Abstract
Sirtuin 3 (SIRT3), a well-known mitochondrial deacetylase, is involved in mitochondrial function and metabolism under various stress conditions. In this study, we found that the expression of SIRT3 was markedly increased by oxidative stress in dopaminergic neuronal cells. In addition, SIRT3 overexpression enhanced mitochondrial activity in differentiated SH-SY5Y cells. We also showed that SIRT3 overexpression attenuated rotenone- or H2O2-induced toxicity in differentiated SH-SY5Y cells (human dopaminergic cell line). We further found that knockdown of SIRT3 enhanced rotenone- or H2O2-induced toxicity in differentiated SH-SY5Y cells. Moreover, overexpression of SIRT3 mitigated cell death caused by LPS/IFN-γ stimulation in astrocytes. We also found that the rotenone treatment increases the level of SIRT3 in Drosophila brain. We observed that downregulation of sirt2 (Drosophila homologue of SIRT3) significantly accelerated the rotenone-induced toxicity in flies. Taken together, these findings suggest that the overexpression of SIRT3 mitigates oxidative stress-induced cell death and mitochondrial dysfunction in dopaminergic neurons and astrocytes.
Keywords: Dopaminergic neuron, Mitochondrial dysfunction, Neurotoxicity, Oxidative stress, SIRT3, Astrocyte/neuron coculture
INTRODUCTION
Dopaminergic neurons are the major source of dopamine (DA) in the CNS. Dopaminergic neurons play an important role in the control of brain functions including movement, cognition, learning, and emotion. Previous studies have suggested that oxidative stress, neuroinflammation, and ageing significantly exacerbate dopaminergic neuronal death [1-3]. Many clinical and experimental studies have suggested that the death of dopaminergic neurons is linked to PD [4, 5]. Interestingly, several proteins related with Parkinson's Disease (PD), such as SNCA, Parkin, PINK1, LRRK2, and DJ-1 are directly linked to the mitochondrial dysfunction and oxidative stress, suggesting that mitochondrial dysfunction may be key pathological mechanism of PD [6]. However, the neuropathological mechanisms underlying the death of dopaminergic neurons have yet to be clearly established.
Mitochondria are major intracellular organelles that modulate various cellular processes, such as cell death, autophagy, differentiation, cell cycle, and proliferation [7]. Many studies have reported that mitochondrial dysfunction is caused by the dysregulation of mitochondrial proteins under various stress conditions [8]. Moreover, mitochondrial dysfunction induces excessive ROS generation and ATP deficits, leading to dopaminergic neuronal death [9]. Recent studies have suggested that mitochondrial dysfunction is associated with many metabolic diseases, including neurodegenerative diseases, immune diseases, cancer, diabetes, and obesity [10-14]. These results demonstrate that mitochondrial dysfunction may play a crucial role in dopaminergic neuronal death.
Sirtuin 3 (SIRT3) is a member of the class III histone deacetylase family, and it exhibits deacetylase activity in mitochondria. Several studies have shown that SIRT3 plays an important role in regulating ATP production, antioxidant mechanisms, inflammation, autophagy, cell death, and metabolism [15-18]. SIRT3 is predominantly localized in the mitochondria and nucleus [19, 20] and is primarily expressed in metabolically active tissues, including the brain, muscle, liver, kidney, and heart [21]. Additionally, the level of SIRT3 is modulated by mitochondrial stress, oxidative stress, proteotoxic stress, and starvation conditions [15, 22, 23]. SIRT3 prevents dopaminergic neuronal loss cause by oxidative stress in in vivo and in vitro models [24, 25]. Intriguingly, Park and colleagues [26] discovered that downregulation of SIRT3 is an essential component of α-synuclein-induced mitochondrial dysfunction. Therefore, a better understanding of the mitochondrial function of SIRT3 in neurons can lead to new therapeutic interventions to prevent and treat mitochondrial dysfunction-associated neurodegenerative diseases.
In the present study, we demonstrated that SIRT3 expression in dopaminergic neurons was upregulated by oxidative stress and that overexpression of SIRT3 attenuated oxidative stress-induced neuronal toxicity. Moreover, we found that SIRT3 regulated the cell death caused by LPS/IFN-γ treatment in astrocytes. By studying the dopaminergic neuronal system in a Drosophila model, we have shown that SIRT2, a Drosophila melanogaster homologue of mammalian SIRT3, protects against the neuronal toxicity induced by oxidative stress.
MATERIALS AND METHODS
Reagents and antibodies
The following reagents were purchased from the indicated providers: dimethyl sulfoxide (DMSO; Sigma, D8418), all-trans retinoic acid (RA; Sigma, R2625), MG132 (Calbiochem/Merck-Millipore, 474791), rotenone (Sigma, R8875), rapamycin (InvivoGen, tlrl-rap), H2O2 (Sigma, 216763), lipopolysaccharide (LPS; Sigma, L4391), and recombinant rat IFN-γ (R&D Systems, 585-IF-100/CF). The following antibodies were used for immunoblotting: anti-SIRT3 (Cell Signaling, 5490), anti-COX-IV (Cell Signaling, 4850), anti-Beta-Actin (Abcam, ab16039), HRP-conjugated anti-Alpha-tubulin (Cell Signaling Technology, 9099), and HRP-conjugated rabbit IgG (Santa Cruz Biotechnology, sc-2004) antibodies.
SH-SY5Y cell culture and differentiation
The SH-SY5Y human neuroblastoma cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, 11995-065) supplemented with 10% heat-inactivated foetal bovine serum (FBS, Gibco, 16000-044) and 50 µg/ml penicillin-streptomycin (Gibco, 15140-122). Cells were grown at 37℃ in a humidified atmosphere containing 5% CO2. For differentiation, SH-SY5Y cells were grown to confluence, and the growth medium (DMEM+10% FBS) was exchanged for differentiation medium (1:1 DMEM/Ham’s F12 supplemented with 1% FBS, 1% MEM-NEAA, 50 μg/ml P/S, and 10 μM all-trans retinoic acid (RA)) every 2 days. The differentiation of SH-SY5Y cells was performed as described previously [27, 28].
Stable transfection
SH-SY5Y cells in 6-well plates (40×104 cells/ml) were transfected with 4 µg of human SIRT3 cDNA using Lipofectamine 3000 reagent (Invitrogen). An empty pCMV6-Flag vector was used as a negative control. Stable transfectants were selected in the presence of 800 µg/ml G418 (Gibco, 10131-027). The expression of transgenes was confirmed by immunoblotting and immunocytochemistry analysis.
siRNA transfection
Differentiated SH-SY5Y cells in 6-well plates (40×104 cells/well) were transfected with control siRNA (Santa Cruz; sc-37007), human SIRT3 siRNA (Santa Cruz; sc-61556), or mouse Sirt3 siRNA (Santa Cruz; sc-61556), using Lipofectamine RNAiMAX reagent (Invitrogen), and then the cells were incubated for 2 days. The downregulation of target protein expression in the transfected cells was confirmed by immunoblot analysis.
Primary cell cultures Primary cultures of dissociated cerebral cortical neurons were prepared from 16-day-old embryonic C57/BL6 mice as described previously [29, 30]. Briefly, mouse embryos were decapitated, and the brains were rapidly removed and placed in a culture dish containing HBSS (Gibco). Cortices were isolated, transferred to a conical tube and washed twice in HBSS (Gibco). Cortical tissues were enzymatically digested with prewarmed papain (20 units/ml) (Worthington Biochemical Corporation) and DNase I (0.005%) for 30 min at 37℃. The tissues were mechanically dissociated (triturated) with 1,000 μl and 200 μl pipette tips to achieve complete tissue homogenization. The cortical cells were centrifuged at 130×g for 10 min at room temperature, and the dissociated cells that were obtained were seeded onto plates coated with poly-D-lysine (Sigma-Aldrich) in neurobasal media containing 2 mM glutamine (Gibco), N2 supplement (Gibco), B27 supplement (Gibco), and 50 μg/ml penicillin-streptomycin (P/S, Gibco). The culture media were changed initially after 5 days and every 3 days thereafter, and the cells were used after being cultured for 14~21 days.
