Abstract
Because identification of the species within the “Streptococcus milleri” group is difficult for the clinical laboratory as the species share overlapping phenotypic characteristics, we wished to confirm biochemical identification with identification by 16S rRNA gene sequence analysis. Ninety-four clinical isolates previously identified as the “Streptococcus milleri” group were reclassified as S. anginosus, S. constellatus, or S. intermedius with the API 20 Strep system (bioMerieux Vikek, Hazelton, Mo.) and the Fluo-card (Key Scientific, Round Rock, Tex.). In addition, we determined the Lancefield group, hemolysis, colony size, colony texture, repetitive extragenic palindromic PCR (rep-PCR) pattern, and cellular fatty acid (CFA) profile (MIDI, Newark, Del.). 16S rRNA gene sequence analysis with 40 selected representative strains showed three distinct groups, with S. constellatus and S. intermedius found to be more closely related to each other than to S. anginosus, and further distinguished a biochemically distinct group of urogenital isolates within the S. anginosus group of isolates. Except for strains unreactive with the Fluo-card (8%), all S. anginosus and S. intermedius strains identified by sequencing were similarly identified by biochemical testing. However, 23% of the selected S. constellatus isolates identified by sequencing (9% of all S. constellatus isolates) would have been identified as S. anginosus or S. intermedius by biochemical tests. Although most S. anginosus strains formed one unique cluster by CFA analysis and most S. constellatus strains showed similar rep-PCR patterns, neither method was sufficiently dependable for identification. Whereas Lancefield group or lactose fermentation did not correspond to sequence or biochemical type, S. constellatus was most likely to be beta-hemolytic and S. intermedius was most likely to have a dry colony type. The most frequent isolate in our population was S. constellatus, followed by S. anginosus. There was an association of S. anginosus with a gastrointestinal or urogenital source, and there was an association of S. constellatus and S. intermedius with both the respiratory tract and upper-body abscesses.
Members of the “Streptococcus milleri” group, which for a brief period were considered a single species loosely synonymous with S. anginosus (5, 15), are now separated into three distinct species: S. anginosus, S. constellatus, and S. intermedius (16, 17). However, because many phenotypic tests for the characterization of the species show considerable overlap, it has been difficult for the clinical laboratory to correctly classify isolates (9). Recently, in an excellent study, Whiley et al. (19) found that seven biochemical tests could separate the “S. milleri” group into the S. anginosus, S. constellatus, and S. intermedius groups. Flynn and Ruoff (6) reported that the Fluo-card (Key Scientific, Round Rock, Tex.), which uses three of these substrates to test for β-d fucosidase, α-d-glucosidase, and β-d-glucosidase activity, when used in conjunction with other tests, correctly identified over 97% of the S. anginosus and S. constellatus species and 88% of the S. intermedius species (6). By use of these better identification schemes, several recent reports associated species with site of isolation or disease. However, the data from different institutions and for different patient populations have shown variations in species distribution and overlapping clinical associations (1, 2, 6, 8, 13, 18). In this paper, we seek a closer association of phenotype, genotype, and site of isolation for “S. milleri” group isolates from a defined population (adult male patients) by confirming the biochemical identification by 16S rRNA gene sequencing.
MATERIALS AND METHODS
Organisms.
All clinical strains were obtained from the Microbiology Laboratory of the Houston Veterans Affairs Medical Center. We examined 101 clinical isolates from our stock collection. The isolates either were identified by the API Rapid Strep test as the S. milleri group or were older isolates identified by nonspecified methods as S. anginosus, S. constellatus, S. intermedius, Streptococcus group F, or small-colony Streptococcus groups C or G. All but seven of these isolates were confirmed to be in the “S. milleri” group (characteristic Streptococcus morphology, catalase negativity, and Lancefield group F or Voges-Proskauer test positive, arginine positive, and sorbitol negative). A second group included strains similar to the “S. milleri” group either by biochemical characterization or by colony morphology, such as S. bovis, S. sanguis, and S. dysgalactiae subsp. equisimilis (which can be Lancefield group C or G). Reference stock strains included four American Type Culture Collection (ATCC) strains (ATCC 27838, ATCC 33397, ATCC 9895, and ATCC 27335) which are described as the S. constellatus type strain, the S. anginosus type strain, S. anginosus, and the S. intermedius type strain, respectively.
