Abstract
The functional receptor for insect ecdysteroid hormones is a heterodimer consisting of two nuclear hormone receptors, ecdysteroid receptor (EcR) and the retinoid X receptor homologue Ultraspiracle (USP). Although ecdysone is commonly thought to be a hormone precursor and 20-hydroxyecdysone (20E), the physiologically active steroid, little is known about the relative activity of ecdysteroids in various arthropods. As a step toward characterization of potential differential ligand recognition, we have analyzed the activities of various ecdysteroids using gel mobility shift assays and transfection assays in Schneider-2 (S2) cells. Ecdysone showed little activation of the Drosophila melanogaster receptor complex (DmEcR-USP). In contrast, this steroid functioned as a potent ligand for the mosquito Aedes aegypti receptor complex (AaEcR-USP), significantly enhancing DNA binding and transactivating a reporter gene in S2 cells. The mosquito receptor also displayed higher hormone-independent DNA binding activity than the Drosophila receptor. Subunit-swapping experiments indicated that the EcR protein, not the USP protein, was responsible for ligand specificity. Using domain-swapping techniques, we made a series of Aedes and Drosophila EcR chimeric constructs. Differential ligand responsiveness was mapped near the C terminus of the ligand binding domain, within the identity box previously implicated in the dimerization specificity of nuclear receptors. This region includes helices 9 and 10, as determined by comparison with available crystal structures obtained from other nuclear receptors. Site-directed mutagenesis revealed that Phe529 in Aedes EcR, corresponding to Tyr611 in Drosophila EcR, was most critical for ligand specificity and hormone-independent DNA binding activity. These results demonstrated that ecdysone could function as a bona fide ligand in a species-specific manner.
Ligand-activated transcription factors in the superfamily of steroid/thyroid/retinoid nuclear hormone receptors play an essential role in regulating the differential expression of genes involved in fundamental processes of animal development and reproduction. In vertebrates, a wide variety of distinct gene-regulatory pathways are realized by the action of chemically diverse ligands, including steroid hormones, thyroid hormones, retinoids, vitamin D, prostaglandins, and oxysterols, in combination with their cognate receptors (38, 39, 66).
In sharp contrast with this diversity of signals in vertebrate animals, the major events in development and reproduction in insects are primarily governed by a small number of known nuclear hormone receptor ligands. Among them, the steroid hormone 20-hydroxyecdysone (20E) is widely accepted as the key hormone regulating a vast array of gene activities (5, 15, 22, 50). The molecular basis of 20E action has been elucidated in great detail (10, 24, 26, 46, 51, 56, 64, 65). The two nuclear hormone receptors that play a central role in the initiation of the 20E-induced gene regulatory hierarchy are the ecdysteroid receptor (EcR) and the retinoid X receptor (RXR) homologue Ultraspiracle (USP) (12, 18, 25, 27, 34, 43, 57, 60). It has been shown that USP is an obligatory heterodimeric partner of EcR, required for both ligand and DNA binding (63, 74, 75). Functional diversity of ecdysteroid receptors within a given species may be achieved in part by differential expression of EcR and USP isoforms. Different EcR isoforms have been identified and cloned from the fruit fly Drosophila melanogaster and from the tobacco hornworm Manduca sexta, and evidence suggests that differential EcR isoform expression contributes to the tissue and stage specificity of 20E action (29, 52, 61). Two USP isoforms have been identified and cloned in the mosquito Aedes aegypti and in M. sexta (30, 31). Both mosquito USP isoforms (USP-A and USP-B) have been shown to form functional heterodimeric complexes with the mosquito EcR, when binding to either ecdysteroid-responsive elements (EcREs) or ecdysteroid ligand (31). The mosquito and tobacco hornworm USP isoforms appear to be functionally distinct, displaying differential response to activation by 20E and thus contributing to the tissue and stage specificity of 20E action (36, 69). Differential ecdysteroid response may also be effected through the differential recognition of a variety of EcREs, including inverted and direct repeats with various spacers (1, 2, 14, 49, 70).
The ecdysteroid 20E is derived via ecdysone-20-monooxygenase-mediated conversion of ecdysone in the peripheral tissues of an insect body (19, 58). However, detailed studies of M. sexta and D. melanogaster suggest a more complex composition of steroid hormones in at least some insects. In M. sexta, the major ecdysteroid secreted by the prothoracic glands is 3-dehydroxyecdysone, which is converted to ecdysone in the hemolymph (33, 54, 71, 72). Furthermore, during the pupal-adult metamorphosis of M. sexta, there are three major hemolymph ecdysteroid peaks: ecdysone, 20E, and 20,26-dihydroxyecdysone (71). In D. melanogaster, the ring gland synthesizes and secretes ecdysone and 20-deoxymakisterone, which are converted in peripheral tissues to 20E and makisterone, respectively (44, 47).
While these findings raise the question of whether ecdysteroids other than 20E can play distinct roles in insect development and reproduction, there are only a few examples of differential action of ecdysteroids. Champlin and Truman (7, 8) have recently presented the most compelling case of ecdysone as an active hormone, demonstrating a role for ecdysone in stimulating cell proliferation during optic lobe neurogenesis in M. sexta.
Study of the direct interaction between physiological ecdysteroids and their receptors has been hampered by the relatively low affinity of receptor-ligand interactions. Previous studies on the interaction between ecdysteroids and the EcR have been conducted primarily using crude receptor-containing cell extracts by analysis of competition for binding of the radiolabeled ecdysteroid ponasterone A (PonA) (11, 37, 40, 53, 76). Recently, a more detailed analysis of the binding of PonA and the nonsteroidal agonist tebufenozide (RH-5992) to the EcR-USP heterodimer has been reported. When tritiated PonA was bound to EcR-USP complexes of D. melanogaster, A. aegypti, and the lepidopteran Choristoneura fumiferana, similar affinities were observed (Kds of 0.8, 2.8, and 3 nM, respectively); the Kds for tebufenozide, a synthetic ecdysteroid agonist, for the same receptors were 336, 28, and 0.5 nM, respectively (16). These data suggest that variability within the hormone binding domain of EcRs may result in substantial difference in the binding of various ligands. Comparatively little is known about the effect of the various endogenous insect ecdysteroids on the DNA binding and transactivating ability of the EcR-USP heterodimer.
Here, we used the Drosophila EcR-B1–USP (DmEcR- DmUSP) and Aedes EcR–USP-B (AaEcR-AaUSP) heterodimers and examined the effect of ecdysone and 20E on their DNA binding and transactivation properties. We show that while 20E activates both DmEcR-DmUSP and AaEcR-AaUSP, ecdysone is an efficient activator only of AaEcR-AaUSP. The differential responsiveness of the mosquito and fly EcR-USP heterodimers to these ecdysteroids was determined by the EcR subunit and not by USP. Furthermore, domain-swapping experiments demonstrated that the high responsiveness to ecdysone was located in the C-terminal portion of the AaEcR ligand binding domain (LBD), within a region previously implicated in receptor heterodimerization. Using site-directed mutagenesis, we have identified a single amino acid, Phe529 in AaEcR, corresponding to Tyr611 in DmEcR, which plays a critical role in ligand specificity and hormone-independent DNA binding activity.
MATERIALS AND METHODS
In vitro protein synthesis and EMSA.
For electrophoretic mobility shift assays (EMSA), nuclear receptor proteins were synthesized in vitro using a coupled transcription-translation (TNT) kit from Promega. The in vitro expression vectors pGEM3Z-AaEcR, pGEM3Z-AaUSP-B, pGEM7Z-DmEcR, and pGEM7Z-DmUSP, with entire open reading frames of indicated nuclear receptor cDNAs, were constructed as described previously (31, 70). TNT-produced protein was quantified by [35S]methionine labeling, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and phosphorimage analysis. Protein yield ranged from 0.1 to 1.6 fmol/μl. A parallel TNT reaction with the same concentration of unlabeled methionine was performed to produce the protein for EMSA. The amounts of receptor proteins were adjusted in order to have comparable levels of DNA-protein complexes for the mosquito and Drosophila EcR-USP after activation with 20E. Under these conditions, the hormone-independent DNA binding of DmEcR-USP was not detectable. Receptor proteins were first incubated with 5 × 10−5M ecdysteroid at room temperature for 30 min in a total volume of 20 μl of HKN buffer containing 20 mM HEPES (pH 7.5), 2 mM dithiothreitol, 100 mM KCl, 7.5% glycerol, 1% NP-40 (Boehringer Mannheim), 2 μg of poly(dI-dC) · poly(dI-dC) (Pharmacia Biotech), and 3 μg of nonspecific single-stranded competitor oligonucleotide (70). Then 50 fmol of 32P-labeled probe IRhsp-1, which is the Drosophila heat shock protein 27 (HSP-27) EcRE (IRhsp-1 [70]), was added to the mixture followed by incubation at room temperature for another 30 min. Bound and free probes were resolved in 5 or 6% native acrylamide gels in 0.5× Tris-borate-EDTA. The gel was vacuum dried and exposed to either X-ray film (Kodak) or a PhosphorImager (Molecular Dynamics) for quantification.
Ecdysteroids and purification.
Muristerone A (MurA), polypodine B (PolB), 20E, 20-hydroxyecdysone 22-acetate (22A), and 2-deoxy-20-hydroxyecdysone (2DE) were purchased from Sigma. PonA was purchased from Invitrogen.