Primary astrocyte cultures were prepared from 1- to 2-day-old C57/BL6 mice as described previously [31]. Briefly, whole brains were homogenized and passed through a 70-μm strainer. The cells were seeded in T75 culture flasks. The cells were grown at 37℃ in a humidified atmosphere containing 5% CO2. The culture medium was changed initially after 5 days and every 2 days thereafter, and the cells were used after being cultured for 14~21 days. Secondary pure astrocyte cultures were obtained by shaking mixed glial cultures at 250 rpm for 4 h; then, the culture medium was discarded. Astrocytes were dissociated using trypsin-EDTA (Life Technologies) and then centrifuged at 800×g for 30 min. The astrocytes obtained were seeded onto plates in DMEM (Life Technologies) supplemented with 10% heat-inactivated FBS and 50 μg/ml P/S. The purity of the cells in culture was determined by immunocytochemistry, which indicated that the cultures contained over 93% GFAP-positive cells. The animals used in the current research were acquired and cared for in accordance with the guidelines published in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Cytotoxicity tests
Differentiated SH-SY5Y cells (8×104 cells/ml) were grown in 96-well plates and treated with rotenone/H2O2 as indicated for 24 h. DMSO was used as a negative control. To measure cytotoxicity, Cell Counting Kit-8 (CCK-8, Enzo Life Science, ALX-850-039-KI02) was used according to the manufacturer’s instructions. Briefly, 10 µl of CCK-8 reagent was added to each well, and the plate was incubated at 37℃ for 2 h. The absorbance at 450 nm was measured by using a microplate reader (Tecan). Cell viability was expressed as a percentage of the control. All experiments were performed in triplicate.
CMFDA staining analysis
CMFDA fluorescence-based assay was used to investigate additional neuronal toxicity. Differentiated SH-SY5Y cells and primary cortical neurons were labeled with a 5 μM concentration of CellTracker Green CMFDA Dye (Invitrogen) for 30 min using the manufacturer’s protocol. Then, the dye solution was aspirated, and the cells were incubated with dye-free medium for 45 min. The samples were mounted and observed with a microscope. Photomicrographs from three randomly chosen fields were obtained and the number of CMFDA-positive cells was counted. The quantification of CMFDA-positive cells was expressed as a percentage of control.
Immunoblot analysis
Cells were homogenized in Cell Lysis Buffer (Cell Signaling, 9803) containing protease and phosphatase inhibitor cocktails. The protein concentrations of the cell lysates were determined by BCA protein assay (Thermo Fisher Scientific, 23225). Next, the protein extracts were mixed with 4× Bolt LDS Sample Buffer (Invitrogen) and 10× Bolt Sample Reducing Agent (Invitrogen), and then they were boiled at 95℃ for 5 min. An equal amount of protein from each sample was separated on Bolt 4~12% Bis-Tris gels (Invitrogen, NW04120BOX) or NuPAGE 3~8% Tris-Acetate gels (Invitrogen, EA0378BOX) and then transferred to polyvinylidene difluoride (PVDF, Invitrogen, LC2005) membranes. After blocking the membranes with 5% skim milk in TBS with 0.025% Tween 20, the blots were probed with antibodies as indicated and detected with an ECL Prime Kit (GE Healthcare, RPN2232). Samples from three independent experiments were used, and the relative expression levels were determined using a Fusion-FX Imaging System (Viber Lourmat).
Quantitative RT-PCR
RNA was extracted from cells and fly heads by using a TRIzol Plus RNA Purification Kit (Invitrogen, 12183-555) according to the manufacturer’s instructions. cDNA synthesis was performed at 37℃ for 120 min from 100 ng of RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368814). Quantitative RT-PCR was performed using a one-step SYBR® PrimeScript RT-PCR Kit (Takara Bio Inc., RR420A) according to the manufacturer’s instructions, which was followed by detection using an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems). Gapdh or 18S rRNA was used as an internal control. The 2−ΔΔCt method was used to calculate the relative changes in gene expression determined by real-time PCR [32].
Mitochondrial oxygen consumption
For assessment of neuronal mitochondrial dysfunction, differentiated SH-SY5Y cells (7×104 cells/ml) were seeded into XF 24-well culture plates (Seahorse Bioscience). The cells were washed twice with XF Base Medium supplemented with 2 mM L-glutamine, 10 mM D-glucose and 1 mM sodium pyruvate (pH 7.4) and incubated at 37℃ in a non-CO2 incubator for 1 h. Mitochondrial dysfunction was evaluated using the XF Cell Mito Stress Test Kit (Seahorse Bioscience) according to the manufacturer’s instructions, followed by measurement using an XF24 Extracellular Flux Analyser (Seahorse Bioscience). The 24-well utility plate was hydrated, treated with 2 µM oligomycin, 2 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), 0.5 µM antimycin A+rotenone, and thenused to calibrate the analyser. . The basal oxygen consumption rate (OCR), ATP production, maximum reserve and respiratory capacity were calculated as previously described [33], with averages calculated from 4 wells per condition in each individual experiment. The OCR was normalized to the total protein concentration (OD). After Seahorse analysis, the plate was centrifuged at 280×g for 5 min. The media was aspirated, and the plate was washed twice with PBS. The cells were lysed in RIPA buffer. Protein concentrations in the cell lysates were determined using a BCA Assay Kit.
Neuron-astrocyte coculture
Primary astrocytes were transfected with human SIRT3 expression construct or mouse Sirt3 siRNA using Lipofectamine 3000 or Lipofectamine RNAiMAX reagent, and then the cells were treated with LPS/IFN-γ for 24 h. The cells were washed twice with PBS, detached, and seeded in the upper compartment of transwell inserts (8 μm pore membrane; Millipore) at a density of 4×104 cells/well. Primary cortical neurons were plated in the lower compartment of 24 transwell plates at a density of 4×104 cells/well. The primary cells were incubated at 37℃ for 24 h and then subjected to a CCK-8 assay and CMFDA staining.
Fly strains
Drosophila stocks were maintained on standard cornmeal agar media at 24℃ unless otherwise noted. UAS-Gfp RNAi, UAS-Sirt2 RNAi, ple-Gal4 driver (also known as TH-Gal4) and the UAS-mCD8::GFP flies were obtained from The Bloomington Stock Center. We crossed female UAS-RNAi flies (Gfp RNAiVALIUM20-EGFP.shRNA.4 and Sirt2 RNAiHMS00485) with male flies containing the ple-Gal4 (dopaminergic neuron-specific) driver to knock down target genes in the dopaminergic neuronal cell population. w;;UAS-mCD8::GFP, ple-Gal4 (ple>mCD8::GFP) was generated using w;; P{UAS-mCD8::GFP.L}LL6 and w*; ; P{ple-Gal4.F}.
Lifespan assays
Adult males (0 to 1 day old) were separated and transferred into experimental vials containing fly media mixed with rotenone (500 µM) at a density of 20 flies per vial (n>80). The number of dead flies was recorded daily, and the flies were transferred to fresh media every other day. All experiments were performed on 80 flies per genotype per time point.
Immunohistochemistry
Adult flies were dissected in PBS and fixed in 4% PFA in PBS for 1 day at 4℃. The brains were then washed six times with PBS and pre-incubated in PBS-T for 1 day at 4℃. After blocking with 5% normal goat serum in PBS-T (0.3% Triton X-100) for 1 hr. The DAPI (4’, 6-diamidino-2-phenylindole dihydrochloride) were incubated with 5% normal goat serum in PBS-T for 2 days at 4℃. The samples were mounted and observed with fluorescence confocal microscope (Leica). Photomicrographs of dopaminergic neurons labeled with mCD8-GFP were acquired through confocal Z-stacks. Numbers of dopaminergic neurons were counted per brain for PPL1clusters.
Statistical analyses
Data were analyzed by unpaired Student’s t-test (Vassar Stats, www.vassarstats.net), and depending on the variables being compared, one-way ANOVA with post hoc analysis was performed as indicated (GraphPad Prism Software). Differences were considered significant when p<0.05 and are indicated as follows: *p<0.05; **p<0.005; ***p<0.001; and n.s., not significant.
