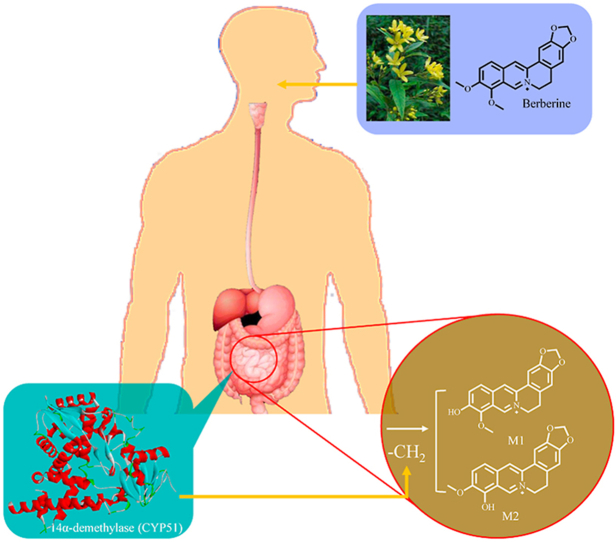
Abstract
Berberine (BBR) is an isoquinoline alkaloid extracted from Coptis chinensis that improves diabetes, hyperlipidemia and inflammation. Due to the low oral bioavailability of BBR, its mechanism of action is closely related to the gut microbiota. This study focused on the CYP51 enzyme of intestinal bacteria to elucidate a new mechanism of BBR transformation by demethylation in the gut microbiota through multiple analytical techniques. First, the docking of BBR and CYP51 was performed; then, the pharmacokinetics of BBR was determined in ICR mice in vivo, and the metabolism of BBR in the liver, kidney, gut microbiota and single bacterial strains was examined in vitro. Moreover, 16S rRNA analysis of ICR mouse feces indicated the relationship between BBR and the gut microbiota. Finally, recombinant E. coli containing cyp51 gene was constructed and the CYP51 enzyme lysate was induced to express. The metabolic characteristics of BBR were analyzed in the CYP51 enzyme lysate system. The results showed that CYP51 in the gut microbiota could bind stably with BBR, and the addition of voriconazole (a specific inhibitor of CYP51) slowed down the metabolism of BBR, which prevented the production of the demethylated metabolites thalifendine and berberrubine. This study demonstrated that CYP51 promoted the demethylation of BBR and enhanced its intestinal absorption, providing a new method for studying the metabolic transformation mechanism of isoquinoline alkaloids in vivo.
Keywords: Berberine, Biotransformation, Gut microbiota, CYP51, Demethylated metabolite
Graphical abstract
Highlights
-
•
The demethylation metabolism of natural drugs difficult to absorb through the gut microbiota was first reported.
-
•
Six different methods were presented to explain the metabolic mechanism of natural isoquinoline alkaloids.
-
•
The findings provided a new idea for studying the mechanism of drug metabolism of gut microbiota.
1. Introduction
The human intestine is the natural host of many microorganisms [1], and the gut microbiota is a complex group containing hundreds of times more genes than the human genome [2]. Referred to as a “hidden organ” of the body [3], the gut microbiota has been reported to be associated with many diseases, including obesity [4], functional bowel disease, colitis [[5], [6], [7], [8]] and cardiovascular disease [9]. There are many metabolic enzymes in the gut microbiota, such as β-glucuronidase, β-glucosidase, nitroreductase and azoreductase [10,11]. These enzymes can interact with oral drugs and produce metabolites that are different from those produced by organs [12]. Therefore, the gut microbiota has good biotransformation ability and can participate in the metabolism of oral natural products in vivo. In addition, studies have shown that regulation of the composition of gut microbiota can contribute to disease treatment [[13], [14], [15]].
Sterol 14α-demethylase (P45014DM, CYP51) is a key biosynthetic enzyme [16] belonging to the cytochrome P450 family of enzymes, which catalyze the 14α-methyl hydroxylation of sterol precursors [17,18]. More than 100 CYP51 sequences have been found in 82 species, some of which contain multiple cyp51 genes. Meanwhile, as the P450 enzyme in bacteria, fungi, lower eukaryotes, higher plants and mammals, CYP51 catalyzes the metabolism of xenobiotics through demethylation. It has been found that CYP51 has five kinds of substrates, including lanosterol, 24,25-dihydrolanosterol, 24-methylene-24,25-dihydrolanosterol, obtusifoliol, and 4β-desmethyllanosterol [19]. These substrates can be demethylated by CYP51. For example, lanosterol can be metabolized by CYP51 to 14-demethyl-14-dehydrolanosterol (FF-MAS). In addition, studies have shown that one of the main metabolic pathways of isoquinoline alkaloids is demethylation, which may be closely related to CYP51, as shown by docking simulation [20].
