Abstract
We developed a scheme for the rapid identification of Mycobacterium species based upon PCR amplification of polymorphic genetic regions with fluorescent primers followed by restriction and analysis by fluorescence capillary electrophoresis. Mycobacterium species were identified by restriction enzyme analysis of a 439-bp segment of the 65-kDa heat shock protein gene (labeled [both strands] at the 5′ end with 4,7,2′,7′-tetrachloro-6-carboxyfluorescein) using HaeIII and BstEII and of a 475-bp hypervariable region of the 16S rRNA gene (labeled [both strands] at the 5′ end with 6-carboxyfluorescein) using HaeIII and CfoI. Samples were analyzed on an automated fluorescence capillary electrophoresis instrument, and labeled fragments were sized by comparison with an internal standard. DNA templates were prepared with pure cultures of type strains. In all, we analyzed 180 strains, representing 22 Mycobacterium species, and obtained distinctive restriction fragment length polymorphism (RFLP) patterns for 19 species. Three members of the Mycobacterium tuberculosis complex had a common RFLP pattern. A computerized algorithm which eliminates subjectivity from pattern interpretation and which is capable of identifying the species within a sample was developed. The convenience and short preparatory time of this assay make it comparable to conventional methodologies such as high-performance liquid chromatography and hybridization assays for identification of mycobacteria.
Public health and clinical laboratories play critical roles in the control of tuberculosis through timely detection, species identification, and drug susceptibility testing to ensure adequate and appropriate treatment. Delays in the diagnosis of tuberculosis seriously impact both patients and tuberculosis control programs. In recent years nucleic acid amplification techniques capable of detecting small numbers of mycobacteria have been proposed for the rapid diagnosis of tuberculosis (14). Several mycobacterial target genes have been investigated with assay systems including commercial kits, which can identify single or multiple mycobacterial species (3, 4, 7, 10, 15, 16). Sensitive and rapid techniques for detecting and identifying Mycobacterium tuberculosis and the nontuberculous mycobacteria targeting polymorphisms in the 65-kDa heat shock protein (HSP) and 16S rRNA genes are among these applications (9, 11, 12, 14–16).
Restriction fragment length polymorphism (RFLP) analysis of DNA amplimers generated by PCR for the identification of mycobacteria provides a rapid means for detecting these species-specific polymorphisms (7, 8, 12, 17). In this method, a gene or gene segment is amplified, the product is cleaved with restriction endonucleases, and the fragments are analyzed electrophoretically, e.g., by agarose or polyacrylamide slab gel or capillary electrophoresis. We developed an assay using PCR amplification with fluorescent primers targeting polymorphic regions of the 65-kDa HSP gene and the 16S rRNA gene followed by restriction digestion and automated fluorescence capillary electrophoresis (FCE). A numerical algorithm was developed to identify 19 Mycobacterium species and the M. tuberculosis complex (MTC).
MATERIALS AND METHODS
Mycobacterium cultures and growth conditions.
We examined 22 Mycobacterium species in this study. Strains were obtained from the American Type Culture Collection, the former Trudeau Mycobacterial Culture (TMC) collection, and the Centers for Disease Control and Prevention (CDC) mycobacterial stock collection (Table 1). The CDC strains and additional isolates were identified in our laboratory. Each strain was cultured at 37°C in complete Middlebrook 7H9 broth (Remel Co., Lenexa, Kan.) until stationary growth phase was achieved or on Lowenstein-Jensen slants (Remel). The identification of all strains was confirmed by high-performance liquid chromatography (HPLC) analysis of their mycolic acids (2, 6). Nonmycobacterial species included Corynebacterium diphtheriae, Tsukamurella sp., Corynebacterium pseudotuberculosis, Nocardia brasiliensis, and Gordona sputi (3).
TABLE 1.