RESULTS
SIRT3 is increased by oxidative or proteotoxic stress
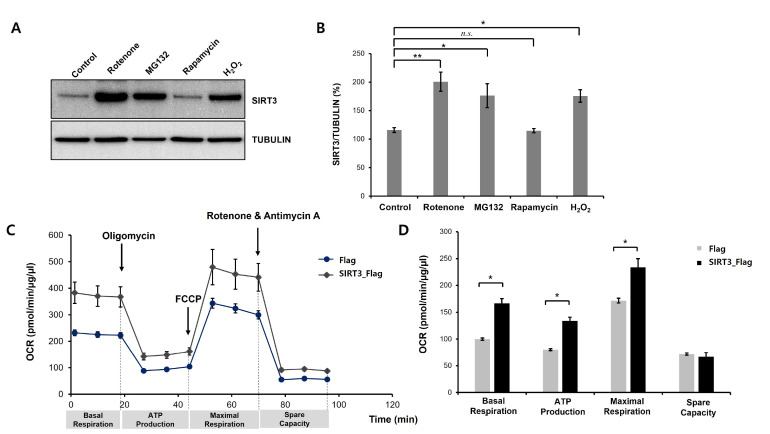
To determine the effects of SIRT3 on oxidative stress-induced neurotoxicity, we first examined the levels of SIRT3 protein in dopaminergic neuronal cells under various stress conditions such as MG132 (ubiquitin proteasome system inhibitor), rapamycin (autophagy inducer), H2O2 (reactive oxygen species) and rotenone (mitochondrial electron transport chain inhibitor). To do this, we induced the differentiation process of SH-SY5Y cells. Previous studies indicated that differentiated SH-SY5Y cells show dopaminergic neuron-like properties, such as neurite extension and the expression of dopaminergic neuronal markers [34-36]. Several studies showed that the protein level of SIRT3 is upregulated by oxidative stress. Moreover, it is already known that MG132 and rapamycin can induce oxidative stress and cell death [37, 38]. In subsequent experiments, we used rotenone and H2O2 as stimuli to induce oxidative stress. To examine the effect of the SIRT3 on the oxidative stress-induced cell death and mitochondrial dysfunction, we generated stable SH-SY5Y cell lines expressing Flag-tagged human SIRT3. We observed whether SIRT3 modulates mitochondrial activity in differentiated SH-SY5Y cells. We monitored the cellular OCR in real time as a measure of mitochondrial respiration and glycolysis using a Seahorse XF24 Extracellular Flux Analyser and a Mitochondrial Stress Test Kit (Seahorse Bioscience). Sequential injections of oligomycin, FCCP, and antimycin A+rotenone enable the measurement of basal respiration, ATP production, maximal respiration, and spare respiratory capacity. Interestingly, we found that the basal respiration, ATP production, and maximal respiration parameters were markedly decreased in Flag-expressing cells compared to SIRT3-overexpressing cells, but the spare respiratory capacity was not altered (Fig. 1C, 1D). Moreover, OCR values were normalized to the total cellular protein concentration for each group. These findings reveal that SIRT3 may play a critical role in the modulation of mitochondrial activity in dopaminergic neuronal cells.
Fig. 1.
Neurotoxic stress increases SIRT3 protein levels in differentiated SH-SY5Y cells. (A) SH-SY5Y cells were treated with all-trans retinoic acid (RA, 10 µM) for 8 days to generate differentiated SH-SY5Y cells. Western blot analysis was performed to measure SIRT3 protein expression. Differentiated SH-SY5Y cells were treated with rotenone (50 µM), MG132 (5 µM), rapamycin (10 µM), and H2O2 (500 µM) for 24 h, and then the cells were harvested for total protein extraction. Rotenone, MG132, and H2O2 significantly increased SIRT3 levels in differentiated SH-SY5Y cells. (B) Quantification of the immunoblots was performed from 3 independent experiments. Tubulin was used for normalization. *p<0.05; **p<0.005; n.s., not significant (one-way ANOVA with Tukey’s post-hoc comparison test). (C) Stable Flag- or human SIRT3-Flag-expressing cell lines were treated with RA (10 µM) for 8 days, and then mitochondrial oxygen consumption was analyzed. Mitochondrial dysfunction of Flag- or SIRT3-Flag-expressing differentiated SH-SY5Y cells was measured by detecting the basal OCR, ATP production, maximum reserve, and respiratory capacity by a Seahorse XF Analyzer. The OCR was normalized to the total protein concentration. (D) Basal respiration, ATP production, maximum respiration, and spare respiratory capacity were quantified and are shown as percentages of the basal values. Data are presented as the mean±SEM. *p<0.05 (unpaired Student’s t-test).
Overexpression of SIRT3 attenuates rotenone- or H2O2-induced neurotoxicity
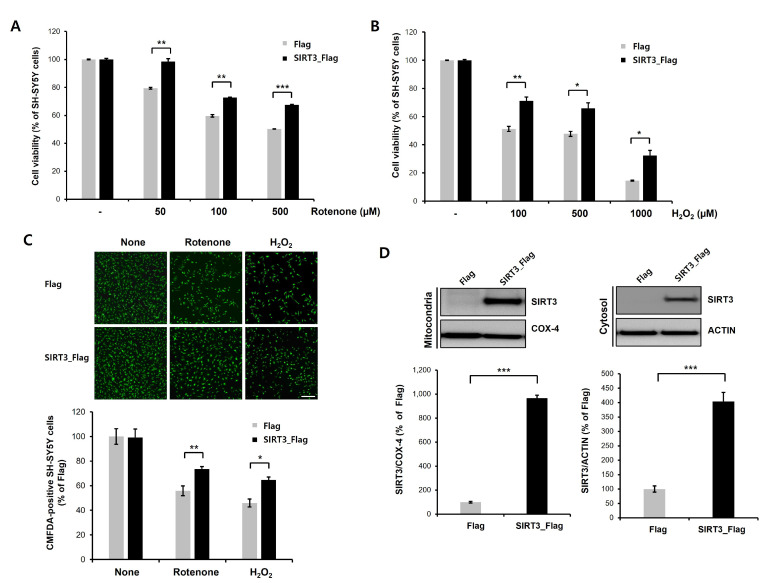
We next investigated whether overexpression of SIRT3 regulates rotenone- or H2O2-induced neuronal toxicity in differentiated SH-SY5Y cells. We showed that rotenone or H2O2 treatment of differentiated SH-SY5Y cells induced cytotoxicity in a dose-dependent manner, and SIRT3 overexpression attenuated rotenone- or H2O2-induced neuronal toxicity (Fig. 2A, 2B). We also confirmed the CCK-8 assay data using CMFDA staining (fluorescent cell tracker). Oxidative stress-induced reduction of CMFDA-labeled cells was notably lower in SIRT3-overexpressiong cells compared to Flag-expressing cells (Fig. 2C). Moreover, we performed subcellular fractionation to measure the SIRT3 protein levels in both the mitochondria and the cytosol. The levels of both mitochondrial and cytosolic SIRT3 protein were significantly increased in SIRT3-overexpressing cells compared to Flag-expressing cells (Fig. 2D). Taken together, these findings suggest that SIRT3 overexpression suppresses rotenone- or H2O2-induced toxicity in dopaminergic neuronal cells.
Fig. 2.
Overexpression of SIRT3 attenuates rotenone- or H2O2-induced neurotoxicity in differentiated SH-SY5Y cells. (A, B) Stable Flag- or human SIRT3-Flag-expressing cell lines were treated with RA (10 µM) for 8 days to generate differentiated SH-SY5Y cells. Flag- or SIRT3-Flag-expressing differentiated SH-SY5Y cells were treated with rotenone (50, 100, or 500 µM; A)/H2O2 (100, 500, or 1000 µM; B) for 24 h, and then CCK-8 analysis was performed. SIRT3-expressing differentiated SH-SY5Y cells exhibited significantly reduced rotenone- or H2O2-induced neuronal toxicity. Data are presented as the mean±SD of 3 independent experiments. *p<0.05; **p<0.005; ***p<0.001 (unpaired Student’s t-test). (C) CMFDA staining of rotenone (100 µM) or H2O2 (500 µM)-treated Flag- or SIRT3-Flag-expressing cells at 24 h. Then, CMFDA-positive neurons were counted under a fluorescence microscope. Data are presented as the mean±SD of 3. *p<0.05; **p<0.005 (unpaired Student’s t-test). Scale bars, 200 µm. (D) Protein levels of mitochondrial and cytosolic SIRT3 in Flag- or SIRT3-Flag-expressing differentiated SH-SY5Y cells. Both mitochondrial and cytosolic SIRT3 protein levels were significantly increased by SIRT3 expression. COX-IV (mitochondrial fraction) and actin (cytosolic fraction) were used for normalization. Data are presented as the mean±SD of 3 independent experiments. ***p<0.001 (unpaired Student’s t-test).
Sirt3 knockdown enhances rotenone- or H2O2-induced neurotoxicity
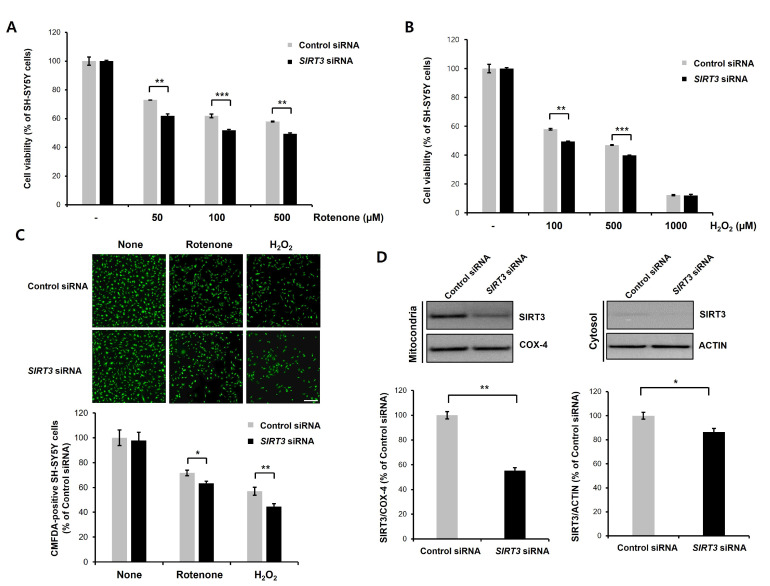
To further confirm that knockdown of SIRT3 contributes to rotenone- or H2O2-induced neurotoxicity, we downregulated the expression of human SIRT3 by RNAi-mediated gene knockdown. Consistent with our previous results, knockdown of SIRT3 greatly aggravated rotenone-induced neurotoxicity (Fig. 3A). H2O2-induced neurotoxicity was also markedly enhanced in SIRT3 knockdown cells compared with control cells (Fig. 3B). Moreover, rotenone- or H2O2-induced reduction of CMFDA-labeled cells were significantly exacerbated by knockdown of SIRT3 (Fig. 3C). As expected, the protein levels of mitochondrial SIRT3 were significantly decreased by SIRT3 knockdown in differentiated SH-SY5Y cells (Fig. 3D). We also showed that the cytosolic SIRT3 protein level was markedly decreased. These data indicate that knockdown of SIRT3 enhances rotenone- or H2O2-induced toxicity in dopaminergic neuronal cells.
