Abstract
Purpose
To develop and evaluate the performance of a radiomic and machine learning model applied to ultrasound images in predicting the risk of malignancy of ovarian masses (OMs).
Methods
Single-center retrospective evaluation of consecutive patients who underwent transvaginal ultrasound (US) with images storage and surgery for ovarian masses. Radiomics methodology was applied to US images according to the International Biomarker Standardization Initiative guidelines. OMs were divided into three homogeneous groups: solid, cystic and motley. TRACE4© radiomic platform was used thus obtaining a full-automatic radiomic workflow. Three different classification systems were created and accuracy, sensitivity, specificity, AUC and standard deviation were defined for each group.
Results
A total of 241 women were recruited. OMs were divided in the three groups: 95 (39.5%) solid, 66 (27.5%) cystic, 80 (33%) motley. For solid OMs, 269 radiomic features were used for the training-validation-testing of the model with accuracy 80%, sensitivity 78%, specificity 83%, AUC 87%. For cystic OMs, 278 radiomic features were used for the training-validation-testing of the model with accuracy 87%, sensitivity 75%, specificity 90%, AUC 88%. For mixed OMs, 306 radiomic features were used for the training-validation-testing of the model with accuracy 81%, sensitivity 81%, specificity 81%, AUC 89%.
Conclusion
Radiomics is a promising tool in improving preoeprative work-up of women diagnosed with OMs. Even in the absence of the subjective impression of expert ultrasound examiner, radiomics allows to easily identify patients with ovarian cancer. Future validation studies on larger series are needed.
Electronic supplementary material
The online version of this article (10.1007/s40477-020-00503-5) contains supplementary material, which is available to authorized users.
Keywords: Radiomics, Ultrasound, Ovarian cancer, Risk of malignancy, Ovarian masses, Predictive model
Introduction
Ovarian cancer (OC) is the fifth most common cancer in women and the most common cause of gynecologic cancer deaths, thus being one of the most lethal cancers in women [1]. It is estimated that over 22,000 new cases of OC were diagnosed and 14,000 women died of this disease in 2019 only in the USA Nearly three cases out of four were diagnosed in the advanced stage of disease [stage IIIC or IV, International Federation of Gynecology and Obstetrics (FIGO)] at presentation [2].
The absence of valid screening and diagnostic programs and the rapid spread of disease through the peritoneal surface represent the main factors driving OC lethality. Growing evidences suggested that clinical examination by qualitative transvaginal ultrasound imaging and CA-125 dosage—currently the available screening examinations—are not enough to detect OC at the early stage in the general population [3] and an accurate protocol to identify high-risk patients is still lacking. Thus, recently, Food and Drug Administration (FDA) recommended against using the currently offered tests for OC screening, suggesting that they might lead to non-negligible inaccuracies [4]. This is of paramount importance especially in women diagnosed with ovarian masses (OMs), often detected incidentally, mostly addressed to surgery and frequently proven to be benign post-surgery at final histology analysis [5]. Therefore, the identification of tools for accurate screening, early diagnosis and prognosis of OC represents a current unmet clinical need.
Recently, radiomics emerged as a new powerful method able to quantify features from radiological medical images [6, 7] containing information that reflect the underlying pathophysiology of cancer tissue [8, 9]. The information extracted might correlate with patient clinical data, leading to the definition of radiomic biomarker profiles that can discriminate cancerous from non-cancerous tissues or different cancer subtypes. Moreover, when radiomics features were used to train machine learning systems to automatically classify tissue images in distinct groups, predictive models were built and proven effective in predicting diagnosis and also prognosis in various types of solid tumors at the level of single patient, representing useful decision support tools for personalized medicine [10]. In the present paper our aim is to test the performance of radiomic analysis on patients diagnosed with OMs. We extracted radiomic features from ultrasonographic images of histologically proven cancerous and non-cancerous OMs, and used selected radiomics features to train machine learning systems to automatically discriminate malignant and benign OMs, not depending on the expertise of ultrasound examiners.
Methods
Study design and study population
This is a single center, retrospective pilot study. The study population includes consecutive women diagnosed with OMs treated at Fondazione IRCCS Istituto Nazionale dei Tumori di Milano from January 1, 2017 to December 31, 2019. Every patient signed written consent for research purpose. Consecutive patients diagnosed with OMs and scheduled to have surgery were included in the study. Inclusion criteria were: (i) diagnosis of OM, (ii) execution of a preoperative ultrasonographic examination within 2 weeks before surgery, (iii) surgery performed. Exclusion criteria were: (i) age <18 years, (ii) absence of ultrasonographic images stored and (iii) consent withdrawn.
