Abstract
Purpose
Birdshot chorioretinopathy (BSCR) is strongly associated with HLA-A29. This study was designed to elucidate the genetic modifiers of BSCR in HLA-A29 carriers.
Methods
We sequenced the largest BSCR cohort to date, including 286 cases and 108 HLA-A29–positive controls to determine genome-wide common and rare variant associations. We further typed the HLA alleles of cases and 45,386 HLA-A29 controls of European ancestry to identify HLA alleles that associate with BSCR risk.
Results
Carrying a second allele that belongs to the HLA-Aw19 broad antigen family (including HLA-A29, -A30, -A31, and -A33) increases the risk for BSCR (odds ratio [OR] = 4.44; P = 2.2e-03). This result was validated by comparing allele frequencies to large HLA-A29-controlled cohorts (n = 45,386; OR > 2.5; P < 1.3e-06). We also confirm that ERAP1 and ERAP2 haplotypes modulate disease risk. A meta-analysis with an independent dataset confirmed that ERAP1 and ERAP2 haplotypes modulate the risk for disease at a genome-wide significant level: ERAP1-rs27432 (OR = 2.46; 95% confidence interval [CI], 1.85–3.26; P = 4.07e-10), an expression quantitative trait locus (eQTL) decreasing ERAP1 expression; and ERAP2-rs10044354 (OR = 1.95; 95% CI, 1.55–2.44; P = 6.2e-09), an eQTL increasing ERAP2 expression. Furthermore, ERAP2-rs2248374 that disrupts ERAP2 expression is protective (OR = 0.56; 95% CI, 0.45–0.70; P = 2.39e-07). BSCR risk is additively increased when combining ERAP1/ERAP2 risk genotypes with two copies of HLA-Aw19 alleles (OR = 13.53; 95% CI, 3.79–54.77; P = 1.17e-05).
Conclusions
The genetic factors increasing BSCR risk demonstrate a pattern of increased processing, as well as increased presentation of ERAP2-specific peptides. This suggests a mechanism in which exceeding a peptide presentation threshold activates the immune response in choroids of A29 carriers.
Keywords: birdshot, ERAP1, HLA-Aw19, HLA-A33, ERAP2
Birdshot chorioretinopathy (BSCR) is a rare bilateral posterior uveitis affecting individuals of European descent with a mean age of 53 years at the time of diagnosis, characterized by a mild vitritis and ovoid cream-colored lesions at the level of the choroid.1–4 The retinal inflammation can include vasculitis, papillitis, and macular edema. Greater than 95% of BSCR patients carry the HLA-A29 allele, corresponding to an odds ratio (OR) of 157.5, the strongest known HLA class I association with any disease.5–9 Indeed, some investigators believe that the diagnosis of the disease can only be made in HLA-A29 carriers.10 Histologic analyses of eyes with BSCR have revealed nongranulomatous nodular infiltrations of the choroid and lymphocytic infiltrates.11,12 BSCR can result in a gradual and severe loss of vision and is usually treated by immunosuppressants or immunomodulating biological therapies.
Despite the association with an HLA class-I protein, expressed on most nucleated cells, the pathology of BSCR is restricted to the posterior ocular tissues. Additionally, the number of estimated BSCR cases in the United States (5000–10,000) is far lower than the HLA-A29 prevalence (7%) in European populations,5,9 suggesting that HLA-A29 is necessary but not sufficient to cause disease and that other factors contribute to the development of BSCR. Previous work identified risk haplotypes and polymorphisms in both ERAP1 (rs2287987, Hap10) and ERAP2 (rs10044354, HapA) that are more prevalent in patients with BSCR relative to healthy controls.8,13 ERAP1 and ERAP2 are endoplasmic reticulum aminopeptidases with complementary substrate preferences that trim peptides to be loaded onto, and presented by, HLA class I proteins.14 Furthermore, expression of ERAP1 and ERAP2 is coordinated, and when ERAP1 expression is decreased ERAP2 expression is increased.15 Alterations in the expression or enzymatic activity levels of ERAP1 and ERAP2 have been shown to alter the peptidome available for presentation by HLA class I proteins.16,17 The ERAP1 risk-associated Hap10 polymorphism is associated with lower expression, as well as reduced peptide trimming activity, of the ERAP1 protein.14 The ERAP2 risk-associated HapA polymorphism produces a full-length functional ERAP2 protein, whereas the protective HapB polymorphism produces no detectable full-length ERAP2 protein.14,18 The combination of a hypoactive ERAP1 and an available ERAP2 is therefore hypothesized to lead to a unique peptidome pool available for presentation by HLA class I proteins. Variation in the canonical peptide motifs in the peptide-binding groove of the HLA class I molecules16,19 further contributes to shaping the peptidome present on the cell surface for immune cell interrogation, recognition, and subsequent immune activation in BSCR.
Here, we study the largest BSCR cohort, to our knowledge, consisting of 286 cases and 108 HLA-A29 controls of European ancestry obtained by the University of Paris (UParis cohort). We investigated the effect of other HLA-A alleles in an HLA-A29–controlled cohort and identified a novel independent risk associated with most members of the Aw19 serotype family (HLA-A29 homozygous, -A30, -A31, or -A33), whereas one member, HLA-A32, is depleted in BSCR patients compared with A29-positive controls. Our analyses also confirmed the BSCR risk associations with both ERAP1 Hap10 and ERAP2 HapA haplotypes. Our results shed light on the underlying mechanisms that support activation of the immune response in A29-positive individuals and resulting in BSCR.
Methods
Study Subjects and Samples
The genomic DNA samples from 286 patients with BSCR and 108 unrelated healthy local French volunteers that exhibited HLA tissue typing common in the French population were included in this study. The patients were recruited at Hôpital Cochin, Paris, France. All patients met the criteria for diagnosis of BSCR as defined both by an international consensus conference held in 2002 and by the Standardization of Uveitis Nomenclature (SUN) Working Group.6,20 In brief, all patients had a posterior bilateral uveitis with multifocal cream-colored or yellow-orange, oval or round choroidal lesions (“birdshot spots”). Although the presence of the HLA-A*29 allele was not a requirement for the diagnosis of BSCR according to the international criteria, all patients included in the current study carried the HLA-A*29 allele. The control DNA samples were collected from volunteer donors recruited by the hematopoietic stem cell donor center of Rheims for France Greffe de Moelle Registry, and local control healthy individuals of the Registry. The Registry has a stringent selection process, eliminating any chronic disease or condition suggestive of at risk for health problems, such as obesity, alcohol or illicit substance use, diabetes, or high blood pressure. The DNA samples were isolated from peripheral blood samples using a standard salting out method or QIAamp Blood Kit (QIAGEN, Chatsworth, CA, USA). The quality and quantity of DNA were determined by ultraviolet spectrophotometry, and the concentration was adjusted to 100 ng/mL. Signed informed consent documentation was obtained from all participants, and all research adhered to the tenets set forth in the Declaration of Helsinki. All study-related data acquisitions were approved by the Paris Cochin institutional review board.
