Abstract
Introduction
In rheumatoid arthritis, time spent using ineffective medications may lead to irreversible disease progression. Despite availability of targeted treatments, only a minority of patients achieve sustained remission, and little evidence exists to direct the choice of biologic disease-modifying antirheumatic drugs in individual patients. Machine learning was used to identify a rule to predict the response to sarilumab and discriminate between responses to sarilumab versus adalimumab, with a focus on clinically feasible blood biomarkers.
Methods
The decision tree model GUIDE was trained using a data subset from the sarilumab trial with the most biomarker data, MOBILITY, to identify a rule to predict disease activity after sarilumab 200 mg. The training set comprised 18 categorical and 24 continuous baseline variables; some data were omitted from training and used for validation by the algorithm (cross-validation). The rule was tested using full datasets from four trials (MOBILITY, MONARCH, TARGET, and ASCERTAIN), focusing on the recommended sarilumab dose of 200 mg.
Results
In the training set, the presence of anti-cyclic citrullinated peptide antibodies, combined with C-reactive protein > 12.3 mg/l, was identified as the “rule” that predicts American College of Rheumatology 20% response (ACR20) to sarilumab. In testing, the rule reliably predicted response to sarilumab in MOBILITY, MONARCH, and ASCERTAIN for many efficacy parameters (e.g., ACR70 and the 28-joint disease activity score using CRP [DAS28-CRP] remission). The rule applied less to TARGET, which recruited individuals refractory to tumor necrosis factor inhibitors. The potential clinical benefit of the rule was highlighted in a clinical scenario based on MONARCH data, which found that increased ACR70 rates could be achieved by treating either rule-positive patients with sarilumab or rule-negative patients with adalimumab.
Conclusions
Well-established and clinically feasible blood biomarkers can guide individual treatment choice. Real-world validation of the rule identified in this post hoc analysis is merited.
Clinical Trial Registration
Supplementary Information
The online version contains supplementary material available at 10.1007/s40744-021-00361-5.
Keywords: Adalimumab, Clinical trial, Machine learning, Precision medicine, Rheumatoid arthritis, Sarilumab
Key Summary Points
| Why carry out this study? |
| In rheumatoid arthritis, despite availability of targeted treatments, only a minority of patients achieve sustained remission. |
| Little evidence exists to direct choice of biologic disease-modifying antirheumatic drugs in individual patients. |
| Our goal was to identify a “rule” based on clinically feasible biomarkers to predict response to sarilumab and discriminate between responses to sarilumab versus adalimumab, using clinical trial data and machine learning. |
| What was learned from the study? |
| The presence of anti-cyclic citrullinated peptide antibodies, combined with C-reactive protein > 12.3 mg/l, emerged as a biomarker “rule” that could potentially predict response to sarilumab. |
| This finding needs to be confirmed in real-world studies. |
Digital Features
This article is published with digital features, including a video abstract to facilitate understanding of the article. To view digital features for this article go to 10.6084/m9.figshare.14512056.
Introduction
Treatment guidelines for rheumatoid arthritis (RA) recognize the importance of attaining clinical improvement within 3 months and remission or low disease activity within 6 months of treatment initiation [1, 2]. Biologic disease-modifying anti-rheumatic drugs (bDMARDs) are recommended in the presence of poor prognostic factors or if response to initial treatment is inadequate. The choice of bDMARD is often based on physician experience, patient preference, and cost, and is complicated by a variety of available agents [3, 4]. Nevertheless, there is a remarkably similar plateau in responder rates for patients achieving 20% (ACR20), 50% (ACR50), and 70% (ACR70) response based on American College of Rheumatology criteria, irrespective of the bDMARD or targeted synthetic DMARD (tsDMARD) studied [5–7].
Given the importance of rapid response to treatment in prevention of irreversible joint damage and improved symptom control, a personalized approach to treatment selection would be preferred over a prolonged and iterative trial-and-error process [8]. However, only a few biomarkers have been identified as candidates for treatment optimization in RA, with at best modest associations with treatment response, and with inconsistent applicability in current clinical practice. For example, the presence of autoantibodies to rheumatoid factor or cyclic citrullinated peptide (CCP) may predict response to rituximab, and genetic factors such as the HLA-DRB1 shared epitope may predict response to tumor necrosis factor inhibitors (TNFi) and tocilizumab, an inhibitor of the interleukin-6 receptor (IL-6R) [3, 9]. It is possible that treatment response would be better predicted by combinations of biomarkers and clinical characteristics [3, 8, 10–13]. However, the multitude of potential clinical and biomarker-based predictors, in combination with their thresholds, and inherent constraints on clinical availability, poses a significant conceptual and computational challenge.
Artificial intelligence techniques such as machine learning are increasingly being used to identify individuals at risk for disease, predict outcome, and optimize treatments [4, 14, 15]. In machine learning, computers apply hypothesis-free algorithms that enable development of data-based mathematical models [4]. To develop machine learning models, a randomly selected subset of data such as that obtained from patients in clinical trials is used to select, among a predefined set of parameters (e.g., clinical or blood biomarkers), those factors that are associated with a certain predefined outcome (e.g., ACR20). Once the parameters are set so that the error in predicting the outcome is minimized, those parameter values (i.e., the “rule”) are validated using the remaining data, or new external data sources [4]. This approach allows the identification of hidden patterns and rules in large datasets, while reducing the risk of overfitting, and having to correctly specify hypotheses a priori [4]. Machine learning has been applied to electronic health records to prognosticate RA disease activity [14, 16] and to define disease phenotypes in RA [17]. However, to the best of our knowledge, it has not yet yielded a robust, clinically feasible rule that would predict treatment response to biologic therapies in patients with RA.
Sarilumab is a human monoclonal antibody to IL-6R approved for the treatment of moderate-to-severe RA [18, 19]. As with other bDMARDs, the characteristics of patients most likely to benefit from sarilumab treatment remain poorly understood. In this post hoc analysis, we used machine learning to identify a simple and clinically feasible rule that could predict favorable response to sarilumab, and in one trial, an incremental response compared with adalimumab.