Ultrasonography images and histopathological data of patients
We retrospectively collected all preoperative ultrasonographic images of patients who underwent surgery for OMs. Ultrasonographic images were stored in.jpg and.DICOM format at the time of preoperative workup. All patients included underwent transvaginal ultrasound eventually completed by transabdominal scan in case of big cysts not completely evaluable with transvaginal approach or in case of suspicion of malignancy (for further abdominal evaluation). Ultrasound examinations were carried out with the same system (General Electrics Voluson™ E8 ultrasound system).
All patients included in the study underwent surgery. Type of surgery performed was tailored on the basis of patients’ and disease characteristics. Intra-operative frozen section was performed in all cases. In case of OC, stage and grading of disease were assessed according to the International Federation of Obstetrics and Gynecologists (FIGO) system [11]. Histological subtypes were reported according to the World Health Organization (WHO) classification [12].
To assess the efficacy of radiomics in a heterogeneous group of patients we categorized OMs in three homogeneous groups: (i) solid, (ii) cystic, and (iii) motley. The solid group included masses without any fluid component; while the cystic group included masses without any solid component except septa (both simple and septated ovarian cysts were considered in this group). The motley group included masses with both fluid and solid components.
We then associated each OM ultrasonographic image to one of the two histopathological classes (malignant/benign). In case of bilateral OMs the lesion with more complex ultrasound appearance (bigger and/or with larger solid component) was considered for the radiomic analysis.
Radiomics study
Radiomics methodology was applied to collected ultrasonographic images of patients, according to the International Biomarker Standardization Initiative (IBSI) guidelines (https://arxiv.org/abs/1612.07003) [13].
For this purpose the TRACE4© radiomic platform was used (http://www.deeptracetech.com/files/TechnicalSheet__TRACE4.pdf) allowing the whole IBSI-compliant radiomic workflow to be obtained in a full-automatic way.
IBSI radiomic workflow included: (i) the segmentation of the lesion region from each patient image, (ii) the preprocessing of image content within the segmented region of interest for the radiomic features extraction, (iii) the extraction of radiomic features from the segmented region of interest, (iv) the selection of radiomic features which remains stable with respect to different segmentations, as may occur by different examiners, and repeatable in test–retest study, (v) the use of such candidate radiomic features to train, validate, and test different systems of machine learning classifiers in the binary classification task of interest (malignant vs benign), by the reduction of such stable and repeatable features to not-redundant features, in a number that is statistically comparable with the number of collected images of patients.