Mycobacterium species tested
| Species | No. of isolates tested | Reference strainsa |
|---|---|---|
| M. tuberculosis | 16 | TMC 119 (ATCC 35810) |
| M. bovis | 10 | TMC 412 (ATCC 35726) |
| M. bovis BCG (BCG Tice) | 10 | TMC 1028 (ATCC 35743) |
| M. africanum | 2 | CDC72-1432 |
| M. microti | 2 | TMC 1608 (ATCC 35782) |
| M. avium | 10 | TMC 716 (ATCC 35717) |
| M. intracellulare | 3 | TMC 1469 (ATCC 35772) |
| M. simiae | 7 | TMC 1226 (ATCC 25275) |
| M. gordonae | 13 | TMC 1324 (ATCC 14470) |
| M. kansasii | 10 | TMC 1214 (ATCC 35777) |
| M. fortuitum | 5 | CDC85-1098 |
| M. peregrinum | 3 | TMC 1547 (ATCC 14467) |
| M. chelonae | 5 | TMC 1524 (ATCC 35749) |
| M. abscessus | 5 | TMC 1542 (ATCC 35751) |
| M. celatum | 9 | ATCC 51131 |
| M. marinum | 10 | 909b |
| M. asiaticum | 8 | CDC88-334 |
| M. mucogenicum | 10 | CDC86-650 |
| M. malmoense | 9 | 01355b |
| M. gastri | 8 | ATCC 25127 |
| M. scrofulaceum | 8 | CDC89-447 |
| M. szulgai | 9 | 954b |
| M. xenopi | 8 | CDC90-TI-723 |
| Total | 180 |
Strain used for development and standardization of the procedure.
Strain provided by W. R. Butler.
DNA extraction and amplifications.
Template DNA was extracted from cultures by agitating cell suspensions with a Mickle cell disrupter (Brinkmann Instruments, Inc., Westbury, N.Y.) in the presence of siliconized glass beads as described previously (13). Extraction of nonmycobacterial nucleic acids was performed as previously described (3). Polymorphic gene regions were amplified with fluorescently labeled oligonucleotide primers. A 439-bp region of the 65-kDa HSP gene (13) was amplified with previously described primers TB11 and TB12 (16) labeled with 4,7,2′,7′-tetrachloro-6-carboxyfluorescein (TET). A hypervariable region of the 16S rRNA gene (475 bp) was amplified with primers BC70 (5′-TAACACATGCAAGTCGAACG-3′) and BC71R (5′-CGTATTACCGCGGCTGCTGG-3′), which were selected with OLIGO, version 5, software (Molecular Biology Insights, Inc., Plymouth, Minn.) and labeled with 6-carboxyfluorescein (FAM). The primers, including the incorporation of 5′ fluorescent tags, were synthesized by ethyl phosphoramidite chemistry at the CDC Biotechnology Core Facility. PCR mixtures contained 1× Taq polymerase buffer containing 2.5 mM MgCl2 (Boehringer Mannheim, Inc., Indianapolis, Ind.), 200 μM concentrations of each of four dNTPs (Perkin-Elmer Applied Biosystems, Inc. [PE-ABI], Foster City, Calif.), 0.5 μM (each) primer, 1 U of Taq polymerase (Boehringer Mannheim), and typically 1 μl of the DNA template in a final volume of 25 μl. Thermocycling of reaction mixtures was performed in a model 9600 thermocycler (PE-ABI) programmed for 35 cycles (30 s at 96°C, 30 s at 61°C, 30 s at 72°C) followed by a 10-min incubation at 72°C. Amplification products were confirmed by electrophoresis on 2% agarose gels followed by staining with ethidium bromide.
Restriction fragment analysis by FCE.