Methods
Data Sources
This post hoc analysis used patient-level data from four phase 3 sarilumab trials: MOBILITY (sarilumab versus placebo in patients with inadequate response to methotrexate; NCT01061736) [20], MONARCH (sarilumab versus adalimumab as monotherapy in patients with inadequate response or intolerant to methotrexate; NCT02332590) [21], TARGET (sarilumab versus placebo in patients intolerant to TNFi; NCT01709578) [22], and ASCERTAIN (comparative safety of sarilumab and tocilizumab; NCT01768572) [23]. Patients in all four trials were ≥ 18 years old and met the ACR 1987 revised classification criteria (MOBILITY) or the ACR 2010/European League Against Rheumatism (EULAR) classification criteria (MONARCH, TARGET, ASCERTAIN) for active RA at baseline [20–23]. Patients had CRP levels ≥ 6 mg/l in MOBILITY, ≥ 8 mg/l in MONARCH and TARGET, and ≥ 4 mg/l in ASCERTAIN [20–23]. At baseline in each trial, at least 65% of patients were seropositive for rheumatoid factor and at least 75% were positive for anti-CCP autoantibodies [20–23]. MOBILITY data were used to train and validate the model (data were randomly split into training and validation sets by the algorithm; henceforth referred to as cross-validation), which was later tested in the full data sets of all four trials.
Variables and Outcome Used for Model Development
Eighteen categorical and 24 continuous baseline variables, including demographics, blood protein biomarkers, and clinical scores from the MOBILITY trial were identified as potential predictors of clinical response (Table 1).
Table 1.
Parameters used in the GUIDE algorithm
| Demographics | Clinical characteristics | Biomarkers |
|---|---|---|
| Continuous | ||
| Age | Duration of RA | C x C motif chemokine ligand 13 |
| SJC (28 joints) | CRP | |
| SJC (68 joints) | IL-6 | |
| TJC (28 joints) | MMP3 | |
| TJC (68 joints) | MMP-degraded type 1 collagen | |
| ACR20 at week 24 | MMP-degraded type 3 collagen | |
| DAS28 at baseline | OC | |
| Joint erosion score | OPG | |
| Joint space narrowing score | RANKL | |
| RANKLF | ||
| RANKL: OPG ratio | ||
| RANKLF: OPG ratio | ||
| Soluble glycoprotein 130 | ||
| Soluble intercellular adhesion molecule | ||
| Soluble IL-6 receptor | ||
| Categorical | ||
| Sex | Prior csDMARDs | Anti-CCP (presence or absence) |
| Alcohol use | Number of csDMARDs | Persistent ADAbs |
| BMI stratum | Number of prior bDMARDs | Rheumatoid factor |
| Race | Type of prior bDMARDs | Transient ADAbs |
| Religion | Duration of RA (3-year strata) | Neutralizing anti-drug antibodies |
| Smoking history | Treatment arm code | |
All variables are baseline except ACR20 at week 24
Anti-CCP anti-cyclic citrullinated peptide, ACR20, American College of Rheumatology 20% response, ADAb anti-drug antibody, BMI body mass index, CRP C-reactive protein, bDMARD biological disease-modifying antirheumatic drug; csDMARD conventional synthetic disease-modifying antirheumatic drug, DAS28 28-joint disease activity score, IL-6 interleukin-6, MMP matrix metalloproteinase, OC osteocalcin, OPG osteoprotegerin, RA rheumatoid arthritis, RANKL receptor activator of NF-kappa B ligand, RANKLF free RANKL, SJC swollen joint count, TJC tender joint count
The clinical endpoint of ACR20 at week 24 was used for the model training, once the machine learning methodology had been chosen (see below).
Choice of the Machine Learning Methodology
Since our goal was identification of a simple, clinically feasible rule with a good prediction performance, we initially tested three decision tree methods, rpart (CRAN library: (https://cran.r-project.org/web/packages/rpart/index.html), C5.0 (https://www.rulequest.com), and the Generalized, Unbiased, Interaction Detection and Estimation (GUIDE; version 27.9 for macOS Mojave 10.14; http://pages.stat.wisc.edu/~loh/guide.html) [24], using either ACR20, ACR50, ACR70, or the 28-joint disease activity score using CRP (DAS28-CRP) as clinical outcomes. Of those, rpart and C5.0 did not provide generalizable approaches for patient stratification. GUIDE provided unbiased variable selection and cross-validation, as well as a final tree that yielded a rule that fits our predefined criteria. The default options of GUIDE were used, including tenfold cross-validation.
Training of the Decision Tree Model
To maximize the information density for model training, a data subset from patients in the MOBILITY trial who had data for all selected biomarkers (n = 163; Table 1) was entered into GUIDE to train the decision tree model where ACR20 ultimately yielded the only cross-validated model. Patient data from the sarilumab 200 mg (n = 63) and 150 mg (n = 100) treatment groups in MOBILITY were pooled. The resulting model was manually reduced by two decision-nodes to achieve greater clinical applicability while maintaining most of its predictive power, and is henceforth referred to as the final model (see “Results”).
Validation of the Model
The final model for predicting sarilumab response was validated using the full dataset for MOBILITY, which included the training subset (Nnew = 1034 plus Ntraining = 163; sarilumab 150 mg or 200 mg, n = 799; placebo, n = 398) and datasets from MONARCH (N = 369; sarilumab 200 mg, n = 184; adalimumab 40 mg, n = 185), TARGET (N = 546; sarilumab 150 mg or 200 mg, n = 365; placebo, n = 181), and ASCERTAIN (N = 202; sarilumab 150 mg or 200 mg, n = 100; tocilizumab 4 mg/kg or 8 mg/kg, n = 102). For each study, the applicable endpoints (Supplementary Material, Table S1) were assessed at baseline and week 24 and placebo-corrected where possible. In addition, we developed a clinical application scenario based on the rule created by the GUIDE algorithm and the MONARCH data [21].
