Abstract
Background
In recent years, farmers have complained that the only way to obtain seeds is to select plants that show good performance under local climate conditions in the region. This study aimed to investigate the diversity of rice accessions grown in the region to build a breeding program.
Methods and results
A total of 62 accessions of rice from farmers and research stations were collected from the Kurdistan region, including short-grain and long-grain types, for molecular genetics and diversity analysis. In this study, 37 polymorphic simple sequence repeat (SSR) markers were selected with several molecular genetics software programs. The results show that these SSR markers are very effective for this investigation, generating a total of 152 observed alleles (Na), 75.166 Effective number of alleles (Ne) and an average of 4.1 and 2.03 alleles per locus, respectively. The average polymorphic information content (PIC) per locus was recorded as 0.404. The research presented here confirms two subpopulations, japonica (C1 and C2) and indica (C3), based on molecular genetics data analysis. Analysis of molecular variance revealed that the 72% variance was due to the variation among populations and 28% within the population.
Conclusions
Altogether, these results indicate that there is very low gene flow. These results show the importance of the study of genetic diversity and relationships for starting breeding and improvement programs for rice in the Kurdistan region.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11033-021-06920-x.
Keywords: Gene flow, Genetic diversity, AMOVA, SSR markers, Rice (Oryza sativa)
Introduction
Rice (Oryza sativa L.) or Asian cultivated species (indica and japonica) are among the top agronomic and nutritionally essential crops worldwide. The indica genotypes are tropical rice cultivars that are grown in lowland conditions. In contrast, the japonica genotypes can be either tropical cultivars adapted to rainfed upland conditions or temperate cultivars adapted to lowland conditions. Based on literary evidence [1], it is believed that rice was introduced into Kurdistan region-Iraq around the 12th century BC.
Rice is the leading consumed food in Iraq and Kurdistan. In the south and middle of Iraq, amber (long-grain) rice is cultivated, and in northern Iraq (the Kurdistan Region), long- and short-grain rice is cultivated [2]. Approximately 70% of rice is imported, and only 30% is produced locally. Improving the quality and quantity of rice production is an important point for farmers in the Kurdistan region (Erbil, Sulaymaniyah and Duhok).In Kurdistan, there are no published data about genetic diversity in rice. The only available information is the local name from the farmers based on the paddy and grain trait variation.
Rice consumption in Iraq was estimated to be 45.7 kg of milled rice per person in year 2017 according to Helgilibrary [3]. The production season “planting start at June–July and harvesting are on September–October” in the middle and south of Iraq [4]. In the Kurdistan Region, the planting season begins in April or May, and the plants are harvested in October [5].
According to Jeong et al. [6], the average of rice import have doubled in the world and Iraq is among the top ten countries of milled rice import. The total global rice trade expected growth of 1.37% annually over the next 10 years because of the high demand from import countries [6]. There is a lack of data for rice production in the Kurdistan region. Regarding the assessment of Ewaid et al. [7], the average rice production from 2007 to 2016 was 271,173 tons in all provinces of Iraq, excluding the Kurdistan region.
Researchers have conducted numerous successful studies worldwide defining the genetic diversity of rice using different molecular markers. The Random Amplified Polymorphic DNA (RAPD) was started by [8, 9]and then combined with the simple sequence repeat (SSR) [10–12]. Amplified fragment length polymorphism (AFLP) was also used for the analysis of the genetic variability of rice [13, 14] in conjunction with SSR [15]. Additionally, the single-nucleotide polymorphism (SNP) marker [16–18] was used along with the SSR [19, 20]. Among these DNA markers, SSR is considered the best marker used to study the genetic diversity of rice in the last two decades [21–23].
There is no published data about genetic diversity and breeding program of rice in the region. The only activity is to select plants that showed good performance under local climate conditions by the farmer. This study will be the first step toward building a breeding program and diversity analysis of rice accessions grown in the region.
Materials and methods
Plant materials
During 2019, samples of approximately 100 rice accessions were collected from the farmer fields and different research institutes of the Iraq and Kurdistan region.The samples were classified based on morphological characteristics, such as seed color, grain size, presence or absence of awns, life cycle, and geographic location (Table S1). A collection of 62 rice accessions (50 from farmers, 9 from the Directory of Agricultural Research in Sulaymaniyah, and 3 from the Al-Mishkhab Research Center) were selected and planted in 2020 for molecular investigation (Fig. 1).Seeds were soaked for 3 days in the lab and on May 20, transferredtofield at Preamagrun-Gaba villagelocation between Latitude 35° 43′ 14.35′′ N Longitude 45° 04′ 32.02′′ and harvested in late October. Cultural practices including irrigation, weed control and fertilizers were conducted during the season.Fresh leaves from each accession were collected from 25-day-old seedlings.