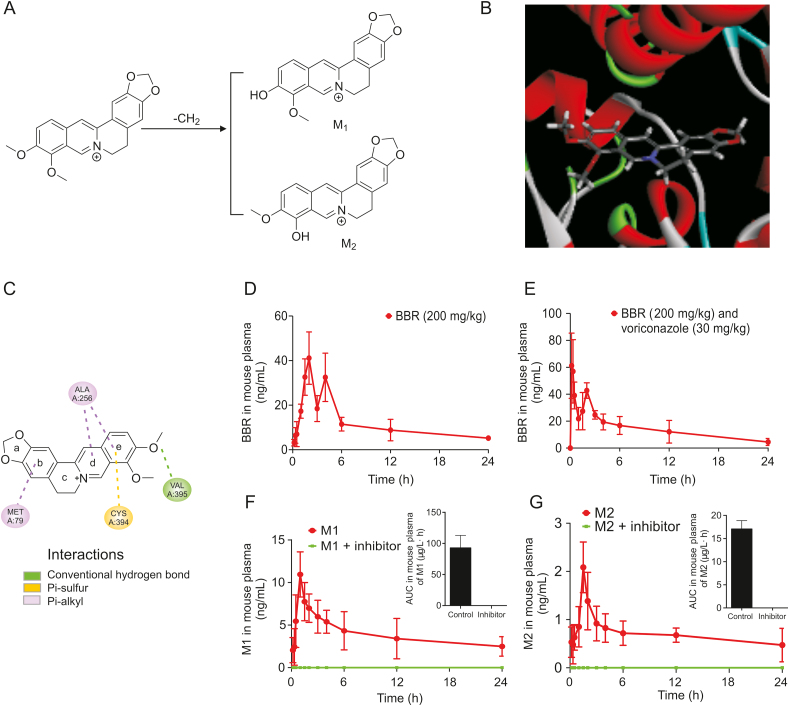
Berberine (BBR), a quaternary ammonium alkaloid isolated from the traditional Chinese medicine Coptis chinensis, is present in six plant families (Oleaceae, Papaveraceae, Ranunculaceae, Rutaceae, Menispermaceae, and Rhamnaceae) that were originally used as antipyretic, antidote and antibacterial drugs in clinical practice [21]. In recent years, studies have shown BBR to be a new lipid-lowering drug, acting by lowering cholesterol, triglyceride and low-density lipoprotein levels, which is different from the mechanism of action of statins [22]. Studies have demonstrated that rats exhibit 16 metabolites in their bile, urine and feces after oral administration of BBR [23] (Fig. 1A), most of which are related to the demethylation process.
Fig. 1.
Molecular docking between berberine (BBR) and CYP51 and pharmacokinetics of BBR in vivo. (A) The major products thalifendine (M1) and berberrubine (M2) demethylated by BBR. (B) Three-dimensional structure showing the molecular docking between BBR and CYP51. (C) The stable binding between BBR and CYP51 showed the ability to promote demethylation by CYP51. (D) Concentration-time curve of BBR in plasma after oral administration in ICR mice (200 mg/kg BBR). (E) Concentration-time curve of BBR in plasma after oral administration in ICR mice (200 mg/kg BBR + 30 mg/kg voriconazole). (F) Concentration-time curve of M1 in plasma after oral administration in ICR mice (200 mg/kg BBR and 200 mg/kg BBR + 30 mg/kg voriconazole). (G) Concentration-time curve of M2 in plasma after oral administration in ICR mice (200 mg/kg BBR and 200 mg/kg BBR + 30 mg/kg voriconazole).
Therefore, the main metabolic process of BBR in vivo called the demethylation reaction was studied. This study would demonstrate that CYP51 promotes the demethylation of BBR and provides a new method for studying the metabolic transformation mechanism of isoquinoline alkaloids in vivo.