Statistical Analysis
Data analyses were performed using R v3.5.2 and Microsoft Excel 2010.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Results
Choice of the Machine Learning Methodology
Retrospectively, we initiated a sensitivity analysis of our methodology by evaluating 14 additional machine learning methods available through PyCaret (https://pycaret.org; Supplementary Material, Table S2), including rule-based (decision trees), regression-based (logistic regression, quadratic discriminant analysis, linear discriminant analysis, ridge classifier), instance-based (k-neighbors classifier), naïve Bayesian, ensemble-based (extra trees classifier, random forest, gradient boosting, light gradient boosting, CatBoost, and Ada Boost), and support vector machines. The PyCaret results confirmed that the decision tree classifier model [25] GUIDE had the best recall, while other performance criteria were comparable. The resulting variable ranking of the PyCaret results (only the four best results are shown) frequently contained the GUIDE-derived variables, which were CRP level and anti-CCP status, but also suggested alternative variables and variable combinations (Supplementary Material, Figure S1).
Development of the Model and Identification of a Predictive Rule
The training and cross-validation dataset included 163 patients who received sarilumab in the MOBILITY trial and had data available for all relevant variables (Table 1).
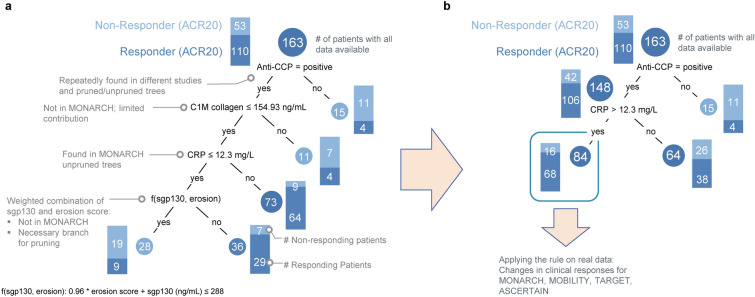
A robust predictive GUIDE model for patient stratification was obtained using ACR20 as the measure of patient response. With ACR50 and ACR70, GUIDE could not generate cross-validated trees, but often included anti-CCP and CRP either as the first decision variable or within the first two branches of the decision tree (data not shown). In the resulting decision tree (Fig. 1A), the model contained, in the following order: anti-CCP, metabolite of type I collagen (C1M), CRP, and a weighted combination of soluble glycoprotein 130 (sgp130) and the erosion score. However, C1M and the weighted combination of sgp130 and erosion score were manually excluded from the resulting model. C1M had only a minor impact on the model performance and the weighted combination of sgp130 and erosion score was only available in MOBILITY. In addition, the exclusion of these variables allowed for greater clinical applicability of the final model.
Fig. 1.
Schematic of the resulting GUIDE decision tree classification approach model (A) and the reduced final model (B). Anti-CCP anti-cyclic citrullinated peptide, C1M metabolite of type I collagen, CRP C-reactive protein, sgp130 soluble glycoprotein 130
Among the 42 candidate variables, the combined presence of anti-CCP (yes/no) and CRP > 12.3 mg/l (chosen by GUIDE) was identified as a predictor (i.e., the rule) of better treatment outcomes with sarilumab. Of the 163 MOBILITY patients included in the training and cross-validation dataset, 84 had a positive status for both anti-CCP and CRP > 12.3 mg/l. The ACR20 response rate in this rule-positive group was 81% (68/84), which was higher than the rate observed in patients without anti-CCP antibodies (27% [4/15]) or in those who had anti-CCP antibodies but their CRP was ≤ 12.3 mg/l (59% [38/64]; Fig. 1B).
Results from the model were similar for both sarilumab doses (150 mg and 200 mg; not shown), so the analysis presented here is focused on the recommended sarilumab dose of 200 mg.
Supplementary file2 (WMV 2,12,490 kb)
Baseline Characteristics of Rule-Positive Patients
Patients who were anti-CCP-positive and had CRP > 12.3 mg/l, i.e., rule-positive patients, accounted for 34–51% of patients in the sarilumab groups across the four trials. On average, rule-positive patients had a more severe disease and more baseline factors suggesting poor prognosis than rule-negative patients (MOBILITY: Table 2; MONARCH, TARGET, ASCERTAIN: Supplementary Material, Tables S3, S4, and S5).
Table 2.
Baseline characteristics of rule-positive and -negative patients in MOBILITY
| Parameter | Sarilumab 200 mg | Placebo | ||
|---|---|---|---|---|
| Rule-positivea patients (n = 213) | Rule-negative patients (n = 210) | Rule-positivea patients (n = 197) | Rule-negative patients (n = 230) | |
| DAS28-CRP | 6.3 ± 0.8 | 5.7 ± 0.8 | 6.2 ± 0.8 | 5.6 ± 0.8 |
| CDAI | 41.9 ± 12.7 | 39.6 ± 11.4 | 40.5 ± 12.4 | 39.1 ± 12.1 |
| HAQ-DI | 1.8 ± 0.7 | 1.6 ± 0.6 | 1.7 ± 0.6 | 1.5 ± 0.7 |
| SJC28 | 12.6 ± 5.9 | 12.0 ± 5.5 | 11.6 ± 5.5 | 11.4 ± 5.0 |
| Presence of RF, n (%) | 197 (92.5) | 156 (74.6) | 179 (90.9) | 179 (77.8) |
| Presence of anti-CCP, n (%) | 213 (100) | 148 (70.5) | 197 (100) | 168 (73.0) |
| RANKL, pmol/l | 2882 ± 6823 | 1301 ± 1772 | 3111 ± 4437 | 2162 ± 3989 |