Genetic Data
We took a comprehensive approach to both sequence the exomes and genotype all samples, to allow for identification of common and rare variants filtered based on high-quality calls. DNA from participants was genotyped on an Illumina Global Screening Array (GSA; Illumina, Inc., San Diego, CA, USA) and imputed to the Haplotype Reference Consortium (HRC) reference panel. Prior to imputation, we retained variants that had a minor allele frequency (MAF) > = 0.1%, missingness < 1%, and Hardy–Weinberg Equilibrium P > 10−15. Imputation using the HRC reference panel yielded 8,385,561 variants with imputation INFO > 0.3 and MAF > 0.5%.
Exome sequencing was performed to a mean depth of 31×, followed by variant calling and quality control as reported previously,21 resulting in 238,942 variants. When integrated, this produced an overall dataset with 8,459,907 variants: 65.5% common (MAF > 5%), 34.5% low-frequency (0.5% < MAF < 5%), and 0.01% rare (MAF < 0.5%).
HLA Genotyping
HLA class I genes (HLA-A, -B, and -C) were amplified in a multiplex PCR reaction with primers encompassing the full genomic loci for each target. The resulting amplicons were enzymatically fragmented to an average size of 250 base pairs and prepared for Illumina sequencing (New England Biolabs, Ipswich, MA, USA). The libraries were sequenced on the Illumina HiSeq 2500 platform on a rapid-run flow cell using paired-end 125 base pair reads with dual 10-base pair indexes. Upon completion of sequencing, raw data from each Illumina HiSeq run was gathered in local buffer storage and uploaded to the DNAnexus platform22 for automated analysis. The FASTQ-formatted reads were converted from the binary base call (BCL) files and assigned to samples identified by specific barcodes using the bcl2fastq conversion software (Illumina). All the reads in sample-specific FASTQ files were subject to HLA typing analysis using an updated version of the PHLAT program23 with the reference sequences consisting of GRCh38 genomic sequences and HLA type reference sequences in the IPD-IMGT/HLA database 3.30.0.24
In addition, HLA allele imputation was performed following SNP2HLA25 with the T1DGC HLA allele reference panel.26 HRC-imputed genotypes in the extended major histocompatibility complex (MHC) region (chr6:25–35 Mb) were filtered for high INFO score (>0.9) and certainty (maximum genotype probability > 0.8 for all genotyped), in order to increase overlap with the T1DGC reference panel; were re-phased along with chromosome 6 array genotypes using SHAPEIT427; and were imputed using Minimac4.28 HLA allele imputation quality was assessed by examining INFO score versus MAF and imputed versus reference panel MAF.
Genetic Association Analyses
Association analyses in each study were performed using the genome-wide Firth logistic regression test implemented in SAIGE.29,30 In this implementation, Firth's approach is applied when P < 0.05 from a standard logistic regression score test. We included for the genomic relationship matrix (GRM) for SAIGE directly genotyped variants with a MAF >1%, <10% missingness, Hardy–Weinberg equilibrium test P > 10−15, and linkage disequilibrium (LD) pruning (1000 variant windows, 100 variant sliding windows, and r2 < 0.1). The association model included as covariates sex and the first 10 ancestry-informative principal components derived from the GRM dataset. Haplotype analyses were performed using PLINK 1.031 –chap and –hap-assoc and –hap-logistic, and in R. High haplotype imputation and phasing quality was indicated by PLINK –hap-phase maximum likelihood haplotype genotypes’ posterior probabilities all equal to one.
HLA-A Allele Association Analyses
Association of HLA-A alleles was performed as follows: For each sample, we typed both HLA-A alleles as described above. Following HLA allele typing, we removed related samples. For the remaining cohort of 282 cases and 106 controls, we next obtained one HLA-A allele that was not A29 (the “second” allele). For samples carrying two copies of A29, we considered A29 as the second allele. We then subjected the cohort to a Fisher's exact test, in which we tested the association of each allele that was identified in three or more BSCR cases, with the case–control status. The threshold of significance for this analysis, considering multiple testing of 14 alleles that are available in three or more cases, was set at P = 0.05/14 = 3.57e-03. To answer the question of whether the A19 allele group was also associated with the case–control status, we combined and tested together the samples in two different ways: (1) carrying all Aw19 alleles (A29, A30, A31, A32, and A33). (2) Because A32 is biologically different than the other Aw19 alleles in its peptide binding domain,32 we also constructed and tested a group made of samples carrying all Aw19 alleles excluding A32. The final ORs and P values are presented in Table 1.
Table 1.
HLA-A Second Allele Frequencies in the French Cohort Compared to UKB and GHS EUR A29 Carriers
| UParis (A29 EUR Carriers) | GHS Cohort 1 (A29 EUR) | GHS Cohort 2 (A29 EUR) | UKB (A29 EUR) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Second HLA-A Allele | Cases (N = 286), n (Frequency) | Controls (N = 108), n (Frequency) | OR (LCI–UCI) | P | Carriers (N = 4014), n (Frequency) | OR (LCI–UCI) | P | Carriers (N = 2829), n (Frequency) | OR (LCI–UCI) | P | Carriers (N = 38,543), n (Frequency) | OR (LCI–UCI) | P |