2. Experimental
2.1. Materials and reagents
BBR was obtained from J&K Scientific Ltd. (Beijing, China). Rotundine (internal standard-1, IS-1) was purchased from the Institute for Food and Drug Control (Beijing, China). Benzylamine (IS-2) was purchased from J&K Scientific, Ltd. (Beijing, China). Thalifendine (M1) was purchased from Shanghai Hekang Biotechnology Co., Ltd. (Shanghai, China). Berberrubine (M2) was purchased from Chengdu Herbipurify Co., Ltd. (Chengdu, China). Voriconazole was purchased from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). Lanosterol was purchased from Chengdu Pufei De Biotech Co., Ltd. (Chengdu, China). Enterobacter cloacae, Enterococcus faecium, Staphylococcus epidermidis, Enterococcus faecalis, Pseudomonas aeruginosa, Acinetobacter baumannii, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Bifidobacterium longum, Bifidobacterium breve, Lactobacillus acidophilus, and Lactobacillus casei were purchased from Nanjing Lezhen Biotechnology Co., Ltd. (Nanjing, China). The sterol 14α-demethylase ELISA Kit was purchased from Nanjing Jin Yibai Biological Technology Co., Ltd. (Nanjing, China).
2.2. Animals
ICR mice (male, 19–21 g) were obtained from SiPeiFu Biotechnology Co., Ltd. (Beijing, China) with the license No. SCXK (Beijing) 2016-0002, and the experimental conditions were as follows: 12 h light/dark cycle, 8 a.m. to 8 p.m.; ambient temperature 20–25 °C; and relative humidity 40%–70%. All animals were fasted for 12 h before starting the experiment. Animal experiments were approved by the Animal Medicine and Experimental Committee of the Chinese Academy of Medical Sciences and Peking Union Medical College, China in accordance with institutional guidelines and ethical guidelines.
2.3. Instruments
The following instruments were used: LC-MS/MS-8050 (Shimadzu Corporation, Kyoto, Japan), LC-MS/MS-8060 (Shimadzu Corporation, Kyoto, Japan) and LC-MS solution workstation for on-line analysis of metabolites (Shimadzu Corporation, Kyoto, Japan); FSH-2 adjustable high-speed homogenizer (Jitan Shenglan Instrument Manufacturing Co., Ltd., Jitan, China); Heraeus Pico21 microcentrifuge (Thermo Fisher Scientific, Dreieich, Germany); MD200-2 nitrogen-blowing instrument (Hangzhou Diansheng Instrument Co., Ltd., Hangzhou, China); ThermoMixer (Eppendorf, Hamburg, Germany); Spectra Max Model 190 microplate reader (Molecular Devices, Silicon Valley, CA, USA).
BBR and its metabolites from each sample in vitro/in vivo and enzymatic reaction studies were analyzed by using a Shimpack XR0ODS II column (75 mm × 3 mm, 2.3 μm, Shimadzu Corporation, Kyoto, Japan) at a column temperature of 40 °C. The mobile phase consisted of water-formic acid (0.5%, V/V) and acetonitrile with a gradient elution (0 min, 90:10; 3.5 min, 75:25; 5.0 min, 70:30; 5.01 min, 80:20; 6.0 min, 90:10); the flow rate was 0.4 mL/min.
Lanosterol and 14-demethyl-14-dehydrolanosterol (FF-MAS) from each sample in the enzymatic reaction study were analyzed by using Alltima TM C18 column (4.6 mm × 150 mm, 5 μm) at a column temperature of 40 °C. The mobile phase consisted of water-ammonia (0.5%, V/V) and acetonitrile, with a gradient elution (0.01 min, 70:30; 2 min, 95:5; 7.0 min, 95:5; 7.01 min, 70:30; 10.0 min, stop); the flow rate was 0.4 mL/min. The ESI+ mode analysis was used, and the specific parameters were as follows: nebulizer gas flow rate, 3.0 L/min; dry gas flow rate, 10.0 L/mL; interface voltage, −4.5 kV; collision-induced dissociation pressure, 230 kPa; desolvation line temperature, 250 °C; and heating block temperature, 400 °C. Multiple reaction monitoring modes were used to quantify the sample. The m/z transitions were m/z 335.70 → 320.10 for BBR, m/z 355.70 → 191.90 for IS-1, m/z 321.65 → 307.15 for M1, m/z 321.65 → 307.15 for M2, m/z 409.15 → 353.30 for lanosterol, m/z 393.20 → 151.10 for FF-MAS, and m/z 108.20 → 91.10 for IS-2.