| OPG, pmol/l | 6.1 ± 3.0 | 6.2 ± 3.4 | 6.0 ± 3.0 | 5.5 ± 1.8 |
| OC, ng/ml | 19.4 ± 8.4 | 21.3 ± 10.3 | 21.6 ± 12.0 | 21.7 ± 11.9 |
| Prior DMARD use, n (%) | 61 (28.6) | 64 (30.5) | 65 (33.0) | 57 (24.8) |
Values are mean ± SD, unless otherwise indicated
aPatients who were anti-CCP-positive and had CRP > 12.3 mg/l
Anti-CCP anti-cyclic citrullinated peptide, CDAI Clinical Disease Activity Index, DAS28-CRP 28-joint Disease Activity Score using C-reactive protein, HAQ-DI Health Assessment Questionnaire-Disability Index, OC osteocalcin, OPG osteoprotegerin, RANKL receptor activator of NF-kappa B ligand, RF rheumatoid factor, SJC swollen joint count
Model Validation and Application of the Rule to Predict Clinical Response
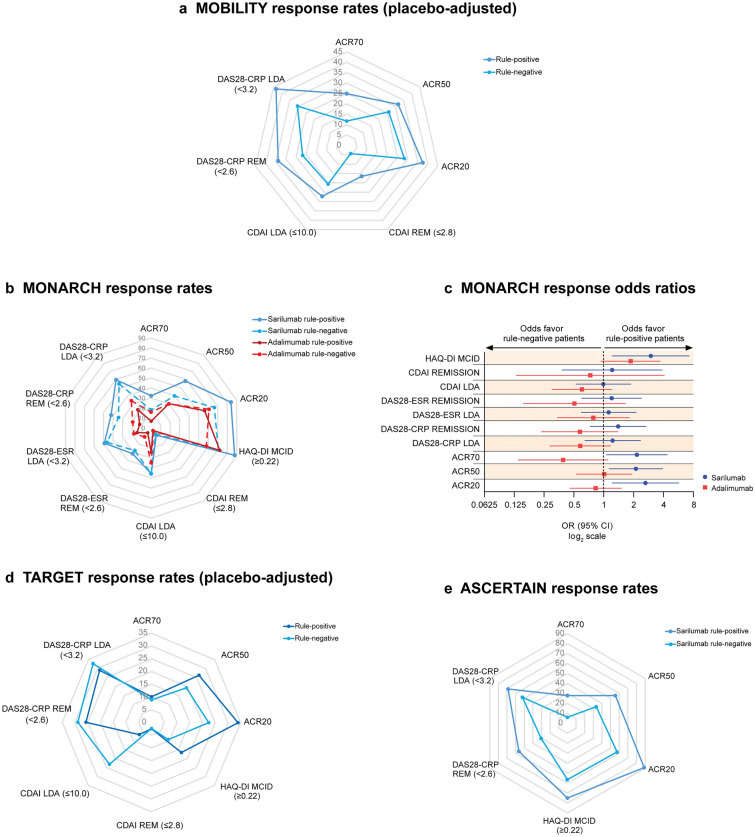
At week 24, response rates in rule-positive sarilumab-treated patients from MOBILITY were superior to those in rule-negative patients, across all endpoints assessed. In the placebo group, responder rates for rule-negative patients were more favorable for CDAI remission, DAS28-CRP remission, and DAS28-CRP LDA, compared with rule-positive patients, whereas response rates for other outcomes were similar (Supplementary Material, Fig. S2A). Across all outcomes, placebo-adjusted response rates in rule-positive patients were higher by approximately 5–15%, compared with rule-negative patients (Fig. 2A).
Fig. 2.
Response rates in rule-positive and rule-negative sarilumab-treated patients. The patient stratification rule was the combined presence of anti-CCP and CRP > 12.3 mg/l. ACR20 ACR 20%, ACR50 ACR 50%, ACR70 ACR 70%, CDAI Clinical Disease Activity Index, DAS28-CRP 28-joint Disease Activity Score using C-reactive protein, DAS28-ESR DAS28 using erythrocyte sedimentation rate, HAQ-DI Health Assessment Questionnaire-Disability Index, LDA low disease activity, MCID minimal clinically important difference, REM remission
In MONARCH, rule-positive patients also had a consistently stronger response to sarilumab than their rule-negative counterparts, which was not observed in adalimumab-treated patients (Fig. 2B, C). Among rule-positive individuals, the ACR70 response rate in those receiving sarilumab was almost five times the rate observed in patients receiving adalimumab (34 vs. 7%), whereas adalimumab treatment resulted in a higher ACR70 response rate among rule-negative patients compared to the overall, unstratified population (16 vs. 12%). Based on MONARCH data, sarilumab treatment of 100 patients (35 of whom were rule-positive) without considering their rule status would result in the ACR70 response in 23 individuals. However, a nearly identical ACR70 response rate (22 individuals) would be achieved if only the 35 rule-positive patients received sarilumab and the remaining 65 rule-negative patients received adalimumab. Adalimumab treatment of 100 patients would result in ACR70 response in 12 individuals (Supplementary Material, Figure S3).
In TARGET, the only trial that enrolled patients with an inadequate response to TNFi treatment, findings were mixed (Fig. 2D). In both the placebo and sarilumab groups, the response rates in rule-positive patients for ACR scores, as well as the DAS28-CRP LDA and remission, were lower than those observed in rule-negative patients (Supplementary Material, Figure S2B).
In ASCERTAIN, sarilumab-treated rule-positive patients had more favorable 24-week response rates compared with rule-negative patients for all endpoints assessed, with a magnitude of difference ranging between 15 and 30% (Fig. 2E). In tocilizumab-treated patients, the rule applied to most clinical endpoints (Supplementary Material, Figure S4).
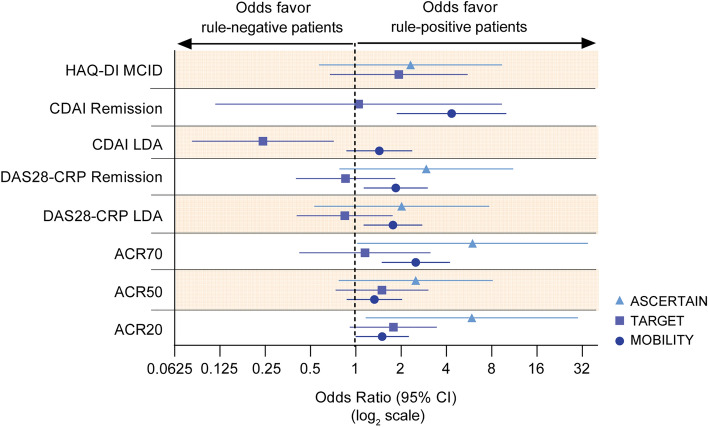
Overall, across MOBILITY, TARGET, and ASCERTAIN, rule-positive patients had higher odds of achieving a clinical response at week 24 than rule-negative patients, with some exceptions observed in TARGET (Fig. 3). The changes from baseline in continuous variables for each trial are shown in Table 3. Across all studies (including TARGET), we calculated a mean improvement in CDAI of 3.4 (± 2.8 [standard error]) and in DAS28-CRP of 0.8 ± 0.4 for rule-positive patients treated with sarilumab compared with rule-negative patients. Conversely, the treatment of rule-negative patients with adalimumab in MONARCH, resulted in a mean CDAI improvement of 3.1 ± 3.0 compared with rule-positive patients (Table 3). Mean change in DAS28-CRP was similar between adalimumab-treated rule-positive and rule-negative patients in MONARCH (Table 3), although response rates for DAS28-CRP remission and LDA showed improvement for rule-negative compared with rule-positive adalimumab-treated patients (Fig. 2B).
