Abstract
The SARS-CoV-2 virus causes a respiratory disease in humans, known as COVID-19. The confirmatory diagnostic of this disease occurs through the real-time reverse transcription and polymerase chain reaction test (RT-qPCR). However, the period of obtaining the results limits the application of the mass test. Thus, chest X-ray computed tomography (CT) images are analyzed to help diagnose the disease. However, during an outbreak of a disease that causes respiratory problems, radiologists may be overwhelmed with analyzing medical images. In the literature, some studies used feature extraction techniques based on CNNs, with classification models to identify COVID-19 and non-COVID-19. This work compare the performance of applying pre-trained CNNs in conjunction with classification methods based on machine learning algorithms. The main objective is to analyze the impact of the features extracted by CNNs, in the construction of models to classify COVID-19 and non-COVID-19. A SARS-CoV-2 CT data-set is used in experimental tests. The CNNs implemented are visual geometry group (VGG-16 and VGG-19), inception V3 (IV3), and EfficientNet-B0 (EB0). The classification methods were k-nearest neighbor (KNN), support vector machine (SVM), and explainable deep neural networks (xDNN). In the experiments, the best results were obtained by the EfficientNet model used to extract data and the SVM with an RBF kernel. This approach achieved an average performance of 0.9856 in the precision macro, 0.9853 in the sensitivity macro, 0.9853 in the specificity macro, and 0.9853 in the F1 score macro.
Keywords: COVID-19; non-COVID-19, Computed tomography images, Transfer learning, Convolutional neural networks, Machine learning
Introduction
The subtype 2 coronavirus, also known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), belongs to a family of viruses that cause respiratory diseases in humans [1]. The outbreak of coronavirus disease 2019 (COVID-19) caused the collapse of health systems in several countries. The infrastructure of public and private health centers did not endure the number of patients with severe symptoms of COVID-19, which contributed to the increase in the number of victims. Thus, some governments have adopted large-scale tests to obtain early diagnosis and reduce the effects of the disease on health systems.
The reverse transcription polymerase chain reaction test (RT-PCR) of the combined oral/nasal swab confirms the diagnosis of COVID-19 by detecting the genetic material of the virus in respiratory samples [1]. However, there are factors that hinder the application of this test, such as the genetic material of the virus becomes detectable generally in the first four or eight days after the onset of symptoms, and undetectable after fourteen days [1]; and due to the high complexity of the test, not all centers have suitable infrastructure and technical expertise to perform [1, 2].
In general, different methods of artificial intelligence have successfully been applied in diverse medical research areas [3]. Medical images and videos have been adopted as a relevant information source for medical diagnostic purposes, in which diverse machine learning algorithms are used to classify images or detect and describe some regions of interest in the analyzed images. Also, different techniques are used to improve image quality [4, 5]. In [6], a denoising filter is designed, which is based on local statistics to improve images quality that is important for providing a proper medical diagnosis of a disease.
The use of radiological medical images, such as chest radiography (x-ray), chest ultrasound and chest computed tomography (CT), has been adopted to assist in the early diagnosis of COVID-19 and to rule out the suspicion of diseases [2, 7], because, in severe cases of the disease, pneumonia is developed [8]. According to [9], the experimental results obtained using CT images were superior than those obtained using x-ray images. In addition, it is important to note that there are several chest CT datasets widely used for research purposes, recently applied in the diagnosis of COVID-19. Some studies demonstrated that chest CT images show some evidences of the development of COVID-19 in patients, as bilateral changes with ground-glass opacity with or without consolidation and interlobular septal thickening [10]. The diagnosis of COVID-19 in symptomatic adult patients, through chest CT images, reaches 90% of the chances of positive evolution of the disease, that is, 90% of sensitivity [10]. According to Jaiswal et al. [11], only CT images cannot be used to identify which virus is causing viral pneumonia. However, when considering these images with the clinical symptoms, travel history and identification of contact with infected people, it can be verified that it is the SARS-CoV-2 [11, 12].
However, during an outbreak of a disease that causes breathing problems, radiologists may be overwhelmed with medical image analysis [2, 8, 13]. In this context, applying different machine learning models [14–17] during a pandemic is very useful, as it performs the automatic analysis of medical images. Deep learning models are the state of the art in identifying COVID-19 and other lung diseases by imaging [2, 11, 18–22]. In [23], a novel deep convolutional neural network method, named VGG16-T, is used to identify lung cancer in early stage using CT images.
In [24], the authors propose a method of deep learning called deep explainable neural networks (xDNN), which is efficient in terms of time and consumption of computational resources. In the first stage of the method, the authors applied a fully connected network layer visual geometry group-16 (VGG-16) [25] to extract the vectors of global characteristics of the images. The results obtained using a SARS-CoV-2 CT-scan dataset [26] and considering accuracy, F1 score and area under the ROC curve (AUC) were 0.9738, 0.9731 and 0.9736, respectively. In [7], authors proposed a model named PatchShuffle stochastic pooling neural network (PSSPNN) for classification between COVID-19, secondary pulmonary tuberculosis, community-captured pneumonia, and healthy subjects. This method is able to show the explainable heatmap, which displays association with lung lesions; their experimental results achieved a microaveraged F1 score of 0.9579.
This work uses transfer learning [27] to extract vectors of global characteristics of image, applying layers of interest from pre-trained convolutional neural networks (CNNs) of the Keras1 application programming interface (API): VGG-16, VGG-19, Inception V3 and EfficientNet-B0. In Pathak et al. [28], experimental results have shown that a COVID-19 classification model built with deep transfer learning achieves better results than other supervised learning models. In addition, the classification models are the result of the application of the xDNN algorithm and Scikit-learn2 API algorithms: k-nearest neighbor (KNN) and support vector machine (SVM) with linear kernels and radial base function (RBF). The main objective is to analyze the impact of features extracted by CNNs, in the design of COVID-19 and non-COVID-19 classification models. The analysis results are estimated by non-parametric statistical tests, specifically Friedman Aligned Ranks [29] and Finner [30].
The main contributions of this work are summarized as follows:
Implementation of different pre-trained CNN models to extract image features and identifying the most appropriated features using Keras API;
Implementation of a high precision classification models, which can assist the diagnostic of COVID-19 and non-COVID-19;
A comparative statistical analysis of the performance of the COVID-19 and non-COVID-19 classification models designed using image features extracted by CNNs;
A framework composed that permits select different combination of pre-trained CNN models and classification models, identifying the best pre-trained CNN and classification models. The implementation code is available to the research community.
Hence, the present research intends to be useful in the classification of CT images that present COVID-19 characteristics. Also, this work presented a detailed methodology in which transfer learning is used, and the best parameters are selected from a pre-trained CNN, and later used as input of a classification algorithm, obtaining high accuracy results. It is worthy to note that the code3 of the solution developed is free available for research purposes.
The article is organized considering the following structure. Section 2 covers studies related to the topic of this work. Section 3 describes the methodology of the objects to create and evaluate the proposed models. In Sect. 4, the results of the model validation are presented and compared. Finally, in Sect. 5, the final considerations of the work are presented.
Theoretical Reference
This section presents the characteristics that help to detect COVID-19 by CT scan and a summary of feature extraction techniques and machine learning applied in this work.
COVID-19 detection based on computed tomography
CT images of the chest with COVID-19 are generated from the emission of radiation beams in the human body [31]. These images show important radiological patterns for the diagnostic of a radiologist and classification based on CT [32, 33]: 1) the initial stage is defined in the period of 0–4 days after the onset of the first symptom, whose ground-glass opacity (GGO) [34, 35] can be seen subpleurally in the lower lobes of the lungs, distributed unilaterally or bilaterally [33]; 2) the progressive stage occurs in the period of 5–8 days with diffuse GGO [36], mosaic paving [37] and consolidation [38] distributed in multiple bilateral lobes [33]; 3) the peak stage appears in the period of 9–13 days with dense consolidation [39] most prevalent [33]; 4) the infection control stage usually occurs after 14 days with the permanence of GGO and the absorption of consolidation and mosaic paving [33].