Fig. 1.
Iraq map showing provinces and locations of rice sampling points
DNA Isolation
Fresh, clean leaves from each accession ground with a pestle and mortar with liquid nitrogen. A total of 150 mg of the ground samples was used for genomic DNA extraction using the Quick-DNA™ Plant/Seed Miniprep Kit, Catalogue No. D6020 (Zymo Research, Irvine, CA, USA). The quantity and quality of DNA were determined by a NanoDrop ND-2000/2000c spectrophotometer (ThermoScientific, USA)and represented by 1% agarose gel in 1X TBE buffer. The gel was viewed using a Labnet gel documentation (LabNet International Inc., Edison, NJ, USA).
PCR amplification of SSR primers
A total of 37 polymorphic SSR markers were obtained from the Gramene database [24] based on their polymorphism information content (PIC) values in previous research (Table S2) after primer screening and used for all 62 rice accessions. The total PCR volume was optimized to 20 µl and included 2 µl of approximately 15 ng DNA template, 6.0 µl PCR master mix of AccuPower® PCR PreMix (Bioneer, Korea), 1.5 µl of each primer (forward and reverse primers), and 9.0 µl of double-distilled water. The PCR protocol was followed, amplification was carried out using a thermocycler (Applied Biosystems), and the PCR conditions were as follows: 5 min at 95 °C; 35 cycles of 50 s at 95 °C; 50 s at annealing temperature (Table S2); and 50 s at 72 °C, followed by 7 min at 72 °C. The amplified products were visualized by ethidium bromide-stained 3% metaphor agarose gels in 1% TBE along with a 100 bp DNA ladder (Add Bio). The gel was viewed using Labnet gel documentation (LabNet International Inc., Edison, NJ, USA), and the gel picture was analyzed using the sample band with a DNA ladder for the band size of each SSR primer. The SSR data were analyzed using POPGENE v1.32 [25] software to determine the allele frequency, Na, Ne and gene diversity per locus in each accession.
Structure analysis
To detect the subpopulations of the genotypes, STRUCTURE 2.3.4 software [26] was used. The run parameters were set up as 100,000 burn-in periods and 100,000 Markov chain Monte Carlo (MCMC) replications. The K value set up from 2 to 10 and 10 replicate runs were performed for each value of K. For selecting the best number of K (subpopulation), Structure Harvester was used [27].
Genetic diversity
A dendrogram of rice genotypes was generated based on the unweighted pair group method with arithmetic mean (UPGMA) method via PowerMarker v3.25 [28] and then visualized using MEGA X [29]. GenAlEx V6.5 [30]was implemented for calculating principal coordinates analysis (PCoA) and analysis of molecular variance (AMOVA).
Results
Allele frequency
Sixty-two rice accessions were investigated using 37 polymorphic SSR markers. All genotypes were collected from Iraq, 50 from farmers in the Kurdistan region (Erbil, Sulaymaniyah and Duhok), nine from the Directory of Agricultural Research in Sulaymaniyah, and three from the Al-Mishkab Rice Research Station near Najaf. A total of 37 polymorphic SSR markers sequence details are available at (Table S2) were selected for genotyping these collections after primer screening. Allele frequency results (Table 1) show Na 152 with an average of 4.1 alleles per locus in this investigation. The maximum Na of 7 alleles was observed for primers RM20, RM257, and RM294.A minimum of 2 alleles were in RM23, RM171, RM172, RM321, and RM7376.The sum of Ne was 75.166 with an average of 2.03 alleles. The highest values of Ne were recorded as 4.544 and 3.827 alleles for RM204 and RM250, respectively, and the lowest value was 1.105 for RM171. Gene diversity ranged from 0.780 (RM204) to 0.095 (RM171), with an average of 0.442. The average of SSR’s PIC was 0.404. Higher PICs of 0.748, 0.694, 0.665 and 0.659 were recorded for loci RM204, RM250, RM507 and RM294, respectively. While the lowest PIC value was 0.090 in locus RM171.
Table 1.