Fig. 3.
Odds ratios of achieving clinical response at week 24 in placebo- (MOBILITY, TARGET) or active-controlled studies (ASCERTAIN): rule-positive versus rule-negative patients. The patient stratification rule was the combined presence of anti-CCP and CRP > 12.3 mg/l. Data presented for MOBILITY and TARGET are placebo-adjusted. ACR20 ACR 20%, ACR50 ACR 50%, ACR70 ACR 70%, DAS28-CRP 28-joint Disease Activity Score using C-reactive protein, HAQ-DI Health Assessment Questionnaire-Disability Index, LDA low disease activity, MCID minimal clinically important difference
Table 3.
Mean ± SE change from baseline to week 24 for applicable endpoints in rule-positive and rule-negative patients in MOBILITY, MONARCH, TARGET, and ASCERTAIN
| MOBILITY | MONARCH | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sarilumab | Placebo | Sarilumab − placebo | Sarilumab | Adalimumab | ||||||
| Rule-positive | Rule-negative | Rule-positive | Rule-negative | Rule-positive | Rule-negative | Rule-positive | Rule-negative | Rule-positive | Rule-negative | |
| CDAI | − 30.0 ± 1.1 | − 25.9 ± 1.0 | − 20.0 ± 1.4 | − 19.4 ± 1.3 | − 10.0 | − 6.4 | − 32.8 ± 1.7 | − 27.4 ± 1.2 | − 24.1 ± 1.7 | − 27.2 ± 1.3 |
| DAS28-CRP | − 3.4 ± 0.1 | − 2.6 ± 0.1 | − 1.7 ± 0.1 | − 1.4 ± 0.1 | − 1.7 | − 1.1 | − 3.5 ± 0.2 | − 2.6 ± 0.1 | − 2.2 ± 0.2 | − 2.0 ± 0.1 |
| DAS28-ESR | N/A | N/A | N/A | N/A | N/A | N/A | − 3.7 ± 0.2 | − 3.1 ± 0.1 | − 2.1 ± 0.2 | − 2.4 ± 0.1 |
| HAQ-DI | N/A | N/A | N/A | N/A | N/A | N/A | − 0.9 ± 0.1 | − 0.5 ± 0.1 | − 0.5 ± 0.1 | − 0.4 ± 0.1 |
| TARGET | ASCERTAIN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sarilumab | Placebo | Sarilumab − placebo | Sarilumab | Tocilizumab | ||||||
| Rule-positive | Rule-negative | Rule-positive | Rule-negative | Rule-positive | Rule-negative | Rule-positive | Rule-negative | Rule-positive | Rule-negative | |
| CDAI | − 30.8 ± 1.8 | − 30.0 ± 1.7 | − 23.7 ± 2.3 | − 24.0 ± 1.5 | − 7.1 | − 6.0 | N/A | N/A | N/A | N/A |
| DAS28-CRP | − 3.3 ± 0.2 | − 3.1 ± 0.2 | − 2.0 ± 0.2 | − 1.9 ± 0.2 | − 1.3 | − 1.1 | − 3.8 ± 0.3 | − 2.4 ± 0.2 | − 3.1 ± 0.2 | − 2.4 ± 0.2 |
| DAS28-ESR | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| HAQ-DI | − 0.7 ± 0.1 | − 0.7 ± 0.1 | − 0.4 ± 0.1 | − 0.6 ± 0.1 | − 0.3 | − 0.1 | − 0.7 ± 0.2 | − 0.5 ± 0.1 | − 0.7 ± 0.1 | − 0.5 ± 0.1 |
The patient stratification rule was the combined presence of anti-CCP and CRP > 12.3 mg/l
CDAI Clinical Disease Activity Index, DAS28-CRP 28-joint Disease Activity Score using C-reactive protein, DAS28-ESR DAS28 using erythrocyte sedimentation rate, HAQ-DI Health Assessment Questionnaire-Disability Index, N/A not applicable
Discussion
In this study, we used machine learning to identify a combination of baseline patient characteristics to predict treatment response to sarilumab and adalimumab. The method found that the presence of anti-CCP antibody and CRP level at a selected cutpoint of > 12.3 mg/l were predictive of a better response to sarilumab, and in one trial where adalimumab data were also available, predicted an incrementally larger response to adalimumab. This approach could facilitate choice of treatment in patients with RA.
Our algorithm identified a simple, clinically applicable rule that considered the large number of combinatorial possibilities between 42 variables and their values or thresholds. Therefore, our study demonstrates the potential of machine learning as a tool for systemic, fast, and deep analysis of the data that can yield rules applicable in clinical practice.
Previously, anti-CCP has been identified as a predictor of response to rituximab and abatacept, and high CRP has been identified as a predictor of response to TNF inhibitors and tocilizumab [26–29]. Our study confirms the predictive potential of the combined presence of these two parameters with data from four independent studies. Of note, biologically plausible parameters that have been identified as predictors of response to sarilumab, such as IL-6 concentration [30], were not included in the rule. This is not unexpected: in patients with RA, IL-6 and CRP levels are highly correlated [31, 32] and machine learning algorithms, which approach data in a non-biased fashion, are set to prefer one of the correlated parameters based on its ability to maximize the predefined outcome (in our case, ACR20). CRP was probably selected because IL-6 varies more between individuals [33] and has a more variable diurnal profile than CRP [34]. The more stable levels of CRP would make it a preferred choice as predictor of response, especially if there was a single biomarker measurement by visit.