Ecdysone was purchased from SIMES (Milan, Italy) and contained approximately 2% 20E. Purification of the ecdysone was carried out on a reversed-phase Nova-Pak C18 cartridge (10 cm by 8 mm; particle size, 4 μm; Waters Associates, Watford, Herts, United Kingdom), using a linear gradient over 30 min of 35 to 100% (vol/vol) methanol-water at 1 ml/min. For each injection, 20 μg of ecdysone was introduced onto the column using a Gilson 234 autoinjector (Gilson, Villiers le Bel, France), and the separation was monitored by UV absorbance at 254 nm. Contaminating 20E eluted at 5 min, with pure ecdysone eluting at 8.5 min. Repeated high-pressure liquid chromatographic runs were carried out; the area around the ecdysone peak was collected over a 3-min period in each case, the fractions were combined, and the solvent was removed by rotary evaporation to yield approximately 1 mg of ecdysone. Analysis of an aliquot of the purified ecdysone under the same chromatographic conditions showed that it was essentially pure.
Reporter and insect expression vectors for transfection assays.
The reporter plasmids ΔMTV-Eip-Luc (Eip-Luc) and ΔMTV-Hsp-Luc (Hsp-Luc) were kind gifts from M. McKeown (Salk Institute, San Diego, Calif.). Partial sequencing indicated Eip-Luc contained four copies of eip-28/29 EcRE, whereas Hsp-Luc contained two copies of hsp-27 EcRE. The expression vector pAc-DmEcR (34) utilizes actin 5C to expressed DmEcR in Schneider-2 (S2) cells. The reporter pAc5-LacZ (Invitrogen) was used to normalize transfection efficiency. The entire AaEcR cDNA was obtained by digesting pcDNA3.1Zeo(+)-AaEcR (70) with BamHI, blunted with Klenow enzyme, and further digested with XbaI. This AaEcR cDNA fragment was then inserted into the EcoRV and XbaI sites of pAc5/V5/HisA (Invitrogen), yielding the expression construct pAc5-AaEcR. AaUSP expression plasmids pAc5-AaUSP-B and pAc5-DmUSP were constructed by inserting the EcoRI cDNA fragments from pcDNA3.1Zeo(+)-AaUSP-B and pcDNA3.1Zeo(+)-DmUSP (70) into the EcoRI site of pAc5/V5/HisA. These expression plasmids utilized the same promoter, actin 5C. All constructs were confirmed by restriction enzyme digestion and partial sequencing.
Cell culture and transient transfection assay.
Drosophila cell line S2 (Invitrogen) was maintained at 22 to 24°C in Schneider Drosophila medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco BRL). Transfection was conducted with LipofectACE (Gibco BRL) with an optimal DNA lipid ratio of 1:20 (wt/wt). Typically, 100 ng of luciferase reporter gene, 25 ng of reporter pAc5-LacZ, 12.5 ng of each receptor, and 3 μg of LipofectACE were mixed in a 24-well plate with a total volume of 20 μl and incubated at room temperature for 30 min. The expression vector pAc5/V5/HisA was used as carrier DNA so that each well received 150 ng of total DNA. The transfection cocktail was overlaid with 500 μl of S2 cells, which were diluted to 106 cells/ml in Drosophila serum-free medium (Gibco BRL). Half of the amount of DNA, LipofectACE, and cells were used for transfection assays in 48-well plates. Transfection was terminated 12 to 15 h later with the addition of 5% fetal bovine serum. After 24 or 36 h of hormone treatment, the medium was aspirated and the cells in suspension and attachment were combined in 100 μl of reporter lysis buffer (Promega) and lysed with three cycles of freezing and thawing. Reporter gene assays were conducted as described for the Promega firefly luciferase reporter and β-galactosidase systems. A luminometer (Turner Designs model TD20e) was used to detect luciferase activity with 10-s delay time and 30-s integration time. The luciferase activity was normalized with β-galactosidase activity. Transfection assays were carried out in duplicates or triplicates, and each independent experiment was repeated at least three times. Although the absolute values of reporter gene activities varied from experiment to experiment, the fold induction of luciferase activity from cells treated with hormone over those treated with control vehicle ethanol remained relatively consistent after normalization with LacZ.
Construction of chimeric receptors.
Five chimeric receptors, AEBsrG1, the reciprocal construct DEBsrG1, DEXma3, DEKpn1, and DEBgl2, were constructed by swapping at the DNA binding domain (DBD) and boundaries between domains C and D, domains D and E, and domains E and F, respectively. A BamHI fragment containing the DmEcR cDNA from pAc5-DmEcR was first subcloned into the BamHI site of pcDNA3.1/Zeo(+), yielding pcDNA3.1/Zeo(+)-DmEcR. pcDNA3.1/Zeo(+)-AaEcR was constructed in a similar way, as described previously (70). The 1,939bp BsrGI-XbaI fragment in pcDNA3.1/Zeo(+)-AaEcR was exchanged with the 2,125bp BsrGI-XbaI fragment in pcDNA3.1/Zeo(+)-DmEcR, yielding two chimeric receptor constructs, pcDNA3.1/Zeo(+)-AEBsrG1 and pcDNA3.1/Zeo(+)-DEBsrG1. pGEM7Z-DEXma3 was created by digesting pGEM7Z-DmEcR with XmaIII and XbaI, and the 3,435-bp fragment containing the vector and 5′ region of the DmEcR cDNA was ligated with a 1,869-bp XmaIII-XbaI fragment bearing the 3′ region of the AaEcR cDNA from pcDNA3.1/Zeo(+)-AaEcR. To make the construct pcDNA3.1/Zeo(+)-DEKpn1, the 1,309-bp KpnI fragment with a 5′ region of AaEcR cDNA in pcDNA3.1/Zeo(+)-AaEcR was replaced with 1,508-bp KpnI fragment with a 5′ region from DmEcR cDNA in pcDNA3.1/Zeo(+)-DmEcR. pcDNA3.1/Zeo(+)-DEBgl2 was created by ligating the 5,959-bp BglII fragment from pcDNA3.1/Zeo(+)-AaEcR with a 3,061-bp BglII fragment from pcDNA3.1/Zeo(+)-DmEcR.
Nine chimeric constructs, AESac1, AENru1, DEBbs1, DETthIII1, DECsp1, DESpe1, DEBsiW1, AENB, and DESS, were prepared by swapping within the LBD using a combination of restriction digestion and PCR amplification techniques. pGEM3Z-AENru1 was constructed by replacing the 1,303-bp NruI-EcoRI fragment in pGEM3Z-AaEcR with the 1,490-bp NruI-EcoRI fragment from pGEM7Z-DmEcR. pGEM7Z-DETthIII1 was constructed by replacing the 1,409-bp fragment in pGEM7Z-DmEcR with the 1,182-bp TthIII1-XbaI fragment from pcDNA3.1/Zeo(+)-AaEcR. To construct pGEM3Z-AESac1, the primer pair DE-Sac1-For and DE-EcoR1-Rev was used to amplify an 836-bp fragment from pGEM7Z-DmEcR. This fragment was digested with SacI and EcoRI to replace the 991-bp SacI-EcoRI fragment in pGEM3Z-AaEcR, yielding the chimera pGEM3Z-AESac1. PCRs were performed with the polymerase Pfu (Promega) with an initial denaturation at 94°C for 2 min followed by 20 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 45 s, and elongation at 72°C for 3 min. To make the chimeric constructs pGEM7Z-DEBbs1, pGEM7Z-DECsp1, pGEM7Z-DESpe1, pGEM7Z-DEBsiW1, and pGEM7Z-DESS, pairs of forward and reverse primers were annealed with the template, either pGEM3Z-AaEcR or pGEM7Z-DmEcR, for PCR amplification, and the amplified fragments were digested with restriction enzymes to allow cloning into the appropriate recipient plasmids. The chimeric plasmid pGEM3Z-AENB was constructed by replacing the 1,284-bp NruI-EcoRI fragment in pGEM3Z-AaEcR with the 729-bp NruI-XbaI fragment in pGEM7Z-DEBsiW1, blunting the EcoRI and XbaI sites.
Construction of site-directed point mutants.
Site-directed mutagenesis of DmEcR was conducted according to the instruction manual for the QuickChange site-directed mutagenesis system (Stratagene). A pair of complementary primers (40 pmol of each) and 10 ng of template plasmid pGEM3Z-AaEcR or pGEM7Z-DmEcR in 100 μl (total volume) were subjected to PCR amplification with Pfu (Promega). PCRs were performed with an initial denaturation at 94°C for 2 min, followed by 15 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 14 min. The PCR products were treated with DpnI (Stratagene) to remove the methylated template DNA then gel purified, and transformed into Escherichia coli. Sixteen site-directed mutants with a single amino acid mutation were constructed this way. Eight of these (pGEM3Z-AEH502A, pGEM3Z-AEA520C, pGEM3Z-AEP523S, pGEM3Z-AEK524M, pGEM3Z-AEC525S, pGEM3Z-AES526L, pGEM3Z-AE1528F, and pGEM3Z-AEF529Y) were AaEcR mutants; the other eight (pGEM7Z-DEA584H, pGEM7Z-DEC602A, pGEM7Z-DES605P, pGEM7Z-DEM606K, pGEM7Z-DES607C, pGEM7Z-DEL608S, pGEM7Z-DEF610I, and pGEM7Z-DEY611F) were DmEcR mutants. These mutants were confirmed by partial sequencing. PCR primer sequences are available upon request.
RESULTS
Differential effects of ecdysone and 20E on EcR-USP DNA binding activity.