Fig. 3.
Knockdown of SIRT3 enhances rotenone- or H2O2-induced neurotoxicity in differentiated SH-SY5Y cells. (A, B) Differentiated SH-SY5Y cells were transfected with control siRNA (100 nM) or human SIRT3-specific siRNA (100 nM) for 2 days, and then a CCK-8 assay and immunoblotting were performed. Control or SIRT3 knockdown cells were treated with rotenone (50, 100, or 500 µM; A)/H2O2 (100, 500, or 1000 µM; B) for 24 h, and then CCK-8 analysis was performed. SIRT3 knockdown cells exhibited significantly increased rotenone- or H2O2-induced neuronal toxicity. Data are presented as the mean±SD of 3 independent experiments. **p<0.005; ***p<0.001 (unpaired Student’s t-test). (C) CMFDA staining of rotenone (100 µM) or H2O2 (500 µM)-treated control siRNA- or SIRT3-downregulating cells at 24 h. Then, CMFDA-positive neurons were counted under a fluorescence microscope. Data are presented as the mean±SD of 3. *p<0.05; **p<0.005 (unpaired Student’s t-test). Scale bars, 200 µm. (D) Protein levels of mitochondrial and cytosolic SIRT3 in control and SIRT3 knockdown cells. SIRT3 protein levels were greatly decreased in the mitochondrial fraction of SIRT3 knockdown cells. COX-IV (mitochondrial fraction) and actin (cytosolic fraction) were used for normalization. Data are presented as the mean±SD of 3 independent experiments. *p<0.05; **p<0.005 (unpaired Student’s t-test).
Sirt2 knockdown enhances rotenone-induced neuronal toxicity in Drosophila
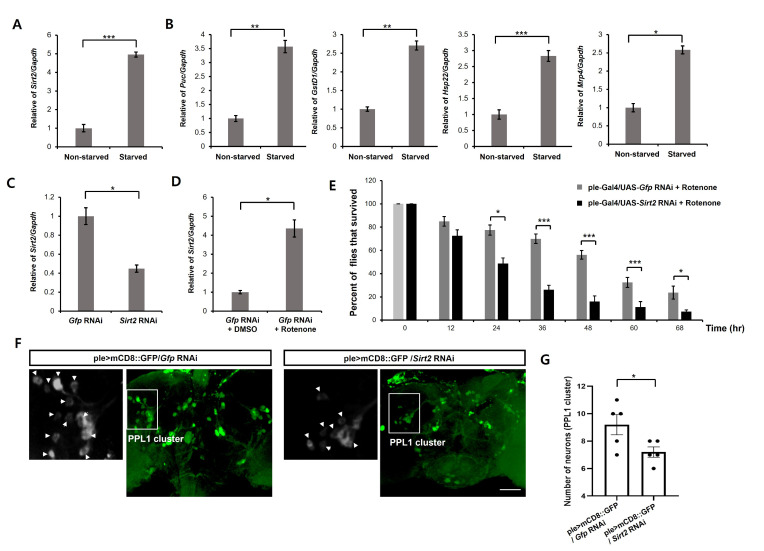
In mammals, calorie restriction induces upregulation of SIRT3 [39]. To check whether Sirt2 function in Drosophila is like those of mammalian SIRT3, we first assessed whether Sirt2 expression is increased by starvation in flies. We found that the levels of Sirt2 mRNA were markedly increased by starvation (Fig. 4A). We also analyzed the expression levels of oxidative stresses-related genes in Drosophila brain. We found that starvation upregulates oxidative stresses-related genes (Puc, GstD1, Hsp22, and Mrp4) (Fig. 4B). These results suggest that starvation induces oxidative stress in fly brain. Next, we downregulated Sirt2 genes by RNAi-mediated knockdown in fly dopaminergic neurons using a ple-Gal4 driver (dopaminergic neuron-specific Gal4 driver). We observed that rotenone treatment increased the level of Sirt2 mRNA in Drosophila brain (Fig. 4D). Then, we examined the effects of Sirt2 downregulation on the rotenone-induced shortened lifespan. Previous studies demonstrated that rotenone leads to a significant reduction in lifespan of Drosophila. Consistent with previous findings, rotenone treatment led to markedly reduced lifespan (Fig. 4E). We also found that downregulation of Sirt2 significantly exacerbated rotenone-induced shortened lifespan in flies (Fig. 4E). We examined the effect of Sirt2 knockdown on rotenone-induced neuronal toxicity in dopaminergic neurons. Not surprisingly, the Sirt2 knockdown markedly increased the rotenone-induced dopaminergic neuronal loss in the PPL1 cluster (Fig. 4F, 4G). These results suggest that Sirt2 is important for suppressing the neurotoxicity of oxidative stress in dopaminergic neurons.
Fig. 4.
Sirt2 knockdown enhances rotenone-induced neuronal toxicity in the Drosophila dopamine system. (A) Levels of dSirt2 mRNA from fly head lysates of 12 h starved wild-type flies were analyzed by real-time PCR. Gapdh was used for normalization. Data are presented as the mean±SD. ***p<0.001 (unpaired Student’s t-test). (B) Levels of oxidative stresses-related genes (dPuc, dGstD1, dHsp22, and dMrp4) mRNA from fly head lysates of 12 h starved wild-type flies. mRNA levels were analyzed by real-time PCR. Gapdh was used to normalize changes in specific gene expression. Genotypes: UAS-Egfp RNAiVALIUM20-EGFP.shRNA.4/+. Data are presented as the mean±SD. *p<0.05; **p<0.005; ***p<0.001 (unpaired Student’s t-test). (C) Levels of Sirt2 mRNA from fly head lysates of ple-Gal4/Gfp RNAi and ple-Gal4/Sirt2 RNAi flies were analyzed by real-time PCR. RNAi-mediated knockdown of Sirt2 in Drosophila reduced the Sirt2 transcription levels. Gapdh was used to normalize changes in specific gene expression. Data are presented as the mean±SD. *p<0.05 (unpaired Student’s t-test). (D) Levels of Sirt2 mRNA from fly head lysates of Ple-Gal4/Gfp RNAi flies fed rotenone (500 μM) were analyzed by real-time PCR. The Sirt2 level was significantly upregulated in the rotenone exposed flies. Gapdh was used to normalize changes in specific gene expression. Data are presented as the mean±SD. *p<0.05 (unpaired Student’s t-test). (E) Survival rate of ple-Gal4/UAS-Gfp RNAi and ple-Gal4/UAS-Sirt2 RNAi flies fed rotenone (500 μM) at the indicated time points. The lifespan of rotenone-treated flies was significantly reduced by the downregulation of Sirt2. The percentage of flies that survived was quantified. Genotypes: Gfp RNAi is ple-Gal4/UAS-Egfp RNAiVALIUM20-EGFP.shRNA.4, and Sirt2 RNAi is ple-Gal4/UAS-Sirt2 RNAiHMS00485. Data are presented as the mean±SEM of 4 independent experiments. *p<0.05; ***p<0.001 (unpaired Student’s t-test). (F, G) Dopaminergic neurons were visualized via expression of UAS-mCD8-GFP under control of the ple-Gal4. The number of dopaminergic neurons in PPL1 cluster was analyzed in ple>mCD8::GFP/Gfp RNAi and ple>mCD8::GFP/Sirt2 RNAi flies after 48 h of exposure to rotenone (500 μM). Arrowheads indicate dopaminergic neurons in PPL1 cluster (left). Genotypes: ple>mCD8::GFP/Gfp RNAi is ple-Gal4,USA-mCD8-GFP/UAS-Egfp RNAiVALIUM20-EGFP.shRNA.4, and ple>mCD8::GFP/Sirt2 RNAi is ple-Gal4,USA-mCD8-GFP/UAS-Sirt2 RNAiHMS00485. Data are presented as the mean±SEM. *p<0.05 (n=5, unpaired Student’s t-test). Scale bars, 50 µm.