SSR markers, chromosome locations and allele frequency across 62 accessions
| Primers ID | Chromosome no. | Na* | Ne* | Gene diversity | PIC | References | |
|---|---|---|---|---|---|---|---|
| RM1 | 1 | 5 | 1.861 | 0.463 | 0.428 | [19, 31–33] | |
| RM4 | 11 | 4 | 2.417 | 0.586 | 0.506 | [33] | |
| RM 20 | 12 | 7 | 2.787 | 0.641 | 0.574 | [33] | |
| RM21 | 11 | 3 | 1.338 | 0.252 | 0.228 | [34, 35] | |
| RM23 | 10 | 2 | 1.33 | 0.248 | 0.217 | [31] | |
| RM159 | 8 | 3 | 1.771 | 0.435 | 0.395 | [31] | |
| RM171 | 10 | 2 | 1.105 | 0.095 | 0.090 | [32, 35] | |
| RM172 | 7 | 2 | 1.330 | 0.248 | 0.217 | [31] | |
| RM195 | 8 | 3 | 1.567 | 0.362 | 0.329 | [31] | |
| RM204 | 6 | 6 | 4.544 | 0.780 | 0.748 | [11] | |
| RM205 | 9 | 4 | 1.795 | 0.443 | 0.410 | [36] | |
| RM207 | 2 | 3 | 1.309 | 0.236 | 0.222 | [35] | |
| RM209 | 11 | 4 | 1.42 | 0.296 | 0.282 | [36] | |
| RM214 | 7 | 3 | 1.612 | 0.379 | 0.347 | [11] | |
| RM223 | 8 | 4 | 2.209 | 0.547 | 0.462 | [11, 34, 35] | |
| RM224 | 11 | 4 | 1.394 | 0.283 | 0.263 | [11, 32, 34] | |
| RM228 | 10 | 4 | 2.409 | 0.585 | 0.52 | [15, 34, 36] | |
| RM241 | 4 | 4 | 1.453 | 0.312 | 0.296 | [15, 35, 37] | |
| RM242 | 9 | 4 | 1.349 | 0.259 | 0.243 | [35, 36] | |
| RM247 | 12 | 5 | 2.766 | 0.638 | 0.599 | [33, 36] | |
| RM250 | 2 | 5 | 3.827 | 0.739 | 0.694 | [15, 31, 34] | |
| RM257 | 9 | 7 | 2.814 | 0.645 | 0.620 | [35] | |
| RM259 | 1 | 6 | 1.907 | 0.476 | 0.455 | [37, 38] | |
| RM294 | 1 | 7 | 3.378 | 0.704 | 0.659 | [31] | |
| RM304 | 10 | 4 | 2.039 | 0.509 | 0.467 | [15] | |
| RM311 | 10 | 5 | 1.946 | 0.486 | 0.454 | [31] | |
| RM314 | 6 | 3 | 1.771 | 0.435 | 0.395 | [11, 15, 31] | |
| RM 316 | 9 | 4 | 2.467 | 0.595 | 0.519 | [11, 19, 31] | |
| RM321 | 9 | 2 | 1.330 | 0.248 | 0.217 | [31] | |
| RM334 | 5 | 3 | 1.343 | 0.255 | 0.235 | [34, 38] | |
| RM339 | 8 | 5 | 1.937 | 0.484 | 0.443 | [15] | |
| RM342 | 9 | 4 | 2.294 | 0.564 | 0.469 | [31] | |
| RM547 | 8 | 4 | 1.14 | 0.122 | 0.118 | [39] | |
| RM507 | 10 | 6 | 3.468 | 0.712 | 0.665 | [36, 37] | |
| RM3134 | 3 | 6 | 2.638 | 0.621 | 0.554 | [31] | |
| RM6250 | 4 | 3 | 1.771 | 0.435 | 0.395 | [31] | |
| RM7376 | 12 | 2 | 1.33 | 0.248 | 0.217 | [31] | |
| Average | – | 4.1 | 2.03 | 0.442 | 0.404 | – | |
Na observed number of alleles, Ne effective number of alleles, Gene Diversity, PIC polymorphism information content
Structure analysis
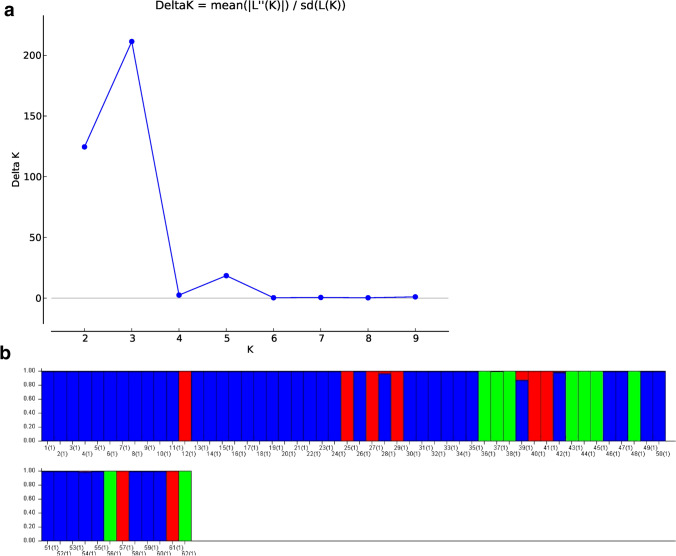
The population genetic structure of 62 rice genotypes was obtained from the STRUCTURE program using SSR genotypic data. The study presents crucial evidence for defining the right population structure of Kurdistan region rice genotypes. The best K (subpopulation) was estimated by Structure Harvester and is presented in Fig. 2, which indicated 3 (K = 3). Consequently, the K = 3 subpopulation results were chosen from STRUCTURE and illustrated in Fig. 2. The genotypes in the blue cluster (C1) represent 45 short-grain rice, the red color cluster (C2) represents eight short-grain rice, and the green color cluster (C3) represents nine long-grain rice. All genotypes assigned to these subpopulations were considered pure because they scored more than 0.80, as shown in Fig. 2.