More specifically:
-
(i)
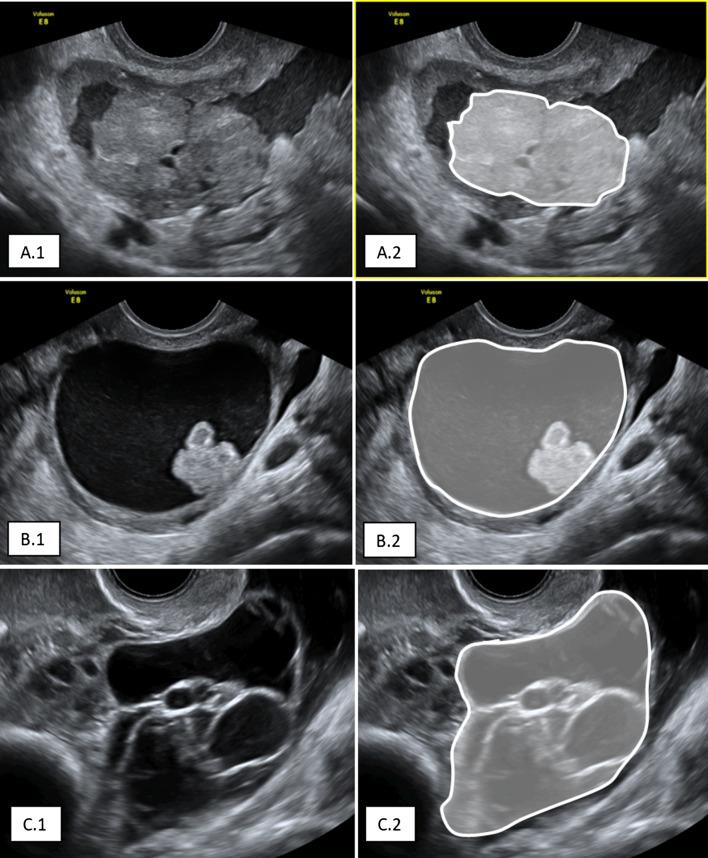
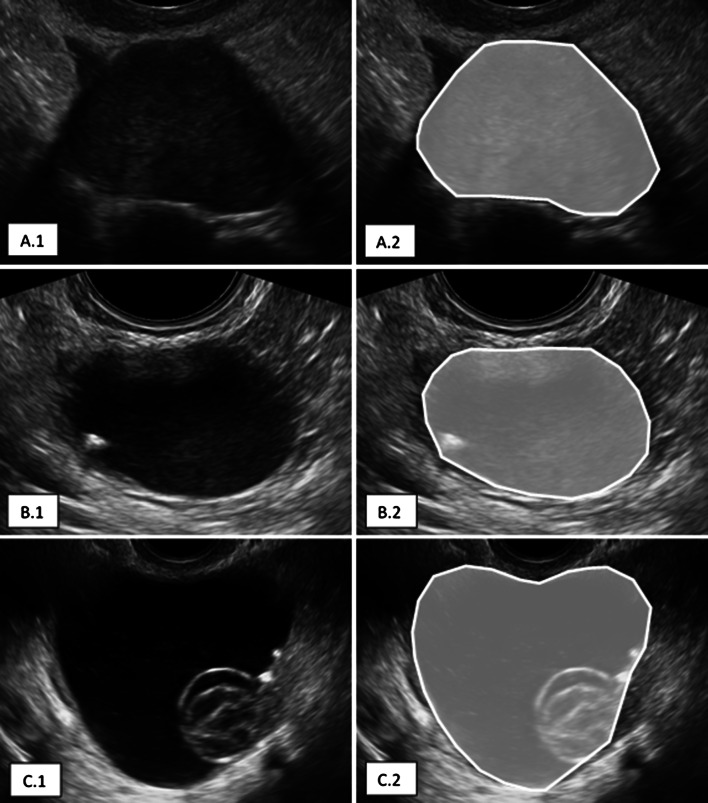
The segmentation of the OMs was performed manually, using the TRACE4 segmentation tool (Figs. 1, 2).
-
(ii)
The preprocessing of image intensities within the segmented region of interest included resampling to isotropic voxel spacing, using a down-sampling scheme by considering image slice thickness of 1 mm and intensity discretization using a fixed number of 64 bins.
-
(iii)
The radiomics features extracted from the segmented region of interest belong to different families: morphology, intensity-based statistics, intensity histogram, gray-level co-occurrence matrix (GLCM), gray-level run length matrix (GLRLM), gray-level size zone matrix (GLSZM), neighborhood gray tone difference matrix (NGTDM), gray-level distance zone matrix (GLDZM), neighboring gray level dependence matrix (NGLDM). Their definition, computation and nomenclature are compliant with the IBSI guidelines, except for the features of the family morphology, originally designed for 3D images, which were replaced with ten 2D equivalent features (e.g., 3D features volume and surface were replaced with 2D features area and perimeter, respectively). Steps from (ii) to (iii) were performed using the TRACE4 Radiomics tool. Radiomic features were reported by TRACE4 according to IBSI standards.
-
(iv)
The selection of radiomic features, stable with respect to different segmentations and repeatable in test–retest study, was performed by ICC (ICC > 0.80) by statistically comparing features obtained by data augmentation strategies, (a) generating random variations of the manual segmentation of the lesion region (performed by the operator), and random rotations of the original images and segmentations to reduce the dependency from the operator and to enrich the dataset, respectively. The selected radiomic features (stable and repeatable) were reported by TRACE4.
-
(v)
For each one of the three homogeneous groups of OMs (solid, cystic, and motley OMs), a different system of machine learning classifier was trained, validated, and tested, for the binary classification task (malignant vs benign, based on histopathology results), reducing the more stable and reproducible features to a signature of not redundant features proper with the number of collected images.
Fig. 1.