Biochemical and phenotypic characterization.
Hemolysis on Columbia agar plates (BBL, Becton Dickinson, Cockeysville, Md.) incubated anaerobically was assessed at 24 to 48 h. A strain that is nonhemolytic around a single colony is often alpha-hemolytic in the heavier areas, and a strain that is alpha-hemolytic around a single colony is often beta-hemolytic in the heavier areas or becomes beta-hemolytic with further incubation. For this study, strains were called beta-hemolytic only if by 48 h beta-hemolysis was seen around a single colony or complete clearing was seen under the heavier areas. Lancefield grouping by latex agglutination (PathoDx; Diagnostic Products Corp. Los Angeles, Calif.) and biochemical testing with the API 20 Strep system (bioMerieux Vikek, Hazelton, Mo.) and the Fluo-card (Key Scientific) were performed according to the manufacturers' instructions. The Fluo-card measures the cleavage of three substrates. We generated a biotype number by assigning numbers according to the substrate's position on the card (1 = β-d-fucosidase activity, 2 = β-d-glucosidase activity, and 3 = α-d-glucosidase activity) and recording only positive reactions to generate a biotype number. The isolate was identified as S. intermedius if it had positive results for 1, 1 and 2, 1 and 3, or 1, 2, and 3, as S. anginosus if it had positive results for 2 or 2 and 3; and S. constellatus if it had positive results for 3.
Cellular fatty acid (CFA) analysis.
Whole-cell fatty acids were extracted and analyzed as reported previously (4), except that to achieve sufficient growth the organisms were grown on Columbia agar or Columbia colistin naladixic acid (CNA) agar (BBL, Becton Dickinson) anaerobically for 48 h. Analysis was performed with an automated Hewlett-Packard HP 5890 II Microbial Identification System (MIDI Systems). Analysis of fatty acid peaks was achieved with the manufacturer's software; however, the dendrogram was made only with data for clinical and reference strains that we grew under similar conditions.
rep-PCR.
Previously described methods with conserved primers corresponding to repetitive extragenic palindromic sequences in a repetitive extragenic palindromic PCR (rep-PCR) were used (4). For our usual protocol, the organisms were harvested with a sterile swab and were resuspended in 0.9% sterile saline to a turbidity that matched that of a 3.0 McFarland standard. The reaction mixture consisted of 5.0 μl of 5× polymerase buffer, 2.5 μl of dimethyl sulfoxide, 0.5 μl of each deoxynucleoside triphosphate (25 mM), 1 μl (50 pmol) of each primer, 4 μl of a sample of the organism suspension, 0.4 μl of Taq polymerase (Perkin-Elmer Cetus), and 10 μl of water, and the mixture was overlaid with 50 μl of light mineral oil. Amplifications were performed on a 96-well Perkin-Elmer thermocycler as described previously (4). The product was separated on a 1.2% agarose (Sigma, St. Louis, Mo.) gel that was stained with ethidium bromide. For analysis of the band patterns, the bands were compared with a DNA molecular size marker.
16S rRNA gene sequencing reactions.
The PCR products were sequenced and purified by using the MicroSeq 500 Gene Kit protocols. This DNA sequencing system uses dRhodamine-labeled dye terminators and provides double-stranded sequence data, and the same primers used in the PCR amplification step described above were used. Sequencing reactions were run on a 4.5% Long Ranger (FMC BioProducts, Rockland, Maine) and 33% urea electrophoresis gel by using an ABI Prism 377 Sequencer (PE Applied Biosystems, Foster City, Calif.). DNA sequence data were analyzed and assembled with Auto Assembler software (PE Applied Biosystems). Bacterial identifications based on 16S rRNA gene sequence data were assigned by using MicroSeq Microbial Identification and Analysis Software (PE Applied Biosystems).
Nucleotide sequence accession numbers.