| Aw19 co-susceptible (29, 30, 31, 33) | 41 (0.14) | 4 (0.04) | 4.44 (1.55–17.53) | 2.20E-03 | 247 (0.06) | 2.63 (1.80–3.78) | 1.29E-06 | 164 (0.06) | 2.69 (1.81–3.92) | 1.07E-06 | 2416 (0.06) | 2.51 (1.75–3.52) | 9.62E-07 |
| A3002 | 13 (0.05) | 1 (0.01) | 5.19 (0.76–222.84) | 0.12 | 45 (0.01) | 4.31 (2.11–8.26) | 6.15E-05 | 20 (0.01) | 6.60 (2.98–14.13) | 3.03E-06 | 396 (0.01) | 4.60 (2.39–8.09) | 1.25E-05 |
| A3301 | 9 (0.03) | 0 (0.00) | Infinite (0.77–infinity) | 0.07 | 39 (0.01) | 3.40 (1.43–7.23) | 3.20E-03 | 32 (0.01) | 2.80 (1.16–6.09) | 0.01 | 255 (0.01) | 4.89 (2.19–9.56) | 1.63E-04 |
| A0301 | 26 (0.09) | 15 (0.14) | 0.63 (0.31–1.35) | 0.20 | 592 (0.15) | 0.59 (0.38–0.90) | 0.01 | 380 (0.13) | 0.63 (0.40–0.97) | 0.03 | 5497 (0.14) | 0.60 (0.38–0.90) | 0.01 |
| A0101 | 71 (0.25) | 17 (0.16) | 1.81 (0.99–3.48) | 0.04 | 662 (0.16) | 1.73 (1.29–2.31) | 2.67E-04 | 448 (0.16) | 1.74 (1.28–2.33) | 3.21E-04 | 7411 (0.19) | 1.39 (1.05–1.83) | 0.02 |
| A3201 | 3 (0.01) | 4 (0.04) | 0.28 (0.04–1.70) | 0.10 | 152 (0.04) | 0.28 (0.06–0.83) | 0.01 | 105 (0.04) | 0.27 (0.05–0.82) | 0.02 | 1323 (0.03) | 0.30 (0.06–0.88) | 0.02 |
| A0201 | 64 (0.22) | 24 (0.22) | 1.03 (0.59–1.85) | 1.00 | 1085 (0.27) | 0.80 (0.59–1.08) | 0.14 | 756 (0.27) | 0.78 (0.57–1.04) | 0.09 | 10680 (0.28) | 0.75 (0.56–1.00) | 0.05 |
| A2902 | 10 (0.03) | 1 (0.01) | 3.95 (0.55–173.12) | 0.30 | 48 (0.01) | 3.07 (1.37–6.24) | 3.65E-03 | 37 (0.01) | 2.70 (1.18–5.61) | 9.43E-03 | 750 (0.02) | 1.83 (0.86–3.43) | 0.08 |
| A2601 | 8 (0.03) | 3 (0.03) | 1.03 (0.24–6.13) | 1.00 | 115 (0.03) | 1.00 (0.42–2.07) | 1.00 | 91 (0.03) | 0.85 (0.35–1.78) | 0.86 | 834 (0.02) | 1.30 (0.55–2.62) | 0.41 |
| A2501 | 3 (0.01) | 2 (0.02) | 0.57 (0.06–6.96) | 0.62 | 101 (0.03) | 0.42 (0.08–1.28) | 0.16 | 47 (0.03) | 0.62 (0.12–1.94) | 0.62 | 673 (0.02) | 0.60 (0.12–1.77) | 0.50 |
| A2402 | 23 (0.08) | 11 (0.10) | 0.79 (0.35–1.86) | 0.55 | 356 (0.09) | 0.92 (0.57–1.44) | 0.83 | 249 (0.09) | 0.89 (0.55–1.40) | 0.74 | 2801 (0.07) | 1.12 (0.69–1.72) | 0.57 |
| A3101 | 9 (0.03) | 2 (0.02) | 1.75 (0.35–16.95) | 0.73 | 115 (0.03) | 1.13 (0.50–2.26) | 0.71 | 75 (0.03) | 1.18 (0.51–2.39) | 0.57 | 1015 (0.03) | 1.20 (0.54–2.33) | 0.58 |
| A1101 | 14 (0.05) | 10 (0.09) | 0.52 (0.21–1.34) | 0.16 | 255 (0.06) | 0.78 (0.41–1.36) | 0.44 | 177 (0.06) | 0.76 (0.40–1.33) | 0.44 | 2278 (0.06) | 0.82 (0.44–1.40) | 0.61 |
| A2301 | 6 (0.02) | 2 (0.02) | 1.16 (0.20–11.91) | 1.00 | 94 (0.02) | 0.92 (0.33–2.10) | 1.00 | 61 (0.02) | 0.96 (0.34–2.24) | 1.00 | 672 (0.02) | 1.21 (0.44–2.68) | 0.65 |
| A6801 | 9 (0.03) | 8 (0.07) | 0.42 (0.14–1.27) | 0.09 | 128 (0.03) | 1.01 (0.45–2.01) | 0.86 | 100 (0.04) | 0.87 (0.38–1.76) | 0.87 | 1142 (0.03) | 1.07 (0.48–2.06) | 0.86 |
| A19 all (29–33) | 44 (0.15) | 8 (0.07) | 2.07 (0.92–4.64) | 0.08 | 399 (0.10) | 1.49 (1.02–2.18) | 0.04 | 269 (0.10) | 1.58 (1.07–2.34) | 0.02 | 3739 (0.10) | 1.51 (1.04–2.18) | 0.03 |
Alleles belonging to the Aw19 broad antigen group that increase risk are A29, A30, A31, and A33; A32 exhibits protection. A Fisher's exact test combining all Aw19 risk alleles presented the strongest enrichment in all comparisons. Only alleles having three or more case carriers were tested. The table is sorted by P values when comparing case frequencies against A29 controls in the UKB.
Results
HLA-Aw19 Broad Antigen Serotype Alleles and BSCR Risk
Our HLA-A29–controlled cohort allowed us to examine the HLA region while controlling for the strong association of HLA-A29 with BSCR and therefore to detect possible additional association signals in the HLA region.
First, we asked whether rare variants on the HLA-A29 background were enriched in BSCR cases. We did not identify significant enrichments (of rare single or aggregated variants) either within or outside the MHC region.
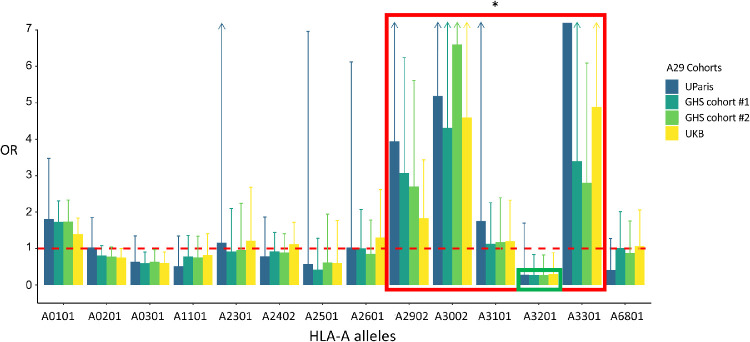
Second, we asked whether other HLA-A alleles in addition to the HLA-A29 allele increased BSCR risk. We constructed an assay to type HLA-A alleles in this cohort (see Methods) and tested the second HLA-A allele (other than the known first HLA-A29) for association with BSCR. We found that additional HLA-A alleles were associated with BSCR, and those with the largest effects belonged to the same HLA-Aw19 broad antigen serotype group: HLA-A29:02, -A30:02, -A31:01, and -A33:01 (Table 1). As a group, HLA-Aw19 alleles were significantly enriched in the second allele of BSCR patients (OR = 4.44; P = 2.2e-03) (Fig. 12, blue bars). This result suggests, for example, that individuals carrying two copies of HLA-A29 would be at a greater risk of developing BSCR compared with those carrying one copy. It also suggests that other Aw19 allele may play a role in BSCR co-susceptibility or pathogenesis in concert with A29. The sole exception within the HLA-Aw19 serotype group is HLA-A32, which has been reported not to share the defining Aw19 binding domain32; HLA-A32 appears to be depleted in BSCR cases and thus protective against BSCR (OR = 0.28; P = 0.1).
Figure 1.
Aw19 enrichment in BSCR cases. Odds ratios for BSCR, comparing frequencies of 14 HLA-A alleles that are present in three or more cases (>1%, x-axis) in 286 UParis cases compared with 108 UParis controls (blue), GHS control cohort 1 (n = 4014, dark green), GHS control cohort 2 (n = 2829, bright green), and UKB controls (n = 38,543, yellow). Aw19 alleles show the highest ORs (red box) that replicates with large A29 control cohorts, with the exception of A32, which is depleted in cases (green box). *P < 0.01.
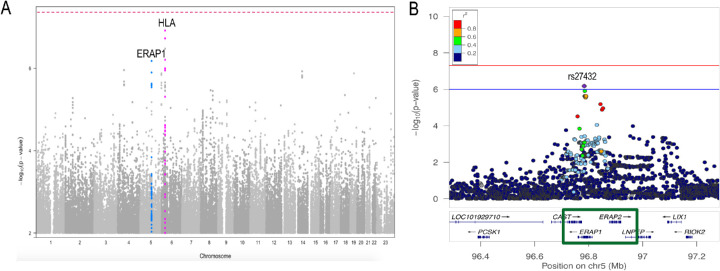
Figure 2.