Fig. 2.
a The number of estimate subgroups (K) ranging from 2 to 10 by Structure Harvester, and b populationstructure of 62 rice for K = 3 revealed by 37 SSR markers and structure program
Genetic diversity
A distance matrix (dissimilarity matrix) result was obtained from PowerMarker [28], and the tree was visualized using MEGA X [29]. The UPGMA clustering method for generating trees was implemented. As shown in Fig. 3, the results of the UPGMA circle tree with three main clusters, blue (45), red (8), and green (9), are the same as the STRUCTURE results. The present study shows a structural analysis of rice genotypes from Iraq with three clusters (C1, C2, and C3) using 37 SSR markers. Additionally, the phylogenetic tree result confirms the STRUCTURE result with the same three clear clusters. However, C1 and C2 were apparent in the same subdivision (Fig. 3), and C3 separated independently. In addition, all grains in the clusters (C1 and C2) are short-grained rice, but the C2 grains are colored, while the C3 samples are long-grained. Furthermore, from C1 and C2, only one genotype (R 49) is registered in the International Rice Genebank Collection under the name of Bazian (IRGC 9506). The Bazian accession is the preferred rice consumer locally among the short grains. In addition, (R42 and R54), which are locally known under the name of Tahalf or Alliance rice, were introduced to the region by the Food and Agriculture Organization (FAO) in the late 1990s. From C3, the genotypes (R43 and R44) are registered under the name Ambar (IRGC 9505). All three varieties from the Al-Mishkhab research center are clustered under the C3 indica subpopulation (R43-Ambar-furat, R44-Ambar-muaazra, and R45-Yasamin).
Fig. 3.

a Circular dendrogram estimated through UPGMA, and b principal co-ordinates analysis of 62genotypes of rice based on 37 SSR markers
The genotypic data were rearranged based on the results obtained from STRUCTURE and PowerMarker. Then, PCoA and AMOVA analysis were performed using GenAlEx. Principal coordinates analysis (PCoA) showed an apparent variance between rice subpopulations (Fig. 3). They were clearly distributed, as shown in the central coordinates (1 vs. 2). Subpopulation C1 was allocated in quadrant 1, subpopulation C2 in quadrant 4, and subpopulation C3 in quadrant 2. Additionally, subpopulations C1 and C2 were more closely related than C3, which agrees with the phylogenetic tree output. The percentage of variation explained by the first 3 axes was 77.31% of the cumulative variation (Table 2).
Table 2.
Percentage of variation explained by the first 3 axes
| Axis | 1 | 2 | 3 |
|---|---|---|---|
| % | 49.22 | 24.29 | 3.79 |
| Cum % | 49.22 | 73.52 | 77.31 |
Table 3 shows the results of AMOVA based on 37 SSR markers (input as allelic distance matrix for F-Statistics analysis). The percentage of molecular variance for the three subpopulation outputs revealed that 71% of the variance was due to the variation among populations and 29% within the individuals (Table 3). The estimated variance among individuals (within single populations) is zero (0). A fixation index (FST) value of 0.726 was recorded at a significant level (P value = 0.001), and the gene flow (Nm) value was 0.095. Furthermore, AMOVA suppresses the source among individuals within populations. A small change was obtained in 72 % (among populations) and 28% (within populations). Finally, the AMOVA results based on PhiPT (Table 4) revealed a large change in the percentage of molecular variance of 75% (among populations) and 15% (within populations).
Table 3.
Analyses of molecular variance for the studied rice accessions
| Source of variation | AMOVA | AMOVA (suppressed) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| df | SS | MS | Est. var. | % | df | SS | MS | Est. var. | % | |
| Among populations | 2 | 562.086 | 281.043 | 10.265 | 71 | 2 | 562.086 | 281.043 | 10.265 | 72 |
| Among individuals within populations | 59 | 208.374 | 3.532 | 0.000 | 0 | |||||
| Within individuals | 62 | 263.000 | 4.242 | 2.242 | 29 | 121 | 471.374 | 3.896 | 3.896 | 28 |
| Total | 123 | 1033.460 | 14.160 | 100 | 123 | 1033.460 | 14.160 | 100 | ||
| FST | 0.726 (p < 0.001) | 0.725 (p < 0.001) | ||||||||
| Nm | 0.095 | 0.094 | ||||||||
Input as allelic distance matrix for F-statistics analysis df degrees of freedom, SS sum of squares, MS mean squared, Est var. estimates of variance, % percentage of variation
Table 4.
