin 8
- IL-10
interleukin 10
- IL-12
interleukin 12
- IL-13
interleukin 13
- TNFa
tumor necrosis factor alpha
- BNP-32
brain natriuretic peptide-32
- CST3
cystatin C
- NGAL
neutrophil gelatinase-associated lipocalin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
CRP has reported receiving consulting fees from Renalytix and has reported serving on the Data Safety and Monitoring Boards of Genfit and Abbott. PG serves on a medical advisory board to SomaLogic, Inc, for which he accepts no salary, honoraria, or any other financial incentives. JVB is cofounder and holds equity in Goldfinch Bio. He is co-inventor on KIM-1 patents assigned to Mass General Brigham, and has received consulting income from Aldeyra, Praxis, Seattle Genetics and Sarepta. JVB’s interests were reviewed and are managed by BWH and MGB Healthcare in accordance with their conflict-of-interest policies.
All other authors declare that no conflicts of interest exist. All authors have read the journal’s policy on conflicts of interest.
Federal demurrer: The opinions expressed herein do not necessarily reflect those of the National Institute of Diabetes Digestive and Kidney Diseases, the National Institutes of Health, the Department of Health and Human Services, or the government of the United States of America.
References
- 1.YALOW RS, BERSON SA. Immunoassay of endogenous plasma insulin in man. J Clin Invest 1960; 39: 1157–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem 2010; 56(2): 186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pertea M, Salzberg SL. Between a chicken and a grape: estimating the number of human genes. Genome Biol 2010; 11(5): 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hathout Y, Brody E, Clemens PR, et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2015; 112(23): 7153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018; 361(6404): 769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gold L, Walker JJ, Wilcox SK, Williams S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol 2012; 29(5): 543–9. [DOI] [PubMed] [Google Scholar]
- 7.Carlyle BC, Trombetta BA, Arnold SE. Proteomic Approaches for the Discovery of Biofluid Biomarkers of Neurodegenerative Dementias. Proteomes 2018; 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raffield LM, Dang H, Pratte KA, et al. Comparison of Proteomic Assessment Methods in Multiple Cohort Studies. Proteomics 2020; 20(12): e1900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund L, Björling E, Oksvold P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 2008; 7(10): 2019–27. [DOI] [PubMed] [Google Scholar]
- 10.Kukova LZ, Mansour SG, Coca SG, et al. Comparison of Urine and Plasma Biomarker Concentrations Measured by Aptamer-Based versus Immunoassay Methods in Cardiac Surgery Patients. J Appl Lab Med 2019; 4(3): 331–42. [DOI] [PubMed] [Google Scholar]
- 11.Tin A, Yu B, Ma J, et al. Reproducibility and Variability of Protein Analytes Measured Using a Multiplexed Modified Aptamer Assay. J Appl Lab Med 2019; 4(1): 30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenen-Stass AM, McClorey G, Manzano R, et al. Identification of novel, therapy-responsive protein biomarkers in a mouse model of Duchenne muscular dystrophy by aptamer-based serum proteomics. Sci Rep 2015; 5: 17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murota A, Suzuki K, Kassai Y, et al. Serum proteomic analysis identifies interleukin 16 as a biomarker for clinical response during early treatment of rheumatoid arthritis. Cytokine 2016; 78: 87–93. [DOI] [PubMed] [Google Scholar]
- 14.Ganz P, Heidecker B, Hveem K, et al. Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients With Stable Coronary Heart Disease. JAMA 2016; 315(23): 2532–41. [DOI] [PubMed] [Google Scholar]
- 15.Parikh CR, Coca SG, Thiessen-Philbrook H, et al. Postoperative biomarkers predict acute kidney injury and poor outcomes after adult cardiac surgery. J Am Soc Nephrol 2011; 22(9): 1748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosner MH, Okusa MD. Acute kidney injury associated with cardiac surgery. Clin J Am Soc Nephrol 2006; 1(1): 19–32. [DOI] [PubMed] [Google Scholar]
- 17.Nephrology ASo. American Society of Nephrology Renal Research Report. J Am Soc Nephrol 2005; 16(7): 1886–903. [DOI] [PubMed] [Google Scholar]
- 18.Belley-Côté EP, Parikh CR, Shortt CR, et al. Association of cardiac biomarkers with acute kidney injury after cardiac surgery: A multicenter cohort study. J Thorac Cardiovasc Surg 2016; 152(1): 245–51.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansour SG, Zhang WR, Moledina DG, et al. The Association of Angiogenesis Markers With Acute Kidney Injury and Mortality After Cardiac Surgery. Am J Kidney Dis 2019; 74(1): 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moledina DG, Isguven S, McArthur E, et al. Plasma Monocyte Chemotactic Protein-1 Is Associated With Acute Kidney Injury and Death After Cardiac Operations. Ann Thorac Surg 2017; 104(2): 613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moledina DG, Parikh CR, Garg AX, et al. Association of Perioperative Plasma Neutrophil Gelatinase-Associated Lipocalin Levels with 3-Year Mortality after Cardiac Surgery: A Prospective Observational Cohort Study. PLoS One 2015; 10(6): e0129619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel UD, Garg AX, Krumholz HM, et al. Preoperative serum brain natriuretic peptide and risk of acute kidney injury after cardiac surgery. Circulation 2012; 125(11): 1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaub JA, Garg AX, Coca SG, et al. Perioperative heart-type fatty acid binding protein is associated with acute kidney injury after cardiac surgery. Kidney Int 2015; 88(3): 576–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams SA, Kivimaki M, Langenberg C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med 2019; 25(12): 1851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta RL, Kellum JA, Shah SV, et al. Acute Kidney Injury Network: report of an initiative to improve outcomes in acute kidney injury. Crit Care 2007; 11(2): R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010; 5(12): e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udovičić M, Baždarić K, Bilić-Zulle L, Petrovečki M. What we need to know when calculating the coefficient of correlation? Biochemia medica: Biochemia medica 2007; 17(1): 10–5. [Google Scholar]
- 28.Wrobel JS, Armstrong DG. Reliability and validity of current physical examination techniques of the foot and ankle. J Am Podiatr Med Assoc 2008; 98(3): 197–206. [DOI] [PubMed] [Google Scholar]
- 29.Diedenhofen B, Musch J. cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One 2015; 10(3): e0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou GY. Toward using confidence intervals to compare correlations. Psychol Methods 2007; 12(4): 399–413. [DOI] [PubMed] [Google Scholar]
- 31.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 2015; 43(Database issue): D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature 2018; 558(7708): 73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker KA, Chen J, Wu A, et al. Large-scale plasma proteomic analysis identifies proteins and biological pathways associated with incident dementia. Alzheimer’s & Dementia 2020; 16(S5): e038307. [Google Scholar]
- 34.SomaLogic. I. Correlation of SOMAmer reagents in the SOMAscan assay and commercially available immunoassays. http://tribe.jhmi.edu/Correlation-of-SOMAmer%C2%AE-reagents-in-the-SOMAscan%C2%AE-assay-and-commercially-available-immunoassays-SS-501-051916-1-1.pdf (Accessed September 2020).
- 35.Chaturvedi AK, Kemp TJ, Pfeiffer RM, et al. Evaluation of multiplexed cytokine and inflammation marker measurements: a methodologic study. Cancer Epidemiol Biomarkers Prev 2011; 20(9): 1902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem 2010; 56(2): 314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