Manhattan and ERAP1 locus zoom plot of the A29-stratified French cohort. (A) Birdshot association analysis of 286 A29 cases and 108 A29 controls showing borderline associations (P < 1e-6) at several loci including HLA on chromosome six (pink) and ERAP1 on chromosome five (blue). (B) Locus zoom plot exhibiting the ERAP1–ERAP2 locus and the top ERAP1–rs27432 risk variant.
The above results presented two issues due to the small numbers of controls in UParis (n = 108): (1) The frequency of alleles might not represent the frequency of HLA-A alleles in general EUR population, and (2) although the high ORs were replicated in several HLA-Aw19 alleles, the numbers are not sufficient to support significant associations. To tackle these concerns, we examined the frequency of HLA-A alleles in three other large European (EUR) ancestry control populations, two cohorts from the Geisinger Health System (GHS cohort 1, n = 77,198; GHS cohort 2, n = 59,072) and the UK Biobank (UKB, n = 463,315). In all three datasets, we selected EUR samples carrying at least one HLA-A29 allele, matching our BSCR cohort: 4014 A29 carriers from GHS cohort 1 (5.2% of all EUR subjects), 2829 A29 carriers from GHS cohort 2 (4.8% of all EUR subjects), and 38,543 A29 carriers from the UKB (8.3% of all EUR subjects). We compared the frequencies of the second HLA-A alleles in these cohorts to those observed in our BSCR cohort (Fig. 1, Table 1). The results support the enrichment of four of the five HLA-Aw19 alleles in BSCR cases, with highest increased risk for HLA-A30:02 (GHS cohort 1, OR = 4.31; GHS cohort 2, OR = 6.6; UKB, OR = 4.6) and HLA-A33 (GHS cohort 1, OR = 3.4; GHS cohort 2, OR = 2.8; UKB, OR = 4.9). When combining samples carrying the four co-susceptibility alleles A29, A30, A31, and A33, we found a highly significant enrichment in BSCR cases when compared with the larger control cohorts (GHS cohort 1, P = 1.29E-06; GHS cohort 2, P = 1.07E-06; UKB, P = 9.62E-07) (Table 1, top row). This analysis excludes A32 because of its biological difference in the sequence of the peptide binding domain as previously reported32 (see Discussion). That said, we performed additional analyses with all Aw19 alleles including A32, and we find that the enrichment in cases was reduced when it was included (Table 1, bottom row). We also tested whether these associations were affected by measurable confounders, conducting logistic regression tests to evaluate the effects of the second HLA-A allele in HLA-A29 carriers, in UParis BSCR cases compared with each control cohort, with covariates included for sex and principal components, calculated based on genetic array data for each analytic set (Supplementary Table S1). We found that these results were consistent with increased risk for the HLA-Aw19 co-susceptibility alleles, A29, A30, A31, and A33.
HLA-A32 Exhibits Protection from BSCR in an HLA-A29 Positive Cohort
HLA-A32 is underrepresented in BSCR cases (3/286, ∼1%) compared with A29 carrier controls (4/108, 3.7%), corresponding to a nominally significant protection from risk (OR = 0.28; P = 0.1) (Table 1). When compared with the larger control cohorts, the trend protection is maintained with both UKB controls (3.4%, OR = 0.3, P = 0.02) and GHS controls (cohort 1: 3.8%, OR = 0.27, P = 0.01; cohort 2: 3.7%, OR = 0.27, P = 0.02). Although nominally significant, this result does not pass the threshold of multiple test correction (P = 3.57e-03) and will have to be further validated with additional case cohorts.
ERAP1 and ERAP2 Are Independently Associated With BSCR
We tested all variants and gene burdens for association with case–control status while controlling for sex and ten principal components using a generalized linear mixed model (SAIGE8; see Methods). Due to the fact that both cases and controls were A29 allele carriers, the expected strong HLA-A signal was at least partially controlled, as evidenced by the strongest HLA P = 8.98E-07, compared with P = 6.6e-74 with 125 cases in the previous BSCR report.8 Overall, no locus passed the genome-wide significance threshold (P < 5e-8). Other than the remnant signal at HLA-A, only the ERAP1/ERAP2-LNPEP locus on chromosome 5 showed an association with disease at P < 1e-6 (Fig. 2).
The top association within the ERAP1/ERAP2-LNPEP locus is the ERAP1 intronic variant rs27432 (OR = 2.58; 95% confidence interval [CI], 1.78–3.76; P = 6.6e-7), a strong expression quantitative trait locus (eQTL) associated with decreased ERAP1 expression,13,15 which also tags the risk-increasing common ERAP1 haplotype.8 We performed a comparable analysis to assess ERAP1 haplotype associations with BSCR status in our data. The results are consistent with three levels of risk differentiated by nonsynonymous ERAP1 variant haplotypes corresponding to Kuiper et al.15: Haps 1+2 (OR = 0.41; case allele frequency (AF) = 0.17; control AF = 0.35; P = 6.7e-06), Hap10 (OR = 1.78; case AF = 0.28; control AF = 0.17; P = 8.0e-03), and haplotypes 3 to 8 (OR = 1.32; case AF = 0.55; control AF = 0.48; P = 0.11) (Supplementary Table S2).
The previously reported top association for BSCR at this locus tags a common variant near ERAP2/LNPEP, rs10044354.8 This reported risk allele is in a strong linkage disequilibrium (D′ = 0.99, R2 = 0.76) with a strong eQTL increasing ERAP2 expression.8 Our results show a nominal association of rs10044354 with increased risk for BSCR (OR = 1.55; 95% CI, 1.13–2.11; P = 5.8e-3). Furthermore, we did not find significant evidence for an interaction of rs10044354 with rs27432-rs2287987 haplotypes (conditional haplotype test P = 0.46).
We next performed a meta-analysis of our results with the published results from Kuiper et al.,8 which yielded genome-wide significant associations for both ERAP1 (rs27432: OR = 2.46; 95% CI, 1.85–3.26; P = 4.07e-10) and ERAP2 (rs10044354: OR = 1.9; 95% CI, 1.55–2.44; P = 6.2e-09) loci with BSCR (Table 2). Both previous and current studies showed consistent directionality for both variants, which, separated by over 201,222 base pairs, show low LD in our cohort (D′ = 0.79, R2 = 0.18).
Table 2.
Top SNPs in ERAP1 and ERAP2 Regions
| Gene | Variant | Variant Type | Study | OR (95% CI) | P | Hom OR | Case MAF | Control MAF | Meta OR (95% CI) | Meta P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| ERAP2-LNPEP * | rs10044354 5:96984791:C:T | Intronic | Kuiper et al.8 | 2.3 (1.69–3.61) | 1.21E-06 | — | 0.63 | 0.42 | ||
| 1.95 (1.55–2.44) | 6.20E-09 | |||||||||
| UParis | 1.55 (1.13–2.11) | 5.80E-03 | 2.6 (1.3–5.15) | 0.52 | 0.41 | |||||
| ERAP1 | rs27432 5:96783569:A:G† | Intronic | Kuiper et al.8 | 2.26 (2.05–2.47) | 7.20E-05 | — | 0.85 | 0.71 | ||
| 2.46 (1.85–3.26) | 4.07E-10 | |||||||||
| UParis | 2.58 (1.78–3.76) | 6.60E-07 | 4.77 (1.98–11.51) | 0.83 | 0.65 |
Variants in ERAP1 and ERAP2 are genome-wide significant when analyzed together with previous results (125 cases and 670 controls.8 Rs10044354 is the top association in the ERAP1–ERAP2 locus in the previous genome-wide association study of Dutch and Spanish cohorts, and rs27432 is the top association in the region in the current French cohort.