The nucleotide sequences of the following isolates have been submitted to GenBank (accession numbers are given in parentheses): 5464 (AF16953), 5302 (AF16954), 5219 (AF16955), 3868 (AF16956), 3276 (AF16957), and 3075 (AF16958).
RESULTS
Figure 1 is the dendrogram based on 16S rRNA gene sequence analysis of a subset of 40 clinical “S. milleri” group isolates selected to include representatives of all phenotypic types. The control strains S. intermedius ATCC 27335T, S. constellatus ATCC 27838T, and S. anginosus ATCC 9895 and 27335 are located in the appropriate clusters. S. constellatus and S. intermedius are more closely related to each other than to S. anginosus and may be more homogeneous than S. anginosus. Table 1 lists some characteristics of these isolates. Larger percentages of S. constellatus and S. intermedius isolates were isolated from abscesses, tissues, or exudates. Within the S. anginosus cluster there was a biochemically distinct group of urinary tract isolates. The distinct numbers for these strains (except strain 4028-3) generated by the API 20 Strep system indicate the ability to ferment raffinose and mannitol and, for strains 1805 and 99M2144, the ability to cleave hippurate. Although all the S. constellatus Lancefield group C isolates have Fluo-card biotype 1,2,3, which would lead to their identification as S. intermedius and most are from a gastrointestinal source, the significance of this finding must be confirmed with additional isolates.
FIG. 1.
Dendrogram of sequence data for 44 strains of “S. milleri” group and other representative clinical and type strains.
TABLE 1.
Summary of characteristics of individual sequenced strainsa
| Stock no. | DNA cluster | Site of isolation | Strep API no. | Fluo-card biotype | Lancefield group | Hemolysis | Original report |
|---|---|---|---|---|---|---|---|
| 5219 | S. anginosus | Bile | 5061410 | 2,3 | F | A/N | |
| 5153 | S. anginosus | ATCC 9895 | 5061410 | 2 | F | A/N | |
| Type strain | S. anginosus | ATCC 33397 | 5061454 | 2 | G | A/N | |
| 5174 | S. anginosus | Eye | 5061411 | 2 | F | A/N | |
| 417-1b | S. anginosus | Peritoneum | 1061410 | 2,3 | F | A/N | |
| 3898 | S. anginosus | Blood | 5061010 | 2 | NG | A/N | |
| 5302c | S. anginosus | Urine | 5065553 | 2,3 | F | A/N | |
| 1805c | S. anginosus | Urine | 7063150 | 2,3 | F | A/N | |
| 4028-3c | S. anginosus | Penile ulcer | 5061010 | 2 | NG | A/N | |
| 4252c | S. anginosus | Urine | 5061553 | 2,3 | F | A/N | |
| 99M2144c | S. anginosus | Decubitus ulcer | 7063152 | 2,3 | NG | A/N | |
| 1432b | S. anginosus | Abscess, arm | 5061410 | 2 | G | A/N | |
| 1942 | S. anginosus | Ulcer, pelvis | 1060004 | 2 | C | B | |
| 5674 | S. anginosus | Blood | 5061010 | 0 | C | A/N | |
| 3075 | S. anginosus | Blood | 5061400 | 2 | C | A/N | |
| 1238b | S. anginosus | Sacral ulcer | 1061014 | 2 | F | A/N, B | |
| 417-2b | S. anginosus | Peritoneum | 1061410 | 2,3 | F | A/N | |
| 902 | S. intermedius | Abscess, head | 5061410 | 1,2,3 | NG | A/N | |
| Type strain | S. intermedius | ATCC 27335 | 5061010 | 1,2,3 | A/N | ||
| 5672 | S. intermedius | Pleural fluid | 1061410 | 1,2,3 | NG | A/N | |
| 3276 | S. intermedius | Abscess and blood | 1061410 | 1,2,3 | F | A/N | |
| 5671 | S. intermedius | Pleural fluid | 1061410 | 1,2,3 | NG | A/N | |
| 5464 | S. constellatus | Abscess, rectum | 1061014 | 3 | F | B | |
| 1422 | S. constellatus | Abscess, head | 5061014 | 3 | F | B | |
| 5243 | S. constellatus | Fluid, chest wall | 1061014 | 3 | F | B | |
| 5255 | S. constellatus | Abscess, arm | 1061004 | 3 | F | B | |
| 3973 | S. constellatus | Abscess | 1071010 | 3 | NG | B | |
| 611 | S. constellatus | Abscess, mouth | 5061431 | 2,3 | NG | A/N | |
| 4119 | S. constellatus | Tissue, thorax | 5061410 | 3 | NG | A/N | |
| 2192 | S. constellatus | Abscess, arm | 1061010 | 3 | F | A/N | |