Analyses of molecular variance (PhiPT) for the studied rice accessions
| Source | df | SS | MS | Est. var. | % |
|---|---|---|---|---|---|
| Among populations | 2 | 1124.172 | 562.086 | 41.113 | 85 |
| Within populations | 59 | 416.747 | 7.064 | 7.064 | 15 |
| Total | 61 | 1540.919 | 48.176 | 100 | |
| FST | 0.853 | (p < 0.001) | |||
| Nm | 0.043 |
Input as genetic distance matrix for calculation of PhiPT
According to the pairwise population FST results (Table 5) below the diagonal, pairwise FST values of 0.685, 0.745, and 0.751 were recorded between subpopulations C1 and C2, C1 and C3, and C2 and C3, respectively. The Nm results (Table 5) above the diagonal showed that the highest Nm occurred (0.115) between C1 and C2 and the lowest (0.083) between C2 and C3.
Table 5.
The pairwise FST (below diagonal), Nm (above diagonal) among rice subpopulations
| Clusters | C1 | C2 | C3 |
|---|---|---|---|
| C1 | 0.000 | 0.115 | 0.085 |
| C2 | 0.685 | 0.000 | 0.083 |
| C3 | 0.745 | 0.751 | 0.000 |
Discussion
Evaluation of genetic diversity is an important factor for rice germplasm collection and breeding program. In this study, 37 SSR markers selected for genotyping 62 accessions of rice. The allele frequency values in the present study are similar to those found by [31, 32, 34]. They reported 2 to 7 alleles per locus in their genetic studies of rice from south Asia and India. The average gene diversity (0.442) in this study agrees with previous results [23, 32]. Most researchers investigating the genetic diversity of rice using SSR markers have utilized PIC as an indicator in their studies for the capability of markers to detect polymorphisms. Singh et al. [20]demonstrated that the PIC value depends on many factors, such as diverse germplasms, population sizes, genotypic methods and oligonucleotide marker loci in the genome. Therefore, different PIC values were reported as 0.240 [34], 0.416 [32], 0.420 [19], 0.483 [23], 0.560 [40], 0.570 [41], 0.630 [31], 0.704 [42], and 0.738 [43].
SSR is considered the best marker used to study the genetic diversity and characterizing of rice germplasm in the last two decades [21–23].The genetic structure analysis of any population is determined by the number of molecular markers that are used in any investigation. Zhang et al. [23] reported that 72 SSR markers are sufficient for population structure analysis of 150 rice varieties. Based on that, 30 SSR markers are sufficient for investigating the population structure analysis of 62 rice genotypes in the present study. However, we have used 37 SSR markers to be more reliable.
The short-grain rice accession clusters grouped under C1 and C2, and the long-grain accessions grouped under C3 based on a circular dendrogram (Fig. 3). The results suggest that clusters C1 and C2 are classified as japonica and cluster C3 indica subpopulations. These results are in agreement with those obtained by [21] in the eastern Himalayan region of northeast India, in Thailand [40], in Egypt [41], and by [32], who indicated two major (japonica and indica) subpopulations. In China, [43] identified three major groups: indica, temperate japonica, and tropical japonica.
In the present study, the allelic form of SSR was used, which is the most standard SSR method to obtain the AMOVA results by GenAlEx. A high level of genetic variation among populations of 71% was found among the studied rice accessions (Table 3). When the allelic form of SSR was used and there were differences among individuals within populations, there was no need to suppress this source of variance. However, when the estimated variance among individuals (within single populations) was set as zero (0) (Table 3), the data estimate was slightly negative (not significantly large differences among individuals were found).Then, within-individual analysis could be suppressed (Table 3). In addition, some researchers use PhiPT, where each diploid genotype is treated as a unit (in quantitative fashion). Because they want to know how different the genes are, from one individual to the next (track of individuals) rather than individual alleles, this will be notified by the degree of freedom (df) in the AMOVA table. In our case, when we used PhiPT to obtain AMOVA results, the percentage of molecular variance results varied. The variation among populations increased to 85, and within the population, it decreased to 15 (Table 4).Finally, all methods of AMOVA revealed high genetic differentiation among the populations, but it was low within the population. Similarly, Ab Razak et al. [16] reported 69 variance among the two populations and 29 variance among individuals in Malaysian rice varieties based on 916 SNP markers. Others reported considerable variance among individuals: 66 % [21], 74% [41], 86% [32].