Examples of manual segmentation of malignant ovarian masses: A.1 solid; A.2 motley; A.3 cystic
Fig. 2.
Examples of manual segmentation of benign ovarian masses: A.1 solid; A.2 motley; A.3 cystic
Each of the three systems is an ensemble of 10 Support Vector Machines, combined with principal components analysis and fisher discriminant ratio with majority vote rule. For each system, nested K-fold cross validation method was used (K = 10). Oversampling technique for the minority class was applied by adaptive synthetic sampling method. Performance of the different classification systems were measured across the different folds (K = 10) in terms of max and mean Accuracy, Sensitivity, Specificity, AUC and standard deviation. Steps (iv) and (v) were performed automatically by the TRACE4 Modeling and Statistics tool.
Results
Ultrasonography images and histopathological data of patients
We retrospectively recruited 241 women with available ultrasonographic images of OMs and histopathological results. Median age was 55 (18–84) years old and median body mass index was 25 (16–39) kg/mq. Most of patients (76%) were postmenopausal. Most of patients (64%) referred symptoms before ultrasound: pelvic or abdominal pain (54%), bloating (22%), increase in abdominal circumference (16%), weight gain or loss (15%), weakness (15%), nausea (13%), irregular menstrual periods (10%), abnormal uterine bleeding (4%). Histological characteristics of the OMs after surgery are presented in Table 1. One hundred patients (87%) with benign pathology at final histology underwent laparoscopic surgery; 15 patients (13%) underwent open surgery because of dimension of OM or suspect of malignancy at preoperative workup. Among patients with malignant disease, 82 patients (65%) had open surgery with surgical staging in case of early stages and optimal cytoreduction in case of advanced stages; 44 patients (35%) had laparoscopy because of preoperative suspect of borderline tumors or advanced stages without possibility to reach complete cytoreduction at preoperative imaging (in this last case, patients underwent multiple laparoscopic biopsies followed by neoadjuvant chemotherapy).
Table 1.
Histopathological characteristics of ovarian masses included in the study
| Variable | Ovarian masses (n = 241) |
|---|---|
| Benign | 115 (48) |
| Malignant | 126 (52) |
| Histological type of benign masses: | n = 115 (100) |
| Serous cystoadenoma/cystoadenofibroma | 45 (39) |
| Mucinous cystoadenoma | 8 (7) |
| Endometrioma | 7 (6) |
| Ovarian fibroma/fibrothecoma | 36 (31) |
| Teratoma | 3 (3) |
| PID | 2 (2) |
| Peritoneal cyst/pseudocysts | 2 (2) |
| Hidrosalpinx | 7 (6) |
| Paraovarian cyst/paratubaric cyst | 5 (4) |
| Histological type of malignant tumors: | n = 126 (100) |
| High grade serous ovarian cancer (HGSOC) | 53 (42) |
| Low grade serous ovarian cancer (LGSOC) | 5 (4) |
| Serous borderline tumor | 12 (10) |
| Mucinous borderline tumor gastrointestinal type | 7 (6) |
| Mucinous borderline tumor endocervical type | 5 (4) |
| Mucinous ovarian cancer | 2 (2) |
| Endometrioid ovarian cancer | 3 (2) |
| Clear cell ovarian cancer | 3 (2) |
| Granulosa cell ovarian tumor | 3 (2) |
| Dysgerminoma | 1 (1) |
| Ovarian metastases from other tumors | 20 (16) |
| Tubal cancer | 1 (1) |
| Yolk sac tumor | 3 (2) |
| Ovarian carcinosarcoma | 5 (4) |
| Sertoli Leydig tumor | 3 (2) |
| FIGO stage of primitive ovarian cancers: | n = 106 (100) |
| IA | 21 (19) |
| IB | 1 (1) |
| IC1 | 4 (4) |
| IC2 | 1 (1) |
| IC3 | 4 (4) |
| IIA | 3 (3) |
| IIB | 6 (6) |
| IIIA1 | 6 (6) |
| IIIA2 | 1 (1) |
| IIIB | 9 (8) |
| IIIC | 33 (31) |
| IVA | 5 (5) |
| IVB | 12 (11) |
Results are presented as n (%)
According to the setting designed for radiomics analysis, the benign/malignant distribution and characteristics of the three groups of OMs are summarized in Table 2.
Table 2.