| 99B136 | S. constellatus | Blood | 5061014 | 3 | F | B | |
| 5245 | S. constellatus | Inguinal abscess | 5061414 | 1,2,3 | C | B | |
| 3197 | S. constellatus | Blood | 1061414 | 3 | NG | B | |
| 5199 | S. constellatus | Abscess, brain | 5041410 | 3 | NG | A/N | |
| 3868 | S. constellatus | Intracranial abscess | 5061415 | 3 | NG | B | |
| 5220 | S. constellatus | Fluid, neck | 5061010 | 2,3 | NG | A/N | |
| Type strain | S. constellatus | ATCC 27823 | 1061010 | 3 | NG | A/N | |
| 5287 | S. constellatus | Fluid, mandible | 1061014 | 3 | NG | B | |
| 5512 | S. constellatus | Blood | 5061411 | 0 | NG | A/N | |
| 1858 | S. constellatus | Abscess | 1061414/6 | 3 | F | B | |
| 2998 | S. constellatus | Blood | 1061410 | 3 | NG | A/N | |
| 3860 | S. constellatus | Abscess, thorax | 1061414 | 3 | NG | B | |
| 1969 | S. constellatus | Abscess, mouth | ND | 1,2,3 | C | B | |
| 5217 | S. constellatus | Perirectal tissue | 5061414 | 1,2,3 | C | B | |
| 5224 | S. gordonii | 1997 CAP survey strain | 4061430 | 2 | NG | A | S. sanguis |
| 5363 | S. bovis-like | Blood | 5050450 | NG | A | S. bovis | |
| 4337 | S. bovis-like | Abscess, brain | 5050450 | NG | A | S. bovis | |
| 4116 | S. uberis-like | Urethral fluid | 5463315 | NG | B | Streptococcus groups E, P, and U | |
| 1959 | S. dysgalactiae subsp. equisimilis | Abscess, heel | 0463015 | 3 | C | B | Streptococcus group C |
| 5254 | S. dysgalactiae subsp. equisimilis | Skin, trachea | 4463015 | 3 | G | B | Streptococcus group G |
| 1429-1b | S. sanguis-S. cristiatus | Abscess, arm | 0260451 | 3 | NG | A | S. sanguis |
| 2916 | S. sanguis-S. cristiatus | Blood | 5/1001410/1 | 0 | NG | A | Streptococcus sp. |
The strains are listed in the same order in which they are listed in Fig. 1. Abbreviations: B, beta-hemolytic; A/N, alpha-hemolytic or nonhemolytic; NG, nongroupable; ND, not determined; CAP, College of American Pathologists.
These isolates were recovered with other “S. milleri” strains.
Genitourinary isolate.
Table 2 shows the concordance of sequence and biochemical identification for isolates within the “S. milleri” group. All S. intermedius and S. anginosus isolates identified by sequencing were similarly identified by biochemical analysis, but five (23%) of the S. constellatus isolates identified by sequencing were identified as either S. anginosus or S. intermedius by our biochemical analysis scheme. As shown in Table 2, for each of these isolates additional enzyme activity was present (they had a Fluo-card biotype of 2,3 or 1,2,3 instead of one of 3 alone, as expected). Part way through the study, we saw that biotypes 2,3 and 1,2,3 might be identified as S. constellatus by sequencing, although according to the Fluo-card instructions and previous work (6) they should have been identified as S. anginosus or S. intermedius, respectively. We therefore sequenced all isolates with those biotypes. We also selected isolates with unusual phenotypes for sequencing, e.g., all Lancefield group G isolates. Overall, 5 of 40 (13%) of the isolates were incorrectly identified and an additional 3 of 40 (7%) of the isolates were unidentified (the isolates did not cleave any of the substrates, and the biotype was recorded as 0). Because all of the sequenced organisms with biotype 3 were S. constellatus and those with biotype 2 were S. anginosus, we assigned 56 additional “S. milleri” group strains with these biotypes accordingly. Three other strains with biotype 1 were assigned to S. intermedius. Table 3 summarizes the phenotypic characteristics associated with the isolates identified by sequencing and all isolates. The results of CFA analysis and Lancefield typing are similar for the same species identified by either scheme. S. anginosus is the only species that exhibited Lancefield type G or A.