According to [33], the related works in the literature are divided into three classification tasks: classification of COVID-19 and non-COVID-19; classification of COVID-19 and pneumonia; and classification of the COVID-19 severity assessment. Unlike the first two tasks, the COVID-19 severity assessment classification seeks to identify the degree of sequelae caused by the disease [40, 41], thus, all patients are infected with SARS-CoV-2. The classification of COVID-19 and pneumonia seeks to separate patients infected with SARS-CoV-2 from those with pneumonia caused by another agent [7, 42–50]. The classification of COVID-19 and non-COVID-19 seeks to distinguish patients diagnosed with infection caused by SARS-Cov-2, from those that present characteristics of classic pneumonia, healthy or with a diagnostic of other lung diseases [51–60]. In addition, some works presented different methodologies [7] that are able to detect the lesion of the image indicating its severity.
In this context, this paper applies the classification task of COVID-19 and non-COVID-19. The validation of the built models is based on the SARS-CoV-2 CT-scan data set. This data set includes CT scans of patients infected with SARS-CoV-2 and other lung diseases.
Feature extraction methods for transfer learning
The feature extraction pre-processor defines a set of data with the ability to influence the performance of a classification model [24, 61]. In traditional machine learning, an expert method is applied to extract a vector of features depending on a certain domain. With deep learning, this pre-processing became inherent to the classification algorithm and to extract vectors from abstract features [24, 62, 63].
According to [28], deep learning models are applied in medical imaging systems, extracting features automatically or using pre-trained networks. The transformation of a pre-trained neural network into a feature extraction method occurs after the removal of some layers. The application of this modified network configures a learning transfer process. The transfer of learning is the enhancement of a new machine learning task, using the transfer of knowledge from an analogous task that has already been perfected [64].
The use of pre-trained networks with the data set ImageNet is widespread in the literature [28, 65]. Thus, pre-trained CNNs with this data set are applied to extract feature vectors in the identification of COVID-19. In [66] pre-trained networks were applied to inception V3 and VGG-19 with the wide and deep neural network (WDNN) and both reached 0.9847 and 0.9904 accuracy, respectively. In [26] the VGG-16 network was applied with xDNN and reached 0.9738 accuracy. In [58] it was applied to network EfficientNet with a vote-based approach and reached 0.9899 accuracy of the test. Due to the good performances, these networks were applied in this work.
A description of each network is presented below:
VGG: is an architecture that uses convolutional layers and the activation function rectified linear unit (ReLU). The VGG-16 and VGG-19 architectures are variations of the VGG with 16 and 19 layers, respectively. The filter dimension of the convolutional layers is 3x3 [25, 67, 68]. Different variations of the VGG network model, suc as 16 and 19, were applied in this work due to the simplicity of this architecture and because they are widely adopted in similar research; for instance, Soares et al. [26] used VGG-16 with xDNN.
Inception: is a CNN architecture published as GoogLeNet, also known as inception V1 [69, 70]. The architecture was improved, and new versions were presented, such as inception V2 and V3. Inception V2 has added batch normalization layers to streamline training [69, 71]. In relation to inception V3, factoring convolutions were added with larger spatial filters to improve computational efficiency [69, 72]. The module inception is the main building block. The convolutional layers have different dimensions of the kernel, 1x1 and 3x3, and an operation of polling [69]. This architecture was utilized in our experimental tests, it was applied in parallel convolutional layers in its main block, as suggested in [73].
EfficientNet: is a CNN architecture proposed by Tan and Quoc [74]. This architecture applies a scaling method that uniformly grows all dimensions of depth, width, and resolution efficiently, using a simple composite coefficient. Thus, a family of 8 convolutional networks was created, named EfficientNet-B0 to EfficientNet-B7. The main building block is the mobile inverted bottleneck convolution (MBConv). This architecture has few parameters, so it consumes less computational resources when compared to other CNNs [67, 74]. This architecture was used in our work, because it contains few parameters, and also because it obtained the best performance in the image classification from the ImageNet dataset, being 8.4 smaller and 6.1 faster on inference, these performance results were obtained in 2019 [74].
Supervised learning algorithms
Supervised learning is a machine learning task. This task uses essential information provided or acquired by human aid [75]. Thus, learning is obtained by classes (categories) identified by human specialists and examples of instances of each class. Classes with their instances make up the data set. The class and instance relationship is maintained in a subset of data called the training set, which is applied to learn a classification function. This classification function is used to classify unlabeled instances [76].
In the literature, different supervised learning techniques are applied in the identification of COVID-19 [24, 77, 78]: convolutional neural network (CNN); support vector machines (SVM); logistic regression (LR); Naive Bayes (NB); linear discriminant analysis (LDA); k-nearest neighbors (KNN); decision tree (DT) and random forest (RF).
In this work, the following algorithms were chosen:
KNN: is a classifier where learning is by analogy. The training set has n-dimensional vectors with their respective classes, and each vector in this set represents a point in n-dimensional space [79]. The classification decision is based on classes of k-nearest neighbors of a new instance [80].
SVM: is a binary vector classifier in an n-dimensional space. The objective of SVM is to find a hyperplane in an n-dimensional space that separates the examples into two distinct classes. The support vectors are the examples in the space (points) closest to the hyperplane. The SVM algorithm seeks to maximize the margin between the support vectors and the hyperplane [77, 81].
xDNN: is a classifier formed by several layers with a clear semantic and functional meaning, according to Plamen and Soares [24]. The use of prototype-based “” rules proposed by the authors makes it easier for users to understand how xDNN works. Prototypes are representative training images that help users visualize, understand, and appreciate similarity to other validation images. In other words, the prototypes are local density peaks and empirically derived probability distributions [26]. Thus, xDNN offers an association between statistical learning and reasoning [24].
Methodology
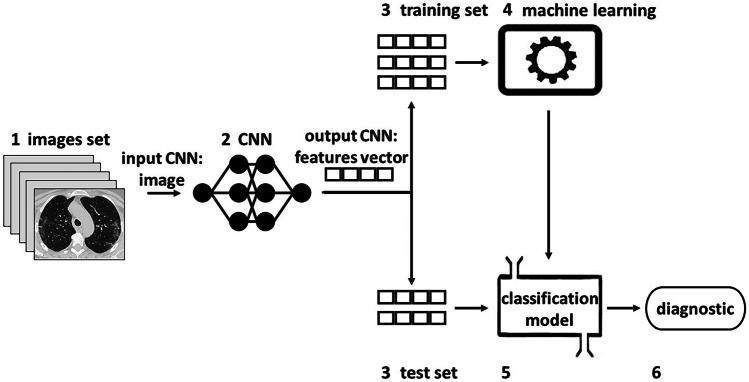
Figure 1 represents the proposed methodology to classify the diagnostic of a patient with suspected COVID-19. The methodology was implemented in python using the Scikit-learn and Keras machine learning algorithms APIs. Below, the steps applied to build the proposed model are presented:
Figure 1.
Proposed methodology to create and evaluate the classification model.
Data set: contains images of COVID-19 and non-COVID-19;
Feature extraction: CNN’s transformed, compressed, and reduced the information in the images. Thus, the Flatten layers transformed data matrices into vectors. The activation functions compressed (coded and transformed) the data to produce a reduced representation of these. In addition, the convolution and pooling operations layers reduced the dimensionality of the features map. The algorithms in this step return a features vector;
Training and test division: the set with feature vectors is randomly divided to perform stratified cross-validation with 10 folds of each model;
Machine learning: trains the model to classify the diagnostic of a patient with suspected COVID-19;
Classification model: it is the final result obtained in machine learning according to the training set. The samples from the test set, without the target classes, are classified by the model;
Diagnostic: it is the result of the classification model, based on an image of the test set with suspected COVID-19. This classification is compared with the target class to measure the performance of each model. Performance is compared in a statistical analysis to identify the best classification models, the best pre-trained CNNs for each model, and the best pre-trained CNN to extract characteristics from images in general.
Data set
The SARS-CoV-2 CT dataset [26] contains 2482 CT images. Of these, 1252 images belong to patients infected with SARS-CoV-2, and 1230 are from patients with other lung diseases. Thus, 50.44% of images belong to the class COVID-19 and 49.46% non-COVID-19. Therefore, the dataset is balanced and did not need any pre-processing.