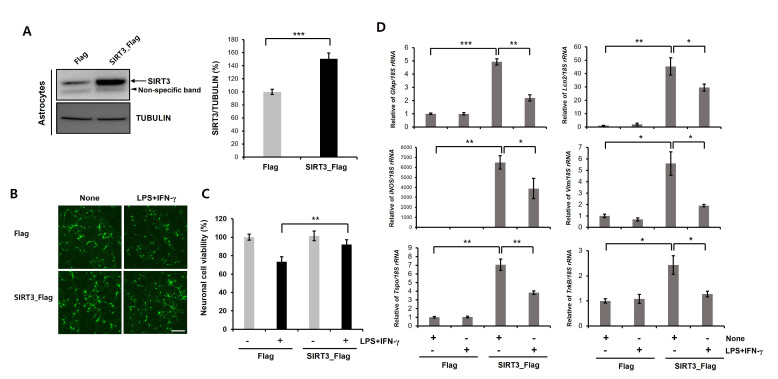
SIRT3 overexpression in astrocytes mitigates reactive astrocyte-induced neurotoxicity
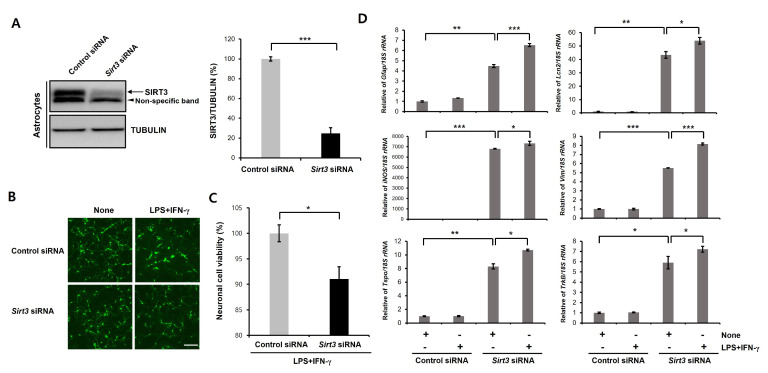
In various neurodegenerative diseases, proinflammatory reactive astrocytes contribute to neuronal death by secreting neurotoxic factors such as proinflammatory cytokines and chemokines. Previous studies demonstrated that reactive astrocyte induced neuronal defect could be a major characteristic of PD [40-42]. Moreover, glial activation is implicated in the dopaminergic neuronal loss in α-synuclein and MPTP-based PD animal models [43, 44]. In addition, reactive astrocytes were accumulated in the substantia nigra pars compacta of PD patients [45, 46]. SIRT3 is expressed both neurons and astrocytes. Thus, SIRT3 may act not only as a cell-intrinsic factor modulating neuronal death but also as a factor regulating non-cell-autonomous neuronal death through glia. To investigate the relevance of SIRT3 to astrocytes mediated neurotoxicity, we used the astrocyte/neuron coculture model. Previous study showed that LPS/IFN-γ-stimulated astrocytes led to a significant neuronal toxicity in a coculture model [47]. We first transfected primary astrocytes with Flag- or Flag-tagged human SIRT3 and then assessed the level of SIRT3 by immunoblotting. SIRT3 expression was significantly higher in the SIRT3-Flag-overexpressing astrocytes than in the Flag-expressing astrocytes (Fig. 5A). Accordingly, we found that LPS/IFN-γ stimulation of primary astrocytes caused neuronal toxicity in transwell cocultures (Fig. 5B). This neuronal toxicity was significantly mitigated in coculture with SIRT3-Flag-overexpressing astrocytes compared to that with Flag-expressing astrocytes (Fig. 5B). We also confirmed these results using ICC of CMFDA dye (fluorescent cell tracker, Fig. 5C). We next performed RT-PCR to examine the level of reactive astrocytic genes (Gfap, Lcn2, iNOS, Vim, Tspo, and TrkB) in SIRT3-overexpressing cells treated with LPS/IFN-γ (Fig. 6D). We found that LPS/IFN-γ-induced upregulation of reactive astrocytic genes was significantly decreased by SIRT3-overexpression (Fig. 5D). Therefore, these results suggest that overexpression of SIRT3 suppresses the activation of astrocytes caused by LPS/IFN-γ. For further confirmation, we downregulated the expression of mouse Sirt3 by RNAi-mediated gene knockdown (Fig. 6A). The siRNA knockdown of Sirt3 enhanced the neuronal toxicity of LPS/IFN-γ-stimulated astrocytes (Fig. 6B). Similar results were also observed in CMFDA staining-based cell viability assay (Fig. 6C). Moreover, the levels of reactive astrocytic genes (Gfap, Lcn2, iNOS, Vim, Tspo, and TrkB) were significantly increased by Sirt3 downregulating astrocytes compared to control astrocytes (Fig. 6D). These data indicate that the SIRT3 modulates neuronal toxicity caused by LPS/IFN-γ treated astrocytes.
Fig. 5.
Astrocytic SIRT3 overexpression mitigates the neurotoxicity caused by LPS/IFN-γ-stimulated astrocytes. (A) Primary astrocytes were transfected with Flag- or human SIRT3-Flag expression constructs for 3 days, and then immunoblotting was performed. Successful transfection of Flag or SIRT3-Flag in cells was confirmed by immunoblotting using an anti-SIRT3 antibody. SIRT3 protein levels were highly increased in SIRT3-Flag-expressing cells compared with Flag-expressing cells. Tubulin was used as loading control. Data are presented as the mean±SD of 3 independent experiments. ***p<0.001 (unpaired Student’s t-test). (B, C) Flag- or SIRT3-Flag-expressing astrocytes were treated with LPS+IFN-γ (100 ng/ml+50 unit/ml) for 24 h and then cocultured with primary cortical neurons in transwell culture inserts. Cell viability was measured using a CMFDA staining (green; B) or CCK-8 assay (C) after a coculture period of 5 days. Data are presented as the mean±SD. **p<0.005 (unpaired Student’s t-test). Scale bars, 20 µm. (D) Flag- or SIRT3-Flag-expressing astrocytes were treated with LPS+IFN-γ (100 ng/ml+50 unit/ml) for 24 h, and then, Real Time-PCR was performed. Transcription levels of target genes are presented as the mean±SD. 18S rRNA was used to normalize changes in specific gene expression. *p<0.05; **p<0.005; ***p<0.001 (one-way ANOVA with Tukey’s post-hoc comparison test).
Fig. 6.
Knock-down of Sirt3 in primary astrocytes enhances the neurotoxicity caused by LPS/IFN-γ-stimulated astrocytes. (A) Primary astrocytes were transfected with control siRNA (50 nM) or mouse Sirt3-specific siRNA (50 nM) for 3 days, and then immunoblotting was performed. Successful knockdown of control siRNA or Sirt3 siRNA in cells was confirmed by immunoblotting using an anti-SIRT3 antibody. SIRT3 protein levels were significantly decreased in Sirt3 siRNA-transfected cells. Tubulin was used as loading control. Data are presented as the mean±SD of 3 independent experiments. ***p<0.001 (unpaired Student’s t-test). (B, C) Control siRNA or Sirt3 siRNA-transfected astrocytes were treated with LPS+IFN-γ (100 ng/ml+50 unit/ml) for 24 h and then cocultured with primary cortical neurons using transwell culture inserts. Neuronal viability was measured using a CMFDA staining (green; B) or CCK-8 assay (C) after a coculture period of 5 days. Data are presented as the mean±SD. *p<0.05 (unpaired Student’s t-test). Scale bars, 20 µm. (D) Control siRNA or Sirt3 siRNA-transfected astrocytes were treated with LPS+IFN-γ (100 ng/ml+50 unit/ml) for 24 h, and then, Real Time-PCR was performed. Transcription levels of target genes are presented as the mean±SD. 18S rRNA was used to normalize changes in specific gene expression. *p<0.05; **p<0.005; ***p<0.001 (one-way ANOVA with Tukey’s post-hoc comparison test).
DISCUSSION
We demonstrated that SIRT3 is a major regulator of oxidative stress-induced neuronal toxicity in dopaminergic neurons. Oxidative stress is currently well known as an inducing factor of neuronal toxicity. In this study, we demonstrated that oxidative stress increased SIRT3 protein levels in differentiated SH-SY5Y cells. We also found that SIRT3 overexpression suppressed oxidative stress-induced neuronal toxicity in differentiated SH-SY5Y cells. Furthermore, downregulation of SIRT3 enhanced rotenone- or H2O2-induced neuronal toxicity in differentiated SH-SY5Y cells. A previous study showed that overexpression of SIRT3 in HT-22 neuronal cells significantly suppressed ROS production and neuronal cell death [48]. We also found that that the rotenone treatment increases the transcription level of Sirt2 in Drosophila brain. Importantly, our data also showed that Sirt2 downregulation significantly enhanced the rotenone-induced toxicity in fly dopaminergic neurons. These findings suggest that SIRT3 is involved in oxidative stress-mediated neurotoxicity in in vivo and in vitro models.