TABLE 2.
Correspondence of cluster by sequencing and Fluo-card identification for selected Voges-Proskauer- and arginine-positive and sorbitol-negative streptococci
| Cluster by sequencing | No. of isolates with the following Fluo-card identification (biotype):
|
||||
|---|---|---|---|---|---|
| S. intermedius (1,2,3) | S. constellatus (3) |
S. anginosus
|
None (0) | ||
| 2,3 | 2 | ||||
| S. intermedius | 5 | ||||
| S. constellatus | 3 | 16 | 2 | 1 | |
| S. anginosus | 7 | 7 | 2 | ||
TABLE 3.
Summary of genotypic and phenotypic characterizationa
| Group (no. of strains) | % Isolates exhibiting the following characteristic:
|
||||||
|---|---|---|---|---|---|---|---|
| Correct Fluo-card ID (% with no ID) | CFA group 7 | CFA group 6 or 9 | Lancefield group
|
||||
| F | C | U | G | ||||
| Sequencing defined S. intermedius (6) | 100 | 0 | 100 | ||||
| All S. intermedius isolates (9) | 100d | 0 | 100 | 25 | 0 | 75 | 0 |
| Sequencing-defined S. constellatus (22) | 72 (5) | 12 | 83 | 32 | 14 | 55 | 0 |
| All S. constellatus isolates (56) | 89 (2)e | 6 | 92 | 36 | 14 | 50 | 0 |
| Sequencing-defined S. anginosus (17) | 94 (6) | 75b | 13 | 53 | 18 | 18 | 12 |
| All S. anginosus isolates (33) | 97 (3)e | 89 | 6 | 44 | 26f | 24 | 6 |
| Non-“S. milleri” isolates (10) | 40g | 0 | 38 | 0 | 20 | 60 | 20 |
| % Isolates exhibiting the following characteristic: | |||||||
| Beta-hemolyticb | Esculin/lactose positiveb | Colony dry, adherentb | Similar rep-PCR pattern (no. of isolates tested) | Blood isolatesc | Upper body abscess, respiratoryd | Lower body abscessd | GI/GU, decubitus ulcerd |
| 11 | 13/100 | 75 | 50 (4) | 11 | 100 | 0 | 0 |
| 64 | 48/52 | ||||||
| 69 | 38/36 | 26 | 82 (22) | 6 | 82 | 16 | 3 |
| 12 | 87/67 | ||||||
| 15 | 75/79 | 0 | 56 (16) | 41 | 20 | 7 | 73 |
| 0 (6) | |||||||
Abbreviations: ID, identification; GI, gastrointestinal; GU, genitourinary.
Clinical strains only.
Percent of total isolates.
Percent of isolates excluding blood.
Assumes that all the unsequenced isolates of Fluo-card biotype 3 are S. constellatus, that all unsequenced isolates of Fluo-card biotype 2 are S. anginosus, and that all unsequenced isolates of Fluo-card biotypes 1 and 1,3 are S. intermedius.
One Lancefield group C isolate was also positive for group A.
These organisms would have been incorrectly identified as one of the “S. milleri” group species with the Fluo-card if they had not been properly screened first.