LD between rs10044354 (ERAP2–LNPEP) and rs27432 (ERAP1): D′ = 0.79, R2 = 0.18.
The reference A-allele is the minor allele, risk is the G-allele.
The expression of ERAP2 has been previously reported to be disrupted by a common splice region variant (rs2248374; AF = 0.53) that causes mis-splicing of intron 10 and eventual transcript degradation via nonsense-mediated decay18,33 and which is in high LD with rs10044354 (D′ = 1; R2 = 0.8). Thus, ∼25% of the population of most ancestries (including European, AF = 0.53; African, AF = 0.57; and South Asian, AF = 0.58) is estimated to be lacking an active ERAP2 protein. We examined both datasets for rs2248374 associations and found that it is protective for BSCR with nominal significance in both datasets (Table 33). We note that, in our data, reciprocal conditional analysis rendered both rs10044354 and rs2248374 non-significant (data not shown), making it difficult to provide support for either variant as causal based on genetics alone. In summary, higher expression of ERAP2 protein increases risk for BSCR and a lower expression is protective.
Table 3.
ERAP2 Splice Region Variant Is Protective for BSCR
| Gene | Variant | Variant Type | Study | OR (95% CI) | P | Hom OR | Case MAF | Control MAF | Meta OR (95% CI) | Meta P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| ERAP2 | rs2248374 5:96900192:A:G | Splice region | Kuiper et al.8 | 0.44 (0.31–0.63) | 6.60E-06 | — | 0.33 | 0.53 | ||
| 0.56 (0.45–0.70) | 2.39E-07 | |||||||||
| UParis | 0.68 (0.5–0.92) | 1.40E-02 | 0.45 (0.23–0.87) | 0.43 | 0.53 |
The common ERAP2 splice region variant rs2248374 that disrupts ERAP2 expression is protective in the current BSCR cohort and the previous Spanish and Dutch cohorts.
Cumulative Effect of HLA-Aw19 Alleles and ERAP1/ERAP2 Haplotypes on BSCR Risk
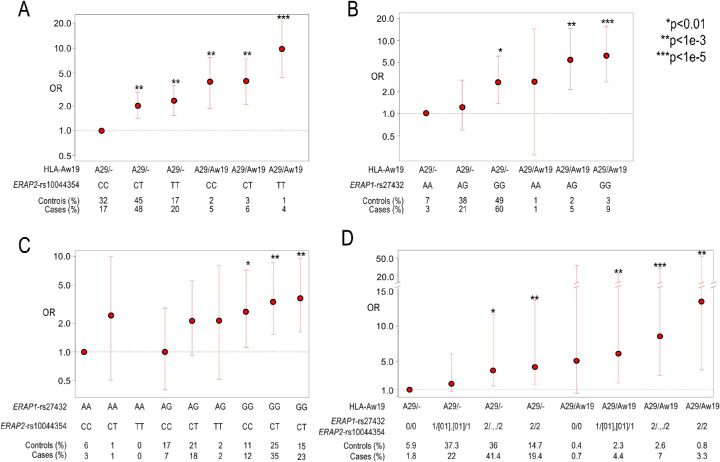
We next examined potential interactions between the ERAP1 and ERAP2 association signals and between HLA-Aw19 and ERAP1/ERAP2 signals by calculating the cumulative effects of HLA-Aw19, ERAP1, and ERAP2 genotypes on BSCR risk using the 286 cases and the 4014 A29 carriers from GHS cohort 1. First, we performed an analysis of ERAP2-rs10044354 risk haplotype, the top non-MHC signal in Kuiper et al.,8 stratified by single (A29/–) versus double (A29/AW19) Aw19 background, which yielded a trend of increased risk with additional ERAP2-rs10044354-T variant alleles, particularly on the double A29/AW19 background (Fig. 3A). We found the highest risk to be the combination of rs10044354-TT and two copies of Aw19 with 12 cases and 34 controls (OR = 9.9; 95% CI, 4.4–21.2; P = 1.66e-07) (Supplementary Table S3). A similar analysis of the ERAP1-rs27432 risk haplotype, our top non-MHC association, stratified by single (A29/–) versus double (A29/AW19) Aw19 background, yielded the same trend of increased risk with additional ERAP1-rs27432-G variant alleles, particularly on the double A29/AW19 background (OR = 6.2; 95% CI, 2.7–15.51; P = 1.54e-06) (Fig. 3B, Supplementary Table S4). We next calculated the combined effects of the ERAP1 risk haplotype tagged by rs27432, and the ERAP2 risk haplotype tagged by rs10044354 (Fig. 3C). We found that the highest risk was conferred by the combination of ERAP1-rs27432-GG and ERAP2-rs10044354-TT (OR = 3.6; 05% CI, 1.62–9.45; P = 4.03e-04) (Supplementary Table S5), and, as mentioned above, our data are consistent with additive effects of the variants/haplotypes. We next combined all risk haplotypes to a single risk analysis. Due to the small number of cases, we combined the genotypes of intermediate genotypes into four main groups: (1) homozygous to the protective alleles in both ERAP1 and ERAP2; (2) homozygous in one and heterozygous in the other; (3) homozygous risk allele in either ERAP1 or ERAP2; and (4) homozygous risk allele in both ERAP1 or ERAP2 (Fig. 3D, Supplementary Table S6). We found a gradual increase in risk with the addition of each risk allele, with the highest risk presented when carrying homozygous risk alleles in both ERAP1 and ERAP2, on top of two copies of A19 alleles (OR = 13.53; 95% CI, 3.79–54.77; P = 1.17e-05). These results suggest that both ERAP1 and/or ERAP2 confer greater BSCR risk, which is further increased in the double Aw19 background.
Figure 3.
The combined risk of ERAP1, ERAP2, and two copies of Aw19. Utilizing 286 BSCR cases and 4014 controls from GHS cohort 1 to calculate additive risk while combining risk factors in ERAP1, ERAP2, and Aw19. (A) An additive genotype model of ERAP2 risk signal tagged by rs10044354 and single (A29/–) or double (A29/Aw19) Aw19 copies relative to lowest risk combination of rs10044354-CC and one copy of Aw19 allele (A29). (B) An additive genotype model of ERAP1 risk signal tagged by rs27432 and single (A29/–) or double (A29/Aw19) Aw19 copies relative to lowest risk combination of rs27432-AA and one copy of Aw19 allele (A29). (C) An additive genotype model of ERAP1 risk signal tagged by rs27432 and ERAP2 signal tagged by rs10044354 relative to lowest risk combination of rs27432-AA and rs10044354-CC. (D) An additive genotype model of ERAP1 and ERAP2 risk signals and single (A29/–) or double (A29/Aw19) Aw19 copies relative to lowest risk combination. The genotypes are combined as follows: 0 = ERAP1 and ERAP2 homozygous for protective allele; 1/[01], [01]/1 = either homozygous protective or heterozygous genotypes of both ERAP1 and ERAP2; 2/., ./2 = homozygous risk allele of either ERAP1 or ERAP2; 2/2 = homozygous risk allele of both ERAP1 and ERAP2.
