Abstract
The TMEM43 has been studied in human diseases such as arrhythmogenic right ventricular cardiomyopathy type 5 (ARVC5) and auditory neuropathy spectrum disorder (ANSD). In the heart, the p.(Ser358Leu) mutation has been shown to alter intercalated disc protein function and disturb beating rhythms. In the cochlea, the p.(Arg372Ter) mutation has been shown to disrupt connexin-linked function in glia-like supporting cells (GLSs), which maintain inner ear homeostasis for hearing. The TMEM43-p.(Arg372Ter) mutant knock-in mice displayed a significantly reduced passive conductance current in the cochlear GLSs, raising a possibility that TMEM43 is essential for mediating the passive conductance current in GLSs. In the brain, the two-pore-domain potassium (K2P) channels are generally known as the “leak channels” to mediate background conductance current, raising another possibility that K2P channels might contribute to the passive conductance current in GLSs. However, the possible association between TMEM43 and K2P channels has not been investigated yet. In this study, we examined whether TMEM43 physically interacts with one of the K2P channels in the cochlea, KCNK3 (TASK-1). Utilizing co-immunoprecipitation (IP) assay and Duolink proximity ligation assay (PLA), we revealed that TMEM43 and TASK-1 proteins could directly interact. Genetic modifications further delineated that the intracellular loop domain of TMEM43 is responsible for TASK-1 binding. In the end, gene-silencing of Task-1 resulted in significantly reduced passive conductance current in GLSs. Together, our findings demonstrate that TMEM43 and TASK-1 form a protein-protein interaction in the cochlea and provide the possibility that TASK-1 is a potential contributor to the passive conductance current in GLSs.
Keywords: TMEM43, KCNK3, Cochlea, Protein interaction domains and motifs, Ion transport
INTRODUCTION
TMEM43 was initially identified in silico as one of the 368 transmembrane (TMEM) proteins with unknown functions [1]. TMEM43 was shown to be expressed throughout the mouse body, including the cochlea, heart, eye, brain, and kidney [2]. Mutations in TMEM43 have been studied in human diseases such as arrhythmogenic right ventricular cardiomyopathy type 5 (ARVC5) in the heart [3-6] and auditory neuropathy spectrum disorder (ANSD) in the cochlea [2]. A p.(Ser358Leu) missense mutation in the TMEM43 has been shown to alter intercalated disc and Cx43 protein expression [4]. Moreover, microelectrode array recordings from HL-1 Cardiac Muscle Cells revealed that TMEM43-p.(Ser358Leu) expressing cells show reduced conduction velocity and asynchronous beating rhythms compared to TMEM43-WT expressing cells [4]. Similarly, a p.(Arg372Ter) nonsense mutation in the TMEM43 disrupts connexin-linked function in the cochlear supporting cells, which is to maintain inner ear homeostasis via recycling and regulating intracellular K+ [2, 7, 8]. Indeed, the TMEM43-p.(Arg372Ter) mutant knock-in mice display a significantly reduced passive conductance current from cochlear glia-like supporting cells (GLSs) [2]. Collectively, TMEM43 is suggested to be essential for mediating background conductance and electrical communication in heart and cochlear cells.
The passive conductance channels, which give rise to a low membrane resistance and linear I-V relationship, are known as the unique property that defines mature astrocytes in the brain [9-11]. Our group has formally revealed the molecular identity of the astrocytic passive conductance channels as a dimer of TWIK-1 and TREK-1 subunits of the two-pore potassium (K2P) channels [12]. In the brain, passive conductance channels are critically involved in K+ ion homeostasis [13] and volume regulation [14, 15]. In the cochlea, the endocochlear potential of +80 mV is generated by maintaining a high K+ concentration in the endolymph, and this potential is critical for the activation of hair cells [16]. Therefore, various channel proteins in GLSs should be thoroughly examined to understand their role in establishing and maintaining K+ ion homeostasis and their associated role in hearing and speech discrimination. Furthermore, the TMEM43-mediated large passive conductance current in GLSs was reminiscent of the typical passive conductance current observed in hippocampal astrocytes [2]. Accordingly, we hypothesized that there is a possibility of TMEM43 protein interacting with one of the K2P channels in the cochlea.
There are 15 known members of the K2P family, each containing two-pore (P) loops and four transmembrane (TM) domains [17]. They have a unique extracellular cap structure formed by two TM1-P1 linkers, containing a cysteine for dimerization [18, 19]. Interestingly, this cysteine residue is not present in the TASK-1/3/5 subfamily [19], suggesting a disulfide-bridge-free binding in the TASK channels. In addition, the K2P channels are regulated by diverse stimuli such as oxygen tension, pH, lipids, mechanical stretch, neurotransmitters, and G protein-coupled receptors [20]. Among the various 15 K2P channels, we searched for candidate K2P subunits that would most likely interact with TMEM43. In the end, we narrowed it down to KCNK3 (TASK-1), which exhibits the most similar properties as the TMEM43-involved currents, sensitive to external acidosis and displaying linear rectification [2, 21]. Although TMEM43 is expected to play critical roles in mammals, studies on this protein are sparse. In this study, we provide a piece of additional information regarding TMEM43 protein that it interacts with one of the K2P channels, KCNK3 (TASK-1), in the cochlea.
MATERIALS AND METHODS
Animals
All experimental procedures were conducted according to protocols approved by the directives of the Animal Care and Use Committee of the Institutional Animal Care and Use Committee of IBS (Daejeon, Republic of Korea). C57BL/6 mouse pups of Postnatal day 5~8 (p5~p8) were purchased from Raon Bio (Youngin, Republic of Korea) or DBL (Eumseong, Republic of Korea) and were sacrificed on the delivered day.