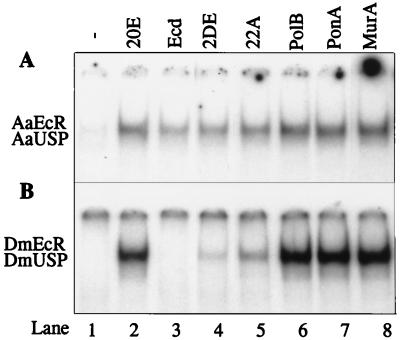
First, we compared the abilities of several ecdysteroids to stimulate the DNA binding activity of the mosquito and Drosophila EcR-USP heterodimers. The following ecdysteroids were tested: ecdysone, 20E, 2DE, 22A, PolB, PonA, and MurA. Each ecdysteroid (5 × 10−5 M) was incubated with receptor proteins prepared by in vitro transcription-translation (see Materials and Methods), and the reaction mix was subjected to EMSA. We first compared the effect of various ecdysteroids on Aedes receptors. A low level of DNA binding by the AaEcR-AaUSP heterodimer was detected in the absence of any ligand (Fig. 1A, lane 1), and this activity was dramatically stimulated by 20E addition (Fig. 1A, lane 2). Ecdysone also significantly stimulated AaEcR-AaUSP DNA binding activity (Fig. 1A, lane 3), although less strongly than 20E. Other ecdysteroids enhanced AaEcR-AaUSP DNA binding activity with the following decreasing potency order: MurA > PonA > PolB > 20E > 22A > 2DE > ecdysone (Fig. 1A).
FIG. 1.
Differential effects of ecdysteroids on receptor DNA binding activities. (A) In vitro-translated AaEcR and AaUSP proteins were incubated with 32P-labeled IRhsp-1 EcRE in the absence of ligand (lane 1) or in the presence of 5 × 10−5 M 20E (lane 2), ecdysone (lane 3), 2DE (lane 4), 22A (lane 5), PolB (lane 6), PonA (lane 7), or MurA (lane 8). The reaction mixtures were subjected to EMSA and autoradiography. (B) Same as panel A except that DmEcR and DmUSP were used as receptor proteins. The molar amount of DmEcR and DmUSP proteins was 50 times more than that of AaEcR and AaUSP so that any trace DNA binding activity of DmEcR-DmUSP could be detected.
We then tested the effect of these ecdysteroids on the DmEcR-DmUSP complex. Unlike the mosquito AaEcR-AaUSP heterodimer, the DmEcR-DmUSP heterodimer exhibited extremely low hormone-independent DNA binding. Even when the molar amounts of DmEcR and DmUSP were 50 times greater than the amounts used for the mosquito receptors, no hormone-independent DNA binding was detected (Fig. 1B, lane 1). Detection of hormone-independent DNA binding by the Drosophila receptor required 100-fold more DmEcR and DmUSP proteins (data not shown). The DmEcR-DmUSP heterodimer exhibited robust activation of its DNA binding in response to 20E, PonA, MurA, and PolB (Fig. 1B, lanes 2 and 6 to 8). The ligand 22A induced appreciable binding of the Drosophila heterodimer, while the effect of 2DE was weak (Fig. 1B, lanes 4 and 5). In stark contrast to what was observed for the mosquito heterodimer, though, ecdysone had no detectable effect on the DNA binding of the Drosophila receptor heterodimer (Fig. 1B, lane 3). Thus, our EMSA results demonstrated a clear differential effect of ecdysone on the DNA binding activity of the AaEcR-AaUSP and DmEcR-DmUSP heterodimers.
To determine the concentration of ecdysone and 20E required to stimulate DNA binding, AaEcR and AaUSP lysates were incubated with increasing concentrations of ecdysone or 20E, ranging from 5 × 10−12 to 5 × 10−5 M, and subjected to EMSA. The effect of 20E on the DNA binding of the AaEcR-AaUSP heterodimer was first evident at 5 × 10−8 M (not shown). DNA binding activity increased proportionally with increasing concentration of 20E and reached its maximal level at 5 × 10−5 M (not shown). The effect of 20E on the Drosophila heterodimer was similar (not shown). Ecdysone was considerably weaker than 20E, with visible stimulation of the AaEcR-AaUSP DNA binding at 5 × 10−6 M hormone (not shown). No stimulation of DNA binding by the DmEcR-DmUSP heterodimer was detected in the EMSA even with 5 × 10−5 M ecdysone (Fig. 1B, lane 3).
EcR protein, not USP protein, conferred specific response to ecdysone.
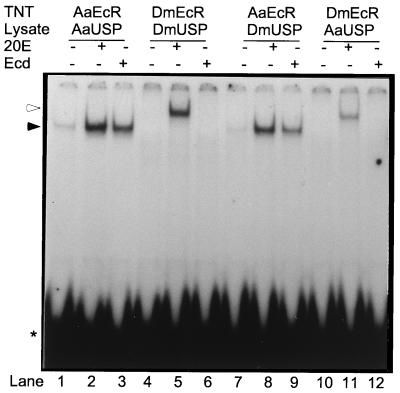
To determine whether EcR or USP dictates the ligand specificity of the heterodimer, we next conducted subunit-swapping experiments. These experiments demonstrated that the behavior of heterodimers with respect to both hormone-independent DNA binding and ligand specificity are determined by the EcR subunit. When AaEcR was paired with DmUSP, the heterodimer exhibited a level of hormone-independent binding similar to that of the AaEcR-AaUSP heterodimer (Fig. 2, lanes 1 and 7). Likewise, the DNA binding activity of the AaEcR-DmUSP heterodimer was highly stimulated by both 20E and ecdysone (Fig. 2, lanes 8 and 9). Testing different concentrations of these two hormones further demonstrated that the AaEcR-DmUSP heterodimer behaved similarly to the AaEcR-AaUSP heterodimer, responding to 10−8 M 20E and 10−6 M ecdysone (not shown). The reciprocal combination of DmEcR and AaUSP had no detectable hormone-independent binding activity (Fig. 2, lane 10). The binding of the DmEcR-AaUSP was stimulated only by 20E and not by ecdysone, as observed for the DmEcR-DmUSP complex (Fig. 2, lanes 11 and 12).
FIG. 2.
AaEcR conferred specific response to ecdysone. In vitro-translated proteins AaEcR and AaUSP (lanes 1 to 3), DmEcR and DmUSP (lanes 4 to 6), AaEcR and DmUSP (lanes 7 to 9), or DmEcR and AaUSP (lanes 10 to 12) were incubated with 50 fmol of 32P-labeled IRhsp-1 EcRE probe either in the absence of hormone (lanes 1, 4, 7, and 10) or in the presence of 5 × 10−5 M 20E (lanes 2, 5, 8, and 11) or ecdysone (lanes 3, 6, 9, and 12). The reaction mixtures were subjected to EMSA and autoradiography. The molar amount of DmEcR and DmUSP proteins was 50 times more than that of AaEcR and AaUSP so that any trace DNA binding activity of DmEcR-DmUSP could be detected. Free probe is indicated by an asterisk. Complexes containing AaEcR and DmEcR proteins are indicated by solid and open arrowheads, respectively.
Differential ecdysteroid-stimulated transactivation by mosquito and Drosophila receptors in Drosophila S2 cells.
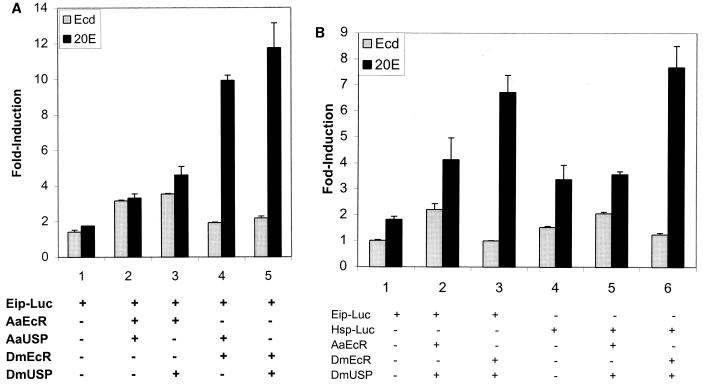
To investigate the effects of 20E and ecdysone on target gene transactivation by the mosquito and Drosophila heterodimers, we used a cell transfection assay. The reporter plasmid Eip-Luc was transfected into Drosophila S2 cells alone or along with pairwise combinations of expression plasmids carrying AaEcR, AaUSP, DmEcR, or DmUSP cDNA. After transfection, cells were incubated either in the absence of hormone or in the presence of 20E or ecdysone at 10−5 M. When challenged with 20E or ecdysone, cells receiving Eip-Luc alone exhibited a low level of activation, which was presumably mediated by the endogenous Drosophila DmEcR (Fig. 3A, column 1). After cotransfection along with AaEcR and AaUSP, the signal was considerably more robust in response to both 20E and ecdysone. Substitution of AaUSP with DmUSP did not change the response (Fig. 3A, columns 2 and 3). The DmEcR-DmUSP and DmEcR-AaUSP heterodimers each responded strongly to 20E and very weakly to ecdysone (Fig. 3A, columns 4 and 5). These results corroborated our finding, from the EMSA subunit swapping experiments, that the specificity of hormonal response is determined by EcR and not by USP. To minimize the variability of conditions, we have used DmUSP as a partner in all subsequent experiments used to characterize functional differences between mosquito and Drosophila EcRs.
FIG. 3.