Genetic differentiation based on FST values was determined by Wright [44]. They were low differentiation (FST = 0.00–0.05), moderate differentiation (FST = 0.05–0.15) and a high level of differentiation (FST of >0.30) [45]. In this study, a high level of genetic differentiation (FST = 0.726, p < 0.001) was indicated between subpopulations C1, C2 and C3 (Table 3). The lowest pairwise FST value of 0.685 was recorded between subpopulations C1 and C2, and the highest of 0.745 was recorded between subpopulations C2 and C3. Similarly, Verma et al. [46]Gouda et al. [47] Suvi et al. [48] reported a very high genetic differentiation FST of 0.827, 0.490 and 0.407 among subpopulations, respectively. The Nm value (0.095) indicates a very low or limited gene flow between subpopulations (C1, C2 and C3) (Table 4). According to [49],a value of Nm less than one will indicate the limitation of gene exchange between populations.
The PCoA, AMOVA, FST and Nm results were the key sources for finding the problem (very low gene flow) in the rice accessions in the region. There is a possibility that most of the individuals within a population are very close, and most farmers are not exchanging seeds. The government is distributing no seeds to farmers. Therefore, each year, the farmers keep a part of their seed for planting the following year.
Conclusions
In conclusion, the molecular diversity of the rice accessions in the region was divided into indica and japonica subpopulations based on a step-by-step analysis of STRUCTURE, PowerMarker, MEGA X, and GenAlEx software. Additionally, SSR proves its effectiveness for identifying a transparent background of rice genotypes in the region and shows that most rice accessions are very close in each subpopulation but under different local names. These findings in the present study are a perfect starting point for a rice breeding program and domestication of new species of rice in the region.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the Kurdistan Institution for Strategic Studies and Scientific Research (KISSR) for letting us work in their labs during the COVID-19 pandemic, 2020 and the Directory of Agricultural Research in Sulaymaniyah for providing nine rice seeds.
Author contributions
EH contributed to conception and design of the study. DH contributed to material preparation, samples collection and genotyping. EH preformed the data analysis and software. DH and EH wrote and edited the manuscript.
Funding
No funding was received for conducting this study.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Both authors have read and agreed to the published version of the manuscript.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Muthukumaran S. Between archaeology and text: the origins of rice consumption and cultivation in the Middle East and the Mediterranean. Pap Inst Archaeol. 2014;24:1–7. doi: 10.5334/pia.465. [DOI] [Google Scholar]
- 2.Ewaid SH, Abed SA, Chabuk A, Al-Ansari N. Water footprint of rice in Iraq. In: Ann R, editor. IOP conference series: earth and environmental science. 1. Bristol: IOP Publishing; 2021. p. p012008. [Google Scholar]
- 3.Helgilibrary (2017) Rice consumption per capita in Iraq. In: Helgilibrary, vol. 2020. https://www.helgilibrary.com/indicators/rice-consumption-per-capita/iraq/. Accessed 23 Oct 2020
- 4.Hameed KA, Mosa A-KJ, Jaber FA. Irrigation water reduction using system of rice intensification compared with conventional cultivation methods in Iraq. Paddy Water Environ. 2011;9(1):121–127. doi: 10.1007/s10333-010-0243-1. [DOI] [Google Scholar]
- 5.Khalil Z, Abdullaev S. Sensitivity of winter crops to climate variability in the irrigated subtropics of Iraq (Al-Diwaniyah) Proced Comput Sci. 2020;167:1066–1079. doi: 10.1016/j.procs.2020.03.405. [DOI] [Google Scholar]
- 6.Jeong J-M, Kim EC, Venkatanagappa S, Lee J-S. Review of rice: production, trade, consumption, and future demand in Korea and worldwide. Korean J Crop Sci. 2017;62(3):157–165. [Google Scholar]
- 7.Ewaid SH, Abed SA, Al-Ansari N. Assessment of main cereal crop trade impacts on water and land security in Iraq. Agronomy. 2020;10(1):98. doi: 10.3390/agronomy10010098. [DOI] [Google Scholar]
- 8.Mazumder SR, Hoque H, Sinha B, Chowdhury WR, Hasan MN, Prodhan SH. Genetic variability analysis of partially salt tolerant local and inbred rice (Oryza sativa L.) through molecular markers. Heliyon. 2020;6(8):e04333. doi: 10.1016/j.heliyon.2020.e04333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rekha T, Martin KP, Sreekumar V, Madassery J (2011) Genetic diversity assessment of rarely cultivated traditional indica rice (Oryza sativa L) varieties. Biotechnol Res Int 10.4061/2011/784719 [DOI] [PMC free article] [PubMed]