Feature extraction by pre-trained CNN Models
From the data sets, the pre-trained models pre-processes the data, which the classification algorithms use to build the models and make the classifications. The CNN-based algorithms of the following functions were used to create the extraction models: VGG16, VGG19 and InceptionV3 (IV3) of the module tf.keras.applications; EfficientNetB0 (EB0) of model efficientnet.tfkeras. Two criteria were fundamental in the definition of the feature extraction layers: position closest to the layer that classifies and experimental tests considering the best performance in classification. The types of the layers chosen in the algorithms were:
FC (Dense) is a fully-connected layer of the VGG16 and VGG19 algorithms. This layer returns a feature vector with 4096 dimensions for each image and is used to extract features [82];
Flatten: it is a leveling layer of the algorithms VGG16 (VGG16 baseline), VGG19 (VGG19 baseline) and was applied to the block6d_add layer of EfficientNetB0 (EB0). This layer converts the data into a vector [83]. The sizes of the vectors returned from the baseline methods VGG16 and VGG19 were 25088 dimensions. Regarding the EB0 method, the returned vector has 9408 dimensions;
GlobalAveragePooling2D: this parameter represents a global pooling operation for spatial representation of the data, which is applied to replace the stage of fully connected layers in conventional CNNs. This operation generates a resource map for each class that will be classified [84]. This operation was used to obtain a drastic reduction of the dimensions of the feature maps in the layers, top_activation, mixed9 and mixed10 of the respective methods of the algorithms: EfficientNetB0 (EB0 baseline) and InceptionV3 (IV3 and IV3 baseline). The sizes of the vectors returned for methods IV3 and IV3 baseline were 2048 dimensions. The vector size returned by the EB0 baseline method was 1280 dimensions Table 1.
Table 1.
Summary of the chosen layer of each applied CNN.
| CNN | Layer type | Layer Name | Output Size |
|---|---|---|---|
| VGG16 | FC (Dense) | FC2 | 4096 |
| VGG16 baseline | Flatten | flatten | 25088 |
| VGG19 | FC (Dense) | FC2 | 4096 |
| VGG19 baseline | Flatten | flatten | 25088 |
| IV3 | GlobalAveragePooling2D | mixed9 | 2048 |
| IV3 baseline | GlobalAveragePooling2D | mixed10 | 2048 |
| EB0 | Flatten | block6d_add | 9408 |
| EB0 baseline | GlobalAveragePooling2D | top_activation | 1280 |
Models
The classification models were generated using data obtained from the feature extraction. Three machine learning algorithms were used to create the classification models: KNeighborsClassifier of the module sklearn.neighbors; SVC module of sklearn.svm; and xDNN4. In general, the standard configuration parameters of the algorithms have not been changed, except for SVC which was applied two kernels, linear and RBF.
Classification measures
The applied measures belong to the module sklearn.metrics and were implemented in python. The metrics of precision (PR), sensitivity (SE), specificity (SP), F1 scores and their respective macro-averages, use the values of the variables in the confusion matrix of each model. Figure 2 presents a confusion matrix.
Figure 2.
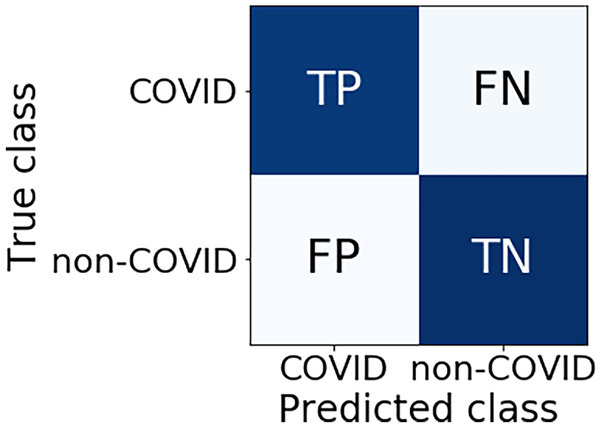
Confusion Matrix.
The values of the variables true positives (TP) and true negatives (TN), quantify the hits. Errors are quantified by the values of the variables false positives (FP) and false negatives (FN).
The metrics are defined in equations. PR calculates the probability of patients with positive results, actually having the disease [85]. The SE calculates the probability of a positive outcome of the disease [86]. SP, on the other hand, calculates the probability of negative results in patients without the disease [86]. The F1 score combines the values of precision and sensitivity, it balances the relative importance of each metric [76]. Finally, the macro averages of the metrics calculate their respective averages across classes [76, 87]. Thus, Eqs. 5, 6, 7, and 8 define the macro precision, macro sensitivity (macro recall), macro specificity, and macro F1 score, respectively.
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
| 8 |
Validation of models and comparative statistical analysis of results
In the validation of the models, the set of features was divided into training and test data by the class StratifiedKFold of the module sklearn.model_se-lection, to apply stratified cross-validation. Stratified cross-validation guarantees the same class distribution in the evaluated sets. This validation divides the set of features into k subsets (folds). Subsets are used as sets of training and testing features, for each k different models.
The results of the F1 macro-average of the models, in each cross-validation fold, were analyzed on the Web platform Statistical Tests for Algorithms Comparison (STAC) [88]. Due to the 32 models created and considering the 10 result values of each, the application Assistant estimated a non-parametric test of aligned rank. Thus, the statistical test Friedman Aligned Ranks was applied with a level of significance , verifying the null hypothesis that the averages of the results of the models are the same. This hypothesis was rejected and then the post-hoc test was applied Finner with a level of significance , verifying the null hypothesis that the average results of each pair of models compared are equal. The level of significance, denoted by indicates a 5% of risk to reject the null hypothesis when it is true. It means that there is a 5% of probability of predicting that the average performances of the algorithms are different when there is not a real difference [89].
Assessment and Results
In this section, the descriptions of the experimental evaluation and results of the pre-trained models of the classification of COVID-19 and non-COVID-19 will be presented, with the SARS-CoV-2 CT set. The objective is to validate the ability to generalize classification models with stratified cross-validation and to identify the best classification model in the applied data set. The experiments measured the performance of the proposed models using the confusion matrix, the metrics of precision, sensitivity, specificity, F1 score and their respective macro-averages, for each cross-validation fold. The average performances of the models in each macro-average and the result of their comparison, referring to the average of the F1 score, are shown in the Table 2.
Table 2.
Average performance of models in the classification of COVID-19 and non-COVID-19 - Cross-validation.