Ultrasonographic and biochemical characteristics of ovarian masses (OMs)
| Group 1, solid OMs (n = 95) | Group 2, cystic OMs (n = 66) | Group 3, motley OMs (n = 80) | ||||
|---|---|---|---|---|---|---|
| Benign | Malignant | Benign | Malignant | Benign | Malignant | |
| N (%) | 38 (40) | 57 (60) | 53 (80) | 13 (20) | 24 (30) | 56 (70) |
| Median max diameter (mm) | 46 | 54 | 44 | 103 | 49 | 82 |
| Bilateral lesions, n (%) | 3 (8) | 18 (32) | 5 (9) | 1 (8) | 2 (8) | 24 (43) |
| Ascites, n (%) | 3 (8) | 17 (30) | 0 (0) | 3 (23) | 0 (0) | 8 (14) |
| Median Ca125 | 17 | 274 | 12 | 92 | 11 | 40 |
Radiomics study
Three hundred and nineteen radiomics IBSI-compliant features were extracted from segmented OMs (see Online Appendix A for a representative patient).
For the first classification task (solid malignant vs benign OMs), 269 radiomics features were found stable with respect to different examiners and to test–retest study (ICC > 0.8) (Online Appendix B1). These features were used for the training-validation-testing (nested tenfold validation) of the first ensemble of support vector machines. Accuracy, Sensitivity, Specificity and AUC of such classifier are shown in Table 3.
Table 3.
Performances of the classifier trained on malignant vs benign ovarian masses in solid, cystic and motley group
| Accuracy | Sensitivity | Specificity | AUC | |
|---|---|---|---|---|
| SOLID OMs | 0.80 ± 0.02 | 0.78 ± 0.02 | 0.83 ± 0.03 | 0.87 ± 0.01 |
| CYSTIC OMs | 0.87 ± 0.03 | 0.75 ± 0.06 | 0.9 ± 0.02 | 0.88 ± 0.04 |
| MOTLEY OMs | 0.81 ± 0.02 | 0.81 ± 0.02 | 0.81 ± 0.02 | 0.89 ± 0.02 |
For the second classification task (cystic malignant vs benign OMs), 278 radiomics features were found stable with respect to different examiners and to test–retest study (ICC > 0.8) (Online Appendix B2). These features were used for the training-validation-testing (nested tenfold validation) of the first ensemble of support vector machines. Accuracy, sensitivity, specificity and AUC of such classifier are shown in Table 3.
For the third classification task (motley malignant vs benign OMs), 306 radiomics features were found stable with respect to different examiners and to test–retest study (ICC > 0.8) (Online Appendix B3). These features were used for the training-validation-testing (nested tenfold validation) of the first ensemble of support vector machines. Accuracy, sensitivity, specificity and AUC of such classifier are shown in Table 3.
Discussion
The present study investigated if the adoption of radiomics could be useful in improving diagnostic work-up of women diagnosed with OM and if, even in absence of the subjective impression of expert ultrasound examiner, radiomics could allow to easily identify patients with OC.
Several investigations tried to identify possible hallmarks for the characterization of patients diagnosed with OM [14, 15], however, no objective parameters work better than the subjective impression of an expert ultrasound examiner [16].
In this study we applied radiomics features to ultrasonographic preoperative images of OMs to build a predictive model to select malignant lesions that should be referred to an oncological center. The performance of our predictive models was >80%, thus suggesting the application of radiomics to ultrasonography to improve the diagnosis of OC. Moreover, the main advantage of our model is that it is not dependent on the experience of the ultrasound examiner except for the categorization of OMs as solid, cystic and motley; the examiner stored the ultrasonographic image of a single patient in DICOM format, sent the image to the TRACE4 software platform and received a probability of classification as “benign” or “malignant” as support to his/her diagnosis.
To the best of our knowledge this is the first study to apply radiomics according to IBSI guidelines and machine learning to ultrasonographic images in gynecology to define the risk of malignancy of OMs.
Few Authors proposed different approaches to automatically identify the presence of ovarian cancer tissue analyzing texture parameters on 2D [17–19] and 3D [20] ultrasonographic images with promising results.