(A) Ecdysone (10−5 M) more potently activated AaEcR than DmEcR in S2 cells. S2 cells were transfected with 25 ng of reporter pAc5-LacZ and 100 ng of reporter plasmid Eip-Luc with no expression plasmid (column 1) or with 12.5 ng each of AaEcR, AaUSP, DmEcR, and DmUSP expression vectors in pairwise combinations: AaEcR and AaUSP (column 2), AaEcR and DmUSP (column 3), DmEcR and AaUSP (column 4), and DmEcR and DmUSP (column 5). After transfection, cells were incubated either in the absence of hormone or in the presence of 5 × 10−5 M 20E or ecdysone for 36 h and harvested for β-galactosidase and luciferase activities. (B) Ecdysone (10−6 M) highly activated only the Aedes receptor, not the Drosophila receptor. S2 cells (2.5 × 105) were transfected with 12.5 ng of reporter pAc5-LacZ and 50 ng of reporter plasmid Eip-Luc (columns 1 to 3) or Hsp-Luc (columns 4 to 6) together with no expression plasmid (columns 1 and 4) or with 6.5 ng each of AaEcR and DmUSP (columns 2 and 5) or DmEcR and DmUSP (columns 3 and 6) expression vectors. After transfection, cells were incubated in the absence of hormone or in the presence of 10−6 M ecdysone (Ecd) or 20E for 24 h and harvested for β-galactosidase and luciferase activities. Luciferase activity was normalized with β-galactosidase activity. The results are expressed as fold induction of the luciferase activity from cells treated with hormone over that from cells treated with control vehicle ethanol.
We also compared transactivation of a reporter construct containing the HSP-27 EcRE with that of EIP. We used 10−6 M 20E and ecdysone, as this amount of ecdysone provided clear discrimination between the strong response of the mosquito receptor and the very weak response of the Drosophila receptor (Fig. 4). These experiments, using the mosquito or Drosophila EcR in combination with DmUSP, showed that responses were, for the most part, similar for different types of reporters. However, both the endogenous receptor and transfected Drosophila receptor did show modestly stronger ecdysone activation of the inverted repeat containing the HSP-27 reporter (Fig. 3B).
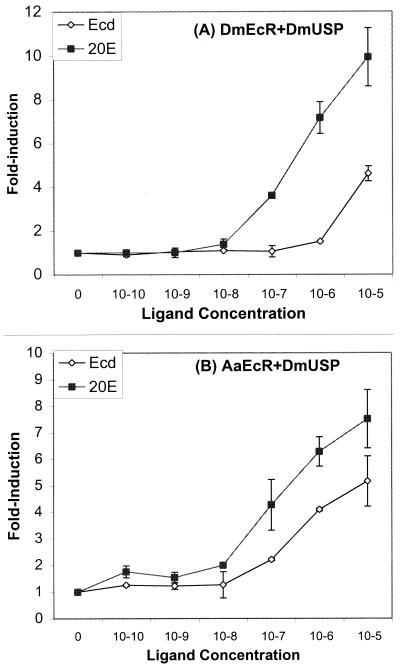
FIG. 4.
Dose-dependent transactivation by 20E and ecdysone in the presence of AaEcR or DmEcR. S2 cells (5 × 105 cells/well) were transfected with 25 ng of reporter pAc5-LacZ and 100 ng of reporter plasmid Eip-Luc with 12.5 ng each of DmEcR and DmUSP (A) or AaEcR and DmUSP (B) expression vectors. After transfection, cells were incubated in the absence of hormone or in the presence of increasing concentrations (from 10−10 to 10−5 M) of 20E or ecdysone for 24 h, and harvested for β-galactosidase and luciferase activities. Luciferase activity was normalized with β-galactosidase activity. The results are expressed as fold induction of the luciferase activity from cells treated with hormone over that from cells treated with control vehicle ethanol. Error bars for some points are too small to be visible on the graph.
When S2 cells were cotransfected with the reporter plasmid along with the DmEcR and DmUSP plasmids, they responded to 10−7 M 20E. However, only a very weak response to ecdysone could be detected at 10−6 M, with strong response seen only at a concentration of 10−5 M (Fig. 4A). When S2 cells were cotransfected with AaEcR and DmUSP plasmids along with the reporter, they responded to 20E at the same concentration, 10−7 M. In contrast, these cells exhibited a markedly enhanced responsiveness to ecdysone relative to cells transfected with DmEcR, showing a clearly detectable level of reporter gene induction at 10−7 M and a relatively high level of transactivation at 10−6 M ecdysone (Fig. 4B).
Taken together, these results established that in transfection assays, DmEcR and AaEcR exhibited similar responsiveness to 20E. In contrast, the Aedes receptor was nearly 2 logs more sensitive than the Drosophila receptor to stimulation with ecdysone.
Mapping the EcR domain responsible for differential recognition of 20E and ecdysone.
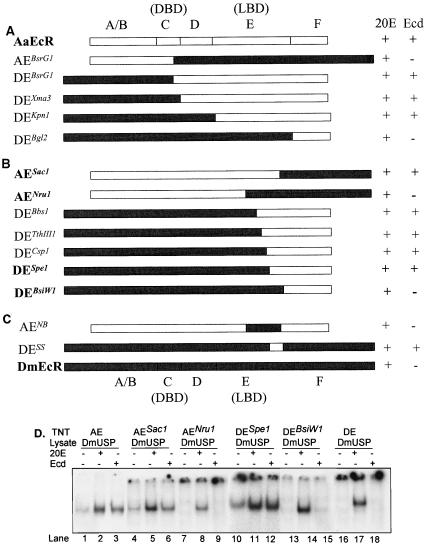
EMSA and transfection assays demonstrated that the EcR protein, and not USP, was responsible for the ligand specificity of the EcR complex. Next, we attempted to identify the EcR domain responsible for differential ligand recognition. The EcR protein possess five functional domains, an N-terminal domain A/B, DBD C, hinge domain D, LBD E, and C-terminal domain F (10, 34). To locate the ligand specificity determinants, we made five chimeric EcR constructs by swapping the appropriate Aedes and Drosophila EcR domains. The AEBsrG1 chimera contained the A/B domain and most of the C domain from AaEcR and the remainder of the C domain along with domains D, E, and F from DmEcR. Reciprocally, DEBsrG1 included the A/B domain and most of the C domain from DmEcR and the rest of domain C along with domains D, E, and F from AaEcR (Fig. 5A). By swapping at the predicted boundaries between domains C and D, domains D and E, and domains E and F, we constructed three additional chimeras with N termini from DmEcR and C termini from AaEcR, namely, DEXma3, DEKpn1, and DEBgl2. These chimeric proteins were produced by in vitro transcription-translation, paired with DmUSP protein, and subjected to EMSA. As seen for DmEcR, the AEBsrG1 and DEBgl2 DNA binding activities were stimulated by 20E, not by ecdysone. These two chimeras did not display any detectable hormone-independent DNA binding activity (not shown). In contrast, the DEBsrG1, DEXma3, and DEKpn1 chimeric proteins, containing AaEcR C termini, exhibited clearly detectable hormone-independent DNA binding activity and significant stimulation by both 20E and ecdysone (Fig. 5A). These results unambiguously mapped the determinants allowing responsiveness to ecdysone to domain E, the LBD. Furthermore, the same region of the AaEcR receptor appeared to be responsible for an increased level of hormone-independent DNA binding activity.
FIG. 5.
(A) Localization of the ecdysone-specific region to the LBD: schematic diagram shows domain-swapping chimeric proteins and their responsiveness to ecdysone. 32P-labeled probe IRhsp-1 EcRE was incubated with in vitro-synthesized DmUSP protein paired with chimeric protein AEBsrG1, DEBsrG1, DEXma3, DEKpn1, or DEBgl2 in the absence of hormone or in the presence of 5 × 10−5 M 20E or ecdysone. Bound and free probes were resolved by EMSA followed by autoradiography. (B) C-terminus of EcR LBD determined ecdysone binding specificity: schematic diagram of subdomain-swapping (within LBD) chimeric EcR proteins and their responsiveness to 20E and ecdysone. 32P-labeled probe IRhsp-1 was incubated with in vitro-synthesized DmUSP protein paired with AESac1, AENru1, DEBbs1, DETthIII1, DECsp1, DESpe1, or DEBsiW1 in the absence of hormone or in the presence of 5 × 10−5 M 20E or ecdysone. Bound and free probes were resolved by EMSA followed by autoradiography. Constructs whose EMSA results are shown in panel D are in bold. (C) Transferable ligand specificity subdomains in AaEcR and DmEcR. 32P-labeled probe IRhsp-1 was incubated with in vitro-synthesized DmUSP protein paired with in vitro-translated AENB or DESS in the absence of hormone or in the presence of 5 × 10−5 M 20E or ecdysone. Bound and free probes were resolved by EMSA followed by autoradiography. Responsiveness to 20E and ecdysone (Ecd) is indicated by a plus sign, while lack of responsiveness is indicated by a minus sign in the schematic diagrams. Solid bars denote DmEcR sequence, and open bars denote AaEcR sequences. Domains A/B, C (DBD), D, E (LBD), and F are pointed out above AaEcR and below DmEcR sequences. (D) Ecdysone responsiveness of critical chimeric proteins revealed by EMSA. The wild-type proteins AaEcR (lanes 1 to 3) and DmEcR (lanes 16 to 18) and chimeric proteins AESac1 (lanes 4 to 6), AENru1 (lanes 7 to 9), DESpe1 (lanes 10 to 12), and DEBsiW1 (lanes 13 to 15) were paired with in vitro-synthesized DmUSP protein and then incubated with 32P-labeled probe IRhsp-1 in the absence of hormone (lanes 1, 4, 7, 10, 13, and 16) or in the presence of 5 × 10−5 M 20E (lane 2, 5, 8, 11, 14, and 17) or ecdysone (lanes 3, 6, 9, 12, 15, and 18). The reaction mixtures were resolved by EMSA followed by autoradiography.