It can be argued that the use of a composite endpoint such as ACR response, which includes the acute phase reactants CRP or ESR, may have influenced the algorithm to select CRP > 12.3 mg/l as one of the components of the rule for an IL-6R inhibitor. However, the rule also predicted CRP-independent endpoints, such as the CDAI and HAQ-DI. ACR scores are based on relative changes and, therefore, unaffected by a potential selection bias. For the other scores associated with low disease activity and remission (e.g., DAS28-CRP remission and LDA), where fixed, relatively low CRP thresholds are required, a selection of high CRP baseline values rather increases the necessary treatment response to achieve these thresholds.
Among the decision tree methods we considered, the GUIDE algorithm was the only one that provided a simple, clinically feasible rule. It also showed the highest precision and competitive accuracy, compared with other methods assessed, as well as a higher transparency and better interpretability, albeit with lower recall.
Since the algorithm was selecting responders regardless of treatment during the model training, patients treated with placebo (MOBILITY and TARGET) and adalimumab (MONARCH) were important controls during the testing phase. We found that rule-positive patients had higher levels of baseline factors associated with poor prognosis and a reduced response to adalimumab treatment or placebo, compared with rule-negative patients. In MOBILITY, the predictive power of the rule was greater for the placebo-adjusted than for the non-adjusted response. The absolute disease state such as CDAI or DAS28-CRP remission/LDA had a relatively low prevalence in these data, and placebo adjusting increases that prevalence, thus improving the performance of the rule. In addition, in settings with active instead of placebo control (e.g., MONARCH trial), our data suggest that the choice of treatment can be improved significantly using this methodology. We explored a clinical scenario and based on the results of the MONARCH trial, sarilumab was clearly favored in rule-positive patients, whereas rule-negative patients could be treated with either adalimumab or sarilumab, based on other priorities (e.g., patient preferences, erosion score, cost).
As noted in the Results section, the rule applied less consistently to patients from TARGET, who had poor tolerance for, or an inadequate response to, TNF inhibitors. Patients with RA who have failed treatment with one drug class are generally less likely to respond to subsequent treatments [26], which may account for some of the inconsistent rule applicability observed in our analysis. However, the overall percentages of TARGET patients achieving remission or low disease activity endpoints were particularly low, making it difficult to demonstrate differences between rule-positive and rule-negative patients in these disease scores.
A less consistent verification to TARGET data suggests that the rule has limits in generalizability such that the rule may not apply to patients who had inadequate response to TNFi. Also, since all data in the training and validation phases came from randomized, controlled clinical trials, which used stringent enrollment criteria, the rule may not apply to a real-world population of patients with RA in the same way. For example, all patients in these trials had to have elevated CRP, and selection of this variable by the machine learning algorithm as an important variable, as well as the exact CRP cutoff value chosen, may have been influenced by the cutoffs required by trials’ inclusion criteria. In addition, radiographic endpoints were only available in the MOBILITY trial, and with the lack of further validation data we excluded this important assessment from the model training. Inclusion of radiographic scores in the rule may be an interesting variable to further increase the accuracy in patient stratification, albeit at the expense of simplicity. Finally, the number of patients in the training set was relatively small. Using a larger set may have resulted in an even more robust rule.
Conclusions
This study used a machine learning approach to identify a simple rule to identify patients with RA who have an increased chance of achieving clinical response to sarilumab, based on laboratory parameters that are readily available in routine practice. In addition, such patients had a lower likelihood of response to placebo and adalimumab, which suggests that the rule can be used for treatment optimization. Real-world validation of this rule, and replication in other clinical trial datasets of other therapies, is merited.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors and Sanofi thank the patients for their participation in the trials, as well as the MOBILITY, MONARCH, TARGET, and ASCERTAIN Steering Committees and Investigators.
Funding
The studies were funded by Sanofi (Cambridge, USA) and Regeneron Pharmaceuticals, Inc. (Tarrytown, USA). This analysis, as well as the Rapid Service Fee for processing of the manuscript in this journal, were funded by Sanofi.
Medical Writing and Editorial Assistance
Medical writing assistance was provided by Richard J. Hogan, PhD, and Vojislav Pejović, PhD, of Elevate Scientific Solutions, a division of Envision Pharma Group. Editorial and graphics assistance was also provided by Elevate Scientific Solutions. Both medical writing and editorial/graphics assistance were funded by Sanofi.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authorship Contributions
Markus Rehberg, Clemens Giegerich, and Hubert van Hoogstraten were involved in the design of the study. Markus Rehberg and Clemens Giegerich were involved in data analysis and in drafting of the manuscript. All authors were involved in interpretation of the results. All authors had full access to the study data, contributed to critical revisions of the manuscript for important intellectual content, and approved the final version of the article to be submitted. Richard J. Hogan, PhD, and Vojislav Pejović, PhD, of Elevate Scientific Solutions, a division of Envision Pharma Group, edited the drafts under the direction of the authors.