Other characteristics in Table 3 are listed for all isolates because of the selection of unusual biotypes for sequencing. S. constellatus isolates were often associated with beta-hemolysis, S. anginosus isolates were often associated with esculin and lactose activity and lack of beta-hemolysis, and S. constellatus and S. intermedius but not S. anginosus might have dry colonies. An important point in Table 3 is the association of specimen type or site of isolation with species. Of our S. anginosus isolates, 43% were from blood, whereas proportionately fewer S. constellatus and S. intermedius isolates were from blood. When we examine the non-blood isolates, most of the S. constellatus and S. intermedius isolates are associated with upper body abscesses or respiratory specimens (82 and 100%, respectively), whereas most S. anginosus are associated with the gastrointestinal or genitourinary tract or lower body abscesses (80%).
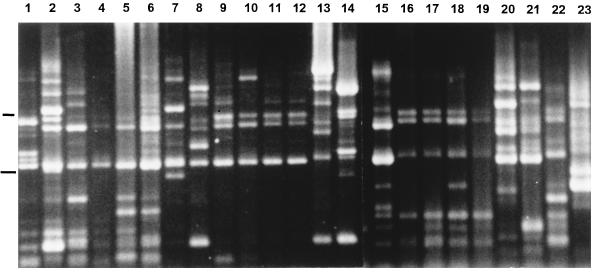
On occasion (8% of specimens), more than one “S. milleri” group phenotype or species was isolated from the same specimen. Two species were isolated from each of three specimens and two hemolytic phenotypes of the same species were isolated from each of two specimens. The sequence data in Fig. 1 show that beta-hemolytic and alpha-hemolytic S. anginosus strains isolated from the same patient (strains 417-1 and 417-2) are different. Figure 2 shows that two strains (one beta-hemolytic strain and one alpha-hemolytic strain) of S. constellatus isolated from the same patient (Fig. 2, lanes 8 and 9) have different rep-PCR patterns.
FIG. 2.
Two agarose gels (combined) showing the PCR products from rep-PCR amplification. Lanes 1 to 12, 16 to 19, and 22, S. constellatus; lanes 15, and 20, and 21, S. anginosus; lanes 13 and 14, S. intermedius. Lower and upper markers, 1 and 2 kb, respectively.
The CFA analysis results were very sensitive to growth conditions. For example, we performed multiple analyses with 15 isolates. If the isolates were grown on the same medium and under the same atmospheric conditions, the runs were reproducible. However, the runs did not match if one subculture was grown on Trypticase soy agar with blood under a CO2 atmosphere and the other was grown anaerobically on Columbia agar. A dendrogram based on CFA analysis of a subset of 85 streptococcal isolates grown anaerobically on Columbia agar or Columbia CNA was made (data not shown). An arbitrary numeric was assigned to each cluster. The number of the cluster(s) associated with each species is shown in Table 3. Most S. constellatus and S. intermedius isolates were found in clusters 5, 6, and 9, while most S. anginosus isolates were found in cluster 7. It is noteworthy that the S. anginosus strains from ATCC and the strains with distinctive colony morphologies (e.g., very mucoid colonies) did not fall in these groups. Table 4 shows that the three species have similar CFA profiles, with S. anginosus being most distinct in the percent 14:0 CFAs. The MIDI database usually assigned the names S. anginosus, S. pyogenes, and S. sanguis, in decreasing probabilities.
TABLE 4.
Major CFAs
| Species | % of total CFAs
|
|||||
|---|---|---|---|---|---|---|
| 14:0 | 16:0 | 18:2 ω6,9c/18:0 ante | 18:1 ω9c | 18:1 ω7c/ω9t/ω12t | 18:0 | |
| S. constellatus | 6–13 | 34–43 | 9–15 | 9–20 | 4–12 | 6–14 |
| S. intermedius | 6–8 | 34–39 | 8–12 | 9–12 | 11–16 | 12–13 |
| S. anginosus | 13–20 | 36–42 | 7–10 | 7–13 | 7–11 | 5–9 |
The rep-PCR analysis was not very useful. Figure 2 shows two gels (combined) with results for representative strains. Although almost none of the strains have exactly the same pattern, some of the S. constellatus (e.g., lanes 9 to 12 and 16 to 19) and S. anginosus (lanes 20 and 21) isolates were similar. The percentage of strains with the predominant pattern is summarized in Table 3.