| Extractor | Model | Macro averages for each metric | Rank | |||
|---|---|---|---|---|---|---|
| PR | SE | SP | F1 | |||
| EB0 | SVC RBF | 0.9856 | 0.9853 | 0.9853 | 0.9853 | a |
| EB0 | SVC linear | 0.9855 | 0.9853 | 0.9853 | 0.9853 | a |
| VGG16 baseline | SVC linear | 0.9815 | 0.9813 | 0.9813 | 0.9813 | ab |
| EB0 | xDNN | 0.9794 | 0.9787 | 0.9787 | 0.9788 | abc |
| VGG19 baseline | SVC linear | 0.9741 | 0.9737 | 0.9737 | 0.9737 | abcd |
| EB0 | KNN | 0.9704 | 0.9696 | 0.9696 | 0.9697 | abcde |
| VGG16 baseline | xDNN | 0.9670 | 0.9661 | 0.9661 | 0.9662 | abcde |
| VGG19 baseline | KNN | 0.9655 | 0.9651 | 0.9651 | 0.9652 | abcdef |
| VGG19 | xDNN | 0.9649 | 0.9646 | 0.9646 | 0.9647 | abcdef |
| VGG16 | xDNN | 0.9630 | 0.9620 | 0.9620 | 0.9621 | bcdefg |
| VGG16 baseline | KNN | 0.9606 | 0.9600 | 0.9600 | 0.9601 | bcdefgh |
| VGG19 baseline | xDNN | 0.9612 | 0.9599 | 0.9599 | 0.9601 | cdefgh |
| IV3 | KNN | 0.9605 | 0.9601 | 0.9601 | 0.9601 | cdefgh |
| EB0 baseline | xDNN | 0.9603 | 0.9590 | 0.9590 | 0.9591 | defgh |
| VGG16 | SVC RBF | 0.9596 | 0.9585 | 0.9585 | 0.9586 | defgh |
| VGG16 | SVC linear | 0.9581 | 0.9570 | 0.9570 | 0.9571 | defgh |
| VGG19 | SVC RBF | 0.9538 | 0.9531 | 0.9531 | 0.9530 | efgh |
| VGG16 | KNN | 0.9529 | 0.9519 | 0.9519 | 0.9520 | efgh |
| IV3 | xDNN | 0.9517 | 0.9509 | 0.9509 | 0.9510 | efgh |
| VGG19 | SVC linear | 0.9515 | 0.9506 | 0.9506 | 0.9505 | efgh |
| VGG19 | KNN | 0.9483 | 0.9481 | 0.9481 | 0.9480 | fgh |
| EB0 baseline | KNN | 0.9461 | 0.9448 | 0.9448 | 0.9450 | fgh |
| EB0 baseline | SVC linear | 0.9428 | 0.9424 | 0.9424 | 0.9425 | ghi |
| IV3 baseline | KNN | 0.9353 | 0.9350 | 0.9350 | 0.9349 | hi |
| EB0 baseline | SVC RBF | 0.9310 | 0.9305 | 0.9305 | 0.9304 | ij |
| IV3 baseline | SVC linear | 0.9253 | 0.9238 | 0.9238 | 0.9238 | ij |
| IV3 baseline | xDNN | 0.9182 | 0.9172 | 0.9172 | 0.9172 | ij |
| IV3 baseline | SVC RBF | 0.9167 | 0.9159 | 0.9159 | 0.9157 | ij |
| VGG19 baseline | SVC RBF | 0.9110 | 0.8938 | 0.8938 | 0.8932 | ij |
| IV3 | SVC linear | 0.8952 | 0.8934 | 0.8934 | 0.8929 | ij |
| IV3 | SVC RBF | 0.8952 | 0.8934 | 0.8934 | 0.8929 | ij |
| VGG16 baseline | SVC RBF | 0.7903 | 0.6317 | 0.6317 | 0.5747 | j |
The averages of the F1 macro with the same letters do not differ at the probability level of 5% by the Finner test
The best performances in the F1 score are with the letter “a”
The worst performances in the F1 score are with the letter “j”
The highest averages for each metric are in bold
The Friedman Aligned Ranks test showed that the average of the F1 macro-average differs between the classification models with p-. Models with the same letters present the same averages in the F1 macro-average, at the probability level of 5% by the Finner test. The models that achieved the best performances are identified with “a”. So, in descending order, the worst models are with the letter “j”. This assessment identified the best CNNs to build each classification model. The best CNNs to build the best KNN models were EB0 and VGG19. Regarding the best SVC models with linear kernel, they were obtained with features provided by EB0, VGG16 baseline and VGG19 baseline. The best xDNN models were built with features provided by EB0, VGG16 baseline and VGG19. The best SVC model with RBF kernel was obtained with EB0 features. Therefore, EB0 is the best algorithm to extract features for the classifiers, as it was the only one that managed to achieve the best performances with each classification algorithm.
In absolute values, the highest performance was for the model with EB0 and SVC RBF, which reached 0.9856 precision and 0.9853 sensitivity, specificity, and F1 score. The lowest performance was the model with VGG16 baseline and SVC RBF, which reached 0.7925 precision, 0.6382 sensitivity and specificity, and 0.5856 F1 scores.
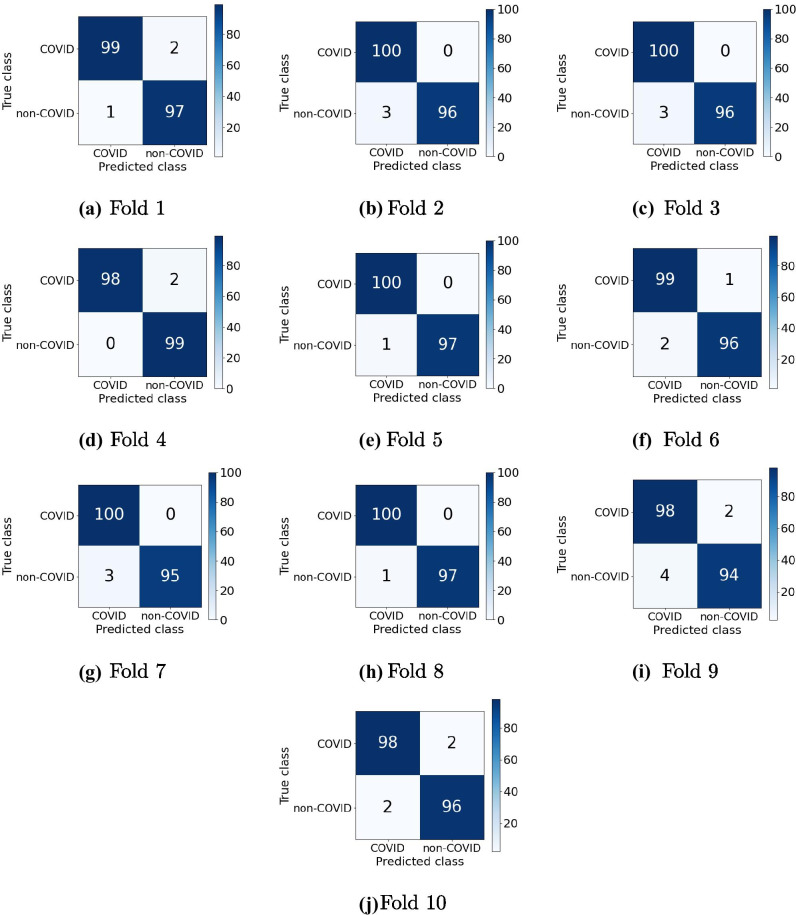
Figure 3 and Table 3 results by cross-validation fold are presented to verify the best performance of the model with EB0 and SVC RBF. The performances measured in the metrics demonstrate that the model can infer a correct diagnosis in both classes of each cross-validation fold.
Figure 3.
Confusion matrices of the model with EB0 and SVC RBF - Cross validation.
Table 3.
Performance of the model with EB0 and SVC RBF in the classification of COVID-19 and non-COVID-19 by cross-validation fold.
| Fold k | PR | SE | SP | F1 | Category |
|---|---|---|---|---|---|
| 1 | 0.9900 | 0.9801 | 0.9897 | 0.9850 | covid |
| 0.9797 | 0.9897 | 0.9801 | 0.9847 | non-covid | |
| 0.9848 | 0.9849 | 0.9849 | 0.9849 | general | |
| 2 | 0.9708 | 1.0000 | 0.9696 | 0.9852 | covid |
| 1.0000 | 0.9696 | 1.0000 | 0.9846 | non-covid | |
| 0.9854 | 0.9848 | 0.9848 | 0.9849 | general | |
| 3 | 0.9708 | 1.0000 | 0.9696 | 0.9852 | covid |
| 1.0000 | 0.9696 | 1.0000 | 0.9846 | non-covid | |
| 0.9854 | 0.9848 | 0.9848 | 0.9849 | general | |
| 4 | 1.0000 | 0.9800 | 1.0000 | 0.9898 | covid |
| 0.9801 | 1.0000 | 0.9800 | 0.9900 | non-covid | |
| 0.9900 | 0.9900 | 0.9900 | 0.9899 | general | |
| 5 | 0.9900 | 1.0000 | 0.9897 | 0.9950 | covid |
| 1.0000 | 0.9897 | 1.0000 | 0.9948 | non-covid | |
| 0.9950 | 0.9948 | 0.9948 | 0.9949 | general | |
| 6 | 0.9801 | 0.9900 | 0.9795 | 0.9850 | covid |
| 0.9896 | 0.9795 | 0.9900 | 0.9846 | non-covid | |
| 0.9849 | 0.9847 | 0.9847 | 0.9848 | general | |
| 7 | 0.9708 | 1.0000 | 0.9693 | 0.9852 | covid |
| 1.0000 | 0.9693 | 1.0000 | 0.9844 | non-covid | |
| 0.9854 | 0.9846 | 0.9846 | 0.9848 | general | |
| 8 | 0.9900 | 1.0000 | 0.9897 | 0.9950 | covid |
| 1.0000 | 0.9897 | 1.0000 | 0.9948 | non-covid | |
| 0.9950 | 0.9948 | 0.9948 | 0.9949 | general | |
| 9 | 0.9607 | 0.9800 | 0.9591 | 0.9702 | covid |
| 0.9791 | 0.9591 | 0.9800 | 0.9690 | non-covid | |
| 0.9699 | 0.9695 | 0.9695 | 0.9696 | general | |
| 10 | 0.9800 | 0.9800 | 0.9795 | 0.9800 | covid |
| 0.9795 | 0.9795 | 0.9800 | 0.9795 | non-covid | |
| 0.9797 | 0.9797 | 0.9797 | 0.9797 | general |
The category values general are the macro-average for each metric
The matrices present a high frequency of correctness in the first diagonal, composed of the frequencies of the correct identifications of COVID-19 (TP) and non-COVID-19 (TN). Each matrix in the Fig. 3 was generated by package functions matplotlib.pyplot in Python.