Absolute BSCR Risk
We calculated what would be the absolute risk of BSCR when considering all risk alleles as presented in Figure 3D. Because the prevalence of BSCR in the general population is estimated at 0.2 to 1.7:100,000,1 we used 1:100,000 as an approximation. We further calculated the absolute risk when carrying one A29 carrier based on the frequency of A29 in the UKB EUR population of 8%, and reached an absolute risk of 1:29,000 (Supplementary Table S6). We found that the absolute risk climbed with each risk genotype presented in Figure 3D, reaching the most prominent risk at 1:2,160 for cases that carry homozygous risk alleles for both ERAP1 and ERAP2 and two copies of Aw19 alleles. Exhibiting a significant increase in absolute risk of disease when carrying all three risk haplotypes. We also estimated the proportion of liability-scale disease variance explained (i.e., heritability), assuming a 1/100,000 prevalence of BSCR, by calculating the expected disease liability for each of our combined-genotype risk estimates and their frequencies in controls. The second HLA-A allele and ERAP1 and ERAP2 variants explain 1.55% of liability-scale disease risk in the A29 carrier population; this is very substantial in comparison with common disease-common variant associations, which typically have OR around 1.1 and explain about 0.1% of disease liability,34,35 and increases by over 10% the BSCR disease variance explained by the HLA-A*29 allele in the general population, as calculated based on results from Kuiper et al.8 (OR = 303, 10.9% heritability).
Discussion
The sequencing of a new large BSCR patient cohort and HLA-A29 controls has confirmed the importance of the ERAP1 and ERAP2 polymorphisms in increasing risk for developing BSCR. ERAP1 and ERAP2 reside back to back on chromosome five in opposite orientation and share the regulatory regions, which upregulate one and downregulate the other, and vice versa.15 The association of both ERAP1 and ERAP2 haplotypes is consistent with a mechanism in which coordinated decreased ERAP1 and increased ERAP2 expression contributes to disease risk. Several studies have reported that the ERAP1 and ERAP2 haplotypes affect their expression, as well as the resulting peptidome.13,15,36
We report the novel finding that several other HLA-Aw19 family alleles (HLA- A29, -A30, -A31, and -A33) contribute additional risk as the second HLA-A allele, in addition to the established HLA-A29 risk allele. HLA-Aw19 family alleles have a similar antigen-binding sequence and therefore would bind similar peptide motifs.32 Hence, the enrichment of Aw19 alleles in cases supports the inferred mechanism underlying activation of the immune response in BSCR: Having two copies of these alleles may increase the cell-surface presentation of specific types of peptides in BSCR cases compared with HLA-A29–positive controls. Furthermore, we found that the HLA-A32 allele within the Aw19 family is potentially protective.
Our results indicate that a decreased expression of ERAP1 and an increased expression of ERAP2 confer stronger risk for BSCR than each one separately. Furthermore, this effect is increased in the presence of two copies of HLA-Aw19. The effect of the second HLA-A allele being one of the HLA-Aw19 alleles, when compared against the largest HLA-A29 positive cohort (UKB), ranges between 1.2 (A31:01) to 4.89 (A33:01), with an OR = 1.83 for a second HLA-A29 allele. Although this is a substantial increase in risk, the effect of the second A*29 (or Aw19) allele is not as high as would be expected under an additive model given the large heterozygous effect of one HLA-A29 allele (OR = 303; P = 2e-63),8 thus exhibiting a partially dominant effect.
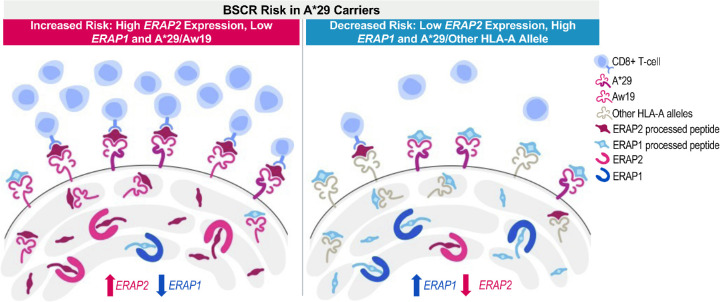
The above combined and additive effects of risk factors associated with peptide processing and presentation are suggestive of a peptide presentation threshold hypothesis as a driving mechanism for the immune response underlying development of BSCR disease (Fig. 4). Results from this and other studies suggest that increased ERAP2 along with decreased ERAP1 expression in BSCR cases would lead to higher availability of ERAP2-processed peptides for presentation onto HLA class I proteins. Additional HLA-Aw19 alleles, with similar peptide-binding properties, would increase the presentation of similar peptides. Therefore, both the production of a unique peptide pool by dominant ERAP2 activity and the increased expression of HLA-Aw19 risk allele proteins presenting these peptides may increase the likelihood that a putative ocular autoantigen would be processed and presented above a certain threshold to activate an immune response (Fig. 4, left panel). On the other hand, having lower expression of ERAP2 (and higher expression of ERAP1), along with a single HLA-A29 allele, lowers the ocular antigenic peptide presentation below the threshold and thus reduces the risk of generating the immunological response leading to BSCR in HLA-A29 healthy control carriers (Fig. 4, right panel). This further highlights the importance of the shaping and generation of the available peptide pool by ERAPs to be presented by specific HLA class I proteins in promoting the generation of an immune response or, in the case of autoimmune disease, an aberrant response to a self-antigen.
Figure 4.
Peptide presentation threshold mechanism in BSCR. An increased expression of ERAP2 and a decreased expression of ERAP1, in addition to two copies of Aw19 alleles, increased the risk for BSCR (left), whereas decreased expression of ERAP2 and increased expression of ERAP1, in the presence of one copy of the A29 allele, decreased the risk for BSCR (right).
ERAP1 and ERAP2 polymorphisms and risk haplotypes have also been reported in other HLA class I-associated autoimmune diseases.37,38 Polymorphisms in ERAP1 increase risk for ankylosing spondylitis in HLA-B*27 carriers, for psoriasis vulgaris in HLA-C*06 carriers, and for Behçet's disease in HLA-B*51 carriers, further supporting the combinatorial impact of peptide trimming and HLA class I allele in initiating autoimmune responses.39–42 Ankylosing spondylitis and Behçet's disease-associated ERAP1 variants have also been experimentally shown to shape the resulting HLA-B*27 and HLA-B*51 peptidomes, respectively.43,44 Therefore, it is possible that the combination of risk ERAP1/ERAP2 haplotypes and specific risk HLA class I alleles can predispose an individual to develop an HLA class I–associated disease in a similar fashion as we hypothesize for BSCR. This implies that the peptide threshold hypothesis may have broader implications as a disease mechanism in HLA class I–associated immunological diseases.