After PCR, each 65-kDa HSP gene amplimer was aliquoted into two tubes and digested with restriction endonucleases HaeIII and BstEII (Roche-Boehringer Mannheim). The 16S rRNA gene amplimers were similarly divided and digested with HaeIII and CfoI (Roche-Boehringer Mannheim). Each digestion reaction mixture consisted of 10 μl of the PCR product, 5 U of enzyme, 2.5 μl of (10×) enzyme buffer, and water to a final reaction volume of 25 μl and was incubated at 37°C (60°C for BstEII) for 1 h. Restriction reactions were inactivated by heating at 95°C for 3 min, and aliquots (5 μl) of each of the four restriction digests were combined into a single tube, diluted 1:50 with deionized water, and stored at −20°C. The samples were prepared for analysis on an FCE instrument (model 310 genetic analyzer; PE-ABI) by combining 1 μl of the diluted sample with 19 μl of deionized formamide and 0.6 μl of TAMARA 500 size standard (PE-ABI). The DNA fragments were denatured at 94°C for 3 min and cooled immediately on ice for 3 min. The samples were electrophoresed through a 47-cm by 50-μm capillary filled with performance-optimized polymer 4C (PE-ABI). Each sample was injected for 5 s and run for 24 min at 15 kV and 60°C.
DNA sequence determination.
Sequences of the 16S rRNA and 65-kDa HSP gene amplimers of all strains were determined by using dye terminator reactions (according to the manufacturer's protocol) followed by electrophoresis on an ABI 373A automated DNA sequencer (PE-ABI). The sequences were analyzed by using DNASIS, version 2.1, for Windows software (Hitachi Corp., San Bruno, Calif.) to determine the locations of restriction sites.
Automated analysis of FCE data.
Algorithms for the identification of species were developed with Excel software, version 7.0, for PC-Windows 95 (Microsoft Corp., Redmond, Wash.) and were based upon sizes (in base pairs) of restriction fragments (Table 2). The presence of FAM- and TET-labeled peaks corresponding to the ends of the restricted polymorphic gene amplimers was considered in the algorithm. Sizes (in base pairs) of the RFLP fragments were compared in the algorithm to the standard values listed in Table 2 (in parentheses). The algorithm was programmed to accept differences of ±4 bp from the standard values. We entered the fragment sizes into the appropriate cells of the Excel spreadsheet, and then the algorithm assigned a species name by using an identification formula according to instructions included with the Excel software. The Excel algorithm assigns the term “other” to patterns not corresponding to the 22 species included in this study.
TABLE 2.
Restriction fragments of polymorphic mycobacterial genes as determined by direct sequencing and FCE
| Species | End fragment size (bp)a
|
|||
|---|---|---|---|---|
| 65-kDa HSP gene
|
16S rRNA gene
|
|||
| BstEII | HaeIII | CfoI | HaeIII | |
| M. tuberculosis | 116 (113), 231 (230) | 127 (126), 152 (150) | 149 (148), 160 (160) | 31 (33), 187 (186) |
| M. bovis | 116 (113), 231 (230) | 127 (126), 152 (150) | 149 (148), 160 (160) | 31 (33), 187 (186) |
| M. bovis BCG | 116 (113), 231 (230) | 127 (126), 152 (150) | 149 (148), 160 (160) | 31 (33), 187 (186) |
| M. africanum | 116 (113), 231 (230) | 127 (126), 152 (150) | 149 (148), 160 (160) | 31 (33), 187 (186) |
| M. microti | 210 (208), 231 (230) | 40 (40), 127 (124) | 138 (139), 146 (147) | 31 (32), 178 (177) |
| M. avium | 210 (208), 231 (230) | 40 (40), 127 (126) | 149 (148), 326 (325) | 31 (32), 178 (179) |
| M. intracellulare | 116 (112), 231 (231) | 40 (41), 127 (126) | 139 (136), 149 (148) | 27 (28), 31 (32) |
| M. simiae | 210 (231), 231 (228) | 126 (123), 185 (182) | 62 (60), 138 (136) | 26 (28), 31 (32) |