Identification of the subdomain responsible for the ligand specificity and hormone-independent DNA binding activity.
Next, using receptor fragments prepared by a combination of restriction digestion and PCR amplification, we prepared nine additional chimeric constructs, swapped at various points within the E domain. Tests with two chimeras, AESac1 and AENru1, containing carboxy-terminal sequences from DmEcR, suggested that the functional differences between the EcRs mapped to the C-terminal portion of the E domain and that the F domain did not play a role in the ligand recognition and heterodimerization (Fig. 5B and D, lanes 4 to 9). These results implied that the region governing ligand specificity and hormone-independent DNA binding activity was located between the NruI and SacI sites in the AaEcR cDNA.
Five more chimeric constructs were produced, proceeding from the NruI site to the SacI site, with N-terminal sequences from DmEcR and C-terminal sequences from AaEcR; these constructs were DEBbs1, DETthIII1, DECsp1, DESpe1, and DEBsiW1 (Fig. 5B). Four of these chimeric proteins, DEBbs1, DETthIII1, DECsp1, and DESpe1, behaved similarly to the mosquito EcR, showing significant hormone-independent DNA binding activity and strong stimulation by either 20E or ecdysone (Fig. 5B and D, lanes 10 to 12). In contrast, the DEBsiW1 chimera failed to show any hormone-independent DNA binding activity, and its DNA binding activity was not enhanced by ecdysone (Fig. 5B and D, lanes 13 to 15).
To further confirm the functionality of the subdomains corresponding to NruI-BsiWI in DmEcR and SacI-SpeI in AaEcR, we constructed two more chimeric receptors, AENB and DESS (Fig. 5C). The chimera AENB, consisting primarily of a AaEcR protein with only the NruI and BsiWI fragment from DmEcR, was highly responsive to 20E, yet this protein displayed no detectable level of hormone-independent DNA binding activity and only trace activity to ecdysone (Fig. 5C). In contrast, the DESS chimera, which was DmEcR containing only the short SacI-SpeI amino acid sequence from AaEcR, exhibited detectable hormone-free DNA binding activity as well as a strong response to 20E and ecdysone (Fig. 6C).
FIG. 6.
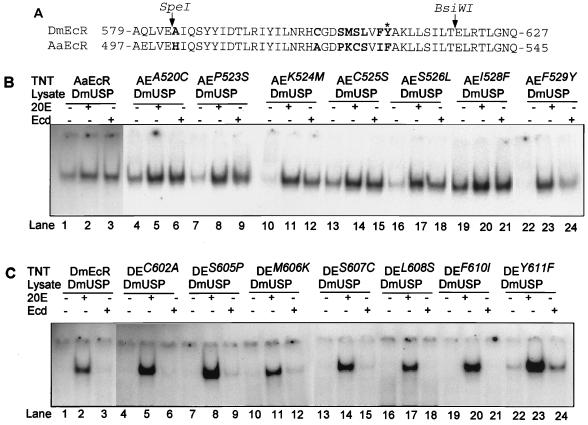
Identification of the critical amino acid affecting heterodimerization and responsiveness to ecdysone. (A) I box in EcR proteins. DmEcR (34) and AaEcR (12) protein sequences are aligned by GCG Bestfit. SpeI and BsiWI sites in AaEcR and DmEcR cDNAs are indicated by arrows. Nonconserved residues which were subjected to site-directed mutagenesis between SpeI and BsiWI sites are in bold. The critical residues F529 in AaEcR and Y611 in DmEcR are indicated by an asterisk. (B) F529 in AaEcR is critical for ligand specificity and hormone-free DNA binding activity. 32P-labeled probe IRhsp-1 was incubated with in vitro-synthesized DmUSP protein paired with the wild-type AaEcR (lanes 1 to 3) or point mutant AEA520C (lanes 4 to 6), AEP523S (lanes 7 to 9), AEK524M (lanes 10 to 12), AEC525S (lanes 13 to 15), AES526L (lanes 16 to 18), AEI528F (lanes 19 to 21), or AEF529Y (lanes 22 to 24) in the absence of hormone (lanes 1, 4, 7, 10, 13, 16, 19, and 22) or in the presence of 5 × 10−5 M 20E (lanes 2, 5, 8, 11, 14, 17, 20, and 23) or ecdysone (lanes 3, 6, 9, 12, 15, 18, 21, and 24). (C) Tyr611 in DmEcR is critical for ligand specificity and hormone-free DNA binding activity. 32P-labeled probe IRhsp-1 was incubated with in vitro-synthesized DmUSP protein paired with the wild-type protein DmEcR (lanes 1 to 3) or point mutant DEC602A (lanes 4 to 6), DES605P (lanes 7 to 9), DEM606K (lanes 10 to 12), DES607C (lanes 13 to 15), DEL608S (lanes 16 to 18), DEF610I (lanes 19 to 21), or DEY611F (lanes 22 to 24) in the absence of hormone (lanes 1, 4, 7, 10, 13, 16, 19, and 22) or in the presence of 5 × 10−5 M 20E (lanes 2, 5, 8, 11, 14, 17, 20, and 23) or ecdysone (lanes 3, 6, 9, 12, 15, 18, 21, and 24). Bound and free probes were resolved by EMSA followed by autoradiography. The molar amount of DmEcR and DE mutants protein was 10 times more than that of the molar amount of AaEcR and AE mutants in the EMSA.
Hence, we concluded that the region of the AaEcR LBD falling between the SpeI and BsiWI restriction sites was responsible for the enhanced levels of hormone-independent DNA binding activity and sensitivity to ecdysone.
AaEcR Phe529/DmEcR Tyr611 is the critical determinant of functional differences in ligand recognition and hormone-independent DNA binding.
Comparing AaEcR and DmEcR protein sequences revealed that 8 out of the 36 amino acids (aa) residing between the SpeI and BsiWI sites were different. These AaEcR/DmEcR amino acid differences were His502/Ala584, Ala520/Cys602, Pro523/Ser605, Lys524/Met606, Cys525/Ser607, Ser526/leu608, Ile528/Phe610, and Phe529/Tyr611 (Fig. 6A). To identify the critical amino acid or amino acids conferring ligand specificity, we created 16 site-directed mutants by replacing each amino acid in AaEcR with the corresponding residue in DmEcR one by one, and vice versa.
First, we constructed eight AaEcR site-directed mutants, in which His502, Ala520, Pro523, Lys524, Cys525, Ser526, Ile528, and Phe529 were mutated to Ala, Cys, Ser, Met, Ser, Leu, Phe, and Tyr, respectively. These mutant proteins were transcribed in vitro, paired with DmUSP, and subjected to EMSA. Four of the site-directed mutants, AEH502A (not shown), AEA520C, AEC525S, and AE1528F, exhibited strong hormone-free DNA binding activity as well as robust responses to 20E and ecdysone, resembling the wild-type AaEcR (Fig. 6B, lanes 1 to 6, 13 to 15, and 19 to 21). Three of these mutants, AEP523S, AEK524M, and AES526L, displayed decreased level of hormone-free DNA binding activity, although they still responded potently to ecdysone (Fig. 6B, lanes 7 to 9, 10 to 12, and 16 to 18). Notably, the mutant AEF529Y exhibited no detectable level of hormone-free DNA binding activity; its response to ecdysone was dramatically reduced compared with other mutants, although its response to 20E was unaltered (Fig. 6B, lanes 22 to 24), indicating that F529 in AaEcR is most critical for conferring high-level hormone-free DNA binding activity as well as a specific response to ecdysone.
We then created eight reciprocal site-directed mutants by converting an amino acid in DmEcR to its corresponding residue in AaEcR; Ala584, Cys602, Ser605, Met606, Ser607, Leu608, Phe610, and Tyr611 in DmEcR were mutated to His, Ala, Pro, Lys, Cys, Ser, Ile, and Phe, respectively, yielding DmEcR mutants DEA584H, DEC602A, DES605P, DEM606K, DES607C, DEL608S, DEF610I, and DEY611F. These mutant constructs were translated in vitro and paired with DmUSP for EMSA. DEA584H (data not shown), DEC602A, DES607C, DEL608S, and DEF610I proteins did not display any hormone-independent heterodimerization, and their DNA binding activity was detected only in the presence of 20E, as observed for the DmEcR parent protein (Fig. 6C, lanes 1 to 6 and 13 to 21). Likewise, the DES605P and DEM606K proteins showed no DNA binding activity in the absence of hormone, strong activity with 20E, and only trace activity with ecdysone (Fig. 6C, lanes 7 to 12), suggesting that the corresponding residues in AaEcR, Pro523 and Lys524, might play a minor role in ligand recognition. Remarkably, a strong effect on both hormone-independent DNA binding and ligand recognition was observed with one of the single amino acid substitutions; the DEY611F protein exhibited significant DNA binding activity in the absence of hormone, and this activity was strongly amplified not only with 20E but also with ecdysone, as observed for the AaEcR protein (Fig. 6C, lanes 22 to 24). These findings suggest a critical role for AaEcR Phe529/DmEcR Tyr611 as a determinant of functional differences between the Drosophila and mosquito EcRs. This single amino acid difference affects both hormone-independent heterodimeric DNA binding and species-specific ligand discrimination.