- 10.Kanawapee N, Sanitchon J, Srihaban P, Theerakulpisut P. Genetic diversity analysis of rice cultivars (Oryza sativa L.) differing in salinity tolerance based on RAPD and SSR markers. Electron J Biotechnol. 2011;14(6):2–2. doi: 10.2225/vol14-issue6-fulltext-4. [DOI] [Google Scholar]
- 11.Ravi M, Geethanjali S, Sameeyafarheen F, Maheswaran M. Molecular marker based genetic diversity analysis in rice (Oryza sativa L.) using RAPD and SSR markers. Euphytica. 2003;133(2):243–252. doi: 10.1023/A:1025513111279. [DOI] [Google Scholar]
- 12.Ren F, Lu B-R, Li S, Huang J, Zhu Y. A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor Appl Genet. 2003;108(1):113–120. doi: 10.1007/s00122-003-1414-x. [DOI] [PubMed] [Google Scholar]
- 13.Sorkheh K, Masaeli M, Chaleshtori MH, Adugna A, Ercisli S. AFLP-based analysis of genetic diversity, population structure, and relationships with agronomic traits in rice germplasm from north region of Iran and world core germplasm set. Biochem Genet. 2016;54(2):177–193. doi: 10.1007/s10528-016-9711-7. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, Gale M, Quarrie S, Jackson M, Bryan G. AFLP markers for the study of rice biodiversity. Theor Appl Genet. 1998;96(5):602–611. doi: 10.1007/s001220050778. [DOI] [Google Scholar]
- 15.Saini N, Jain N, Jain S, Jain RK. Assessment of genetic diversity within and among Basmati and non-Basmati rice varieties using AFLP, ISSR and SSR markers. Euphytica. 2004;140(3):133–146. doi: 10.1007/s10681-004-2510-y. [DOI] [Google Scholar]
- 16.Ab Razak S, Azman NHEN, Kamaruzaman R, Saidon SA, Yusof MFM, Ismail SN, Jaafar MA, Abdullah N. Genetic diversity of released Malaysian rice varieties based on single nucleotide polymorphism markers. Czech J Genet Plant Breed. 2020;56(2):62–70. doi: 10.17221/58/2019-CJGPB. [DOI] [Google Scholar]
- 17.Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3(9):e163. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao K, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, Tyagi W, Ali ML, Tung C-W, Reynolds A. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE. 2010;5(5):e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courtois B, Frouin J, Greco R, Bruschi G, Droc G, Hamelin C, Ruiz M, Clément G, Evrard JC, Van Coppenole S. Genetic diversity and population structure in a European collection of rice. Crop Sci. 2012;52(4):1663–1675. doi: 10.2135/cropsci2011.11.0588. [DOI] [Google Scholar]
- 20.Singh N, Choudhury DR, Singh AK, Kumar S, Srinivasan K, Tyagi R, Singh N, Singh R. Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS ONE. 2013;8(12):e84136. doi: 10.1371/journal.pone.0084136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhury B, Khan ML, Dayanandan S. Genetic structure and diversity of indigenous rice (Oryza sativa) varieties in the Eastern Himalayan region of Northeast India. SpringerPlus. 2013;2(1):1–10. doi: 10.1186/2193-1801-2-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169(3):1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS ONE. 2011;6(12):e27565. doi: 10.1371/journal.pone.0027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tello-Ruiz MK, Naithani S, Stein JC, Gupta P, Campbell M, Olson A, Wei S, Preece J, Geniza MJ, Jiao Y. Gramene 2018: unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 2018;46(D1):D1181–D1189. doi: 10.1093/nar/gkx1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh F, Yang R, Boyle T (1999) POPGENE (version 1.31). Microsoft window-bases freeware for population genetic analysis. University of Alberta and the Centre for International Forestry Research. https://sites.ualberta.ca/~fyeh/index.html
- 26.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Earl DA. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 28.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6(1):288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jasim Aljumaili S, Rafii M, Latif M, Sakimin SZ, Arolu IW, Miah G (2018) Genetic diversity of aromatic rice germplasm revealed by SSR markers. BioMed Res Int 10.1155/2018/7658032 [DOI] [PMC free article] [PubMed]