| M. gordonae | 116 (113), 231 (229) | 40 (38), 161 (158) | NF | 28 (28), 31 (32) |
| M. kansasii | 210 (207), 231 (229) | 127 (123), 185 (181) | 139 (136), 149 (148) | 31 (32), 178 (177) |
| M. fortuitum | 116 (113), 231 (231) | 41 (43), 145 (142) | 62 (59), 134 (130) | 31 (32), 178 (165) |
| M. peregrinum | 210 (208), 231 (233) | 98 (102), 139 (136) | 62 (59), 134 (130) | 31 (30), 178 (165) |
| M. chelonae | 131 (128), 310 (310) | 40 (44), 197 (195) | 137 (137), 325 (325) | 31 (30), 184 (182) |
| M. abscessus | 210 (207), 231 (232) | 40 (40), 145 (142) | 137 (136), 326 (326) | 31 (31), 185 (183) |
| M. celatum | 210 (208), 231 (231) | 40 (39), 127 (123) | 138 (139), 324 (328) | 31 (30), 176 (180) |
| M. marinum | 210 (208), 231 (231) | 40 (40), 106 (103) | 149 (148), 326 (325) | 31 (32), 178 (178) |
| M. asiaticum | 210 (208), 231 (231) | 40 (40), 106 (103) | 139 (139), 149 (148) | 31 (30), 178 (179) |
| M. mucogenicum | 116 (113), 310 (307) | 40 (36), 145 (141) | 31 (30), 136 (133) | 101 (97), 178 (177) |
| M. malmoense | 116 (119), 231 (229) | 40 (42), 106 (105) | 139 (138), 149 (148) | 31 (30), 178 (177) |
| M. gastri | 131 (127), 231 (229) | 40 (38), 127 (127) | 139 (136), 149 (148) | 31 (31), 178 (178) |
| M. scrofulaceum | 210 (206), 231 (230) | 40 (40), 127 (123) | 138 (136), 146 (144) | 26 (29), 31 (33) |
| M. szulgai | NF | 40 (40), 127 (123) | 149 (150), 160 (168) | 27 (28), 31 (32) |
| M. xenopi | 116 (113), 231 (229) | 40 (40), 161 (158) | 133 (135), 139 (143) | 26 (29), 96 (99) |
FCE values are in parentheses. NF, no restriction fragments.
RESULTS
Amplimers for the 65-kDa HSP gene (439 bp) and the 16S rRNA gene (475 bp) were produced for all Mycobacterium species but for none of the nonmycobacterial species. Up to four fluorescent restriction fragments for each polymorphic gene (two fragments per restriction enzyme except for those instances in which the amplimer was not cut due to the absence of restriction sites) were generated for the 65-kDa HSP and 16S rRNA gene amplimers. Among the 22 Mycobacterium species, five patterns were observed when the 65-kDa HSP amplimers were digested with BstEII (Table 2). Specific patterns generated by BstEII were observed for three species: M. mucogenicum (116- and 310-bp fragments), M. gastri (131 and 231 bp), and M. chelonae (131 and 310 bp). No BstEII digestion fragments were obtained for M. szulgai. Two BstEII patterns, fragments of 116 plus 231 bp and 210 plus 230 bp, were common among the remaining 19 species. The HaeIII digestions of the 65-kDa HSP gene amplimers produced eight different patterns (Table 2). One pattern (126- plus 150-bp fragments) was common to the MTC species M. tuberculosis, M. bovis (including the M. bovis BCG strains), and M. africanum, but not to M. microti. Although M. microti is included in the complex, its HaeIII pattern (40 plus 126 bp) was shared with M. avium and M. intracellulare. Of the remaining six patterns, two were unique to a single species and four were observed in groups of between two and seven species each. The CfoI digests of the 16S rRNA gene amplimer produced patterns unique for M. mucogenicum (31- plus 136-bp fragments) and M. xenopi (133 plus 139 bp). No CfoI fragments were observed for M. gordonae, and the remaining patterns were observed among groups of between two and seven species each. Six different patterns resulting from HaeIII digestion of the 16S rRNA gene amplimer were observed among the 22 species. Unique patterns were observed for M. xenopi (26- plus 96-bp fragments), M. mucogenicum (101 plus 178 bp), and the MTC species except M. microti (31 plus 187 bp). The three remaining patterns were observed in 2 to 10 species each.