Primary neuron culture
C57BL/6 pups of embryonic day 15 were used. Embryos were dissociated from amniotic sac, and fetuses were put into a sterile petri dish with HBSS. They were decapitated, and skull and meninges were removed on ice. Only cortex was collected in 50 ml tube with HBSS and washed 3 times with 20 ml of HBSS to clear the debris. The cortex was incubated in 5 ml HBSS containing 0.1% trypsin for 5 min in a 37℃ water bath (shaking every 1 min). Afterward, 5 ml FBS (1X volume) was added to the tube, and the cortex was dissociated by pipetting. The cells were collected using a cell strainer (40 μm) and centrifuge (800 g, 3 min). The supernatant was removed, and the cell pellet was resuspended in Neurobasal A medium (10888022, Gibco). 1×104 cells were plated on 0.1 mg/ml Poly-L-Ornithine hydrobromide (PLO) (P3655, Sigma-Aldrich)-coated 24 well plates and incubated in the incubator (37℃, 5% CO2). The neuron culture was maintained by replacing the half of medium with fresh medium: Neurobasal A medium+2% B27 (17504-044, Gibco)+1% Glutamax (35050-061, Thermofisher Scientific)+0.5% P/S (250 ul/50 ml) (SV30010, Hyclone) every 2~3 days.
Plasmids
TMEM43 (Myc-DDK-tagged)-Human transmembrane protein 43 (TMEM43) (NM_024334.2) was purchased from OriGene (RC200998), KCNK3 (Myc-DDK-tagged) (TASK-1) (NM_002246.3) was purchased from OriGene (RC215155), GJB2 (NM_004004) Human Tagged ORF Clone was purchased from OriGene (RC202092), and Cx30-msfGFP was purchased from Addgene (69019). The coding sequences of these genes were cloned into CMV-MCS-IRES2-EGFP or CMV-MCS-IRES2-dsRed plasmid vector using BglII/XmaI sites. TMEM43 truncation mutations were made using the EZchange site-directed mutagenesis kit (EZ004S, Enzynomics).
Heterologous expression of TMEM43 on HEK293T cell lines
Human embryonic kidney (HEK) 293T cells were purchased from ATCC (CRL-3216). The cell line has been tested for mycoplasma contamination. HEK293T cell was cultured in DMEM (10-013, Corning) supplemented with 10% heat-inactivated fetal bovine serum (10082-147, Gibco) and 10,000 units/ml penicillin-streptomycin (15140-122, Gibco) at 37℃ in a humidified atmosphere of 95% air and 5% CO2. According to the manufacturer’s protocol, the transfection of expression vectors was performed with Effectene Transfection Reagent (Effectene, 301425, Qiagen). One day before performing the experiments, HEK293T cells were transfected with each DNA 1 μg per 35 mm dish.
Co-immunoprecipitation (IP) and western blot
DNA transfected HEK293T cells, or cochlear tissues of an average of 20 mice (p5~p8) per sample were extracted and homogenized with PierceTM IP Lysis Buffer (87787, Thermo Fisher Scientific) and HaltTM Protease and Phosphatase.
Inhibitor Cocktail (100X) (78446, Thermo Fisher Scientific). Equal amounts of precleared cell lysates were incubated with mouse anti-TASK-1 (KCNK3) monoclonal (1 μg, ab186352, Abcam) antibody or control IgG (1 μg) overnight. Protein A/G-Agarose beads (20422, Thermo Fisher Scientific) were added to the mixtures and further incubated with rotation for 1 h at 4℃, followed by a wash with lysis buffer. Bound proteins were eluted from the beads with SDS-PAGE sample buffer, and western blotting was performed with rabbit anti-FLAG (1:500, 2368s, Cell Signaling) or rabbit anti-TMEM43 (1:100, NBP1-84132, Novus).
Duolink® proximity ligation assay (PLA)
Duolink® In Situ Red Starter Kit Mouse/Rabbit (DUO92101, Sigma-Aldrich) and far-red detection kit (DUO92013, Sigma-Aldrich) were used. On the experimental day, cochlea tissue (p5~p8) was obtained, fixed with 4% paraformaldehyde, and washed with 0.3% Triton-X100 containing PBS. After blocking, the sample was incubated with rabbit anti-TMEM43 polyclonal (1:100, NBP1-84132, Novus) and mouse anti-TASK-1 (KCNK3) monoclonal (1:100, ab186352, Abcam) at 4℃, overnight. The next day, the sample was incubated with anti-rabbit MINUS and anti-mouse PLUS probe, ligase, and polymerase sequentially. DNA strands participate in rolling circle DNA synthesis only when two probes are in close proximity (<40 nm). Fluorescent-labeled complementary oligonucleotide probes were observed under Zeiss confocal microscopy.
Construction and packaging of shRNA–lentiviral vectors
For Task-1 gene silencing, three candidates for mouse Task-1 shRNA (GenBank accession no. NM_010608) sequences were cloned into lentiviral pSicoR vector using XhoI/XbaI sites, as previously described [22]. The candidate shRNA containing pSicoR vector was packaged into high-titer lentivirus by the IBS virus facility (Republic of Korea). The target regions of the 3 candidate Task-1 shRNAs are: 5’-ggtgaacgcatcaacaccttc-3’, 5’-ggactttcttccaggcctatt-3’ and 5’- gccttcagcttcgtgtacatc-3’. Candidate #2 (5’-ggactttcttccaggcctatt-3’) was used in the study. For control shRNA, scrambled sequence 5’-tcgcatagcgtatgccgtt-3’ was inserted in place of shRNA sequence. Knockdown efficiency was determined by RT-PCR data analysis with ImageJ. We analyzed the transcripts of Task-1 and Gapdh by primer pairs as follows: the forward primer (5'- atcaccgtcatcaccaccat-3') and the reverse primer (5'-acgaaggtgttgatgcgttc-3') for Task-1, and the forward primer (5'-tgatgacatcaagaaggtggtgaag-3') and the reverse primer (5'-tccttggaggccatgtaggccat-3') for Gapdh. When calculating the knockdown efficiency, the virus infection efficiency was considered by dividing the knockdown efficiency with infection efficiency.