2.4. Method of molecular docking
The interaction between BBR and CYP51 was simulated using Discovery Studio software (v16.1.0.15350). The crystal structure of CYP51 was obtained from the Protein Database (PDB) with PDB ID 2W09.
The BBR structure was constructed using ChemDraw15.0, and the structure of the target compound was subjected to energy minimization by the minimization module in Discovery Studio Client software (v16.1.0.15350) to obtain an optimized structure. The CDOCKER method was selected to simulate the interaction between the target compound and the protein after optimization. The pose cluster radius parameter was set to 0.5, and the rest of the parameters were set to default values.
2.5. Analytical methods of BBR pharmacokinetics in vivo
Animals were divided into BBR group and voriconazole group, and the ICR mice were given 200 mg/kg BBR and 200 mg/kg BBR +30 mg/kg voriconazole in single oral administrations. After administration for 0.17, 0.33, 0.5, 1, 1.5, 2, 3, 4, 6, 12, and 24 h, blood was taken from the retrobulbar venous plexus and transferred to a sample tube to which heparin had been added. Samples were stored at −20 °C prior to analysis. After 3 h of administration, the mice were free to eat. All animal programs were approved by the Animal Care Welfare Committee of the Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). In addition, all animal experiments were conducted in strict accordance with the Laboratory Animal Care and Use Guidelines issued by the Animal Protection and Welfare Institute.
The plasma (100 μL) was added to 300 μL of methanol (containing 50 ng/mL IS) to precipitate the protein. After vortexing, the tubes were centrifuged at 10,800 rpm for 10 min, the supernatant was extracted, and 2 μL was injected. The concentrations used for the standard curve were as follows: 1, 2, 10, 100, and 1000 ng/mL.
The two-compartment model was used to simulate pharmacokinetic parameters, and the data were analyzed by using DAS 3.0.
2.6. Preparation of intestinal bacterial incubation in vitro
An anaerobic medium for intestinal bacteria was prepared according to literature reports [11]. The obtained medium was autoclaved at 0.1 MPa and 121 °C for 20 min and used after being cooled.
Intestinal bacterial cultures were prepared according to literature reports [24]. After being anesthetized, the mice were sacrificed by cervical dislocation. The abdomen was incised, the colon was removed, and the colon contents were homogenized in an anaerobic chamber. After mixing 1 g of the contents in 20 mL of anaerobic medium, the cultures were incubated at 37 °C for 60 min under anaerobic conditions (N2 environment) and set aside.
The samples were divided into five groups: negative control group; the 10, 50, and 100 mg/mL BBR groups; and the 50 mg/mL BBR + 3.5 μg/mL voriconazole group. BBR and voriconazole were dissolved in methanol; BBR was formulated at concentrations of 10, 50, and 100 mg/mL; and voriconazole was formulated at a concentration of 3.5 μg/mL. Then, 10 μL of the sample solution was added to each sample tube, 1 mL of the intestinal culture medium was added in an anaerobic chamber, and the sample tubes were sealed with sealing films and incubated at 37 °C for 12, 24, 36, 48, 60, and 72 h while using methanol as a negative control. When the incubation was completed, the samples were removed, added to 1 mL of methanol, vortexed for 30 s and centrifuged at 14,800 rpm for 15 min. The supernatant was diluted in groups as follows: 10 times for the 10 mg/mL BBR group; 50 times for the 50 mg/mL BBR group; 100 times for the 100 mg/mL BBR group, and 50 times for the 50 mg/mL BBR + 3.5 μg/mL voriconazole group. After dilution, 100 μL of each sample was added to 300 μL of methanol (containing 50 ng/mL IS), vortexed and centrifuged at 14,800 rpm for 10 min. The supernatant was extracted, and 2 μL was injected. The concentrations used for the standard curve were as follows: 1, 2, 10, 100, and 1000 ng/mL.
2.7. Preparation of liver and kidney homogenates for BBR metabolism in vitro
The mice were sacrificed by cervical dislocation, and the abdomen was incised to remove the liver and kidneys. With a ratio of 1 g of organ to 5 mL of physiological saline, the samples were homogenized for use.