When fragment sizes obtained by FCE analysis were compared with those determined by DNA sequence analysis, it was found that differences ranged from 1 to 4 bp for 180 (98%) of the fragments (except for one HaeIII fragment each for M. fortuitum and M. peregrinum) (Table 2). The average difference in sizes determined by FCE compared with those determined by sequence analyses was 3 bp. Each strain listed in Table 1 was evaluated by FCE analysis at least twice, and peak sizes varied by no more than 2 bp on any successive run. Fluorescence RFLP chromatograms for six species are shown in Fig. 1. Likewise, peak sizes for all isolates of each species varied by no more than 2 bp, and all 180 isolates were correctly identified when fragment sizes were entered into the algorithm. The algorithm was programmed to accept the variations in fragment sizes within these limits. Although six and nine species could be identified after digestion of the 65-kDa HSP and 16S rRNA gene amplimers, respectively, by a single enzyme, identification of all species required analysis of all eight fragment sizes. The order of entry of the four fragment sizes into fields of the Excel algorithm for each of the polymorphic genes was by ascending size (i.e., smallest to largest fragments). The use of two fluorescent labels allowed identification of restriction fragments of each of the two polymorphic gene amplimers since digests were run simultaneously.
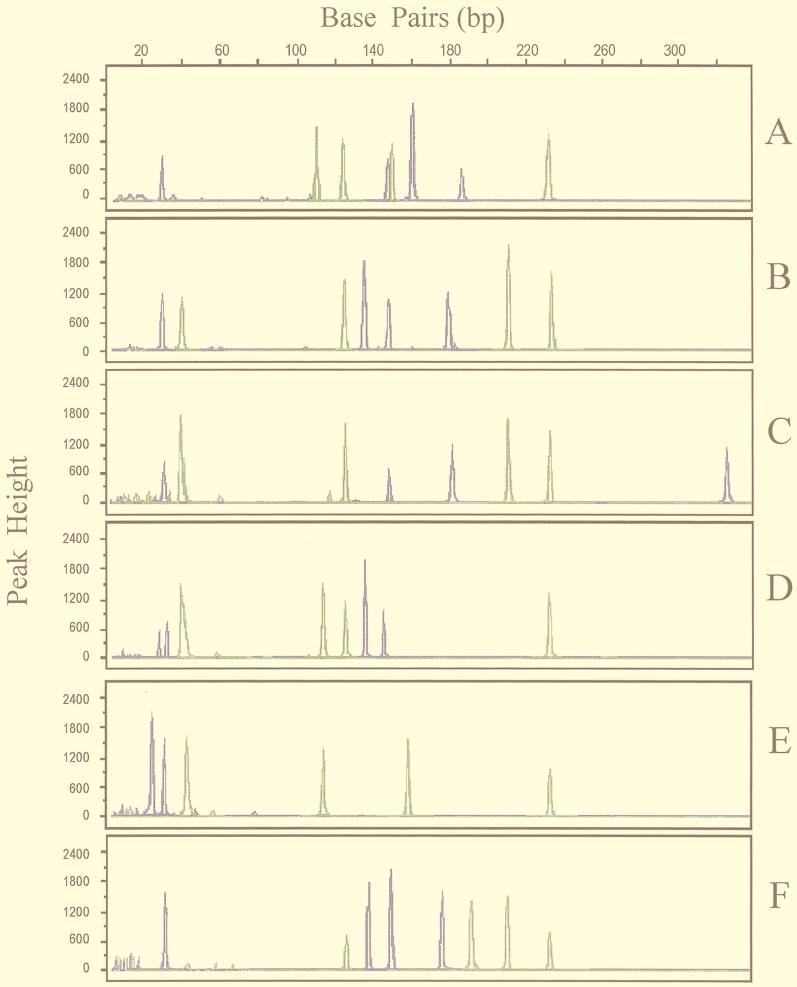
FIG. 1.