DISCUSSION
In contrast to most nuclear receptors, EcRs are characterized by a relatively low level of amino acid conservation (10, 18, 23). While the DBD is highly conserved among EcRs of different insects, the rest of the EcR protein shows substantial divergence relative to other members of nuclear hormone receptor superfamily. It is particularly surprising that even the LBD/heterodimerization domain, presumably recognizing the same hormone, 20E, varies from 87% identity between Drosophila and Aedes to 67% between Drosophila and Bombyx. Although no dramatic differences in the binding affinities for 20E or PonA have been observed among EcRs, the nonsteroidal agonist tebufenozide (RH-5992) binds different EcRs with strikingly different affinities (16, 17). Using gel mobility shift and transactivation assays, Suhr et al. (59) identified determinants in EcR of the silkworm Bombyx mori (BmEcR) which are responsible for activation of this receptor by tebufenozide. Construction of chimeric BmEcR-DmEcRs revealed that the tebufenozide sensitivity of BmEcR was correlated with a high level of hormone-independent heterodimer complex formation and DNA binding relative to that of the DmEcR. Discrete determinants within the hinge region (D) and the middle and C-terminal portions of the LBD (E2 and E3) of the BmEcR protein are presumably involved in both heterodimerization and increased affinity to tebufenozide (59).
In this report, we present the first molecular evidence of the differential effect of two natural insect steroid hormones on EcRs. Gel mobility shift and transfection assays indicated that the mosquito EcR was considerably more sensitive to ecdysone than was the Drosophila receptor; we show here that while 20E activated both DmEcR-DmUSP and AaEcR-AaUSP with equal efficiency, ecdysone activated AaEcR-AaUSP with significantly higher efficiency. The differential responsiveness of the mosquito and fly EcR-USP heterodimers to ecdysteroids was determined by the EcR subunit and not by USP. Furthermore, domain-swapping experiments mapped the high responsiveness to a 36-aa region in the C-terminal portion of the EcR LBD. Interestingly, we found that the increased responsiveness to a natural insect steroid hormone, ecdysone, also correlates with the enhanced hormone-independent DNA binding activity of the mosquito EcR. This specific region lies in E3, one of the regions involved in high level hormone-independent DNA binding activity of the BmEcR (59). However, in contrast to what was observed for BmEcR, our detailed domain swapping did not show any significant involvement of the hinge (domain D) or the middle region of the LBD (E2) in hormone-independent DNA binding activity and ligand sensitivity of the mosquito EcR.
EMSA indicated that ecdysone failed to stimulate discernible DNA binding activity of DmEcR, although this steroid exhibited only moderately lower activity than 20E in experiments using AaEcR. In transfection assays in S2 cells, ecdysone was capable of activating both receptors, DmEcR and AaEcR, but ecdysone was still much weaker than 20E in activating DmEcR. One possibility is that the effect of ecdysone on DmEcR in cell transfection assays could result from conversion of ecdysone to 20E or other more active ecdysteroids. However, neither time courses of ecdysone response nor preincubation experiments show changes in the relative efficacy of ecdysone over time (data not shown). Thus, it seems plausible that in vivo, other factors may act to stabilize the receptor-ligand complex on at least some targets enough to allow detection of weak interactions between ecdysone and DmEcR.
Analysis of Aedes-Drosophila EcR chimeras revealed that the region responsible for significantly augmented hormone-independent DNA binding activity and ecdysone responsiveness is located between aa 502 and 529 in AaEcR, corresponding to aa 584 to 611 in DmEcR. This region of EcR corresponds to helices 9 and 10 within other nuclear hormone receptors for which crystal structure data are available (4, 6, 48, 62, 68, 73). In estrogen receptor and RXR homodimers, helices 9 and 10 are located at the dimerization interface (4, 62). Using domain-swapping techniques, Perlmann et al. (45) localized a dimerization box, designated the identity (I) box, within this interval and demonstrated its critical role in the formation of RXR-retinoic acid receptor (RAR) and RXR-thyroid hormone receptor heterodimers. We aligned the I-box sequence of EcRs with that of human RARγ, which displays highest identity with EcRs among those receptors with available crystal structures. This region contains the predicted helices 9 and 10 as well as the loop connecting these two helices. Between the SpeI and BsiWI sites flanking the region responsible for functional differences in the Drosophila and mosquito EcRs, there are eight amino acid differences between these two proteins, seven of which are clustered within the loop region of the I box (Fig. 6). Interestingly, the predicted loop region in the I box of BmEcR, which has been shown to differ significantly from DmEcR with respect to its hormone-independent heterodimerization and agonist recognition (59), has seven amino acids different from those of DmEcR. Indeed, alignment of other cloned EcRs shows that the loop region is the most diverged portion of the putative I boxes in these receptors (Fig. 7). It remains to be tested whether the synergistic action of several amino acids or the critical Phe/Tyr residue determines the ligand specificity in EcRs of different arthropods.
FIG. 7.
Putative I boxes in EcR proteins. I boxes of EcR protein sequences from 15 arthropod species are aligned by GCG Pileup. Helices 9 and 10 in human RARγ (HsRARγ) (48) are indicated by dotted lines. Residues affecting hormone-free DNA binding activity and ecdysone responsiveness in AaEcR and DmEcR are in bold. The most critical residue, Phe/Tyr, is in bold italics. DmEcR, CfEcR, BaEcR, and BmEcR are underlined as their proteins contain a Tyr at the critical ligand specificity site. Data bank search yielded EcR protein sequences from 16 species: 6 Diptera species, the Mediterranean fruit fly Ceratitis capitata (CcEcR [67]), the sheep blowfly Lucilia cuprina (LcEcR [23]), the yellow fever mosquitoes A. aegypti (AaEcR, 12) and A. albopictus (not shown, as its EcR I box is 100% identical to AaEcR [28]), the midge Chironomus tentans (CtEcR [27]), and D. melanogaster (DmEcR [34]); 6 Lepidoptera species, the spruce budworm C. fumiferana (CfEcR [35]), squinting bush brown Bicyclus anynana (BaEcR [R. K. Reinhardt, P. Weber, and P. B. Koch, submitted to GenBank, accession no. CAB63236]), the silkworm B. mori (BmEcR [32, 60]), the tobacco budworm Heliothis virescens (HvEcR [41]), the tobacco hornworm M. sexta (MsEcR [18]), and the buckeye Junonia coenia (JcEcR [R. K. Reinhardt, P. Weber, and P. B. Koch submitted to GenBank, accession no. CAB63485]); 1 Orthoptera species, the migratory locust Locusta migratoria (LmEcR [55]); 1 Coleoptera species, the yellow mealworm Tenebrio moliter (TmEcR [42]); 1 crustacean species, the Atlantic sand fiddler crab Celuca pugilator (CpEcR [13]); and 1 Ixodidae species, the tick Amblyomma americanum (AamEcR [20]).
EMSA analysis of the effects of single amino acid substitutions revealed that substitution of a single residue, corresponding to Phe529 of AaEcR, for Tyr611 of DmEcR renders the Drosophila receptor responsive to ecdysone. Reciprocally, converting the Phe529 to Tyr dramatically reduces the mosquito receptor responsiveness to ecdysone while concurrently abolished its ligand-free DNA binding activity. The ability of the less polar residue, Phe, to confer enhanced sensitivity to the less polar ligand, ecdysone, is consistent with the possibility of direct interaction between Phe529/Tyr611 and ecdysteroids; crystal structure data for related receptors suggest, however, that this residue is unlikely to lie within the ligand binding pocket. In light of the correlation between ecdysone sensitivity and hormone-independent DNA binding, it therefore seems more likely that the primary effect of Phe529 is in establishing a more stable hydrophobic dimerization interface. Stabilization of the heterodimer would in turn facilitate ligand binding and kinetically favor the establishment of the ternary complex containing receptor, ligand, and DNA (75). While 20E is capable of driving the formation of complexes involving either the Drosophila or mosquito EcR-USP-ecdysone appears to require a prestabilized dimer to ensure productive ligand-receptor interaction.
Hagedorn et al. (21) have shown that in the adult mosquito, neuroendocrine signals triggered by a blood meal cause the ovaries to secrete ecdysone, which is presumably converted into 20E by peripheral tissues. The secretion of ovarian ecdysone reaches in maximal levels at 16 h post-blood meal, with hemolymph ecdysteroid levels closely following with a peak at 18-h post-blood meal (21). Further analysis of hemolymph ecdysteroids showed that they consist of ecdysone and 20E at a 1:1.5 ratio, with the ratio of ecdysone to 20E in the hemolymph remaining approximately constant throughout the vitellogenic cycle (3). Taken together with our findings of a high level of AaEcR sensitivity to ecdysone, these observations suggest the possibility that ecdysone is an active hormone in the mosquito. Further studies will be required to determine whether ecdysone is involved in distinct physiological responses, as appears to be the case in Manduca eye development (7, 8), and to characterize those responses.
In conclusion, our study revealed at the molecular level, for the first time, that ecdysone could act as a potent ligand for an EcR. Furthermore, we have established differential responses of insect EcRs to natural ecdysteroids, previously demonstrated only for synthetic nonsteroid agonists (59). We have identified the molecular determinants defining hormone-independent DNA binding activity/heterodimerization as well as differential ecdysteroid responses. By furnishing new insights into the structural and functional properties of insect EcRs, these studies are expected to pave the way for the development of new EcR ligand-based pesticides for use in the control of important disease vectors and other pest species.