Understanding the mitochondrial function of SIRT3 in neurons can lead to new therapeutic agents to prevent and treat mitochondria-related neurodegenerative diseases. Previous studies have suggested that SIRT3 deficiency significantly induces mitochondrial dysfunction and neuronal damage [49, 50]. SIRT3 is associated with proteotoxic- and mitochondrial stress-mediated cell death in cancer cells [22]. In this study, SIRT3 were significantly increased by oxidative stress in mammalian neuronal cell and Drosophila. We also found that the rotenone- or H2O2-induced neuronal toxicity in differentiated SH-SY5Y cells was suppressed by SIRT3 overexpression (Fig. 2). Consistently, rotenone- or H2O2-induced neuronal toxicity were significantly enhanced by SIRT3 downregulation (Fig. 3). We also observed that mitochondrial oxygen consumption was increased in SIRT3-overexpressing cells compared to Flag-expressing control cells (Fig. 1C, 1D). The SIRT3 has NAD+-dependent deacetylase activity, and the activity of SIRT3 regulates mitochondrial function. Previous studies have reported that SIRT3 was significantly decreased in frontal cortex and hippocampus of post-mortem human and mouse AD brains [51-53]. Moreover, viniferin increases the levels of SIRT3 and antioxidant enzyme Mn-SOD, leading to protects Huntingtin protein-induced neurotoxicity. The reduction of SIRT3 is also associated with loss of PGC-1α, acetylation of SOD2 and ATP synthase β in MPTP-administrated mouse PD model [54, 55]. Furthermore, SOD1G93A-induced mitochondrial fragmentation in motor neurons was dramatically reduced by SIRT3 [56]. In addition, previous studies suggest that alpha-synuclein or oxidative stress-induced mitochondrial defect in PD mouse model is associated with SIRT3 [24, 26]. These results indicate that SIRT3 might play an important role in mitochondria-related neurodegenerative diseases.
Several studies have shown that starvation can induce the oxidative stress [57]. Starvation-induced oxidative stress is closely related to starvation-induced cell toxicity [58-61]. Furthermore, Sirt3 deficiency leads to mitochondrial dysfunction and increase the vulnerability of cells to oxidative stress [62-65]. Thus, we investigated the effects of starvation on the oxidative stresses-related genes in Drosophila brain. We found that starvation can increase the expression of oxidative stresses-related genes (Puc, GstD1, Hsp22, and Mrp4) (Fig. 4B). We also showed that rotenone treatment clearly increased the levels of Sirt2 transcription in Drosophila (Fig. 4D). Sirt2 knockdown markedly increased the rotenone-induced dopaminergic neuronal loss in the PPL1 cluster and enhanced the rotenone-induced shortened life span in Drosophila (Fig. 4E~4G). Taken together, our findings suggest that SIRT2 regulates oxidative stress-induced toxicity in Drosophila dopaminergic neuron.
Many studies have shown that LPS/IFN-γ-induced proinflammatory cytokines in astrocytes lead to neuronal death in astrocyte/neuron coculture models. Interestingly, these proinflammatory cytokines are significantly elevated in the plasma of PD patients [66, 67]. Moreover, a recent meta-analysis revealed that IL-6, TNF-α, and IL-1β levels were significantly increased in the peripheral blood of PD patients [68]. In this study, we found that the SIRT3 overexpression mitigated neuronal toxicity caused by LPS/IFN-γ stimulated astrocytes (Fig. 5). The number of CMFDA-positive neurons was notably decreased in co-cultures with LPS/IFN-γ-treated astrocytes, and SIRT3 overexpression significantly attenuated the neurotoxicity of LPS/IFN-γ-stimulated astrocytes (Fig 5). Furthermore, we found that the neurotoxicity of LPS/IFN-γ-stimulated astrocytes was enhanced by knock-down of Sirt3 (Fig. 6). Accordingly, adjudin, a potential SIRT3 activator, has shown to reduce LPS- and stroke-induced neuroinflammation [69]. Interestingly, we found that LPS/IFN-γ-induced reactive astrocytic genes (Gfap, Lcn2, iNOS, Vim, Tspo, and TrkB) are significantly downregulated by SIRT3 overexpression in primary astrocytes, suggesting that SIRT3 may play a critical role in the activation of astrocytes caused by LPS/IFN-γ treatment. SIRT3 overexpression can also inhibit ischemic stroke-mediated astrocyte activation and glial scar formation [70]. These findings suggest that overexpression of SIRT3 in astrocytes may be linked to decreased levels of proinflammatory cytokines and that these cytokines are mediators of neuronal death in PD.
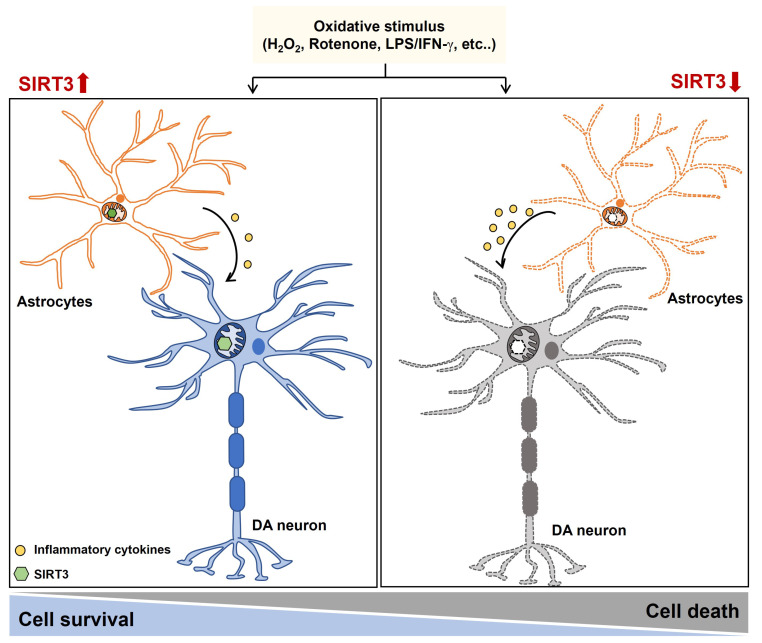
In the present study, we demonstrated that SIRT3 expression in dopaminergic neurons was upregulated by oxidative stress. We identified that SIRT3 played a critical role in the neurotoxicity induced by oxidative stress by regulating mitochondrial function in mammalian cells as well as a Drosophila. Moreover, we found that SIRT3 is a major regulator of neuronal toxicity caused by LPS/IFN-γ stimulated astrocytes (Fig. 7). Therefore, our data suggest that targeting SIRT3 may represent a novel therapeutic intervention for mitochondria-related neurodegenerative diseases.
Fig. 7.
Schematic representation of the effect of SIRT3 on the oxidative stress. SIRT3 plays a protective role in the oxidative stress-induced cell death and inflammatory responses by regulating mitochondrial function in neurons and astrocytes.
ACKNOWLEDGEMENTS
This work was supported by the KBRI Research Program of the Ministry of Science, ICT & Future Planning (21-BR-02-15); the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (NRF-2020R1A2C4002366 and NRF-2021R1C1C1008688); and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, South Korea (grant number: HI14C1135).