Similar experiences have been recently described in literature applied to hepatocellular carcinoma ultrasound images [21] where the authors concluded that radiomics could help in liver tumor evaluations, including diagnosis, differential diagnosis, and clinical prognosis. The most important experiences in literature described radiomics applied to MRI, PET/CT or CT-scan; a few studies have been conducted in patients with adnexal lesions with different purposes: to categorize ovarian masses (benign/malignant and in case of malignant type I/type II ovarian tumors) [22] and to relate tumor heterogeneity to prognosis and risk or recurrence [23–26].
A strength of our study is the radiomic analysis according to IBSI guidelines, that allows better comparison with further analysis by other research groups. Moreover, we were able to correlate radiomics features with histologic specimen, allowing the development of artificial intelligence (AI) models predictive of the risk of malignancy based on the sole in vivo measurement of radiomic features. Another strength point is the number of cases available for radiomics analysis and to develop the models (241), allowing the use of k-fold cross validation to properly train, validate and test the AI models on separate folds of image cases.
However, our work has some limitations, in particular the manual segmentation of the lesions, impacting on the work of clinicians and potentially including a certain percentage of intrinsic error. An advantage over the segmentation offered by the TRACE4 platform is that the selected features are independent of the contour traced by the operator because this contour is manipulated by TRACE4 functions with a multiplicity of small variations (small random deformations) that generate as many contours as if they were traced by a multiplicity of operators. The TRACE4 platform selects stable radiomic features with respect to these different contours.
The main weakness of the study is related to the inherit biases of the retrospective, single center—single ultrasonographic system study design. Additionally, another possible limitation of our study is the limited sample size for some subgroups of lesions due to the rarity of diseases (malignant cystic lesions and benign motley lesions), but this is a pilot study to build a predictive model that will need to be validated on larger series.
Nowadays, transvaginal ultrasound represents the first-choice imaging technique to approach the female pelvis, and it’s widely used to complete clinical examination even in an office setting by non-experienced examiners who often are not able to categorize incidental OMs.
Compared to the other models described in literature, our model has the advantage to be not dependent by the experience of the examiner, requiring very few skills including image manual segmentation.
The most important score actually in use to categorize OMs is ADNEX [27–29], built and validated by International Ovarian Tumor Analysis (IOTA) group. This is a very useful model because it defines not only the risk of malignancy with a performance similar to the subjective impression of the experienced ultrasound examiner, but also stratifies the risk of the OM to be a borderline tumor, an early or advanced ovarian cancer or a metastasis; on the other side, the variables used to calculate the risk according to ADNEX assume that the examiner knows at least IOTA terminology [30] and it requires a minimum of experience to correctly apply the terminology and run the algorithm.
ADNEX provides a percentage of risk of malignancy that needs a further interpretation to define the management of the lesion and the performance of the model changes according to chosen cut-off to define malignancy; for the most used cut off of 10% the model shows a sensitivity of 96.5% and a specificity of 71.3%. External validations of the model showed similar results to the original paper [31, 32].
To apply our model, the examiner should only define the OM as solid, cystic or motley and trace the outline of the lesion, without any other info required, thus allowing a wide possibility of applications, even by basic ultrasound examiners. A further advantage of our model is to eliminate uncertainty thus providing the classification of the OM into benign/malignant and not an estimation of the risk of malignancy which needs interpretation to drive the subsequent management; this could facilitate the application of the model in a basic setting to select patients for second level evaluation and referral to oncological center.
Our future perspectives are to optimize the model, avoiding the division of the OMs into the three groups by the examiners, thus building a unique model where the software automatically recognizes the type of mass, that could be embedded into ultrasound machines allowing an immediate real time classification of ovarian lesions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
Conceptualization, V.C., G.B., I.C., F.R.; methodology and formal analysis, I.C., M.I., C.S.; data curation, I.C., M.I., C.S., F.B., G. S., M.S.; software C.S., writing, V.C., G.B., M.I., C.S.; review and editing, I.C., F.R., M.S.; supervision, I.C., F.R.