HLA-A32 is the only HLA-Aw19 member that is found at lower rates in BSCR patients compared with controls, suggesting that it could be protective. We considered whether this is a dominant protection effect or rather the effect of the increased frequency of the HLA-Aw19 alleles in cases. Because 15% of cases carry two copies of HLA-A29, this will diminish the case numbers by 0.85 compared to the large three to four times difference in frequency when compared to the control sets (UParis frequency is 1%, UKB frequency = 3%, and GHS frequency = 4%). For this reason, we do not expect the effect of the damaging HLA-Aw19 alleles to create a false protection effect of that size for HLA-A32. To answer the question why HLA-A32 has a reverse effect than the other HLA-Aw19 alleles, we considered that the HLA-Aw19 serotype was initially identified by antibody binding to related family members; however, this identifies the HLA-A proteins based on structure outside of the peptide-binding groove. Serofamilies have since been re-analyzed by overall and peptide-binding region sequences.32 Comparison of the sequences in the peptide-binding region reveals that HLA-A32 is more distantly related than the other Aw19 alleles that are identified as novel risk factors in this present study: HLA-A29, -A30, -A31, and -A33. When examining the differences in sequence between these Aw19 alleles, two main differences are evident: at position 9, which is part of the peptide-binding domain, and a stretch of amino acids at positions 79 to 83 that are only found in HLA-A32 and not the other Aw19 alleles (Supplementary Fig. S1). Theoretically, the peptide pool bound by HLA-A32 would differ from the remaining members of the Aw19 family and would not activate the same subset of responding CD8 T cells. This adds further evidence supporting the hypothesis of the threshold requirement of an increased concentration of the driving autoantigenic peptide pool presented on high-risk HLA-A proteins as a driving component for development of BSCR uveitis.
HLA class I alleles not only present peptides to CD8 T cells that can initiate an adaptive immune response but they are also recognized by killer immunoglobulin receptors on natural killer (NK) cells involved in innate immune surveillance. The amino acids at positions 79 to 83 codes in HLA-A32 contain the Bw4 epitope, shared by a subset of HLA-B and HLA-A alleles known to bind KIR3DL1, which delivers an inhibitory signal to the NK cell.45 Interestingly, HLA-A32 is the only allele in the Aw19 serotype family that contains an HLA-Bw4 motif and thus is a ligand for the inhibitory KIR3DL1. There are reports of autoimmune HLA class I–associated diseases with protective KIR3DL1 or Bw4 genetic associations.46–49
The new data presented in this investigation do not provide an answer to the question of the origin of the ocular specificity of BSCR, in distinction to other autoimmune disorders that are strongly HLA associated, which manifest more widely. We speculate that an undefined antigen specifically and exclusively expressed in the choroid, retinal pigment epithelium, or perhaps outer retina, is the pathogenic driver of the disease. This molecule must (1) have a very restricted distribution (exclusively ocular) or have an alternatively expressed isoform in the eye and (2) have an alternative structure or be aberrantly processed, presented, or sensed by T cells, in patients with BSCR. The current study has not found a clear candidate as for the antigen, which may indicate that the critical difference between BSCR patients and unaffected HLA-A29+ individuals involves antigen processing and/or presentation.
Regarding the consideration of the additional HLA-Aw19 alleles as additional diagnosis criteria for BSCR, all of our patients with BSCR are HLA-A29 positive, and we are not aware of reports of HLA-A29–negative patients who are homozygous or heterozygous for A30, A31, or A33. We expect that following this report of the additional risk in other HLA-A alleles, more extensive serotyping will be performed for the diagnosis of BSCR that may help us shed more light on this important question.
In summary, the combinatorial impact of ERAP1/ERAP2 shaping the immunopeptidome along with differential peptide selection by the key residues in HLA-A29 and HLA-Aw19 family members supports the immunological hypothesis of a peptide pool that is generated by this combination and available for immune cell recognition and activation initiating an inflammatory cascade. Avenues to reduce the expression and recognition of ERAP2-processed and HLA-Aw19-presented peptides in the eye may be beneficial against BSCR disease and/or progression.
Supplementary Material
Acknowledgments
The authors thank Jonas Kuiper for his expertise and assistance providing results from previous BSCR genome-wide association analysis, as well as performing additional quality control and fine mapping. We also thank Rachel Kirschner for her expertise constructing the scientific illustration pertaining to the hypothesis of quantitative threshold of peptide presentation.
Disclosure: S. Gelfman, Regeneron Genetics Center (E); D. Monnet, None; A.J. Ligocki, Regeneron Pharmaceuticals (E); T. Tabary, None; A. Moscati, Regeneron Genetics Center (E); X. Bai, Regeneron Genetics Center (E); J. Freudenberg, Regeneron Genetics Center (E); B. Cooper, Regeneron Pharmaceuticals (E); J.A. Kosmicki, Regeneron Genetics Center (E); S. Wolf, Regeneron Genetics Center (E); M.A.R. Ferreira, Regeneron Genetics Center (E); J. Overton, Regeneron Genetics Center (E); J. Weyne, Regeneron Pharmaceuticals (E); E.A. Stahl, Regeneron Genetics Center (E); A. Baras, Regeneron Genetics Center (E); C. Romano, Regeneron Pharmaceuticals (E); J.H.M. Cohen, None; G. Coppola, Regeneron Genetics Center (E); A. Brézin, None
References
- 1. Minos E, Barry RJ, Southworth S, et al.. Birdshot chorioretinopathy: current knowledge and new concepts in pathophysiology, diagnosis, monitoring and treatment. Orphanet J Rare Dis. 2016; 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shao EH, Menezo V, Taylor SR. Birdshot chorioretinopathy. Curr Opin Ophthalmol. 2014; 25: 488–494. [DOI] [PubMed] [Google Scholar]
- 3. Menezo V, Taylor SR. Birdshot uveitis: current and emerging treatment options. Clin Ophthalmol. 2014; 8: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ryan SJ, Maumenee AE. Birdshot retinochoroidopathy. Am J Ophthalmol. 1980; 89: 31–45. [DOI] [PubMed] [Google Scholar]
- 5. Nussenblatt RB, Mittal KK, Ryan S, Green WR, Maumenee AE. Birdshot retinochoroidopathy associated with HLA-A29 antigen and immune responsiveness to retinal S-antigen. Am J Ophthalmol. 1982; 94: 147–158. [DOI] [PubMed] [Google Scholar]
- 6. Levinson RD, Brezin A, Rothova A, Accorinti M, Holland GN. Research criteria for the diagnosis of birdshot chorioretinopathy: results of an international consensus conference. Am J Ophthalmol. 2006; 141: 185–187. [DOI] [PubMed] [Google Scholar]
- 7. Brezin AP, Monnet D, Cohen JH, Levinson RD. HLA-A29 and birdshot chorioretinopathy. Ocul Immunol Inflamm. 2011; 19: 397–400. [DOI] [PubMed] [Google Scholar]
- 8. Kuiper JJW, Van Setten J, Ripke S, et al.. A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum Mol Genet. 2014; 23: 6081–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. LeHoang P, Ozdemir N, Benhamou A, et al.. HLA-A29.2 subtype associated with birdshot retinochoroidopathy. Am J Ophthalmol. 1992; 113: 33–35. [DOI] [PubMed] [Google Scholar]