References
- 1.Qian L, Flood PM, Hong JS. Neuroinflammation is a key player in Parkinson's disease and a prime target for therapy. J Neural Transm (Vienna) 2010;117:971–979. doi: 10.1007/s00702-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeve AK, Krishnan KJ, Turnbull DM. Age related mitochondrial degenerative disorders in humans. Biotechnol J. 2008;3:750–756. doi: 10.1002/biot.200800066. [DOI] [PubMed] [Google Scholar]
- 3.Dias V, Junn E, Mouradian MM. The role of oxidative stress in Parkinson's disease. J Parkinsons Dis. 2013;3:461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surmeier DJ. Determinants of dopaminergic neuron loss in Parkinson's disease. FEBS J. 2018;285:3657–3668. doi: 10.1111/febs.14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michel PP, Hirsch EC, Hunot S. Understanding dopaminergic cell death pathways in Parkinson disease. Neuron. 2016;90:675–691. doi: 10.1016/j.neuron.2016.03.038. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Wu Q, Shi J, Zhou S. Regulation of SIRT3 on mitochondrial functions and oxidative stress in Parkinson's disease. Biomed Pharmacother. 2020;132:110928. doi: 10.1016/j.biopha.2020.110928. [DOI] [PubMed] [Google Scholar]
- 7.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramaniam SR, Chesselet MF. Mitochondrial dysfunction and oxidative stress in Parkinson's disease. Prog Neurobiol. 2013;106-107:17–32. doi: 10.1016/j.pneurobio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bose A, Beal MF. Mitochondrial dysfunction in Parkinson's disease. J Neurochem. 2016;139 Suppl 1:216–231. doi: 10.1111/jnc.13731. [DOI] [PubMed] [Google Scholar]
- 10.Walker MA, Volpi S, Sims KB, Walter JE, Traggiai E. Powering the immune system: mitochondria in immune function and deficiency. J Immunol Res. 2014;2014:164309. doi: 10.1155/2014/164309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Lin TY, Lee G, Paddock MN, Momb J, Cheng Z, Li Q, Fei DL, Stein BD, Ramsamooj S, Zhang G, Blenis J, Cantley LC. Mitochondrial one-carbon pathway supports cytosolic folate integrity in cancer cells. Cell. 2018;175:1546–1560.e17. doi: 10.1016/j.cell.2018.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burbulla LF, Song P, Mazzulli JR, Zampese E, Wong YC, Jeon S, Santos DP, Blanz J, Obermaier CD, Strojny C, Savas JN, Kiskinis E, Zhuang X, Krüger R, Surmeier DJ, Krainc D. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson's disease. Science. 2017;357:1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolson GL. Mitochondrial dysfunction and chronic disease: treatment with natural supplements. Integr Med (Encinitas) 2014;13:35–43. [PMC free article] [PubMed] [Google Scholar]
- 14.West AP. Mitochondrial dysfunction as a trigger of innate immune responses and inflammation. Toxicology. 2017;391:54–63. doi: 10.1016/j.tox.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J, Shi L, Liang F, Xu W, Li T, Gao L, Sun Z, Yu J, Zhang J. Sirt3 ameliorates oxidative stress and mitochondrial dysfunction after intracerebral hemorrhage in diabetic rats. Front Neurosci. 2018;12:414. doi: 10.3389/fnins.2018.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koyama T, Kume S, Koya D, Araki S, Isshiki K, Chin-Kanasaki M, Sugimoto T, Haneda M, Sugaya T, Kashiwagi A, Maegawa H, Uzu T. SIRT3 attenuates palmitate-induced ROS production and inflammation in proximal tubular cells. Free Radic Biol Med. 2011;51:1258–1267. doi: 10.1016/j.freeradbiomed.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 17.Jablonski RP, Kim SJ, Cheresh P, Williams DB, Morales-Nebreda L, Cheng Y, Yeldandi A, Bhorade S, Pardo A, Selman M, Ridge K, Gius D, Budinger GRS, Kamp DW. SIRT3 deficiency promotes lung fibrosis by augmenting alveolar epithelial cell mitochondrial DNA damage and apoptosis. FASEB J. 2017;31:2520–2532. doi: 10.1096/fj.201601077R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Chen T, Xiao M, Li N, Wang S, Su H, Guo X, Liu H, Yan F, Yang Y, Zhang Y, Bu P. Mouse Sirt3 promotes autophagy in AngII-induced myocardial hypertrophy through the deacetylation of FoxO1. Oncotarget. 2016;7:86648–86659. doi: 10.18632/oncotarget.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.e05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dittenhafer-Reed KE, Richards AL, Fan J, Smallegan MJ, Fotuhi Siahpirani A, Kemmerer ZA, Prolla TA, Roy S, Coon JJ, Denu JM. SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metab. 2015;21:637–646. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34:699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcus JM, Andrabi SA. SIRT3 regulation under cellular stress: making sense of the ups and downs. Front Neurosci. 2018;12:799. doi: 10.3389/fnins.2018.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henn V. [Neck pain from the neurological viewpoint] Schweiz Rundsch Med Prax. 1988;77:340–342. German. [PubMed] [Google Scholar]
- 25.Zeng R, Wang X, Zhou Q, Fu X, Wu Q, Lu Y, Shi J, Klaunig JE, Zhou S. Icariin protects rotenone-induced neurotoxicity through induction of SIRT3. Toxicol Appl Pharmacol. 2019;379:114639. doi: 10.1016/j.taap.2019.114639. [DOI] [PubMed] [Google Scholar]
- 26.Park JH, Burgess JD, Faroqi AH, DeMeo NN, Fiesel FC, Springer W, Delenclos M, McLean PJ. Alpha-synuclein-induced mitochondrial dysfunction is mediated via a sirtuin 3-dependent pathway. Mol Neurodegener. 2020;15:5. doi: 10.1186/s13024-019-0349-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambjørn M, Dubreuil V, Miozzo F, Nigon F, Møller B, Issazadeh-Navikas S, Berg J, Lees M, Sap J. A loss-of-function screen for phosphatases that regulate neurite outgrowth identifies PTPN12 as a negative regulator of TrkB tyrosine phosphorylation. PLoS One. 2013;8:e65371. doi: 10.1371/journal.pone.0065371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Deng YN, Zhang JY, Liu J, Li YB, Su H, Qu QM. SIRT3 protects rotenone-induced injury in SH-SY5Y cells by promoting autophagy through the LKB1-AMPK-mTOR pathway. Aging Dis. 2018;9:273–286. doi: 10.14336/AD.2017.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enokido Y, Akaneya Y, Niinobe M, Mikoshiba K, Hatanaka H. Basic fibroblast growth factor rescues CNS neurons from cell death caused by high oxygen atmosphere in culture. Brain Res. 1992;599:261–271. doi: 10.1016/0006-8993(92)90400-4. [DOI] [PubMed] [Google Scholar]
- 30.Araki W, Yuasa K, Takeda S, Shirotani K, Takahashi K, Tabira T. Overexpression of presenilin-2 enhances apoptotic death of cultured cortical neurons. Ann N Y Acad Sci. 2000;920:241–244. doi: 10.1111/j.1749-6632.2000.tb06929.x. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Landar A, Darley-Usmar VM. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes FM, Schröder R, da Frota ML, Jr, Zanotto-Filho A, Müller CB, Pires AS, Meurer RT, Colpo GD, Gelain DP, Kapczinski F, Moreira JC, Fernandes Mda C, Klamt F. Comparison between proliferative and neuron-like SH-SY5Y cells as an in vitro model for Parkinson disease studies. Brain Res. 2010;1337:85–94. doi: 10.1016/j.brainres.2010.03.102. [DOI] [PubMed] [Google Scholar]
- 35.Korecka JA, van Kesteren RE, Blaas E, Spitzer SO, Kamstra JH, Smit AB, Swaab DF, Verhaagen J, Bossers K. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One. 2013;8:e63862. doi: 10.1371/journal.pone.0063862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forster JI, Köglsberger S, Trefois C, Boyd O, Baumuratov AS, Buck L, Balling R, Antony PM. Characterization of differentiated SH-SY5Y as neuronal screening model reveals increased oxidative vulnerability. J Biomol Screen. 2016;21:496–509. doi: 10.1177/1087057115625190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan WH, Hou Y, Meng FK, Wang XF, Luo YN, Ge PF. Proteasome inhibitor MG-132 induces C6 glioma cell apoptosis via oxidative stress. Acta Pharmacol Sin. 2011;32:619–625. doi: 10.1038/aps.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kofman AE, McGraw MR, Payne CJ. Rapamycin increases oxidative stress response gene expression in adult stem cells. Aging (Albany NY) 2012;4:279–289. doi: 10.18632/aging.100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barger JL, Vann JM, Cray NL, Pugh TD, Mastaloudis A, Hester SN, Wood SM, Newton MA, Weindruch R, Prolla TA. Identification of tissue-specific transcriptional markers of caloric restriction in the mouse and their use to evaluate caloric restriction mimetics. Aging Cell. 2017;16:750–760. doi: 10.1111/acel.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGeer PL, McGeer EG. Glial reactions in Parkinson's disease. Mov Disord. 2008;23:474–483. doi: 10.1002/mds.21751. [DOI] [PubMed] [Google Scholar]