- 32.Nachimuthu VV, Muthurajan R, Duraialaguraja S, Sivakami R, Pandian BA, Ponniah G, Gunasekaran K, Swaminathan M, Suji K, Sabariappan R. Analysis of population structure and genetic diversity in rice germplasm using SSR markers: an initiative towards association mapping of agronomic traits in Oryza sativa. Rice. 2015;8(1):1–25. doi: 10.1186/s12284-015-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S, Xu W, Vijayakumar C, Ali J, Fu B, Xu J, Jiang Y, Marghirang R, Domingo J, Aquino C. Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theor Appl Genet. 2003;108(1):131–140. doi: 10.1007/s00122-003-1400-3. [DOI] [PubMed] [Google Scholar]
- 34.Anandan A, Anumalla M, Pradhan SK, Ali J. Population structure, diversity and trait association analysis in rice (Oryza sativa L.) germplasm for early seedling vigor (ESV) using trait linked SSR markers. PLoS ONE. 2016;11(3):e0152406. doi: 10.1371/journal.pone.0152406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy S, Banerjee A, Mawkhlieng B, Misra A, Pattanayak A, Harish G, Singh S, Ngachan S, Bansal K. Genetic diversity and population structure in aromatic and quality rice (Oryza sativa L.) landraces from North-Eastern India. PLoS ONE. 2015;10(6):e0129607. doi: 10.1371/journal.pone.0129607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohapatra S, Pandit E, Barik S, Patra B, Meher J, Pradhan S. Genetic diversity and population structure using linked SSR markers for heat stress tolerance in rice. ORYZA-Int J Rice. 2017;54(2):158–168. doi: 10.5958/2249-5266.2017.00021.2. [DOI] [Google Scholar]
- 37.Becerra V, Paredes M, Ferreira ME, Gutiérrez E, Díaz LM. Assessment of the genetic diversity and population structure in temperate japonica rice germplasm used in breeding in Chile, with SSR markers. Chil J Agric Res. 2017;77(1):15–26. doi: 10.4067/S0718-58392017000100002. [DOI] [Google Scholar]
- 38.Babu BK, Meena V, Agarwal V, Agrawal P. Population structure and genetic diversity analysis of Indian and exotic rice (Oryza sativa L.) accessions using SSR markers. Mol Biol Rep. 2014;41(7):4329–4339. doi: 10.1007/s11033-014-3304-5. [DOI] [PubMed] [Google Scholar]
- 39.Mahalingam A, Saraswathi R, Ramalingam J (2013) Simple sequence repeat (SSR) markers for assessing genetic diversity among the parental lines of hybrid rice (Oryza sativa L.). Afr J Biotechnol 10.5897/AJB12.557
- 40.Pathaichindachote W, Panyawut N, Sikaewtung K, Patarapuwadol S, Muangprom A. Genetic diversity and allelic frequency of selected Thai and exotic rice germplasm using SSR markers. Rice Sci. 2019;26(6):393–403. doi: 10.1016/j.rsci.2018.11.002. [DOI] [Google Scholar]
- 41.Salem KF, Sallam A. Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. CR Biol. 2016;339(1):1–9. doi: 10.1016/j.crvi.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Dang X, Thi TGT, Edzesi WM, Liang L, Liu Q, Liu E, Wang Y, Qiang S, Liu L, Hong D. Population genetic structure of Oryza sativa in East and Southeast Asia and the discovery of elite alleles for grain traits. Sci Rep. 2015;5(1):1–13. doi: 10.1038/srep11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Zheng X, Xu Q, Yuan X, Huang L, Zhou H, Wei X, Ge S. Genetic diversity and classification of Oryza sativa with emphasis on Chinese rice germplasm. Heredity. 2014;112(5):489–496. doi: 10.1038/hdy.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. doi: 10.1111/j.1558-5646.1965.tb01731.x. [DOI] [Google Scholar]
- 45.Wright S. Evolution and the genetics of populations: a treatise in four volumes: vol 4: variability within and among natural populations. Chicago: University of Chicago Press; 1978. [Google Scholar]
- 46.Verma H, Borah J, Sarma R. Variability assessment for root and drought tolerance traits and genetic diversity analysis of rice germplasm using SSR markers. Sci Rep. 2019;9(1):1–19. doi: 10.1038/s41598-019-52884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gouda AC, Ndjiondjop MN, Djedatin GL, Warburton ML, Goungoulou A, Kpeki SB, N’Diaye A, Semagn K. Comparisons of sampling methods for assessing intra-and inter-accession genetic diversity in three rice species using genotyping by sequencing. Sci Rep. 2020;10(1):1–14. doi: 10.1038/s41598-020-70842-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suvi WT, Shimelis H, Laing M, Mathew I, Shayanowako AIT. Assessment of the genetic diversity and population structure of rice genotypes using SSR markers. Acta Agric Scand B. 2020;70(1):76–86. doi: 10.1080/09064710.2019.1670859. [DOI] [Google Scholar]
- 49.Slatkin M. Rare alleles as indicators of gene flow. Evolution. 1985;39(1):53–65. doi: 10.1111/j.1558-5646.1985.tb04079.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.