ACKNOWLEDGMENTS
We thank H. H. Rees for help in purification of ecdysone and M. McKeown for providing reporter constructs; we thank Michael J. Mienaltowski and Richard J. Miksicek for critical reading of the manuscript.
This work was supported by grant AI 36959 from the National Institutes of Health.
REFERENCES
- 1.Antoniewski C, Laval M, Hahan A, Lepesant J-A. The ecdysone response enhancer of the Fbp 1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol Cell Biol. 1994;14:4465–4474. doi: 10.1128/mcb.14.7.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniewski C, O'Grady M S, Edmondson R G, Lassieur S M, Benes H. Characterization of an EcR/USP heterodimer target site that mediates ecdysone responsiveness of the Drosophila Lsp-2 gene. Mol Gen Genet. 1995;249:545–556. doi: 10.1007/BF00290580. [DOI] [PubMed] [Google Scholar]
- 3.Borovsky D, Whisenton L R, Thomas B R, Fuchs M S. Biosynthesis and distribution of ecdysone and 20-OH-ecdysone in Aedes-aegypti. Arch Insect Biochem Physiol. 1986;3:19–30. [Google Scholar]
- 4.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 5.Bownes M. Expression of the genes coding for vitellogenin (yolk protein) Annu Rev Entomol. 1986;31:507–531. [Google Scholar]
- 6.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 7.Champlin D T, Truman J W. Ecdysteroids govern two phases of eye development during metamorphosis of the moth, Manduca sexta. Development. 1998;125:2009–2018. doi: 10.1242/dev.125.11.2009. [DOI] [PubMed] [Google Scholar]
- 8.Champlin D T, Truman J W. Ecdysteroid control of cell proliferation during optic lobe neurogenesis in the moth Manduca sexta. Development. 1998;125:269–277. doi: 10.1242/dev.125.2.269. [DOI] [PubMed] [Google Scholar]
- 9.Cherbas L, Lee K, Cherbas P. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- 10.Cherbas P, Cherbas L. Molecular aspects of ecdysteroid hormone action. In: Gilbert L I, Tata J R, Atkinson B G, editors. Metamorphosis, postembryonic reprogramming of gene expression in amphibian and insect cells. San Diego, Calif: Academic Press; 1996. pp. 175–222. [Google Scholar]
- 11.Cherbas P, Cherbas L, Lee S S, Nakanishi K. 26-[125I]iodoponasterone A is a potent ecdysone and a sensitive radioligand for ecdysone receptors. Proc Natl Acad Sci USA. 1988;85:2096–2100. doi: 10.1073/pnas.85.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho W-L, Kapitskaya M Z, Raikhel A S. Mosquito ecdysteroid receptor: analysis of the cDNA and expression during vitellogenesis. Insect Biochem Mol Biol. 1995;25:19–27. doi: 10.1016/0965-1748(94)00045-j. [DOI] [PubMed] [Google Scholar]
- 13.Chung A C, Durica D S, Clifton S W, Roe B A, Hopkins P M. Cloning of crustacean ecdysteroid receptor and retinoid-X receptor gene homologs and elevation of retinoid-X receptor mRNA by retinoic acid. Mol Cell Endocrinol. 1998;139:209–227. doi: 10.1016/s0303-7207(98)00056-2. [DOI] [PubMed] [Google Scholar]
- 14.D'Avino P P, Crispi S, Cherbas L, Cherbas P, Furia M. The moulting hormone ecdysone is able to recognize target elements composed of direct repeats. Mol Cell Endocrinol. 1995;113:1–9. doi: 10.1016/0303-7207(95)03584-t. [DOI] [PubMed] [Google Scholar]
- 15.Dhadialla T S, Raikhel A S. Endocrinology of mosquito vitellogenesis. In: Davey K G, Peter R E, Tobe S S, editors. Perspectives in comparative endocrinology. Toronto, Canada: National Research Council; 1994. pp. 275–281. [Google Scholar]
- 16.Dhadialla T S, Carlson G R, Le D P. New insecticides with ecdysteroidal and juvenile hormone activity. Annu Rev Entomol. 1998;3:545–569. doi: 10.1146/annurev.ento.43.1.545. [DOI] [PubMed] [Google Scholar]
- 17.Dhadialla T S, Tzertzinis G. Characterization and partial cloning of ecdysteroid receptor from a cotton boll weevil embryonic cell line. Arch Insect Biochem Physiol. 1997;35:45–57. doi: 10.1002/(SICI)1520-6327(1997)35:1/2<45::AID-ARCH5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara H, Jindra M, Newitt R, Palli S R, Hiruma K, Riddiford L M. Cloning of an ecdysone receptor homolog from Manduca sexta and the developmental profile of its mRNA in wings. Insect Biochem Mol Biol. 1995;25:845–856. doi: 10.1016/0965-1748(95)00023-o. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert L I, Rybczynski R, Tobe S S. Endocrine cascade in insect metamorphosis. In: Gilbert L I, Tata J R, Atkinson B G, editors. Metamorphosis, postembryonic reprogramming of gene expression in amphibian and insect cells. San Diego, Calif: Academic Press; 1996. pp. 60–108. [Google Scholar]
- 20.Guo X, Harmon M A, Laudet V, Mangelsdorf D J, Palmer M J. Isolation of a functional ecdysteroid receptor homologue from the ixodid tick Amblyomma americanum (L.) Insect Biochem Mol Biol. 1997;27:945–962. doi: 10.1016/s0965-1748(97)00075-1. [DOI] [PubMed] [Google Scholar]
- 21.Hagedorn H H, O'Connor J D, Fuchs M S, Sage B, Schlaeger D A, Bohm M K. The ovary as a source of alpha-ecdysone in an adult mosquito. Proc Natl Acad Sci USA. 1975;72:3255–3259. doi: 10.1073/pnas.72.8.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagedorn H H. Physiological roles of hemolymph ecdysteroids in adult insect. In: Koolman J, editor. Ecdysone, from chemistry to mode of action. New York, N.Y: Thieme Medical Publishers, Inc.; 1989. pp. 279–289. [Google Scholar]
- 23.Hannan G N, Hill R J. Cloning and characterization of LcEcR: a functional ecdysone receptor from the sheep blowfly Lucilia cuprina. Insect Biochem Mol Biol. 1997;27:479–488. doi: 10.1016/s0965-1748(97)00019-2. [DOI] [PubMed] [Google Scholar]
- 24.Henrich V C, Brown N E. Insect nuclear receptors: a developmental and comparative perspective. Insect Biochem Mol Biol. 1995;25:881–897. doi: 10.1016/0965-1748(95)00030-y. [DOI] [PubMed] [Google Scholar]
- 25.Henrich V C, Sliter T J, Lubahn D B, MacIntyre A, Gilbert L I. A steroid/thyroid hormone receptor superfamily member in Drosophila melanogaster that shares extensive sequence similarity with a mammalian homologue. Nucleic Acids Res. 1990;18:4143–4148. doi: 10.1093/nar/18.14.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henrich V C, Rybczynski R, Gilbert L I. Peptide hormones, steroid hormones, and puffs: mechanisms and models in insect development. Vitam Horm. 1999;55:73–125. doi: 10.1016/s0083-6729(08)60934-6. [DOI] [PubMed] [Google Scholar]
- 27.Imhof M O, Rusconi S, Lezzi M. Cloning of a Chironomus tentans cDNA encoding a protein (cEcR) homologous to the Drosophila melanogaster ecdysteroid receptor (dEcR) Insect Biochem Mol Biol. 1993;23:115–124. doi: 10.1016/0965-1748(93)90089-b. [DOI] [PubMed] [Google Scholar]
- 28.Jayachandran G, Fallon A M. Evidence for expression of EcR and USP components of the 20-hydroxyecdysone receptor by a mosquito cell line. Arch Insect Biochem Physiol. 2000;43:87–96. doi: 10.1002/(SICI)1520-6327(200002)43:2<87::AID-ARCH5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Jindra M, Malone F, Hiruma K, Riddiford L M. Developmental profiles and ecdysteroid regulation of the mRNAs for two ecdysone receptor isoforms in the epidermis and wings of the tobacco hornworm, Manduca sexta. Dev Biol. 1996;180:258–272. doi: 10.1006/dbio.1996.0299. [DOI] [PubMed] [Google Scholar]
- 30.Jindra M, Huang J Y, Malone F, Asahina M, Riddiford L M. Identification and mRNA developmental profiles of two ultraspiracle isoforms in the epidermis and wings of Manduca sexta. Insect Mol Biol. 1997;6:41–53. doi: 10.1046/j.1365-2583.1997.00153.x. [DOI] [PubMed] [Google Scholar]
- 31.Kapitskaya M, Wang S-F, Cress D E, Dhadialla T S, Raikhel A S. The mosquito ultraspiracle homologue, a partner of ecdysteroid receptor heterodimer: cloning and characterization of isoforms expressed during vitellogenesis. Mol Cell Endocrinol. 1996;121:119–132. doi: 10.1016/0303-7207(96)03847-6. [DOI] [PubMed] [Google Scholar]
- 32.Kamimura M, Tomita S, Fujiwara H. Molecular cloning of an ecdysone receptor (B1 isoform) homologue from the silkworm, Bombyx mori, and its mRNA expression during wing disc development. Comp Biochem Physiol B. 1996;113:341–347. doi: 10.1016/0305-0491(95)02032-2. [DOI] [PubMed] [Google Scholar]
- 33.Kiriishi S, Rountree D B, Sakurai S, Gilbert L I. Prothoracic gland synthesis of 3-dehydroecdysone and its hemolymph 3 beta-reductase mediated conversion to ecdysone in representative insects. Experientia. 1990;46:716–721. doi: 10.1007/BF01939944. [DOI] [PubMed] [Google Scholar]