The framework proposed using the model with EB0 and SVC RBF a-chieved similar performance to those obtained by the works and reported in [26, 90–92]. In [90] a cross-validation with ten folds was used, as well as this work, and applied a method based on generative adversarial network (GAN), obtaining 0.9879 of F1 score. The other works used different methods to analyse the data obtained in relation to the considerations applied in this work. In [26], the model with VGG-16 and xDNN achieved a performance of 0.973 in the F1 score. In [91], an optimized convolutional neural network model, called ADECO-CNN, reached 0.996 of sensitivity, 0.992 of precision, and 0.997 of specificity. In [92], a semi-supervised shallow neural network structure, called Parallel Quantum-Inspired Self-supervised Network (PQIS-Net), achieved a performance of 0.948 considering the F1 score. It is worthy to note that some recent methods also have the additional advantage of identifying the region of lesion and its severity using a heatmap [7].
Conclusion
This work implemented pre-trained CNNs to extract the vectors of global features from CT images. The vectors were used to design classification models for COVID-19 and non-COVID-19. The Algorithms of CNNs applied were VGG16, VGG19, InceptionV3 (IV3) and EfficientNetB0 (EB0). The classification algorithms applied were KNeighborsClassifier (KNN), SVC and Explainable Deep Neural networks (xDNN).
In the experimental evaluation, the performances of each model were compared in a statistical analysis using the Friedman Aligned Ranks and Finner tests. This analysis identified the best classification models, the best pre-trained CNNs to extract characteristics from images for each model, and the best pre-trained CNN in general. Thus, the KNN classifier achieved the best performance with EB0 and VGG19. The SVC linear classifier obtained the best performance with EB0, VGG16 baseline, and VGG19 baseline. The SVC RBF classifier achieved the best performance with the features extracted by the EB0 algorithm. Finally, the XDNN classifier performed best with the features provided by EB0, VGG16 baseline, and VGG19. The SVC RBF with data extracted by EB0 was the model that obtained the highest average performances: 0.9856 in the precision macro, 0.9853 in the sensitivity macro, 0.98553 in the specificity macro, and 0.9853 in the F1 score macro. In this scenario, the EB0 algorithm is the best to extract features for the classifiers, as it was the only one that achieved the best performances in each classification model.
Therefore, the use of algorithms based on transfer learning to extract characteristics from CT images was beneficial to identify COVID-19 and non-COVID-19, in the cross-validation tests. With the exception of one model, the models achieved excellent results. The inferiority of the model with VGG16 baseline and SVC RBF can be attributed to the mapping to another larger space, carried out by the kernel. Another justification is the lack of variation in the parameters of the classification algorithm. In addition, the better performances of the models with EB0, when compared with the others, can be justified by the lack of variation in the parameters of the feature extraction algorithms.
In future works, the parameters variation of the feature extraction and classification algorithms will be carried out to try to reach the maximum performance in the identification of COVID-19 and non-COVID-19. In addition, the same approach will be applied to classify the stages of COVID-19. In addition, different methods to improve image quality will be used in future experiments.
Acknowledgements
Sattam Al Otaibi would like to thank Taif University Researchers Supporting Project number (TURSP2020/228), Taif University, Taif, Saudi Arabia. Authors also wish to thank the Brazilian National Council for Scientific and Technological Development (CNPq), the SINE-CNPq research group and Multimedia Signal Processing laboratory at UFLA.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arthur A. M. Teodoro, Email: arthur.teodoro@estudante.ufla.br
Douglas H. Silva, Email: douglas.silva2@estudante.ufla.br
Muhammad Saadi, Email: muhammad.saadi@ucp.edu.pk.
Ogobuchi D. Okey, Email: ogobuchi.okey@estudante.ufla.br
Renata L. Rosa, Email: renata.rosa@ufla.br
Sattam Al Otaibi, Email: srotaibi@tu.edu.sa.
Demóstenes Z. Rodríguez, Email: demostenes.zegarra@ufla.br
References
- 1.Magno, L., Rossi, T. A., Mendonça-Lima, F. W. D., Santos, C. C. D., Campos, G. B., Marques, L. M., & Dourado, I. (2020). Challenges and proposals for scaling up COVID-19 testing and diagnosis in Brazil. Ciencia & Saude Coletiva, 25, 3355-3364. [DOI] [PubMed]
- 2.Barbosa RC, Rosa RL, da Silva KCN, Rodriguez DZ. Ct-fastnet: Detection of covid-19 from chest computedtomography (ct) images using artificial intelligence. Brazilian Journal of Development. 2020;6(7):50315–50330. doi: 10.34117/bjdv6n7-619. [DOI] [Google Scholar]
- 3.Bao, Y., Qiu, W., Tang, P., & Cheng, X. (2021). Efficient, revocable and privacy-preserving fine-grained data sharing with keyword search for the cloud-assisted medical iot system. IEEE Journal of Biomedical and Health Informatics, 1. 10.1109/JBHI.2021.3100871 [DOI] [PubMed]
- 4.Singh, P., Diwakar, M., Cheng, X., Shankar, A. (2021). A new wavelet-based multi-focus image fusion technique using method noise and anisotropic diffusion for real-time surveillance application, Journal of Real-Time Image Processing 1–18.
- 5.Chen S, Shi D, Sadiq M, Cheng X. Image denoising with generative adversarial networks and its application to cell image enhancement. IEEE Access. 2020;8:82819–82831. doi: 10.1109/ACCESS.2020.2988284. [DOI] [Google Scholar]
- 6.Singh K, Sharma B, Singh J, Srivastava G, Sharma S, Aggarwal A, Cheng X. Local statistics-based speckle reducing bilateral filter for medical ultrasound images. Mobile Networks and Applications. 2020;25(6):2367–2389. doi: 10.1007/s11036-020-01615-2. [DOI] [Google Scholar]
- 7.Wang, S. H., Zhang, Y., Cheng, X., Zhang, X., & Zhang, Y. D. (2021). Psspnn: Patchshuffle stochastic pooling neural network for an explainable diagnosis of covid-19 with multiple-way data augmentation. Computational and Mathematical Methods in Medicine 2021. [DOI] [PMC free article] [PubMed] [Retracted]
- 8.Yang, X., He, X., Zhao, J., Zhang, Y., Zhang, S., & Xie, P. (2020). Covid-ct-dataset: A ct scan dataset about covid-19. arXiv:2003.13865.