Compliance with ethical standards
Conflict of interest
No potential conflicts of interest were disclosed. No funding source was obtained.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bagnoli M, Canevari S, Califano D, Losito S, Maio MD, Raspagliesi F, Multicentre Italian Trials in Ovarian cancer (MITO) translational group et al. Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: a cohort study. Lancet Oncol. 2016;17:1137–1146. doi: 10.1016/S1470-2045(16)30108-5. [DOI] [PubMed] [Google Scholar]
- 3.Pinsky PF, Yu K, Kramer BS, Black A, Buys SS, Partridge E, et al. Extended mortality results for ovarian cancer screening in the PLCO trial with median 15 years follow-up. Gynecol Oncol. 2016;143:270–275. doi: 10.1016/j.ygyno.2016.08.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.FDA. The FDA recommends against using screening tests for ovarian cancer screening: FDA Safety Communication. 2016 Available from: https://www.fda.gov/MedicalDevices/Safety/AlertsandNotices/ucm519413.htm. Accessed 16 Mar 2020
- 5.Froyman W, Landolfo C, De Cock B, Wynants L, Sladkevicius P, Testa AC, et al. Risk of complications in patients with conservatively managed ovarian tumours (IOTA5): a 2-year interim analysis of a multicentre, prospective, cohort study. Lancet Oncol. 2019;20(3):448–458. doi: 10.1016/S1470-2045(18)30837-4. [DOI] [PubMed] [Google Scholar]
- 6.Nougaret S, Tardieu M, Vargas HA, Reinhold C, Vande Perre S, Bonanno N, et al. Ovarian cancer: an update on imaging in the era of radiomics. Diagn Interv Imaging. 2019;100(10):647–655. doi: 10.1016/j.diii.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Kumbhare D, Shaw S, Ahmed S, Noseworthy MD. Quantitative ultrasound of trapezius muscle involvement in myofascial pain: comparison of clinical and healthy population using texture analysis. J Ultrasound. 2020;23(1):23–30. doi: 10.1007/s40477-018-0330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh AC, Li H, Zhu Y, Zhang J, Khramtsova G, Drukker K, et al. Radiogenomics of breast cancer using dynamic contrast enhanced MRI and gene expression profiling. Cancer Imaging. 2019;19(1):48. doi: 10.1186/s40644-019-0233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12(8):862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prat J; FIGO Committee on Gynecologic Oncology Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2014;124(1):1–5. doi: 10.1016/j.ijgo.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Kurman RJ, Carcangiu ML, Herrington CS, Young RH, editors. WHO Classification of tumours of female reproductive organs. 4. Geneva: WHO; 2014. [Google Scholar]
- 13.Zwanenburg A, Leger S, Valli`eres M, Lock S. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003
- 14.Kaijser J. Towards an evidence-based approach for diagnosis and management of adnexal masses: findings of the International Ovarian Tumour Analysis (IOTA) studies. Facts Views Vis Obgyn. 2015;7(1):42–59. [PMC free article] [PubMed] [Google Scholar]
- 15.Sconfienza LM, Perrone N, Delnevo A, Lacelli F, Murolo C, Gandolfo N, et al. Dignostic value of contrast-enhanced ultrasonography in the caracterization of ovarian tumors. J Ultrasound. 2010;13(1):9–15. doi: 10.1016/j.jus.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meys EM, Kaijser J, Kruitwagen RF, Slangen BF, Van Calster B, Aertgeerts B, et al. Subjective assessment versus ultrasound models to diagnose ovarian cancer: a systematic review and meta-analysis. Eur J Cancer. 2016;58:17–29. doi: 10.1016/j.ejca.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Zimmer Y, Tepper R, Akselrod S. An automatic approach for morphological analysis and malignancy evaluation of ovarian masses using B-scans. Ultrasound Med Biol. 2003;29(11):1561–1570. doi: 10.1016/j.ultrasmedbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Acharya UR, Molinari F, Sree SV, Swapna G, Saba L, Guerriero S, et al. Ovarian tissue characterization in ultrasound: a review. Technol Cancer Res Treat. 2015;14(3):251–261. doi: 10.1177/1533034614547445. [DOI] [PubMed] [Google Scholar]
- 19.Lucidarme O, Akakpo JP, Granberg S, Sideri M, Levavi H, Schneider A, et al. A new computer-aided diagnostic tool for non-invasive characterisation of malignant ovarian masses: results of a multicentre validation study. Eur Radiol. 2010;20(8):1822–1830. doi: 10.1007/s00330-010-1750-6. [DOI] [PubMed] [Google Scholar]