Cochlear organotypic culture and shRNA treatment
Neonatal mouse (C57/BL6, P5) cochlear turns were isolated in ice-cold sterile Hanks’ Balanced Salt Solution (HBSS). Cochlear segments were attached on Cell-Tak (354240, Corning) coated coverslips and incubated overnight in DMEM/F12 medium containing 1% fetal bovine serum, 5 µg/ml ampicillin, B27 and N2 (37°C, 5% CO2 humidified incubator). Upon confirming stable attachment, the tissues were treated with either control or shRNA carrying lentivirus diluted in culture medium (1:1,000) for 48 h. The medium was then replaced with a fresh one. After an additional 24 h of incubation, the tissues were subjected to further examination.
Immunocytofluorescence on cochlear tissue
The virus infection on cultured cochlea tissue was confirmed via the presence of mCherry signal. For immunostaining, cochlea tissue was fixed in ice-cold 4% paraformaldehyde, for 10 min and incubated in blocking/permeabilizing buffer (PBS with 5% goat serum and 0.25% Triton-X100). Then, the preparations were incubated overnight at 4°C with rabbit anti-TMEM43 polyclonal (1:100, NBP1-84132, Novus) and mouse anti-TASK-1 (KCNK3) monoclonal (1:100, ab186352, Abcam) diluted in the blocking/permeabilizing buffer. After 3 washes, the cochlear turns were reacted with donkey anti-rabbit Alexa Fluor 594 (1:500, 711-585-152, Jackson ImmunoResearch) donkey anti-mouse Alexa Fluor 488 (1:500, 715-545-150, Jackson ImmunoResearch) in blocking/permeabilizing buffer for 1 h at room temperature. Samples were then rinsed once with blocking/permeabilizing buffer and twice with PBS. Using Fluorsave reagent (345789, Calbiochem), the tissues were mounted on glass slides and covered with a coverslip. Specific immunolabeling was examined under Zeiss confocal microscope. No immunoreactivity was found when the primary antibodies were omitted.
Electrophysiological recording in the cochlear supporting cells
Cultured cochlea tissue infected with lentivirus was used. Recordings were done in HEPES buffer containing (mM): 144 NaCl, 5.8 KCl, 1.3 CaCl2, 2 MgCl2, 10 HEPES, 0.7 NaH2PO4, and 5.6 D-glucose (pH 7.4 was adjusted with NaOH). Glia-like supporting cells located below the inner hair cell layer were whole-cell patch clamped, and current traces were elicited by 1-sec ramps descending from +100 mV to -100 mV with -60 mV holding potential. Recording electrodes (7~11 MΩ) supplemented with (mM): 126 K-Gluconate, 5 HEPES, 0.5 MgCl2, and 10 BAPTA (pH adjusted to 7.3 with KOH) were advanced through tissue under positive pressure. Slice chamber was mounted on the stage of an upright Hamamatsu digital camera viewed with a 60X water immersion objective with infrared differential interference contrast optics using Imaging Workbench Software. Electrical signals were digitized and sampled with Digidata 1320A and Multiclamp 700B amplifier (Molecular Devices) using pCLAMP 10.2 software. Data were sampled at 10 kHz and filtered at 2 kHz. Glass pipettes were pulled from a micropipette puller (P-97, Sutter Instrument), and all experiments were conducted at room temperature 20~22℃.
Data analysis and statistical analysis
Off-line analysis was carried out using Clampfit version 10.4.1.10 and GraphPad Prism version 7.02 software. When comparing between two samples, the significance of data was assessed by Student’s two-tailed unpaired t-test. One-way ANOVA with Kruskal-Wallis test with Dunn's posthoc test was used to compare 3 samples, as the data did not pass the normality test. Significance levels were given as: N.S. p>0.05, *p<0.05, **p<0.01, ***p<0.001 and #p<0.0001.