- 41.An H, Lee H, Yang S, Won W, Lee CJ, Nam MH. Adenovirus-induced reactive astrogliosis exacerbates the pathology of Parkinson's disease. Exp Neurobiol. 2021;30:222–231. doi: 10.5607/en21013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heo JY, Nam MH, Yoon HH, Kim J, Hwang YJ, Won W, Woo DH, Lee JA, Park HJ, Jo S, Lee MJ, Kim S, Shim JE, Jang DP, Kim KI, Huh SH, Jeong JY, Kowall NW, Lee J, Im H, Park JH, Jang BK, Park KD, Lee HJ, Shin H, Cho IJ, Hwang EM, Kim Y, Kim HY, Oh SJ, Lee SE, Paek SH, Yoon JH, Jin BK, Kweon GR, Shim I, Hwang O, Ryu H, Jeon SR, Lee CJ. Aberrant tonic inhibition of dopaminergic neuronal activity causes motor symptoms in animal models of Parkinson's disease. Curr Biol. 2020;30:276–291.e9. doi: 10.1016/j.cub.2019.11.079. [DOI] [PubMed] [Google Scholar]
- 43.Fellner L, Irschick R, Schanda K, Reindl M, Klimaschewski L, Poewe W, Wenning GK, Stefanova N. Toll-like receptor 4 is required for α-synuclein dependent activation of microglia and astroglia. Glia. 2013;61:349–360. doi: 10.1002/glia.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberatore GT, Jackson-Lewis V, Vukosavic S, Mandir AS, Vila M, McAuliffe WG, Dawson VL, Dawson TM, Przedborski S. Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease. Nat Med. 1999;5:1403–1409. doi: 10.1038/70978. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch EC, Breidert T, Rousselet E, Hunot S, Hartmann A, Michel PP. The role of glial reaction and inflammation in Parkinson's disease. Ann N Y Acad Sci. 2003;991:214–228. doi: 10.1111/j.1749-6632.2003.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 46.Teismann P, Tieu K, Cohen O, Choi DK, Wu DC, Marks D, Vila M, Jackson-Lewis V, Przedborski S. Pathogenic role of glial cells in Parkinson's disease. Mov Disord. 2003;18:121–129. doi: 10.1002/mds.10332. [DOI] [PubMed] [Google Scholar]
- 47.Jang E, Kim JH, Lee S, Kim JH, Seo JW, Jin M, Lee MG, Jang IS, Lee WH, Suk K. Phenotypic polarization of activated astrocytes: the critical role of lipocalin-2 in the classical inflammatory activation of astrocytes. J Immunol. 2013;191:5204–5219. doi: 10.4049/jimmunol.1301637. [DOI] [PubMed] [Google Scholar]
- 48.Dai SH, Chen T, Wang YH, Zhu J, Luo P, Rao W, Yang YF, Fei Z, Jiang XF. Sirt3 attenuates hydrogen peroxide-induced oxidative stress through the preservation of mitochondrial function in HT22 cells. Int J Mol Med. 2014;34:1159–1168. doi: 10.3892/ijmm.2014.1876. [DOI] [PubMed] [Google Scholar]
- 49.Tyagi A, Nguyen CU, Chong T, Michel CR, Fritz KS, Reisdorph N, Knaub L, Reusch JEB, Pugazhenthi S. SIRT3 deficiency-induced mitochondrial dysfunction and inflammasome formation in the brain. Sci Rep. 2018;8:17547. doi: 10.1038/s41598-018-35890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, Kim Y, Liu T, Hwang YJ, Hyeon SJ, Im H, Lee K, Alvarez VE, McKee AC, Um SJ, Hur M, Mook-Jung I, Kowall NW, Ryu H. SIRT3 deregulation is linked to mitochondrial dysfunction in Alzheimer's disease. Aging Cell. 2018;17:e12679. doi: 10.1111/acel.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin J, Han P, Song M, Nielsen M, Beach TG, Serrano GE, Liang WS, Caselli RJ, Shi J. Amyloid-β increases Tau by mediating Sirtuin 3 in Alzheimer's disease. Mol Neurobiol. 2018;55:8592–8601. doi: 10.1007/s12035-018-0977-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin J, Li S, Nielsen M, Carcione T, Liang WS, Shi J. Sirtuin 3 attenuates amyloid-β induced neuronal hypometabolism. Aging (Albany NY) 2018;10:2874–2883. doi: 10.18632/aging.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lutz MI, Milenkovic I, Regelsberger G, Kovacs GG. Distinct patterns of sirtuin expression during progression of Alzheimer's disease. Neuromolecular Med. 2014;16:405–414. doi: 10.1007/s12017-014-8288-8. [DOI] [PubMed] [Google Scholar]
- 54.Liu L, Peritore C, Ginsberg J, Kayhan M, Donmez G. SIRT3 attenuates MPTP-induced nigrostriatal degeneration via enhancing mitochondrial antioxidant capacity. Neurochem Res. 2015;40:600–608. doi: 10.1007/s11064-014-1507-8. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Ren X, Zhang Q, Li Z, Ma S, Bao J, Li Z, Bai X, Zheng L, Zhang Z, Shang S, Zhang C, Wang C, Cao L, Wang Q, Ji J. PGC-1α/ERRα-Sirt3 pathway regulates DAergic neuronal death by directly deacetylating SOD2 and ATP synthase β. Antioxid Redox Signal. 2016;24:312–328. doi: 10.1089/ars.2015.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song W, Song Y, Kincaid B, Bossy B, Bossy-Wetzel E. Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: neuroprotection by SIRT3 and PGC-1α. Neurobiol Dis. 2013;51:72–81. doi: 10.1016/j.nbd.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livingstone DR. Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar Pollut Bull. 2001;42:656–666. doi: 10.1016/S0025-326X(01)00060-1. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Ren S, Liu Y, Gao K, Liu Z, Zhang Z. Inhibition of starvation-triggered endoplasmic reticulum stress, autophagy, and apoptosis in ARPE-19 cells by taurine through modulating the expression of Calpain-1 and Calpain-2. Int J Mol Sci. 2017;18:2146. doi: 10.3390/ijms18102146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White EZ, Pennant NM, Carter JR, Hawsawi O, Odero-Marah V, Hinton CV. Serum deprivation initiates adaptation and survival to oxidative stress in prostate cancer cells. Sci Rep. 2020;10:12505. doi: 10.1038/s41598-020-68668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ayers MC, Sherman ZN, Gallagher JEG. Oxidative stress responses and nutrient starvation in MCHM treated saccharomyces cerevisiae. G3 (Bethesda) 2020;10:4665–4678. doi: 10.1534/g3.120.401661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee KK, Ayyub C, Sengupta S, Kolthur-Seetharam U. dSir2 deficiency in the fatbody, but not muscles, affects systemic insulin signaling, fat mobilization and starvation survival in flies. Aging (Albany NY) 2012;4:206–223. doi: 10.18632/aging.100435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banerjee KK, Ayyub C, Ali SZ, Mandot V, Prasad NG, Kolthur-Seetharam U. dSir2 in the adult fat body, but not in muscles, regulates life span in a diet-dependent manner. Cell Rep. 2012;2:1485–1491. doi: 10.1016/j.celrep.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 63.Banerjee KK, Ayyub C, Sengupta S, Kolthur-Seetharam U. Fat body dSir2 regulates muscle mitochondrial physiology and energy homeostasis nonautonomously and mimics the autonomous functions of dSir2 in muscles. Mol Cell Biol. 2013;33:252–264. doi: 10.1128/MCB.00976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei T, Huang G, Gao J, Huang C, Sun M, Wu J, Bu J, Shen W. Sirtuin 3 deficiency accelerates hypertensive cardiac remodeling by impairing angiogenesis. J Am Heart Assoc. 2017;6:e006114. doi: 10.1161/JAHA.117.006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y, Chung ACK, Fan R, Lee HM, Xu G, Tomlinson B, Chan JCN, Kong APS. Sirt3 deficiency increased the vulnerability of pancreatic beta cells to oxidative stress-induced dysfunction. Antioxid Redox Signal. 2017;27:962–976. doi: 10.1089/ars.2016.6859. [DOI] [PubMed] [Google Scholar]
- 66.Rocha NP, Teixeira AL, Scalzo PL, Barbosa IG, de Sousa MS, Morato IB, Vieira EL, Christo PP, Palotás A, Reis HJ. Plasma levels of soluble tumor necrosis factor receptors are associated with cognitive performance in Parkinson's disease. Mov Disord. 2014;29:527–531. doi: 10.1002/mds.25752. [DOI] [PubMed] [Google Scholar]
- 67.Scalzo P, Kümmer A, Cardoso F, Teixeira AL. Increased serum levels of soluble tumor necrosis factor-alpha receptor-1 in patients with Parkinson's disease. J Neuroimmunol. 2009;216:122–125. doi: 10.1016/j.jneuroim.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 68.Qin XY, Zhang SP, Cao C, Loh YP, Cheng Y. Aberrations in peripheral inflammatory cytokine levels in Parkinson disease: a systematic review and meta-analysis. JAMA Neurol. 2016;73:1316–1324. doi: 10.1001/jamaneurol.2016.2742. [DOI] [PubMed] [Google Scholar]
- 69.Liu T, Zhang T, Yu H, Shen H, Xia W. Adjudin protects against cerebral ischemia reperfusion injury by inhibition of neuroinflammation and blood-brain barrier disruption. J Neuroinflammation. 2014;11:107. doi: 10.1186/1742-2094-11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang X, Geng K, Zhang J, Zhang Y, Shao J, Xia W. Sirt3 mediates the inhibitory effect of adjudin on astrocyte activation and glial scar formation following ischemic stroke. Front Pharmacol. 2017;8:943. doi: 10.3389/fphar.2017.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]