- 20.Acharya UR, Sree VS, Saba L, Molinari F, Guerriero S, Suri JS. Ovarian tumor characterization and classification: a class of GyneScan™ systems. Conf Proc IEEE Eng Med Biol Soc. 2012;2012:4446–4449. doi: 10.1109/EMBC.2012.6346953. [DOI] [PubMed] [Google Scholar]
- 21.Yao Z, Dong Y, Wu G, Zhang Q, Yang D, Yu JH, et al. Preoperative diagnosis and prediction of hepatocellular carcinoma: radiomics analysis based on multi-modal ultrasound images. BMC Cancer. 2018;18(1):1089. doi: 10.1186/s12885-018-5003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Mao Y, Chen X, Wu G, Liu X, Zhang P, et al. Magnetic resonance imaging radiomics in categorizing ovarian masses and predicting clinical outcome: a preliminary study. Eur Radiol. 2019;29:3358–3371. doi: 10.1007/s00330-019-06124-9. [DOI] [PubMed] [Google Scholar]
- 23.Vargas HA, Veeraraghavan H, Micco M, Nougaret S, Lakhman Y, Meier AA, et al. A novel representation of inter-site tumour heterogeneity from pre-treatment computed tomography textures classifies ovarian cancers by clinical outcome. Eur Radiol. 2017;27:3991–4001. doi: 10.1007/s00330-017-4779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzo S, Botta F, Raimondi S, Origgi D, Buscarino V, Colarieti A, et al. Radiomics of high-grade serous ovarian cancer: association between quantitative CT features, residual tumour and disease progression within 12 months. Eur Radiol. 2018;28:4849–4859. doi: 10.1007/s00330-018-5389-z. [DOI] [PubMed] [Google Scholar]
- 25.Nougaret S, Tardieu M, Vargas HA, Reinhold C, Vande Perre S, Bonanno N, et al. Ovarian cancer: an update on imaging in the era of radiomics. Diagn Intervent Imaging. 2019;100(10):647–655. doi: 10.1016/j.diii.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Meier A, Veeraraghavan H, Nougaret S, Lakhman Y, Sosa R, Soslow RA, et al. Association between CT-texture-derived tumor heterogeneity, outcomes, and BRCA mutation status in patients with high-grade serous ovarian cancer. Abdom Radiol. 2019;44:2040–2047. doi: 10.1007/s00261-018-1840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Calster B, Van Hoorde K, Valentin L, Testa AC, Fischerova D, Van Holsbeke C, International Ovarian Tumour Analysis Group et al. Evaluating the risk of ovarian cancer before surgery using the ADNEX model to differentiate between benign, borderline, early and advanced stage invasive, and secondary metastatic tumours: prospective multicentre diagnostic study. BMJ. 2014;349:5920. doi: 10.1136/bmj.g5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Calster B, Van Hoorde K, Froyman W, Kaijser J, Wynants L, Landolfo C, et al. Practical guidance for applying the ADNEX model from the IOTA group to discriminate between different subtypes of adnexal tumors. Facts Views Vis Obgyn. 2015;7(1):32–41. [PMC free article] [PubMed] [Google Scholar]
- 29.Van Calster B. External validation of ADNEX model for diagnosing ovarian cancer: evaluating performance of differentiation between tumor subgroups. Ultrasound Obstet Gynecol. 2017;50(3):406–407. doi: 10.1002/uog.17391. [DOI] [PubMed] [Google Scholar]
- 30.Timmerman D, Valentin L, Bourne TH, Collins WP, Verrelst H, Vergote I, International Ovarian Tumor Analysis (IOTA) Group Terms, definitions and measurements to describe the sonographic features of adnexal tumors: a consensus opinion from the International Ovarian Tumor Analysis (IOTA) Group. Ultrasound Obstet Gynecol. 2000;16(5):500–505. doi: 10.1046/j.1469-0705.2000.00287.x. [DOI] [PubMed] [Google Scholar]
- 31.Szubert S, Wojtowicz A, Moszynski R, Zywica P, Dyczkowski K, Stachowiak A, et al. External validation of the IOTA ADNEX model performed by two independent gynecologic centers. Gynecol Oncol. 2016;142(3):490–495. doi: 10.1016/j.ygyno.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 32.Sayasneh A, Ferrara L, De Cock B, Saso S, Al-Memar M, Johnson S, et al. Evaluating the risk of ovarian cancer before surgery using the ADNEX model: a multicentre external validation study. Br J Cancer. 2016;115(5):542–548. doi: 10.1038/bjc.2016.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.