RESULTS AND DISCUSSION
TMEM43 interacts with TASK-1 in the in vitro system
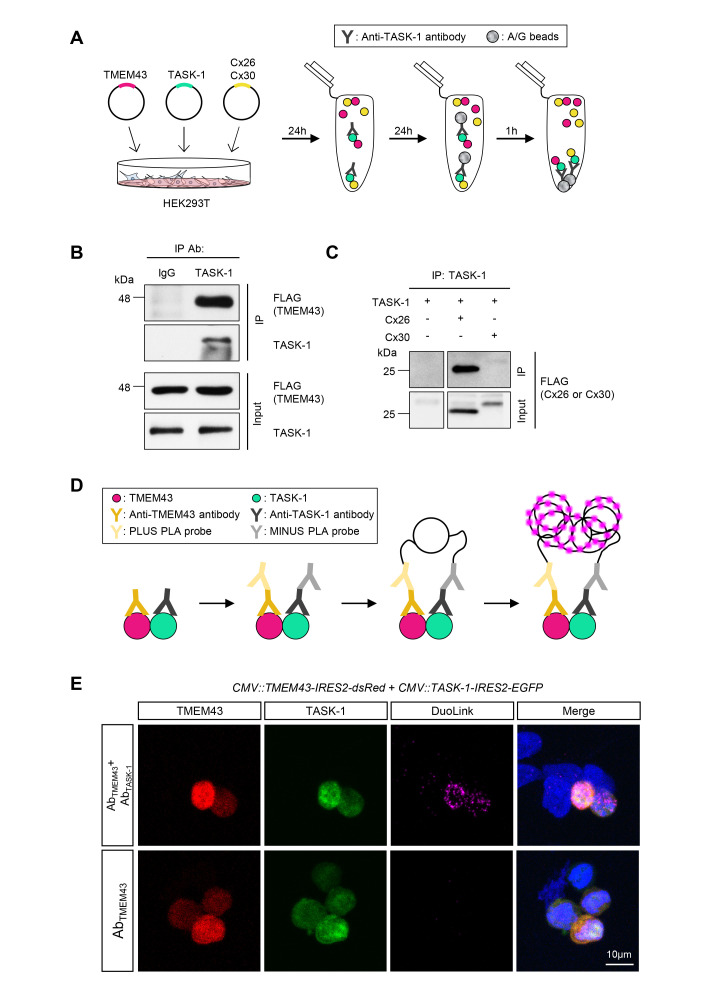
In order to examine whether TMEM43 interacts with TASK-1, human TMEM43 and TASK-1 genes were cloned into an internal ribosome entry site (IRES2) vector. The IRES sequences can translate two independent proteins under the same promoter in a cap-independent mechanism via recruiting the eukaryotic ribosome to the mRNA [23]. Unlike fluorescent-tagged proteins, which may have disturbed protein trafficking or interaction, the fluorescent reporter molecules are expressed without affecting TMEM43 or TASK-1 protein in the IRES system. Using this tool, we heterologously expressed TMEM43 and TASK-1 together in the HEK293T cell line and performed a co-immunoprecipitation (IP) assay (Fig. 1A). The cell lysate was pulled down with TASK-1 antibody and blotted with anti-FLAG to detect TMEM43 protein. A positive band appeared at the size of TMEM43 protein, indicating that TMEM43 protein was immunoprecipitated with TASK-1 (Fig. 1B). As the previous study has shown interaction of TMEM43 with Cx26 and Cx30 proteins [2], we next examined if TASK-1 also interacts with the connexins (Fig. 1A). The positive western blot band for connexin protein was only present when the TASK-1 was co-expressed with Cx26 but not with Cx30 (Fig. 1C). This result demonstrates that TASK-1 interacts with Cx26 protein.
Fig. 1.
TMEM43 interacts with TASK-1 in the heterologous overexpression system. (A) Schematic diagram illustrating the heterologous DNA expression in HEK293T cell line and procedures of the co-IP assay. Tmem43, Cx26, and Cx30 coding sequences tagged with FLAG and Task-1 coding sequences were cloned into IRES2 vectors. Anti-TASK-1 antibody was used for immunoprecipitation, and the pulled-down protein was blotted with an anti-FLAG antibody. (B) Co-IP result, performed with normal IgG or TASK-1 antibody using lysates of HEK293T cells co-expressing TMEM43-FLAG and TASK-1. (C) Co-IP results of TASK-1 with Cx26-FLAG and Cx30-FLAG. Anti-FLAG antibody was used to detect the connexin channels. The first lane is a negative control. (D) A cartoon depicting the principle of the Duolink PLA. If the proteins of interest are in close proximity, the DNA probes hybridize to make circular DNA. This DNA can be amplified and visualized by fluorescently labeled complementary oligonucleotide probes. (E) Duolink PLA with anti-TMEM43 and anti-TASK-1 antibodies. Duolink PLA signal was recognized as far-red fluorescent (λex 644 nm, λem 669 nm), indicative of proximity of TMEM43 and TASK-1 (<40 nm). The lower panel is a negative control, using only an anti-TMEM43 antibody.
To further examine the direct interaction of TMEM43 and TASK-1 in the in vitro system, we implemented a Duolink proximity ligation assay (PLA). This tool allows visualizing protein-protein interaction with high specificity and sensitivity; hybridization of oligonucleotide arms of the PLA probes creates a template for rolling circle amplification when the epitopes of the target proteins are in a close proximity (<40 nm) (Fig. 1D) [24]. Consistent with the co-IP results, we observed a positive PLA signal with TMEM43 and TASK-1 antibodies in the TMEM43 and TASK-1 co-expressed cells, but not with TMEM43 antibody alone (Fig. 1E), highly indicating close proximity of the two proteins. Taken together, these results demonstrate that TMEM43 and TASK-1 can bind in the heterologous expression system.