- 34.Koelle M R, Talbot W S, Segraves W A, Bender M T, Cherbas P, Hogness D S. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 35.Kothapalli R, Palli S R, Ladd T R, Sohi S S, Cress D, Dhadialla T S, Tzertzinis G, Retnakaran A. Cloning and developmental expression of the ecdysone receptor gene from the spruce budworm, Choristoneura fumiferana. Dev Genet. 1995;17:319–330. doi: 10.1002/dvg.1020170405. [DOI] [PubMed] [Google Scholar]
- 36.Lan Q, Hiruma K, Hu X, Jindra M, Riddiford L M. Activation of a delayed-early gene encoding MHR3 by the ecdysone receptor heterodimer EcR-B1-USP-1 but not by EcR-B1-USP-2. Mol Cell Biol. 1999;19:4897–4906. doi: 10.1128/mcb.19.7.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Amin J, Voellmy R. Ecdysterone receptor is a sequence-specific transcription factor involved in the developmental regulation of heat shock genes. Mol Cell Biol. 1991;11:3660–3675. doi: 10.1128/mcb.11.7.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 39.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maroy P, Dennis R, Beckers C, Sage B A, O'Connor J D. Demonstration of an ecdysteroid receptor in a cultured cell line of Drosophila melanogaster. Proc Natl Acad Sci USA. 1978;75:6035–6038. doi: 10.1073/pnas.75.12.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez A, Scanlon D, Gross B, Perara S C, Palli S R, Greenland A J, Windass J, Pongs O, Broad P, Jepson I. Transcriptional activation of the cloned Heliothis virescens (Lepidoptera) ecdysone receptor (HvEcR) by muristeroneA. Insect Biochem Mol Biol. 1999;29:915–930. doi: 10.1016/s0965-1748(99)00067-3. [DOI] [PubMed] [Google Scholar]
- 42.Mouillet J F, Delbecque J P, Quennedey B, Delachambre J. Cloning of two putative ecdysteroid receptor isoforms from Tenebrio molitor and their developmental expression in the epidermis during metamorphosis. Eur J Biochem. 1997;248:856–863. doi: 10.1111/j.1432-1033.1997.00856.x. [DOI] [PubMed] [Google Scholar]
- 43.Oro A E, McKeown M, Evans R M. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature. 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- 44.Pak M D, Gilbert L I. A developmental analysis of ecdysteroids during the metamorphosis of Drosophila melanogaster. J Liq Chromatogr. 1987;10:2591–2611. [Google Scholar]
- 45.Perlmann T, Umesono K, Rangarajan P N, Forman B M, Evans R M. Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol Endocrinol. 1996;10:958–966. doi: 10.1210/mend.10.8.8843412. [DOI] [PubMed] [Google Scholar]
- 46.Raikhel A S, Miura K, Segraves W A. Nuclear receptors in mosquito vitellogenesis. Am Zool. 1999;39:722–735. [Google Scholar]
- 47.Redfern C P F. Evidence for the presence of makisterone-a in Drosophila larvae and the secretion of 20-deoxymakisterone-a by the ring gland. P Natl Acad Sci USA. 1984;81:5643–5647. doi: 10.1073/pnas.81.18.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 49.Riddihough G, Pelham H R B. An ecdysone response element in the Drosophila hsp27 promoter. EMBO J. 1987;6:3729–3734. doi: 10.1002/j.1460-2075.1987.tb02707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riddiford L M. Hormone and Drosophila development. In: Bate M, Martinez A, editors. The development of Drosophila melanogaster. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 899–939. [Google Scholar]
- 51.Riddiford L M, Hiruma K, Lan Q, Zhou B H. Regulation and role of nuclear receptors during larval molting and metamorphosis of lepidoptera. Am Zool. 1999;39:736–746. [Google Scholar]
- 52.Robinow S, Talbot W S, Hogness D S, Truman J W. Programmed cell death in the Drosophila CNS is ecdysone-regulated and coupled with a specific ecdysone receptor isoform. Development. 1993;119:1251–1259. doi: 10.1242/dev.119.4.1251. [DOI] [PubMed] [Google Scholar]
- 53.Sage B A, Tanis M A, O'Connor J D. Characterization of ecdysteroid receptors in cytosol and naive nuclear preparations of Drosophila-Kc cells. J Biol Chem. 1982;257:6373–6379. [PubMed] [Google Scholar]
- 54.Sakurai S, Williams C M. Short-loop negative and positive feedback on ecdysone secretion by prothoracic gland in the tobacco hornworm, Manduca sexta. Gen Comp Endocrinol. 1989;75:204–216. doi: 10.1016/0016-6480(89)90072-5. [DOI] [PubMed] [Google Scholar]
- 55.Saleh D S, Zhang J, Wyatt G R, Walker V K. Cloning and characterization of an ecdysone receptor cDNA from Locusta migratoria. Mol Cell Endocrinol. 1998;143:91–99. doi: 10.1016/s0303-7207(98)00131-2. [DOI] [PubMed] [Google Scholar]
- 56.Segraves W A. Steroid receptors and orphan receptors in Drosophila development. Semin Cell Biol. 1994;5:105–113. doi: 10.1006/scel.1994.1014. [DOI] [PubMed] [Google Scholar]
- 57.Shea M J, King D L, Conboy M J, Mariani B D, Kafatos F C. Proteins that bind to Drosophila chorion cis-regulatory elements: a new C2H2 zinc finger protein and a C2C2 steroid receptor-like component. Genes Dev. 1990;4:1128–1140. doi: 10.1101/gad.4.7.1128. [DOI] [PubMed] [Google Scholar]
- 58.Smith S L, Bollenbacher W E, Gilbert L I. Ecdysone 20-monooxygenase activity during larval-pupal development of Manduca sexta. Mol Cell Endocrinol. 1983;31:227–251. doi: 10.1016/0303-7207(83)90151-x. [DOI] [PubMed] [Google Scholar]
- 59.Suhr S T, Gil E B, Senut M C, Gage F H. High level transactivation by a modified Bombyx ecdysone receptor in mammalian cells without exogenous retinoid X receptor. Proc Natl Acad Sci USA. 1998;95:7999–8004. doi: 10.1073/pnas.95.14.7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swevers L, Drevet J R, Junke M D, Iatrou K. The silkmoth homolog of the Drosophila ecdysone receptor (B1 isoform): cloning and analysis of expression during follicular cell differentiation. Insect Biochem Mol Biol. 1995;25:857–866. doi: 10.1016/0965-1748(95)00024-p. [DOI] [PubMed] [Google Scholar]
- 61.Talbot W S, Swyryd E A, Hogness D S. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 1993;73:1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- 62.Tanenbaum D M, Wang Y, Williams S P, Sigler P B. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas H E, Stunnenberg H G, Stewart A F. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature. 1993;362:471–475. doi: 10.1038/362471a0. [DOI] [PubMed] [Google Scholar]
- 64.Thummel C S. Files on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- 65.Thummel C S. Dueling orphans—interacting nuclear receptors coordinate Drosophila metamorphosis. Bioessays. 1997;19:669–672. doi: 10.1002/bies.950190806. [DOI] [PubMed] [Google Scholar]
- 66.Tsai M J, O'Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 67.Verras M, Mavroidis M, Kokolakis G, Gourzi P, Zacharopoulou A, Mintzas A C. Cloning and characterization of CcEcR. An ecdysone receptor homolog from the mediterranean fruit fly ceratitis capitata. Eur J Biochem. 1999;265:798–808. doi: 10.1046/j.1432-1327.1999.00788.x. [DOI] [PubMed] [Google Scholar]
- 68.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 69.Wang S-F, Li C, Zhu J, Miura K, Miksicek R J, Raikhel A S. Differential expression and regulation by 20-hydroxyecdysone of mosquito Ultraspiracle isoforms. Dev Biol. 2000;218:99–113. doi: 10.1006/dbio.1999.9575. [DOI] [PubMed] [Google Scholar]
- 70.Wang S-F, Miura K, Miksicek R J, Segraves W A, Raikhel A S. DNA binding and transactivation characteristics of the mosquito ecdysone receptor-ultraspiracle complex. J Biol Chem. 1998;273:27531–27540. doi: 10.1074/jbc.273.42.27531. [DOI] [PubMed] [Google Scholar]
- 71.Warren J T, Sakurai S, Rountree D B, Gilbert L I. Synthesis and secretion of ecdysteroids by the prothoracic glands of Manduca-sexta. J Insect Physiol. 1988;34:571–576. [Google Scholar]
- 72.Warren J T, Sakurai S, Rountree D B, Gilbert L I, Lee S S, Nakanishi K. Regulation of the ecdysteroid titer of Manduca sexta: reappraisal of the role of the prothoracic glands. Proc Natl Acad Sci USA. 1988;85:958–962. doi: 10.1073/pnas.85.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams S P, Sigler P B. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 74.Yao T-P, Segraves W A, Oro A E, McKeown M, Evans R M. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 75.Yao T-P, Forman B M, Jlang Z, Cherbas L, Chen J-D, McKeown M, Cherbas P, Evans R M. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;336:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 76.Yund M A, King D S, Fristrom J W. Ecdysteroid receptors in imaginal discs of Drosophila melanogaster. Proc Natl Acad Sci USA. 1978;75:6039–6043. doi: 10.1073/pnas.75.12.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]