Analysis of PCR-RFLP patterns of 65-kDa HSP and 16S rRNA genes of six Mycobacterium species using FCE. Terminal BstEII and HaeIII restriction fragments of the 439-bp 65-kDa HSP gene amplimer are labeled with TET (green), and CfoI and HaeIII fragments of the 475-bp 16S rRNA gene amplimer are labeled with FAM (blue). A, M. tuberculosis; B, M. microti; C, M. avium; D, M. intracellulare; E, M. gordonae; F, M. kansasii.
DISCUSSION
These data indicate that at least 19 Mycobacterium species and the MTC can be identified by fluorescence RFLP analysis. The eight data points obtained after amplification, restriction, and electrophoresis of the 65-kDa HSP and 16S rRNA genes can be identified by GeneScan analysis and may be entered into the algorithm manually or by direct computer downloading. We have recently performed additional evaluations using polymicrobic samples containing various mixtures of the 22 species, as well as species other than these, and the “other” designation has been assigned in every instance (data not shown). Although automated fluorescence sequence analysis of the polymorphic genes provides the most definitive genotypic data to differentiate Mycobacterium species or complexes, the method is considerably more expensive and time-consuming than fluorescence PCR-RFLP (15). There was good overall agreement among fragment sizes determined by FCE and sequence analysis, and acceptable size ranges for each data point were adjusted within the algorithm accordingly.
In laboratories that do not have an FCE instrument (e.g., PE-ABI model 310), the samples may be analyzed on vertical polyacrylamide gel electrophoresis (PAGE) gels. Under these circumstances, electrophoresis, especially for restriction digests of the polymorphic gene amplimers, should be performed in a standardized fashion, e.g., by using the same PAGE gel formulation and electrophoretic conditions. These gels may be analyzed by fluorescence scanners, which should yield banding patterns like those obtained by FCE, or by staining with ethidium bromide, which will reveal the total banding pattern for the restricted gene amplimers. We maintain transparent overlays of banding patterns for each species to facilitate identification of samples when the latter method is performed. Telenti et al. (16) previously showed that restriction analysis of the 65-kDa HSP gene amplimer alone with BstEII and HaeIII provided sufficient information to identify most clinically important Mycobacterium species and subspecies. However, we often encountered difficulty in interpreting patterns by total-band restriction endonuclease analysis of the 65-kDa HSP gene only when the analysis was performed with PAGE gels. This difficulty is due to similarities in some band sizes that are critical for discrimination of species and to the presence of nonspecific gel bands, such as those for partially digested fragments. In the first automated analysis of labeled end fragments to identify Mycobacterium species, Avaniss-Aghajani et al. (1) used five restriction enzymes to digest amplimers of the 16S rRNA gene hypervariable region that were fluorescently labeled at one end. The single-band fluorescent products were detected in polyacrylamide slab gels and sized by GeneScan analysis. Although this method quantitates the sizes of restriction fragments, it is somewhat cumbersome since it requires that large polyacrylamide slab gels be poured and that five enzymes be used. Moreover, it does not identify some clinically important mycobacterial species. We found that the most convenient and definitive genotypic method to identify 22 Mycobacterium species was to determine the sizes of the endmost restriction fragments of both polymorphic genes.
We have found it useful to precede this RFLP analysis with similar FCE analyses to identify the presence of insertion element IS6110 (5) or IS1245 (7) when preliminary identification of the MTC or M. avium is necessary. Oligonucleotide primers for these amplifications are labeled with a different fluorescent tag, 4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein. Alternative labeling enables simultaneous FCE examination of the insertion element amplimers along with the polymorphic genes.