DISCUSSION
Our sequence data separate the “S. milleri” group into three species, with S. constellatus and S. intermedius being most closely related (distance matrix, 1.7%) and S. anginosus being farther away (distance matrix, 6.8%). This confirms the relationship found by some investigators (3, 11, 14) but not by others (7, 17). In addition, our sequences clustered but did not distinguish at the species level a subgroup of urinary tract isolates of S. anginosus, some of which were biochemically more active (e.g., they fermented raffinose) and which may correspond to the isolates described as separate species by Bergman et al. (3). A subgroup of S. constellatus Lancefield group C isolates may also be associated with gastrointestinal specimens.
We showed good agreement between the identification obtained by sequencing and that obtained with the Fluo-card and API Strep system combined for S. anginosus and S. intermedius (100% for those strains that utilize the sugars in the Fluo-card). However, in contrast to Flynn and Ruoff (6), we found discrepancies when identifying S. constellatus. All of the S. constellatus isolates possessed α-d-glucosidase activity, as expected, but 23% of those selected for sequencing had additional activity which led to a lack of agreement of results. Because of these discrepant results, overall agreement was 91%. The strains with discrepant results are similar to those characterized as intermediate between S. constellatus and S. intermedius (19). If the Fluo-card interpretation criteria were changed so that isolates of Fluo-card biotype 1,2,3 which are both beta-hemolytic and Lancefield group C were identified as S. constellatus, then the overall discrepancies would drop to 2%. Identification with a 32-well API strip not commercially available in the United States may also hold potential (1, 10).
The reported frequency of isolation of each of the species within the “S. milleri” group is variable and seems to be at least partially a function of how the isolates were selected and how carefully they were identified. In most of the series in which species have been assigned to the “S. milleri” group, S. anginosus is the most common isolate (1, 2, 6, 12, 13, 18). S. constellatus could be the most common isolate in our study because of our population (essentially no women or children and few genitourinary tract specimens), because we may have inadvertently selected for the presence of beta-hemolysis, and/or because fewer blood isolates were examined. All of these factors would decrease the proportion of S. anginosus isolates. In addition, if our data can be extended to other populations, some S. constellatus isolates would be misidentified as S. anginosus or S. intermedius. Our data confirmed the predominance of S. anginosus in the blood and S. constellatus and S. intermedius in abscesses (6, 12) and the distributions of hemolysis and Lancefield type among species (13). There was a strong association of S. anginosus with a gastrointestinal or urogenital source and a strong association of S. constellatus and S. intermedius with specimens from both upper body abscesses and the respiratory tract.
The greater understanding achieved by correlating the 16S rRNA gene sequence genotypic grouping with the source of isolation and careful phenotypic description has promoted a better recognition of the niche of S. constellatus, S. intermedius, and S. anginosus and should ultimately clarify their clinical significance.
REFERENCES
- 1.Ahmet Z, Warren M, Houang E T. Species identification of the Streptococcus milleri group isolated from the vagina by ID 32 Strep System and differential phenotypic characteristics. J Clin Microbiol. 1995;33:1592–1595. doi: 10.1128/jcm.33.6.1592-1595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantar C, Fernandez Canigia L, Relloso S, Lanza A, Bianchini H, Smayevsky J. Species belonging to the “Streptococcus milleri” group: antimicrobial susceptibility and comparative prevalence in significant clinical specimens. J Clin Microbiol. 1996;34:2020–2022. doi: 10.1128/jcm.34.8.2020-2022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman S, Selig M, Collins M D, Farrow J A E, Baron E J, Dickersin G R, Ruoff K L. “Streptococcus milleri” strains displaying a gliding type of motility. Int J Syst Bacteriol. 1995;45:235–239. doi: 10.1099/00207713-45-2-235. [DOI] [PubMed] [Google Scholar]