The intracellular loop domain of TMEM43 is necessary for TASK-1 interaction
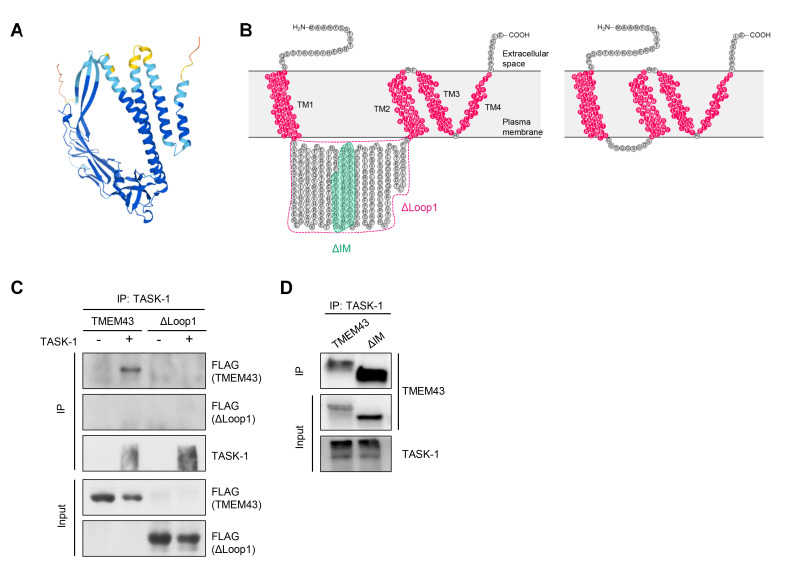
We next investigated the interacting domain of TMEM43 that is required for interaction with TASK-1. We predicted protein structure of TMEM43 with AlphaFold, the state-of-the-art deep neural network-based method with atomic level accuracy (Fig. 2A) [25]. The bioinformatic analyses predicted TMEM43 protein with 4 TM domains and a large intracellular loop (Loop1) containing an intramembrane (IM) domain [2, 3, 25]. Based on the predictions, we firstly made a deletion mutation of Loop1, which lies between TM1 and TM2 region, and linked the two TM with three GGS linkers (Fig. 2B). The co-IP assay revealed that the interaction between TMEM43-ΔLoop1 and TASK-1 was disrupted (Fig. 2C). We next targeted the IM domain (Fig. 2B) and found that the interaction between TMEM43-ΔIM with TASK-1 was intact (Fig. 2D). These findings indicate that TMEM43 intracellular loop (Loop1) is necessary for TASK-1 interaction. The exact binding domain at TMEM43 Loop1, other than IM domain, should be further examined.
Fig. 2.
The binding domain of TMEM43 for TASK-1 interaction. (A) TMEM43 protein structure prediction based on AlphaFold. (B) Left: amino acids of TMEM43 protein and its predicted topology based on AlphaFold. TM domains are colored in pink. The truncated domains are dotted-lined with different colors. Right: amino acids of TMEM43-ΔLoop1 with three GGS linkers. (C) Co-IP results performed with TASK-1 antibody using lysates of HEK293T cells expressing TASK-1 and TMEM43-FLAG full length and Loop1 truncation mutant (ΔLoop1). Expected band size for TMEM43-ΔLoop1 is 20 kDa. (D) Co-IP data showing that both TMEM43-WT and TMEM43-ΔIM are immuno-pulled down with anti-TASK-1 antibody. The expected band size for TMEM43-ΔIM is 41 KDa.
TMEM43 interacts with TASK-1 in the mouse cochlea
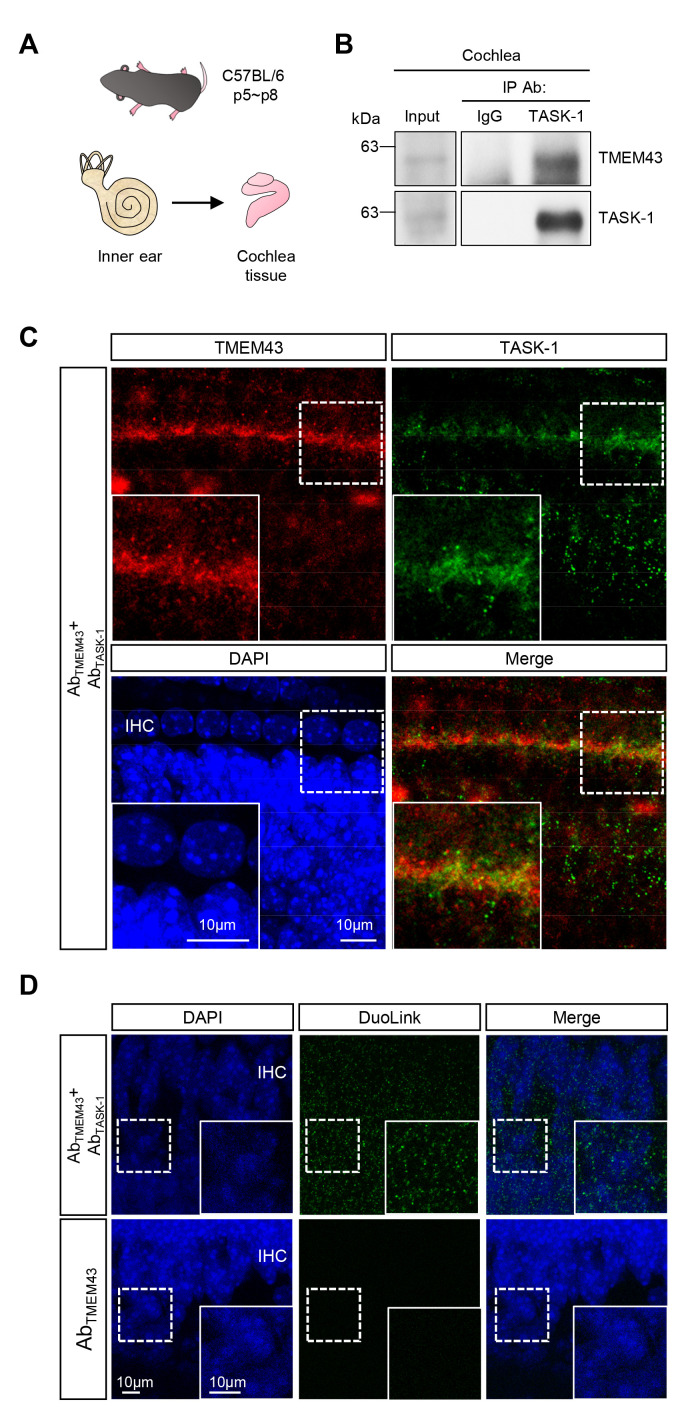
We subsequently examined TMEM43 interaction with TASK-1 in the cochlea tissue. The cochlea tissue was obtained from the inner ear of C57BL/6 pups (Fig. 3A). Consistent with the heterologous expression system results, immunoprecipitates from cochlear homogenates with anti-TASK-1 antibody showed a positive TMEM43 signal (Fig. 3B). Interestingly, the co-IP band size for TMEM43 was at around 60 kDa in the cochlea tissue (Fig. 3B), which was different from 47 kDa in the heterologous system (Fig. 1B). This discrepancy of size gap may arise from the absence of many proteins required for proper protein processing (such as chaperone and trafficking proteins) in cell lines but not in native environments. Furthermore, it has been demonstrated that co-translational and post-translational modification, in particular glycosylation, can also affect many proteins in membrane trafficking and functional gating properties [26, 27]. In fact, we have observed two TMEM43 bands, at size 43 kDa and 60 kDa from the cochlea lysates. However, after co-IP with anti-TASK-1 antibody, the major TMEM43 band appeared at 60 kDa but not at 43 kDa. This result is different from the result of the previous study [2], which showed TMEM43 band size at 43 kDa after co-IP with anti-Cx30 antibody. Therefore, these results together suggest that Cx30 likely interacts with the native TMEM43 (43 kDa) while TASK-1 preferentially associates with the post-translationally modified form of TMEM43 (60 kDa). Identification of these possible chaperones and membrane trafficking proteins or post-translational modifications of TMEM43 in GLSs awaits future investigations.