Disclosures
Markus Rehberg, Clemens Giegerich, Amy Praestgaard, Hubert van Hoogstraten, Melitza Iglesias-Rodriguez: Employees of Sanofi and may hold stock and/or stock options in the company. Jeffrey R. Curtis: Research grants and consulting fees (AbbVie, Amgen, Bristol-Myers Squibb, Corrona, Janssen, Lilly, Myriad, Pfizer, Regeneron, Roche, and UCB). Jacques-Eric Gottenberg: Research grants (Bristol-Myers Squibb, Pfizer, Roche); speaking/consulting fees (AbbVie, Bristol-Myers Squibb, Gilead, Janssen, Lilly, MSD, Pfizer, Roche, UCB). Andreas Schwarting: Grant/research support and honoraria from GSK, Pfizer, Janssen, Roche, BMS, MSD. Santos Castañeda: Grants/research support from MSD and Pfizer, consultation fees/participation in company-sponsored speaker’s bureau from Amgen, Celgene, Lilly, MSD, Pfizer, Sanofi, SOBI, and travel aids for congresses from BMS, MSD, Lilly, Pfizer, Roche. Andrea Rubbert-Roth: Speakers’ bureaus for AbbVie, Bristol-Myers Squibb, Chugai, Hexal/Novartis, Janssen, Lilly, Merck Sharp & Dohme, Pfizer, Roche, and Sanofi, and has provided consultancy for Chugai, Lilly, Roche, and Sanofi. Ernest HS Choy: Received research grants and/or consulting fees from AbbVie, Amgen, AstraZeneca, Biogen, Bio-Cancer, Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Chelsea Therapeutics, Chugai Pharma, Daiichi Sankyo, Eli Lilly, Ferring Pharmaceuticals, GlaxoSmithKline, Hospita, Ionis, Janssen, Jazz Pharmaceuticals, MedImmune, Merck Sharp & Dohme, Merrimack Pharmaceutical, Napp, Novartis, Novimmune, ObsEva, Pfizer, R-Pharm, Regeneron Pharmaceuticals, Inc., Roche, Sanofi, SynAct Pharma, Tonix and UCB, and participated in speakers’ bureaus for Amgen, Boehringer Ingelheim, Bristol-Myers Squibb, Chugai Pharma, Eli Lilly, Hospira, Merck Sharp & Dohme, Novartis, Pfizer, Regeneron Pharmaceuticals, Inc., Roche, Sanofi-Aventis, and UCB.
Prior presentation
These data were presented in part as an oral presentation at the American College of Rheumatology (ACR) Convergence Meeting, 4–10 November 2020.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Qualified researchers can request access to patient-level data and related study documents including the clinical study report, study protocol with any amendments, blank case report form, statistical analysis plan, and dataset specifications. Patient-level data will be anonymized and study documents will be redacted to protect the privacy of trial participants. Further details on Sanofi’s data sharing criteria, eligible studies, and process for requesting access can be found online.
Footnotes
The original online version of this article was revised due to correction in the supplementary information.
Change history
8/1/2022
The original article is updated to correct the error in the supplementary video
Change history
10/29/2021
A Correction to this paper has been published: 10.1007/s40744-021-00389-7
Contributor Information
Markus Rehberg, Email: markus.rehberg@sanofi.com.
Clemens Giegerich, Email: clemens.giegerich@sanofi.com.
Amy Praestgaard, Email: amy.praestgaard@sanofi.com.
Hubert van Hoogstraten, Email: hubert.vanhoogstraten@sanofi.com.
Melitza Iglesias-Rodriguez, Email: melitza.iglesias-rodriguez@sanofi.com.
Jeffrey R. Curtis, Email: jrcurtis@uabmc.edu
Jacques-Eric Gottenberg, Email: jegotten@gmail.com.
Andreas Schwarting, Email: schwarting@uni-mainz.de.
Santos Castañeda, Email: scastas@gmail.com.
Andrea Rubbert-Roth, Email: andrea.rubbert-roth@kssg.ch.
Ernest H. S. Choy, Email: choyeh@cardiff.ac.uk
References
- 1.Singh JA, Saag KG, Bridges SL, Jr, et al. 2015 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res. 2016;68(1):1–25. doi: 10.1002/acr.22783. [DOI] [PubMed] [Google Scholar]
- 2.Smolen JS, Landewe RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2019-216655. [DOI] [PubMed] [Google Scholar]
- 3.Karsdal MA, Bay-Jensen AC, Henriksen K, et al. Rheumatoid arthritis: a case for personalized health care? Arthritis Care Res (Hoboken) 2014;66(9):1273–1280. doi: 10.1002/acr.22289. [DOI] [PubMed] [Google Scholar]
- 4.Hugle M, Omoumi P, van Laar JM, Boedecker J, Hugle T. Applied machine learning and artificial intelligence in rheumatology. Rheumatol Adv Pract. 2020;4(1):rkaa005. doi: 10.1093/rap/rkaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venerito V, Lopalco G, Cacciapaglia F, Fornaro M, Iannone F. A Bayesian mixed treatment comparison of efficacy of biologics and small molecules in early rheumatoid arthritis. Clin Rheumatol. 2019;38(5):1309–1317. doi: 10.1007/s10067-018-04406-z. [DOI] [PubMed] [Google Scholar]
- 6.Migliore A, Bizzi E, Egan CG, Bernardi M, Petrella L. Efficacy of biological agents administered as monotherapy in rheumatoid arthritis: a Bayesian mixed-treatment comparison analysis. Ther Clin Risk Manag. 2015;11:1325–1335. doi: 10.2147/TCRM.S89678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis G, Jr, Holweg CT, Kummerfeld SK, et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res Ther. 2014;16(2):R90. doi: 10.1186/ar4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson KJ, Sanchez HN, Schoenbrunner N. Defining response to TNF-inhibitors in rheumatoid arthritis: the negative impact of anti-TNF cycling and the need for a personalized medicine approach to identify primary non-responders. Clin Rheumatol. 2019;38(11):2967–2976. doi: 10.1007/s10067-019-04684-1. [DOI] [PubMed] [Google Scholar]
- 9.Plant D, Maciejewski M, Smith S, et al. Profiling of gene expression biomarkers as a classifier of methotrexate nonresponse in patients with rheumatoid arthritis. Arthritis Rheumatol. 2019;71(5):678–684. doi: 10.1002/art.40810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahler M, Martinez-Prat L, Sparks JA, Deane KD. Precision medicine in the care of rheumatoid arthritis: focus on prediction and prevention of future clinically-apparent disease. Autoimmun Rev. 2020;19(5):102506. doi: 10.1016/j.autrev.2020.102506. [DOI] [PubMed] [Google Scholar]
- 11.Aletaha D. Precision medicine and management of rheumatoid arthritis. J Autoimmun. 2020;110:102405. doi: 10.1016/j.jaut.2020.102405. [DOI] [PubMed] [Google Scholar]
- 12.Giacomelli R, Afeltra A, Alunno A, et al. Guidelines for biomarkers in autoimmune rheumatic diseases - evidence based analysis. Autoimmun Rev. 2019;18(1):93–106. doi: 10.1016/j.autrev.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Lequerré T, Rottenberg P, Derambure C, Cosette P, Vittecoq O. Predictors of treatment response in rheumatoid arthritis. Joint Bone Spine. 2019;86(2):151–158. doi: 10.1016/j.jbspin.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Norgeot B, Glicksberg BS, Trupin L, et al. Assessment of a deep learning model based on electronic health record data to forecast clinical outcomes in patients with rheumatoid arthritis. JAMA Netw Open. 2019;2(3):e190606. doi: 10.1001/jamanetworkopen.2019.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan Y, Zhang H, Quang D, et al. Machine learning to predict anti-tumor necrosis factor drug responses of rheumatoid arthritis patients by integrating clinical and genetic markers. Arthritis Rheumatol. 2019;71(12):1987–1996. doi: 10.1002/art.41056. [DOI] [PubMed] [Google Scholar]
- 16.Yoo J, Lim MK, Ihm C, Choi ES, Kang MS. A study on prediction of rheumatoid arthritis using machine learning. Int J Appl Engineering Res. 2017;12(20):9858–9862. [Google Scholar]
- 17.Zhou SM, Fernandez-Gutierrez F, Kennedy J, et al. Defining disease phenotypes in primary care electronic health records by a machine learning approach: a case study in identifying rheumatoid arthritis. PLoS ONE. 2016;11(5):e0154515. doi: 10.1371/journal.pone.0154515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.KEVZARA (sarilumab) [Prescribing Information] 2017. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/761037s001lbl.pdf.