- 4.Clarridge J E, III, Raich T J, Pirwani D, Simon B B, Tsai L, Rodriguez M, Regnery R, Zollo A, Rambo C. A strategy to detect Bartonella infections in a routine clinical laboratory yields Bartonella henselae from a human immunodeficiency virus-positive patient and a unique Bartonella sp. from his cat. J Clin Microbiol. 1995;33:2107–2113. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coykendall A L, Wesbecher P M, Gustafson K B. Streptococcus milleri, Streptococcus constellatus, and Streptococcus intermedius are later synonyms of Streptococcus anginosus. Int J Syst Bacteriol. 1987;37:222–228. [Google Scholar]
- 6.Flynn C, Ruoff K. Identification of “Streptococcus milleri” group isolates to the species level with a commercially available rapid test system. J Clin Microbiol. 1995;33:2704–2708. doi: 10.1128/jcm.33.10.2704-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnier F, Gerbaud G, Courvalin P, Galimand M. Identification of clinically relevant viridans group streptococci to the species level by PCR. J Clin Microbiol. 1997;35:2337–2341. doi: 10.1128/jcm.35.9.2337-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Garces Jose-Luis, Alos J, Cogollos R. Bacteriologic characteristics and antimicrobial susceptibility of 70 clinically significant isolates of Streptococcus milleri group. Diagn Microbiol Infect Dis. 1994;19:69–73. doi: 10.1016/0732-8893(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 9.Hinnebusch C J, Nikolai D M, Bruckner D A. Comparison of API Rapid STREP, Baxter Microscan Rapid Pos ID panel, BBL Minitek Differential Identification system, IDS RapID STR system, and Vitek GPI to conventional biochemical tests for identification of viridans streptococci. Am J Clin Pathol. 1991;45:459–463. doi: 10.1093/ajcp/96.4.459. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs J A, Stobberignh E E. Species identification of “Streptococcus milleri” with the Rapid ID 32 Strep system. Med Microbiol Lett. 1994;3:315–322. [Google Scholar]
- 11.Jacobs J A, Schot C S, Bunschoten A E, Schouls L M. Rapid species identification of Streptococcus milleri strains by line blot hybridization: identification of a distinct 16S rRNA population closely related to Streptococcus constellatus. J Clin Microbiol. 1996;34:1717–1721. doi: 10.1128/jcm.34.7.1717-1721.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs J A, Pietersen H G, Stobbringh E E, Soeters P B. Bacteremia involving the Streptococcus milleri group: analysis of 19 cases. Clin Infect Dis. 1994;19:704–713. doi: 10.1093/clinids/19.4.704. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs J A, Pietersen H G, Stobbringh E E, Soeters P B. Streptococcus anginosus, Streptococcus constellatus, and Streptococcus intermedius: clinical relevance, hemolytic, and serologic characteristics. Am J Clin Pathol. 1995;104:547–553. doi: 10.1093/ajcp/104.5.547. [DOI] [PubMed] [Google Scholar]
- 14.Knight R G, Shlaes D M. Physiological characteristics and deoxyribonucleic acid relatedness of Streptococcus intermedius strains. Int J Syst Bacteriol. 1988;38:19–24. [Google Scholar]
- 15.Ruoff K L. Streptococcus anginosus (“Streptococcus milleri”): the unrecognized pathogen. Clin Microbiol Rev. 1988;1:102–108. doi: 10.1128/cmr.1.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiley R A, Beighton D. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus angiosus as distinct species. Int J Syst Bacteriol. 1991;41:1–5. doi: 10.1099/00207713-41-1-1. [DOI] [PubMed] [Google Scholar]
- 17.Whiley R A, Duke B, Hardie J M, Hall L M C. Heterogeneity among 16S-23S rRNA intergenic spacers of species within the Streptococcus milleri group. Microbiology. 1995;141:1461–1467. doi: 10.1099/13500872-141-6-1461. [DOI] [PubMed] [Google Scholar]
- 18.Whiley R A, Beighton D, Winstanley T G, Fraser H Y, Hardie J M. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J Clin Microbiol. 1992;30:243–244. doi: 10.1128/jcm.30.1.243-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whiley R A, Frazer H, Hardie J M, Beighton D. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the Streptococcus milleri group. J Clin Microbiol. 1990;28:1497–1501. doi: 10.1128/jcm.28.7.1497-1501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]