- 9.Benmalek, E., Elmhamdi, J., & Jilbab, A. (2021). Comparing ct scan and chest x-ray imaging for covid-19 diagnosis. Biomedical Engineering Advances,1, 100003. [DOI] [PMC free article] [PubMed]
- 10.Park, J. Y., Freer, R., Stevens, R., Soneji, N., & Jones, N. (2021). The accuracy of chest ct in the diagnosis of covid-19: An umbrella review. https://www.cebm.net/covid-19/the-accuracy-of-chest-ct-in-the-diagnosis-of-covid-19-an-umbrella-review
- 11.Jaiswal, A., Gianchandani, N., Singh, D., Kumar, V., & Kaur, M. (2020). Classification of the covid-19 infected patients using densenet201 based deep transfer learning. Journal of Biomolecular Structure and Dynamics 1–8. PMID: 32619398. 10.1080/07391102.2020.1788642 [DOI] [PubMed]
- 12.Xie, X., Zhong, Z., Zhao, W., Zheng, C., Wang, F., & Liu, J. (2020). Chest ct for typical coronavirus disease 2019 (covid-19) pneumonia: Relationship to negative rt-pcr testing. Radiology 296, 2, 200343. PMID: 32049601. 10.1148/radiol.2020200343 [DOI] [PMC free article] [PubMed]
- 13.Carrillo D, Lam DN, Nardelli PHJ, Pournaras E, Morita P, Rodríguez DZ, Dzaferagic M, Siljak H, Jung A, Laurent H-D, Macaluso I, Ullah M, Fraidenraich G, Popovski P. Containing future epidemics with trustworthy federated systems for ubiquitous warning and response. Frontiers in Communications and Networks. 2021;2:11. doi: 10.3389/frcmn.2021.621264. [DOI] [Google Scholar]
- 14.Mendonça RV, Teodoro AAM, Rosa RL, Saadi M, Melgarejo DC, Nardelli PHJ, Rodríguez DZ. Intrusion detection system based on fast hierarchical deep convolutional neural network. IEEE Access. 2021;9:61024–61034. doi: 10.1109/ACCESS.2021.3074664. [DOI] [Google Scholar]
- 15.Ribeiro DA, Silva JC, Lopes Rosa R, Saadi M, Mumtaz S, Wuttisittikulkij L, et al. Light field image quality enhancement by a lightweight deformable deep learning framework for intelligent transportation systems. Electronics. 2021;10(10):1136. doi: 10.3390/electronics10101136. [DOI] [Google Scholar]
- 16.Saadi, M., Saeed, Z., Ahmad, T., Saleem, M. K., & Wuttisittikulkij, L. (2019). Visible light-based indoor localization using k-means clustering and linear regression. Transactions on Emerging Telecommunications Technologies,30(2), e3480.
- 17.Terra Vieira S, Lopes Rosa R, Zegarra Rodríguez D, Arjona Ramírez M, Saadi M, Wuttisittikulkij L. Q-meter: Quality monitoring system for telecommunication services based on sentiment analysis using deep learning. Sensors. 2021;21(5):1880. doi: 10.3390/s21051880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardila, D., Kiraly, A. P., Bharadwaj, S., Choi, B., Reicher, J. J., Peng, L., Tse, D., Etemadi, M., Ye, W., Corrado, G., Naidich, D. P., & Shetty, S. (2019). End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature Medicine 25, 6, 954–961. 10.1038/s41591-019-0447-x [DOI] [PubMed]
- 19.Rosa RL, De Silva MJ, Silva DH, Ayub MS, Carrillo D, Nardelli PHJ, Rodríguez DZ. Event detection system based on user behavior changes in online social networks: Case of the covid-19 pandemic. IEEE Access. 2020;8:158806–158825. doi: 10.1109/ACCESS.2020.3020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, G., Liu, Z., van der Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. In 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, July 21-26, 2017, IEEE Computer Society, pp. 2261–2269. 10.1109/CVPR.2017.243
- 21.Li, L., Qin, L., Xu, Z., Yin, Y., Wang, X., Kong, B., Bai, J., Lu, Y., Fang, Z., Song, Q., Cao, K., Liu, D., Wang, G., Xu, Q., Fang, X., Zhang, S., Xia, J., & Xia, J. (2020). Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary ct: Evaluation of the diagnostic accuracy. Radiology 296, 2, 201178. PMID: 32191588. 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed]
- 22.Suzuki, K. (2017). Overview of deep learning in medical imaging. Radiological Physics and Technology 10, 3, 257–273. 10.1007/s12194-017-0406-5 [DOI] [PubMed]
- 23.Pang S, Meng F, Wang X, Wang J, Song T, Wang X, Cheng X. Vgg16-t: A novel deep convolutional neural network with boosting to identify pathological type of lung cancer in early stage by ct images. International Journal of Computational Intelligence Systems. 2020;13:771–780. doi: 10.2991/ijcis.d.200608.001. [DOI] [Google Scholar]
- 24.Angelov PP, Soares EA. Towards explainable deep neural networks (xdnn) Neural Networks. 2020;130:185–194. doi: 10.1016/j.neunet.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Simonyan, K., & Zisserman, A. (2015). Very deep convolutional networks for large-scale image recognition. In 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings, Y. Bengio and Y. LeCun, Eds.
- 26.Soares, E., Angelov, P., Biaso, S., Froes, M. H., & Abe, D. K. (2020). Sars-cov-2 ct-scan dataset: A large dataset of real patients ct scans for sars-cov-2 identification. medRxiv.
- 27.Jain R, Jain N, Aggarwal A, Hemanth DJ. Convolutional neural network based alzheimerâ??s disease classification from magnetic resonance brain images. Cognitive Systems Research. 2019;57:147–159. doi: 10.1016/j.cogsys.2018.12.015. [DOI] [Google Scholar]
- 28.Pathak, Y., Shukla, P., Tiwari, A., Stalin, S., Singh, S., & Shukla, P. (2020). Deep transfer learning based classification model for covid-19 disease. IRBM. 10.1016/j.irbm.2020.05.003 [DOI] [PMC free article] [PubMed]
- 29.Hodges JL, Lehmann EL. Rank methods for combination of independent experiments in analysis of variance. In: Lehmann L, Rojo J, editors. Selected Works of E. Boston, MA: Springer, US; 2012. pp. 403–418. [Google Scholar]
- 30.Finner H. On a monotonicity problem in step-down multiple test procedures. Journal of the American statistical association. 1993;88(423):920–923. doi: 10.1080/01621459.1993.10476358. [DOI] [Google Scholar]
- 31.Pavan Kumar, M. R., & Jayagopal, P. (2020). Generative adversarial networks: a survey on applications and challenges. International Journal of Multimedia Information Retrieval. [DOI] [PMC free article] [PubMed]
- 32.Pan, F., Ye, T., Sun, P., Gui, S., Liang, B., Li, L., Zheng, D., Wang, J., Hesketh, R. L., Yang, L., & Zheng, C. (2020). Time course of lung changes on chest ct during recovery from 2019 novel coronavirus (covid-19) pneumonia. Radiology 295, 3, 715–721. PMID: 32053470. [DOI] [PMC free article] [PubMed]
- 33.Shi, F., Wang, J., Shi, J., Wu, Z., Wang, Q., Tang, Z., He, K., Shi, Y., & Shen, D. (2020). Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. CoRR abs/2004.02731. [DOI] [PubMed]
- 34.Hansell, D. M., Bankier, A. A., MacMahon, H., McLoud, T. C., Muller, N. L., & Remy J. (2008) Fleischner society: Glossary of terms for thoracic imaging. Radiology 246 (3), 697–722, pMID: 18195376. 10.1148/radiol.2462070712 [DOI] [PubMed]
- 35.Kobayashi, Y., & Mitsudomi, T. (2013). Management of ground-glass opacities: should all pulmonary lesions with ground-glass opacity be surgically resected? Translational Lung Cancer Research,2, 5. https://tlcr.amegroups.com/article/view/1608 [DOI] [PMC free article] [PubMed]
- 36.Santos, M. L. A. d. O., Marchiori, E., Vianna, A. D., Souza Jr., A. S., & Moraes, H. P. D. (2003). Ground-glass opacity in diffuse lung diseases: high-resolution computed tomography-pathology correlation. Radiologia Brasileira 36, 329–338. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0100-39842003000600003&nrm=iso