- 19.KEVZARA (sarilumab) [Summary of Product Characteristics] 2017. Available at: https://www.ema.europa.eu/en/documents/product-information/kevzara-epar-product-information_en.pdf.
- 20.Genovese MC, Fleischmann R, Kivitz AJ, et al. Sarilumab plus methotrexate in patients with active rheumatoid arthritis and inadequate response to methotrexate: results of a phase III study. Arthritis Rheumatol. 2015;67(6):1424–1437. doi: 10.1002/art.39093. [DOI] [PubMed] [Google Scholar]
- 21.Burmester GR, Lin Y, Patel R, et al. Efficacy and safety of sarilumab monotherapy versus adalimumab monotherapy for the treatment of patients with active rheumatoid arthritis (MONARCH): a randomised, double-blind, parallel-group phase III trial. Ann Rheum Dis. 2017;76(5):840–847. doi: 10.1136/annrheumdis-2016-210310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischmann R, van Adelsberg J, Lin Y, et al. Sarilumab and nonbiologic disease-modifying antirheumatic drugs in patients with active rheumatoid arthritis and inadequate response or intolerance to tumor necrosis factor inhibitors. Arthritis Rheumatol. 2017;69(2):277–290. doi: 10.1002/art.39944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emery P, Rondon J, Parrino J, et al. Safety and tolerability of subcutaneous sarilumab and intravenous tocilizumab in patients with rheumatoid arthritis. Rheumatology (Oxford) 2019;58(5):849–858. doi: 10.1093/rheumatology/key361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh W-Y. Classification and regression trees. Wiley Interdiscip Rev Data Min Knowl Discov. 2011;1(1):14–23. doi: 10.1002/widm.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan JR. Induction of decision trees. Mach Learn. 1986;1:81–106. [Google Scholar]
- 26.Wijbrandts CA, Tak PP. Prediction of response to targeted treatment in rheumatoid arthritis. Mayo Clin Proc 2017;92(7):1129–43. [DOI] [PubMed]
- 27.Harrold LR, Litman HJ, Connolly SE, et al. Effect of anticitrullinated protein antibody status on response to abatacept or antitumor necrosis factor-α therapy in patients with rheumatoid arthritis: a US national observational study. J Rheumatol. 2018;45(1):32–39. doi: 10.3899/jrheum.170007. [DOI] [PubMed] [Google Scholar]
- 28.Pers YM, Fortunet C, Constant E, et al. Predictors of response and remission in a large cohort of rheumatoid arthritis patients treated with tocilizumab in clinical practice. Rheumatology (Oxford) 2014;53(1):76–84. doi: 10.1093/rheumatology/ket301. [DOI] [PubMed] [Google Scholar]
- 29.Harrold L, Wittstock K, Kelly S, et al. The comparative effectiveness of abatacept versus TNF inhibitors in patients who are ACPA positive and have the shared epitope: results from a US national observational study. Arthritis Rheumatol. 2020;72(suppl 10):0801. doi: 10.1186/s42358-024-00352-4. [DOI] [PubMed] [Google Scholar]
- 30.Boyapati A, Schwartzman S, Msihid J, et al. Association of high serum interleukin-6 levels with severe progression of rheumatoid arthritis and increased treatment response differentiating sarilumab from adalimumab or methotrexate in a post hoc analysis. Arthritis Rheumatol. 2020;72(9):1456–1466. doi: 10.1002/art.41299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boss B, Neeck G. Correlation of IL-6 with the classical humoral disease activity parameters ESR and CRP and with serum cortisol, reflecting the activity of the HPA axis in active rheumatoid arthritis. Z Rheumatol 2000;59 Suppl 2:II/62–4. [DOI] [PubMed]
- 32.Cronstein BN. Interleukin-6–a key mediator of systemic and local symptoms in rheumatoid arthritis. Bull NYU Hosp Jt Dis. 2007;65(Suppl 1):S11–S15. [PubMed] [Google Scholar]
- 33.van Leeuwen MA, Westra J, Limburg PC, van Riel PL, van Rijswijk MH. Clinical significance of interleukin-6 measurement in early rheumatoid arthritis: relation with laboratory and clinical variables and radiological progression in a three year prospective study. Ann Rheum Dis. 1995;54(8):674–677. doi: 10.1136/ard.54.8.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izawa S, Miki K, Liu X, Ogawa N. The diurnal patterns of salivary interleukin-6 and C-reactive protein in healthy young adults. Brain Behav Immun. 2013;27(1):38–41. doi: 10.1016/j.bbi.2012.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Qualified researchers can request access to patient-level data and related study documents including the clinical study report, study protocol with any amendments, blank case report form, statistical analysis plan, and dataset specifications. Patient-level data will be anonymized and study documents will be redacted to protect the privacy of trial participants. Further details on Sanofi’s data sharing criteria, eligible studies, and process for requesting access can be found online.