Fig. 3.
Endogenous TMEM43 interacts with TASK-1 in mouse cochlea tissue. (A) An illustration of materials used for ex vivo study. C57BL/6 pups of p5~p8 were used, and the cochlea tissue was acutely dissected from the inner ear. (B) Co-IP data performed with cochlea tissue. Tissue lysates were pulled down with anti-TASK-1 antibody and blotted with anti-TMEM43 antibody. (C) Immunostaining of cochlea tissue with anti-TMEM43 and anti-TASK-1 antibodies. Inset images show magnified GLSs with TMEM43 and TASK-1 signals. (D) Duolink PLA with anti-TMEM43 and -TASK-1 antibodies, performed with acutely dissected mouse cochlea tissue. Duolink PLA signal was positive at GLSs (green). The amplified PLA red fluorescent signal was pseudo-colored as green for better data display. Inset: magnified images. The lower panel is a negative control, using only an anti-TMEM43 antibody.
We next performed immunohistochemistry with the two antibodies to analyze TMEM43 and TASK-1 protein expression in the intact cochlea tissue. The confocal micrographs of the organs of Corti demonstrated a strong co-localization of TMEM43 and TASK-1 immunoreactivities in the cochlear GLSs (Fig. 3C). Furthermore, the Duolink PLA was done in the cochlea tissue using anti-TMEM43 and anti-TASK-1 antibodies. Similar to the in vitro results (Fig. 1E), a positive PLA signal was observed in the cochlea only when both antibodies were present (Fig. 3D). Thus, from the above ex vivo cochlea data, we could conclude that the endogenous TMEM43 protein can interact with the native TASK-1 protein in the mouse cochlea tissue.
TASK-1 is involved in the cochlear passive conductance current
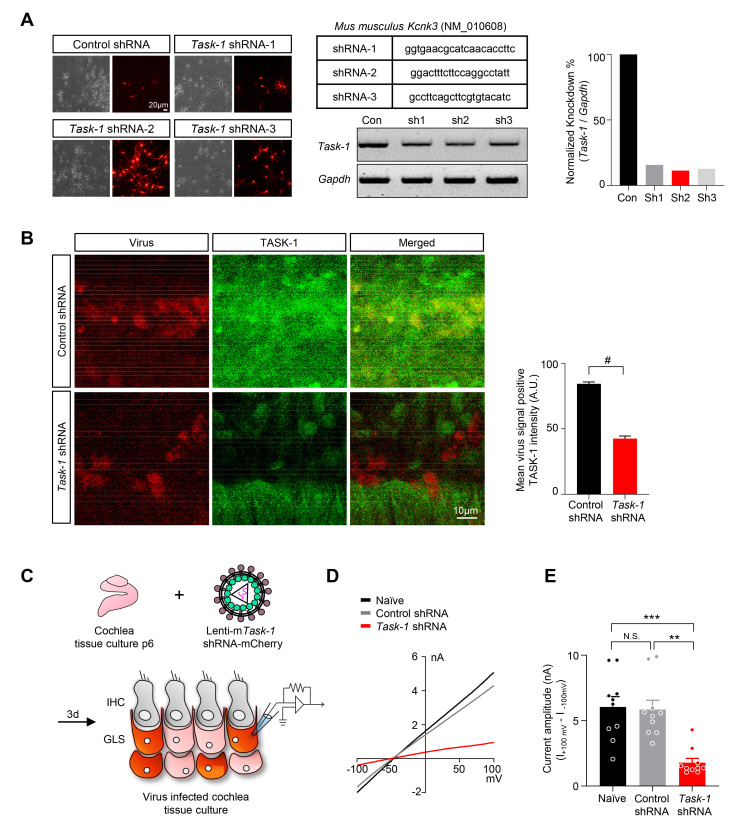
TMEM43 has been reported to mediate the passive conductance current in the cochlear GLSs [2]. Accordingly, we tested if TASK-1 is also involved in the cochlear passive conductance current. To begin with, we developed a mouse Task-1 shRNA to genetically knockdown Task-1 mRNA. A total of 3 candidate shRNAs to target Task-1 were developed and packaged into lentivirus (Fig. 4A). We then infected the cultured neurons with each candidate shRNAs to reduce Task-1 expression (Fig. 4A). Among the 3 candidates, we chose candidate number 2, as both knockdown efficiency and virus expression were highest compared to the other two candidates (Fig. 4A). The Task-1 shRNA-carrying lentivirus could effectively infect cultured cochlea tissue and successfully ablate TASK-1 protein expression (Fig. 4B). In order to measure TASK-1 mediated current in the cochlear GLSs, we cultured cochlea tissue, infected with Task-1 shRNA-carrying lentivirus and did whole-cell patch-clamp recordings on the virus-infected GLSs (Fig. 4C). As a result, gene-silencing of Task-1 led to a significant loss of passive conductance current in the cochlear GLSs (Fig. 4D, E). These results provide possibility that TASK-1 is a potential contributor to the passive conductance current in GLSs.