Liver and kidney homogenates were divided into two groups: BBR group (50 mg/mL) and voriconazole (3.5 μg/mL) group. BBR was formulated to a concentration of 50 mg/mL, and voriconazole was formulated to a concentration of 3.5 μg/mL. Then, 10 μL of the sample solution was added to each sample tube, and 1 mL of the organ homogenate was added and incubated at 37 °C for 0, 15, 30, 60, 90, and 120 min. When the incubation was completed, the samples were removed, treated with 1 mL of methanol, vortexed for 30 s and centrifuged at 14,800 rpm for 15 min. Then, 100 μL of the supernatant of each sample was added to 300 μL of methanol (containing 50 ng/mL IS). Subsequently, the samples were vortexed and centrifuged at 14,800 rpm for 10 min. The supernatant was extracted, and 2 μL was injected. The concentrations used for the standard curve were as follows: 1, 2, 10, 100, and 1000 ng/mL.
2.8. Preparation of liver and kidney microsomal for BBR metabolism in vitro
The mice were sacrificed by cervical dislocation, and the liver and kidneys were removed into beakers containing physiological saline at 4 °C. Then, the organs were washed until no blood remained; filter paper was used to blot the remaining liquid; the excess tissues were removed and weighed; Tris-KCl was added at 2 mL/g according to the weight of the organ so that the mixture was homogenized. Then, the homogenate was centrifuged at 10,000 g and 4 °C for 25 min to extract the supernatant. After the addition of Tris solution, sample centrifugation was continued at 105,000 g and 4 °C for 1 h. Then, the supernatant was discarded, and the precipitate was resuspended with an appropriate amount of Tris-KCl and centrifuged at 105,000 g and 4 °C for 1 h. Finally, the supernatant was discarded again, 8 mL of Tris-KCl was added and the sample was suspended to obtain the prepared liver and kidney microsomes.
Liver and kidney microsomes were divided into two groups: BBR group (50 mg/mL) and voriconazole group (50 mg/mL BBR + 3.5 μg/mL voriconazole). BBR was formulated to a concentration of 50 mg/mL, voriconazole was formulated to a concentration of 3.5 μg/mL and the NADPH generation system was formulated as follows: 1.3 mmol/L NADP, 3.3 mmol/L G-6-P, 0.4 U/mL G-6-PD, and 3.3 mmol/L MgCl2. The prepared mouse liver and kidney microsomes were diluted with 4 °C Tris buffer (pH 7.4) until the protein concentration was 0.5 mg/mL. After incubation for 3 min at 37 °C in advance, 10 μL of sample solution and NADPH solution were added and incubated at 37 °C for 0, 15, 30, 60, 90, and 120 min. Then, 100 μL of the supernatant of each sample was added to 300 μL of methanol (containing 50 ng/mL IS). Subsequently, the samples were vortexed and centrifuged at 14,800 rpm for 10 min. The supernatant was extracted, and 2 μL was injected. The concentration of the standard curve was as follows: 1, 2, 10, 100, and 1000 ng/mL.
2.9. Preparation of standard bacterial strains for BBR metabolism in vitro
The number of colonies of 14 standard bacterial strains on culture medium was counted by the plate method after resuscitating the cells. According to the results after counting, the culture media for the standard bacterial strains were diluted until the bacterial concentrations were the same.
Then, 0.9 mL of sterilized anaerobic medium and 100 μL of the diluted 14 standard bacterial cultures were added to each sample tube in an anaerobic chamber. Then, each tube was blown with nitrogen and sealed. After incubation at 37 °C for 1 h in advance, 10 μL of BBR (50 mg/mL) was added to each sample, which was then incubated at 37 °C for 24 h. Then, 1 mL of methanol (containing 50 ng/mL IS) was added to each sample, which was then vortexed and centrifuged at 14,800 rpm for 10 min. The supernatant was extracted, and 2 μL was injected. The concentrations used for the curve were as follows: 1, 2, 10, 100, 500, and 1000 ng/mL.
Samples of each standard bacterial strain were divided into three groups after four single strains were screened: negative control group (methanol), BBR group (50 mg/mL BBR), and voriconazole group (50 mg/mL BBR + 3.5 μg/mL voriconazole). The four standard bacterial strains, namely, E. faecalis, S. epidermidis, E. cloacae, and E. faecium, were mixed with 50 mg/mL BBR and incubated to determine the concentration of BBR and the activity of CYP51 at 48 and 72 h. Then, the percentage of BBR metabolism was calculated.
The 0.9 mL of sterilized anaerobic culture medium and 100 μL of the diluted single-culture medium were added to each sample in an anaerobic chamber. Then, each tube was blown with nitrogen, sealed, incubated at 37 °C for 24 h and ultrasonically disrupted. Then, the activity of CYP51 was determined by the CYP51 ELISA Kit.