- 37.Vabo, K. A. D., & Damato, S. D. (2011). Tomographic and pathological findings of crazy-paving pattern. Radiologia Brasileira 44, 215 – 219. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0100-39842011000400005&nrm=iso
- 38.Elicker, B., Pereira, C. A. D. C., Webb, R., & Leslie, K. O. (2008). High-resolution computed tomography patterns of diffuse interstitial lung disease with clinical and pathological correlation. Jornal Brasileiro de Pneumologia 34, 715 – 744. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1806-37132008000900013&nrm=iso [DOI] [PubMed]
- 39.Marchiori, E., Zanetti, G. A., & Hochhegger, B. (2015). Dense consolidations. Jornal Brasileiro de Pneumologia 41, 388–388. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1806-37132015000400388&nrm=iso [DOI] [PMC free article] [PubMed]
- 40.Tang, Z., Zhao, W., Xie, X., Zhong, Z., Shi, F., Liu, J., & Shen, D. (2020). Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. CoRR abs/2003.11988. arXiv: 2003.11988. https://arxiv.org/abs/2003.11988
- 41.Xiao, L. S., Li, P., Sun, F., Zhang, Y., Xu, C., Zhu, H., Cai, F. Q., He, Y. L., Zhang, W. F., Ma, S. C., Hu, C., Gong, M., Liu, L., Shi, W., & Zhu, H. (2020). Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Frontiers in bioengineering and biotechnology 8, 898. PMCID: PMC7411489. 10.3389/fbioe.2020.00898 [DOI] [PMC free article] [PubMed]
- 42.Shi, F., Xia, L., Shan, F., Wu, D., Wei, Y., Yuan, H., Jiang, H., Gao, Y., Sui, H., & Shen, D. (2020). Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. arXiv:2003.09860. [DOI] [PubMed]
- 43.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary ct: Evaluation of the diagnosticaccuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai, H. X., Wang, R., Xiong, Z., Hsieh, B., Chang, K., Halsey, K., Tran, T. M. L., Choi, J. W., Wang, D. C., Shi, L. B., Mei, J., Jiang, X. L., Pan, I., Zeng, Q. H., Hu, P. F., Li, Y. H., Fu, F. X., Huang, R. Y., Sebro, R., Yu, Q. Z., Atalay, M. K., & Liao, W. H. (2020). Artificial intelligence augmentation of radiologist performance in distinguishing covid-19 from pneumonia of other origin at chest ct. Radiology 296, 3, E156–E165. PMID: 32339081. 10.1148/radiol.2020201491. https://pubmed.ncbi.nlm.nih.gov/32339081 [DOI] [PMC free article] [PubMed]
- 45.Ko, H., Chung, H., Kang, W. S., Kim, K. W., Shin, Y., Kang, S. J., Lee, J. H., Kim, Y. J., Kim, N. Y., Jung, H., & Lee, J. (2020). Covid-19 pneumonia diagnosis using a simple 2d deep learning framework with a single chest ct image: Model development and validation. Journal of medical Internet research 22, 6, e19569–e19569. PMCID: PMC7332254. 10.2196/19569 [DOI] [PMC free article] [PubMed]
- 46.Ni, Q., Sun, Z. Y., Qi, L., Chen, W., Yang, Y., Wang, L., Zhang, X., Yang, L., Fang, Y., Xing, Z., Zhou, Z., Yu, Y., Lu, G. M., & Zhang, L. J. (2020). A deep learning approach to characterize 2019 coronavirus disease (covid-19) pneumonia in chest ct images. European Radiology 30, 12, 6517–6527. 10.1007/s00330-020-07044-9 [DOI] [PMC free article] [PubMed]
- 47.Hu, R., Ruan, G., Xiang, S., Huang, M., Liang, Q., & Li, J. (2020). Automated diagnosis of covid-19 using deep learning and data augmentation on chest ct. medRxiv. arXiv:https://www.medrxiv.org/content/early/2020/05/03/2020.04.24.20078998.full.pdf, 10.1101/2020.04.24.20078998. https://www.medrxiv.org/content/early/2020/05/03/2020.04.24.20078998
- 48.Ying, S., Zheng, S., Li, L., Zhang, X., Zhang, X., Huang, Z., Chen, J., Zhao, H., Wang, R., Chong, Y., Shen, J., Zha, Y., & Yang, Y. (2020). Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images. medRxiv. arXiv:https://www.medrxiv.org/content/early/2020/02/25/2020.02.23.20026930.full.pdf, 10.1101/2020.02.23.20026930 [DOI] [PMC free article] [PubMed]
- 49.Xu, X., Jiang, X., Ma, C., Du, P., Li, X., Lv, S., Yu, L., Ni, Q., Chen, Y., Su, J., et al. (2020). A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 6, 10, 1122–1129. 10.1016/j.eng.2020.04.010 [DOI] [PMC free article] [PubMed]
- 50.Han Z, Wei B, Hong Y, Li T, Cong J, Zhu X, et al. Accurate screening of covid-19 using attention-based deep 3d multiple instance learning. IEEE Transactions on Medical Imaging. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 51.Ragab, D. A., & Attallah, O. (2020). Fusi-cad: Coronavirus (covid-19) diagnosis based on the fusion of cnns and handcrafted features. PeerJ Computer Science,6, e306. [DOI] [PMC free article] [PubMed]
- 52.Tetila, E., Bressem, K., Astolfi, G., Sant’Ana, D. A., Pache, M. C., & Pistori, H. (2020) System for quantitative diagnosis of covid-19-associated pneumonia based on superpixels with deep learning and chest ct. 10.21203/rs.3.rs-123158/v1
- 53.Alshazly H, Linse C, Barth E, Martinetz T. Explainable covid-19 detection using chest ct scans and deep learning. Sensors. 2021;21:2. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panwar, H., Gupta, P., Siddiqui, M. K., Morales-Menendez, R., Bhardwaj, P., & Singh, V. (2020). A deep learning and grad-cam based color visualization approach for fast detection of covid-19 cases using chest x-ray and ct-scan images. Chaos, Solitons & Fractals,140, 110190. 10.1016/j.chaos.2020.110190 [DOI] [PMC free article] [PubMed]
- 55.Liang, L., Ma, L., Qian, L., Chen, J. (2020). An algorithm for out-of-distribution attack to neural network encoder. arXiv:2009.08016.
- 56.Kechagias-Stamatis, O., Aouf, N., & Koukos, J. A. (2020). Deep learning fusion for covid-19 diagnosis. medRxiv. arXiv:https://www.medrxiv.org/content/early/2020/12/14/2020.12.11.20246546.full.pdf, 10.1101/2020.12.11.20246546
- 57.Silva, P., Luz, E., Silva, G., Moreira, G., Silva, R., Lucio, D., & Menottu, D. (2020). Efficient deep learning model for covid-19 detection in large ct images datasets: A cross-dataset analysis. 10.21203/rs.3.rs-41062/v1 [DOI] [PMC free article] [PubMed]
- 58.Silva, P., Luz, E., Silva, G., Moreira, G., Silva, R., Lucio, D., & Menotti, D. (2020). Covid-19 detection in ct images with deep learning: A voting-based scheme and cross-datasets analysis. Informatics in Medicine Unlocked,20, 100427. 10.1016/j.imu.2020.100427 [DOI] [PMC free article] [PubMed]
- 59.Wang Z, Liu Q, Dou Q. Contrastive cross-site learning with redesigned net for covid-19 ct classification. IEEE J. Biomed. Health Informatics. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen, S., Saha, S., Chatterjee, S., Mirjalili, S., & Sarkar, R. (2021). A bi-stage feature selection approach for covid-19 prediction using chest ct images. Applied Intelligence. 10.1007/s10489-021-02292-8 [DOI] [PMC free article] [PubMed]
- 61.Rodríguez DZ, Rosa RL, Almeida FL, Mittag G, Müller S. Speech quality assessment in wireless communications with mimo systems using a parametric model. IEEE Access. 2019;7:35719–35730. doi: 10.1109/ACCESS.2019.2902798. [DOI] [Google Scholar]
- 62.Lasmar EL, de Paula FO, Rosa RL, Abrahão JI, Rodríguez DZ. Rsrs: Ridesharing recommendation system based on social networks to improve the user’s qoe. IEEE Transactions on Intelligent Transportation Systems. 2019;20(12):4728–4740. doi: 10.1109/TITS.2019.2945793. [DOI] [Google Scholar]
- 63.Rosa, R. L., Rodriguez, D. Z., Bressan, G. (2013). Sentimeter-br: A new social web analysis metric to discover consumers’ sentiment, in: 2013 IEEE International Symposium on Consumer Electronics (ISCE), IEEE, 2013, pp. 153–154.
- 64.Torrey, L., & Shavlik, J. (2010). Transfer learning. In Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI global, pp. 242–264.