Fig. 4.
Gene-silencing of mTask-1 and TASK-1-regulated cochlear passive conductance current. (A) Mouse primary neuron culture infected with lentivirus packaged with mTask-1 shRNA candidates. The target sequences for 3 shRNA candidates are shown in the table. The knockdown efficiency was tested with RT-PCR and normalized by Gapdh level for comparison. The virus infection efficiency was measured and reflected when calculating the knockdown percentage. (B) Cultured cochlea tissue infected with control or Task-1 shRNA-carrying lentivirus and immunostained with anti-TASK-1 antibody. Mean TASK-1 intensity of virus-infected cells is quantified. (C) A diagram describing cultured cochlea tissue infected with Task-1 shRNA-carrying lentivirus and the infected GLSs patch-clamped with a glass pipette containing a high K+ internal solution. (D) Representative I-V curve measured from naïve (black), control shRNA treated (gray), and Task-1 shRNA treated (red) cochlea tissue. (E) Summary bar graph of current amplitude from (D).
In conclusion, our findings provide molecular and physiological lines of evidence manifesting that TMEM43 protein interacts with a K2P channel, TASK-1. The congruent results from Co-IP and Duolink PLA in both in vitro overexpression systems and ex vivo cochlea tissue strengthens our hypothesis that the TMEM43 protein may interact with a K2P channel in the cochlea. Furthermore, the electrophysiological recordings from cochlear GLSs revealed that TASK-1 also has chance to mediate the large K+ current in the cochlea. One of the major functions of cochlear GLSs, is to regulate cochlear K+ homeostasis via leak K+ channels and gap junction channels [28]. As this function of K+ homeostasis by GLSs is critical for hearing, we expect to see further studies regarding cochlear K2P channels. Lastly, because TMEM43 is ubiquitously expressed in mammals and has detrimental dominant-negative effects on human diseases, detailed research on this protein is required. With the new information provided from this study, we anticipate active investigations concerning TMEM43 protein on background passive conductance currents and human diseases.
ACKNOWLEDGEMENTS
This work was supported by the Institute for Basic Science (IBS), Center for Cognition and Sociality (IBS-R001-D2) to C.J.L. The graphical abstract was created with BioRender.com.
References
- 1.Stelzer G, Dalah I, Stein TI, Satanower Y, Rosen N, Nativ N, Oz-Levi D, Olender T, Belinky F, Bahir I, Krug H, Perco P, Mayer B, Kolker E, Safran M, Lancet D. In-silico human genomics with GeneCards. Hum Genomics. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang MW, Oh DY, Yi E, Liu X, Ling J, Kim N, Sharma K, Kim TY, Lee S, Kim AR, Kim MY, Kim MA, Lee M, Han JH, Han JJ, Park HR, Kim BJ, Lee SY, Woo DH, Oh J, Oh SJ, Du T, Koo JW, Oh SH, Shin HW, Seong MW, Lee KY, Kim UK, Shin JB, Sang S, Cai X, Mei L, He C, Blanton SH, Chen ZY, Chen H, Liu X, Nourbakhsh A, Huang Z, Kang KW, Park WY, Feng Y, Lee CJ, Choi BY. A nonsense TMEM43 variant leads to disruption of connexin-linked function and autosomal dominant auditory neuropathy spectrum disorder. Proc Natl Acad Sci U S A. 2021;118:e2019681118. doi: 10.1073/pnas.2019681118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merner ND, Hodgkinson KA, Haywood AF, Connors S, French VM, Drenckhahn JD, Kupprion C, Ramadanova K, Thierfelder L, McKenna W, Gallagher B, Morris-Larkin L, Bassett AS, Parfrey PS, Young TL. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82:809–821. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siragam V, Cui X, Masse S, Ackerley C, Aafaqi S, Strandberg L, Tropak M, Fridman MD, Nanthakumar K, Liu J, Sun Y, Su B, Wang C, Liu X, Yan Y, Mendlowitz A, Hamilton RM. TMEM43 mutation p.S358L alters intercalated disc protein expression and reduces conduction velocity in arrhythmogenic right ventricular cardiomyopathy. PLoS One. 2014;9:e109128. doi: 10.1371/journal.pone.0109128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskin B, Skinner JR, Sanatani S, Terespolsky D, Krahn AD, Ray PN, Scherer SW, Hamilton RM. TMEM43 mutations associated with arrhythmogenic right ventricular cardiomyopathy in non-Newfoundland populations. Hum Genet. 2013;132:1245–1252. doi: 10.1007/s00439-013-1323-2. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez F, Zorio E, Jimenez-Jaimez J, Salguero-Bodes R, Zwart R, Gonzalez-Lopez E, Molina P, Bermúdez-Jiménez F, Delgado JF, Braza-Boïls A, Bornstein B, Toquero J, Segovia J, Van Tintelen JP, Lara-Pezzi E, Garcia-Pavia P. Clinical characteristics and determinants of the phenotype in TMEM43 arrhythmogenic right ventricular cardiomyopathy type 5. Heart Rhythm. 2020;17:945–954. doi: 10.1016/j.hrthm.2020.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Zhao L, Liu J, Hu Y, Sun J, Yang S. Supporting cells-a new area in cochlear physiology study. J Otol. 2008;3:9–17. doi: 10.1016/S1672-2930(08)50002-X. [DOI] [Google Scholar]