- 10. Herbort CP Jr, Pavésio C, LeHoang P, et al.. Why birdshot retinochoroiditis should rather be called ‘HLA-A29 uveitis’? Br J Ophthalmol. 2017; 101: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuiper JJ, Rothova A, Schellekens PAW, Ossewaarde-van Norel A, Bloem AC, Mutis T. Detection of choroid- and retina-antigen reactive CD8(+) and CD4(+) T lymphocytes in the vitreous fluid of patients with birdshot chorioretinopathy. Hum Immunol. 2014; 75: 570–577. [DOI] [PubMed] [Google Scholar]
- 12. Pulido JS, Canal I, Salomão D, Kravitz D, Bradley E, Vile R. Histological findings of birdshot chorioretinopathy in an eye with ciliochoroidal melanoma. Eye (Lond). 2012; 26: 862–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuiper JJW, van Setten J, Devall M, et al.. Functionally distinct ERAP1 and ERAP2 are a hallmark of HLA-A29-(Birdshot) uveitis. Hum Mol Genet. 2018; 27: 4333–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lopez de Castro JA. How ERAP1 and ERAP2 shape the peptidomes of disease-associated MHC-I proteins. Front Immunol. 2018; 9: 2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paladini F, Fiorillo MT, Vitulano C, et al.. An allelic variant in the intergenic region between ERAP1 and ERAP2 correlates with an inverse expression of the two genes. Sci Rep. 2018; 8: 10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanz-Bravo A, et al. Allele-specific alterations in the peptidome underlie the joint association of HLA-A*29:02 and endoplasmic reticulum aminopeptidase 2 (ERAP2) with birdshot chorioretinopathy. [DOI] [PMC free article] [PubMed]
- 17. Alvarez-Navarro C, Martin-Esteban A, Barnea E, Admon A, López de Castro JA. Endoplasmic reticulum aminopeptidase 1 (ERAP1) polymorphism relevant to inflammatory disease shapes the peptidome of the birdshot chorioretinopathy-associated HLA-A*29:02 antigen. Mol Cell Proteomics. 2015; 14: 1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andres AM, Dennis MY, Kretzschmar WW, et al.. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010; 6: e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarkizova S, Klaeger S, Le PM, et al.. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol. 2020; 38: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. The Standardization of Uveitis Nomenclature Sun Working, Group. Classification criteria for birdshot chorioretinitis. Am J Ophthalmol. 2021; 228: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Hout CV, Tachmazidou I, Backman JD, et al.. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020; 586: 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reid JG, Carroll A, Veeraraghavan N, et al.. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014; 15: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai Y, Ni M, Cooper B, Wei Y, Fury W. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics. 2014; 15: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson J, Soormally AR, Hayhurst JD, Marsh SGE. The IPD-IMGT/HLA database - new developments in reporting HLA variation. Hum Immunol. 2016; 77: 233–237. [DOI] [PubMed] [Google Scholar]
- 25. Jia X, Han B, Onengut-Gumuscu S, et al.. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013; 8: e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rich SS, Concannon P, Erlich H, et al.. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006; 1079: 1–8. [DOI] [PubMed] [Google Scholar]
- 27. Delaneau O, Zagury JF, Robinson MR, Marchini JL, Dermitzakis ET. Accurate, scalable and integrative haplotype estimation. Nat Commun. 2019; 10: 5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Das S, Forer L, Schönherr S, et al.. Next-generation genotype imputation service and methods. Nat Genet. 2016; 48: 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mbatchou J, Barnard L, Backman J, et al.. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021; 53: 1097–1103. [DOI] [PubMed] [Google Scholar]
- 30. Zhou W, Nielsen JB, Fritsche LG, et al.. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018; 50: 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McKenzie LM, Pecon-Slattery J, Carrington M, O'Brien SJ. Taxonomic hierarchy of HLA class I allele sequences. Genes Immun. 1999; 1: 120–129. [DOI] [PubMed] [Google Scholar]
- 33. Coulombe-Huntington J, Lam KC, Dias C, Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 2009; 5: e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrett JC, Hansoul S, Nicolae DL, et al.. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008; 40: 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stahl EA, Raychaudhuri S, Remmers EF, et al.. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010; 42: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanz-Bravo A, Martín-Esteban A, Kuiper JJW, et al.. Allele-specific alterations in the peptidome underlie the joint association of HLA-A*29:02 and endoplasmic reticulum aminopeptidase 2 (ERAP2) with birdshot chorioretinopathy. Mol Cell Proteomics. 2018; 17: 1564–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Babaie F, Hosseinzadeh R, Ebrazeh M, et al.. The roles of ERAP1 and ERAP2 in autoimmunity and cancer immunity: new insights and perspective. Mol Immunol. 2020; 121: 7–19. [DOI] [PubMed] [Google Scholar]
- 38. Yao Y, Liu N, Zhou Z, Shi L. Influence of ERAP1 and ERAP2 gene polymorphisms on disease susceptibility in different populations. Hum Immunol. 2019; 80: 325–334. [DOI] [PubMed] [Google Scholar]
- 39. Evans DM, Spencer CCA, Pointon JJ, et al.. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011; 43: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiśniewski A, Matusiak Ł, Szczerkowska-Dobosz A, Nowak I, Łuszczek W, Kuśnierczyk P. The association of ERAP1 and ERAP2 single nucleotide polymorphisms and their haplotypes with psoriasis vulgaris is dependent on the presence or absence of the HLA-C*06:02 allele and age at disease onset. Hum Immunol. 2018; 79: 109–116. [DOI] [PubMed] [Google Scholar]
- 41. Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Consortium 2, Strange A, Capon F, et al.. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010; 42: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takeuchi M, Ombrello MJ, Kirino Y, et al.. A single endoplasmic reticulum aminopeptidase-1 protein allotype is a strong risk factor for Behçet's disease in HLA-B*51 carriers. Ann Rheum Dis. 2016; 75: 2208–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanz-Bravo A, Alvarez-Navarro C, Martín-Esteban A, Barnea E, Admon A, López de Castro JA. Ranking the contribution of ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 (ERAP1) polymorphisms to shaping the HLA-B*27 peptidome. Mol Cell Proteomics. 2018; 17: 1308–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guasp P, Barnea E, González-Escribano MF, et al.. The Behçet's disease-associated variant of the aminopeptidase ERAP1 shapes a low-affinity HLA-B*51 peptidome by differential subpeptidome processing. J Biol Chem. 2017; 292: 9680–9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood. 2008; 112: 708–710. [DOI] [PubMed] [Google Scholar]
- 46. Vendelbosch S, Heslinga SC, John M, et al.. Study on the protective effect of the KIR3DL1 gene in ankylosing spondylitis. Arthritis Rheumatol. 2015; 67: 2957–2965. [DOI] [PubMed] [Google Scholar]
- 47. Berinstein J, Pollock R, Pellett F, Thavaneswaran A, Chandran V, Gladman DD. Association of variably expressed KIR3dl1 alleles with psoriatic disease. Clin Rheumatol. 2017; 36: 2261–2266. [DOI] [PubMed] [Google Scholar]
- 48. Petrushkin H, Norman PJ, Lougee E, et al.. KIR3DL1/S1 allotypes contribute differentially to the development of Behçet disease. J Immunol. 2019; 203: 1629–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levinson RD, Du Z, Luo L, et al.. Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes Immun. 2008; 9: 249–258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.