- 65.Barbosa RC, Ayub MS, Rosa RL, Rodríguez DZ, Wuttisittikulkij L. Lightweight pvidnet: a priority vehicles detection network model based on deep learning for intelligent traffic lights. Sensors. 2020;20(21):6218. doi: 10.3390/s20216218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hassan, E., Hikal, N. A., & Elmuogy, S. (2020). An efficient technique for ct scan images classification of covid-19. Journal of Intelligent & Fuzzy Systems Preprint, 1–14. Preprint. 10.3233/JIFS-201985
- 67.Shoeibi, A., Khodatars, M., Alizadehsani, R., Ghassemi, N., Jafari, M., Moridian, P., Khadem, A., Sadeghi, D., Hussain, S., Zare, A., Sani, Z. A., Bazeli, J., Khozeimeh, F., Khosravi, A., Nahavandi, S., Acharya, U. R., & Shi, P. (2020). Automated detection and forecasting of COVID-19 using deep learning techniques: A review. CoRR abs/2007.10785. arXiv:2007.10785. https://arxiv.org/abs/2007.10785
- 68.Alom, M. Z., Taha, T. M., Yakopcic, C., Westberg, S., Sidike, P., Nasrin, M. S., Esesn, B. C. V., Awwal, A. A. S., & Asari, V. K. (2018). The history began from alexnet: A comprehensive survey on deep learning approaches. CoRR abs/1803.01164. arXiv:1803.01164. http://arxiv.org/abs/1803.01164
- 69.Alshazly, H., Linse, C., Barth, E., & Martinetz, T. (2021). Explainable covid-19 detection using chest ct scans and deep learning. Sensors,21, 2. arXiv:2011.05317. https://arxiv.org/abs/2011.05317 [DOI] [PMC free article] [PubMed]
- 70.Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S. E., Anguelov, D., Erhan, D., Vanhoucke, V., & Rabinovich, A. (2015). Going deeper with convolutions. In IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2015, Boston, MA, USA, June 7-12, 2015, IEEE Computer Society, pp. 1–9. 10.1109/CVPR.2015.7298594
- 71.Ioffe, S., & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, ICML 2015, Lille, France, 6-11 July 2015, F. R. Bach and D. M. Blei, Eds., vol. 37 of JMLR Workshop and Conference Proceedings, JMLR.org, pp. 448–456. http://proceedings.mlr.press/v37/ioffe15.html
- 72.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna, Z. (2015). Rethinking the inception architecture for computer vision. CoRR abs/1512.00567. arXiv:1512.00567. http://arxiv.org/abs/1512.00567
- 73.Adewole, S., Yeghyayan, M., Hyatt, D., Ehsan, L., Jablonski, J., Copland, A., Syed, S., & Brown, D. (2021). Deep learning methods for anatomical landmark detection in video capsule endoscopy images. In Proceedings of the Future Technologies Conference (FTC) 2020, Volume 1 (Cham, 2021), Springer International Publishing, pp. 426–434. [DOI] [PMC free article] [PubMed]
- 74.Tan, M., & Le, Q. V. (2019). Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, ICML 2019, 9-15 June 2019, Long Beach, California, USA, K. Chaudhuri and R. Salakhutdinov, Eds., vol. 97 of Proceedings of Machine Learning Research, PMLR, pp. 6105–6114. http://proceedings.mlr.press/v97/tan19a.html
- 75.Guimarães, R., Rodríguez, D. Z., Rosa, R. L., & Bressan, G. (2016). Recommendation system using sentiment analysis considering the polarity of the adverb. In 2016 IEEE International Symposium on Consumer Electronics (ISCE), IEEE, pp. 71–72.
- 76.Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval: The concepts and technology behind search. 2. Boston, MA: Addison-Wesley Publishing Company; 2011. [Google Scholar]
- 77.Mohamadou, Y., Halidou, A., & Kapen, P. T. (2020). A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of covid-19. Applied Intelligence 50, 11, 3913–3925. 10.1007/s10489-020-01770-9 [DOI] [PMC free article] [PubMed]
- 78.Affonso, E. T., Rodríguez, D. Z., Rosa, R. L., Andrade, T., & Bressan, G. (2016). Voice quality assessment in mobile devices considering different fading models. In 2016 IEEE International Symposium on Consumer Electronics (ISCE), pp. 21–22. 10.1109/ISCE.2016.7797329
- 79.Gomes, I. R., Gomes, C. R., Gomes, H. S., & Cavalcante, G. P. D. S. (2018). Empirical radio propagation model for dtv applied to non-homogeneous paths and different climates using machine learning techniques. PloS one 13, 3, e0194511–e0194511. 10.1371/journal.pone.0194511 [DOI] [PMC free article] [PubMed]
- 80.Aha D, Kibler D. Instance-based learning algorithms. Machine Learning. 1991;6:37–66. doi: 10.1007/BF00153759. [DOI] [Google Scholar]
- 81.Vapnik V. Universal learning technology: Support vector machines. NEC Journal of Advanced Technology. 2005;2(2):137–144. [Google Scholar]
- 82.Zhou, W., Newsam, S., Li, C., & Shao, Z. (2017) Learning low dimensional convolutional neural networks for high-resolution remote sensing image retrieval. Remote Sensing 9, 5, 489. 10.3390/rs9050489
- 83.Shekar, G., Revathy, S., & Goud, E. K. (2020). Malaria detection using deep learning. In 2020 4th International Conference on Trends in Electronics and Informatics (ICOEI)(48184), pp. 746–750. 10.1109/ICOEI48184.2020.9143023
- 84.Bouchabou, D., Nguyen, S. M., Lohr, C., Leduc, B., & Kanellos, I. (2020). Fully convolutional network bootstrapped by word encoding and embedding for activity recognition in smart homes. CoRR abs/2012.02300. arXiv:2012.02300. https://arxiv.org/abs/2012.02300
- 85.Pourseidi, B., & Khorram-Manesh, A. (2007). Triple non-invasive diagnostic test for exclusion of common bile ducts stones before laparoscopic cholecystectomy. World journal of gastroenterology 13, 43, 5745–5749. PMID: 17963302. 10.3748/wjg.v13.i43.5745 [DOI] [PMC free article] [PubMed]
- 86.Sizova L. Diagnostic accuracy of the 2002 eular criteria for the clinical suspicion of rheumatoid arthritis and the sensitivity of the new 2010 acr/eular classification criteria in â??early arthritisâ? Archives of Rheumatology. 2011;26(1):007–012. [Google Scholar]
- 87.Cardoso, I., Almeida, E., Allende-Cid, H., Frery, A. C., Rangayyan, R. M., Azevedo-Marques, P. M., & Ramos, H. S. (2018). Evaluation of deep feedforward neural networks for classification of diffuse lung diseases. In Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications (Cham, 2018), M. Mendoza and S. Velastín, Eds., Springer International Publishing, pp. 152–159.
- 88.Rodríguez-Fdez, I., Canosa, A., Mucientes, M., & Bugarín, A. (2015). STAC: A web platform for the comparison of algorithms using statistical tests. In 2015 IEEE International Conference on Fuzzy Systems, FUZZ-IEEE 2015, Istanbul, Turkey, August 2-5, 2015, pp. 1–8. 10.1109/FUZZ-IEEE.2015.7337889
- 89.Liu Z, Blasch E, John V. Statistical comparison of image fusion algorithms: Recommendations. Information Fusion. 2017;36:251–260. doi: 10.1109/FUZZ-IEEE.2015.7337889. [DOI] [Google Scholar]
- 90.Goel, T., Murugan, R., Mirjalili, S., & Chakrabartty, D. K. (2021). Automatic screening of covid-19 using an optimized generative adversarial network. Cognitive Computation. 10.1007/s12559-020-09785-7 [DOI] [PMC free article] [PubMed]
- 91.Castiglione, A., Vijayakumar, P., Nappi, M., Sadiq, S., & Umer, M. (2021). Covid-19: Automatic detection of the novel coronavirus disease from ct images using an optimized convolutional neural network. IEEE Transactions on Industrial Informatics, 1. 10.1109/TII.2021.3057524 [DOI] [PMC free article] [PubMed]
- 92.Konar D, Panigrahi BK, Bhattacharyya S, Dey N, Jiang RM. Auto-diagnosis of COVID-19 using lung CT images with semi-supervised shallow learning network. IEEE Access. 2021;9:28716–28728. doi: 10.1109/ACCESS.2021.3058854. [DOI] [Google Scholar]