- 8.Kikuchi T, Kimura RS, Paul DL, Takasaka T, Adams JC. Gap junction systems in the mammalian cochlea. Brain Res Brain Res Rev. 2000;32:163–166. doi: 10.1016/S0165-0173(99)00076-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhou M, Schools GP, Kimelberg HK. Development of GLAST(+) astrocytes and NG2(+) glia in rat hippocampus CA1: mature astrocytes are electrophysiologically passive. J Neurophysiol. 2006;95:134–143.:mature. doi: 10.1152/jn.00570.2005. [DOI] [PubMed] [Google Scholar]
- 10.D'Ambrosio R, Wenzel J, Schwartzkroin PA, McKhann GM, 2nd, Janigro D. Functional specialization and topographic segregation of hippocampal astrocytes. J Neurosci. 1998;18:4425–4438. doi: 10.1523/JNEUROSCI.18-12-04425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinhäuser C, Berger T, Frotscher M, Kettenmann H. Heterogeneity in the membrane current pattern of identified glial cells in the hippocampal slice. Eur J Neurosci. 1992;4:472–484. doi: 10.1111/j.1460-9568.1992.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 12.Hwang EM, Kim E, Yarishkin O, Woo DH, Han KS, Park N, Bae Y, Woo J, Kim D, Park M, Lee CJ, Park JY. A disulphide-linked heterodimer of TWIK-1 and TREK-1 mediates passive conductance in astrocytes. Nat Commun. 2014;5:3227. doi: 10.1038/ncomms4227. [DOI] [PubMed] [Google Scholar]
- 13.Walz W. Role of astrocytes in the clearance of excess extracellular potassium. Neurochem Int. 2000;36:291–300. doi: 10.1016/S0197-0186(99)00137-0. [DOI] [PubMed] [Google Scholar]
- 14.O'Connor ER, Kimelberg HK. Role of calcium in astrocyte volume regulation and in the release of ions and amino acids. J Neurosci. 1993;13:2638–2650. doi: 10.1523/JNEUROSCI.13-06-02638.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson JE, Evers JA. Correlations between energy metabolism, ion transport, and water content in astrocytes. Can J Physiol Pharmacol. 1992;70 Suppl:S350–S355. doi: 10.1139/y92-282. [DOI] [PubMed] [Google Scholar]
- 16.Nin F, Hibino H, Doi K, Suzuki T, Hisa Y, Kurachi Y. The endocochlear potential depends on two K+ diffusion potentials and an electrical barrier in the stria vascularis of the inner ear. Proc Natl Acad Sci U S A. 2008;105:1751–1756. doi: 10.1073/pnas.0711463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein SA, Bayliss DA, Kim D, Lesage F, Plant LD, Rajan S. International Union of Pharmacology. LV. Nomenclature and molecular relationships of two-P potassium channels. Pharmacol Rev. 2005;57:527–540. doi: 10.1124/pr.57.4.12. [DOI] [PubMed] [Google Scholar]
- 18.Zúñiga L, Zúñiga R. Understanding the cap structure in K2P channels. Front Physiol. 2016;7:228. doi: 10.3389/fphys.2016.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein M, Rinné S, Kiper AK, Ramírez D, Netter MF, Bustos D, Ortiz-Bonnin B, González W, Decher N. Functional mutagenesis screens reveal the 'cap structure' formation in disulfide-bridge free TASK channels. Sci Rep. 2016;6:19492. doi: 10.1038/srep19492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesage F, Lazdunski M. Molecular and functional properties of two-pore-domain potassium channels. Am J Physiol Renal Physiol. 2000;279:F793–F801. doi: 10.1152/ajprenal.2000.279.5.F793. [DOI] [PubMed] [Google Scholar]
- 21.Patel AJ, Honoré E. Properties and modulation of mammalian 2P domain K+ channels. Trends Neurosci. 2001;24:339–346. doi: 10.1016/S0166-2236(00)01810-5. [DOI] [PubMed] [Google Scholar]
- 22.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 24.Alam MS. Proximity ligation assay (PLA) Curr Protoc Immunol. 2018;123:e58. doi: 10.1002/cpim.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazniewska J, Weiss N. Glycosylation of voltage-gated calcium channels in health and disease. Biochim Biophys Acta Biomembr. 2017;1859:662–668. doi: 10.1016/j.bbamem.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe I, Zhu J, Recio-Pinto E, Thornhill WB. Glycosylation affects the protein stability and cell surface expression of Kv1.4 but Not Kv1.1 potassium channels. A pore region determinant dictates the effect of glycosylation on trafficking. J Biol Chem. 2004;279:8879–8885. doi: 10.1074/jbc.M309802200. [DOI] [PubMed] [Google Scholar]
- 28.Wangemann P. Supporting sensory transduction: cochlear fluid homeostasis and the endocochlear potential. J Physiol. 2006;576(Pt 1):11–21. doi: 10.1113/jphysiol.2006.112888. [DOI] [PMC free article] [PubMed] [Google Scholar]