The genotypic FCE method for the identification of Mycobacterium species may be a useful alternative to more widely accepted methodologies, particularly HPLC. Although both methods require expensive instrumentation, these instruments can be applied to other tasks in clinical and public health laboratories. Reagent costs and hands-on time requirements are somewhat higher for the genotypic method, related primarily to the performance of PCR, especially PCR involving fluorescent primers, restriction enzymes, and electrophoresis reagents. An advantage is the lack of subjectivity in pattern interpretation. Used with pure mycobacterial cultures, both methods can provide identification results in one work day. Since the genotypic method involves amplification of target DNA, it offers greater sensitivity than HPLC; both targets could be amplified with DNA extracted from as few as 100 bacilli (data not shown). This sensitivity, coupled with the specificity of amplifications for Mycobacterium species, may support the use of the genotypic assay for direct identification of mycobacteria in patient specimens.
REFERENCES
- 1.Avaniss-Aghajani E, Jones K, Holtzman A, Aronson T, Glover N, Boian M, Froman S, Brunk C. Molecular technique for rapid identification of mycobacteria. J Clin Microbiol. 1996;34:98–102. doi: 10.1128/jcm.34.1.98-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler W R, Jost K C, Jr, Kilburn J O. Identification of mycobacteria by high-performance liquid chromatography. J Clin Microbiol. 1991;29:2468–2472. doi: 10.1128/jcm.29.11.2468-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooksey R C, Crawford J T. Detection of the 10 kDa antigen structural gene in mycobacteria by using the polymerase chain reaction. Diagn Microbiol Infect Dis. 1994;18:215–218. doi: 10.1016/0732-8893(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 4.Del Portillo P, Thomas M C, Martinez E, Maranon C, Valladares B, Patarroyo M F, Lopes C M. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J Clin Microbiol. 1996;34:324–328. doi: 10.1128/jcm.34.2.324-328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenach K D, Cave M D, Bates J H, Crawford J T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161:977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 6.Floyd M, Silcox V, Jones W, Butler W, Kilburn J. Separation of Mycobacterium tuberculosis and Mycobacterium bovis by using high-performance liquid chromatography of mycolic acids. J Clin Microbiol. 1992;30:1327–1330. doi: 10.1128/jcm.30.5.1327-1330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janssen K. The polymerase chain reaction. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1994. pp. 15.1.1–15.1.4. [Google Scholar]
- 9.Kox L F F, Jansen H M, Knijper S, Kolk A H J. Multiplex PCR assay for immediate identification of the infecting species in patients with mycobacterial disease. J Clin Microbiol. 1997;35:1492–1498. doi: 10.1128/jcm.35.6.1492-1498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kyriakopoulos A M, Tassios P T, Matsiota-Bernard P, Marinis E, Tsauousidou S, Legakis N J. Characterization to species level of Mycobacterium avium complex strains from human immunodeficiency virus-positive and -negative patients. J Clin Microbiol. 1997;35:3001–3003. doi: 10.1128/jcm.35.11.3001-3003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S, Wall S, Saunders N A. Heminested inverse PCR for IS6110 fingerprinting of Mycobacterium tuberculosis strains. J Clin Microbiol. 1996;34:1686–1690. doi: 10.1128/jcm.34.7.1686-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plikaytis B B, Plikaytis B D, Yakrus M A, Butler W A, Woodley C L, Silcox V A, Shinnick T M. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J Clin Microbiol. 1992;30:1815–1822. doi: 10.1128/jcm.30.7.1815-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plikaytis B B, Gelber R H, Shinnick T M. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J Clin Microbiol. 1990;28:1913–1917. doi: 10.1128/jcm.28.9.1913-1917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinnick T M, Jonas V. Molecular approaches to the diagnosis of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: American Society for Microbiology; 1994. pp. 517–530. [Google Scholar]
- 15.Springer B, Stockman L, Teschner K, Roberts G D, Bottger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Totsch M, Bromomelkamp E, Stucker A, Fille M, Gross R, Wiesner P, Schmid K W, Bocker W, Dockhorn-Dworniczak B. Identification of mycobacteria to the species level by automated restriction enzyme fragment length polymorphism analysis. Virchows Arch. 1995;427:85–89. doi: 10.1007/BF00203742. [DOI] [PubMed] [Google Scholar]