2.10. 16S rRNA analysis of ICR mouse feces
By using 16S V3-V4:340–805R specific primers, the V3-V4 region of 16S rRNA was targeted, and the 16S rRNA gene was amplified. The PCR products were mixed in equal proportions. Then, the PCR products were purified by the OMEGA Gel Extraction Kit. The sequencing library was constructed using the NEXTflex Rapid Illumina DNA-seq Kit, while the quality of the library was tested by using a Qube 2 fluorometer and an Agilent Bioanalyzer 2100. Finally, the library was sequenced on the HISEQ 2500 platform, and 250-bp paired-end reads were generated.
Sequences were analyzed using Quantitative Insights Into Microbial Ecology (QIIME) software. First, the reads were classified by using the quality filters module of QIIME. Then, each operational taxonomic unit (OTU) was selected for a representative sequence, and the classification information for each representative sequence was annotated by using the Ribosomal Database Project classifier. Sequences with similarity >97% were assigned to the same OTU.
2.11. Expression and functional verification of CYP51
The cyp51 gene of Saccharomyces cerevisiae S288C (NC_022591.1) was synthesized by Sangon Biotech (Shanghai, China). The cyp51 gene was amplified using primer-F/primer-R. The primers for the cyp51 gene were designed by Primer 5.0 as follows: cyp51-F: AGCAAATGGGTCGCGGATCCAGCGCGACCAAAAGCATCG; cyp51-R: TCGAGTGCGGCCGCAAGCTTGATTTTCTGTTCCGGGTTACGT. Thirty-five cycles of PCR were performed with Phanta polymerase (Vazyme Biotech Co., Ltd., Nanjing, China) in a 50-μL system. Then, 1% agarose gel electrophoresis and gene sequencing were performed to verify the gene amplification. The cyp51 fragment was cloned between the HindIII/BamHI sites of the expression vector pET28a (New England Biolabs, Ipswich, MA, USA) using Gibson Assembly Master Mix (New England Biolabs, Ipswich, MA, USA) to obtain pET28a-cyp51.
The plasmid pET28a-cyp51 was introduced into E. coli BL21 cells (Vazyme Biotech Co., Ltd., Nanjing, China). After verifying the sequence, clones were used to express the cyp51 protein (ONH80457.1). E. coli BL21 colonies with pET28a-cyp51 were grown in 100 mL of Luria-Bertani broth containing 15 μg/mL kanamycin at 37 °C. When the amplified bacterial solution reached an OD600 of 0.6–0.8, isopropyl β-d-1-thiogalactopyranoside was added at a final concentration of 0.1 mM to induce expression, and the cells were grown at 16 °C for 20 h. After centrifugation, the bacterial cells were suspended in 10 mL of phosphate buffered saline (PBS) solution, and E. coli BL21 with a pet28a plasmid lacking the CYP51 gene was used as a control. Supernatants and pellets were prepared for SDS gel electrophoresis. After concentrating to 1 mL through a microporous membrane (MWCO: 30,000), the concentrated solution was used for functional verification.
The enzymatic reaction system was composed of PBS solution (1×) containing NADPH (0.5 mM), enzyme lysate (10 μL), and lanosterol (10 μg/mL), and incubated for 0, 1, 2, and 4 h. The products obtained after incubation were used to analyze lanosterol and FF-MAS by LC-MS/MS. The enzymatic reaction system was composed of 1×PBS solution containing NADPH (0.5 mM), enzyme lysate (10 μL), and substrate BBR (10 μg/mL), and incubated for 0, 1, 2, and 4 h. The products obtained after incubation were used to analyze M1 and M2 by LC-MS/MS.
2.12. Statistical analysis
Statistical analysis was performed using ANOVA and Student's t-test in GraphPad Prism version 5 (GraphPad, San Diego, CA, USA). The data are expressed as mean ± standard deviation (SD), and P values less than 0.05 are considered statistically significant.
3. Results and discussion
3.1. Molecular docking between BBR and CYP51
The putative chemical mechanism for the docking of BBR alkaloids by CYP51 (PDB ID: 2W09) is shown in Fig. 1B. Based on the results of molecular docking, BBR can stably bind to CYP51 with a binding energy of −23.1 kcal/mol (Fig. 1C). In addition, the form of action demonstrated that the BBR structure can form various bonds with CYP51 for tight binding to promote the demethylation effect of CYP51 (ring b of BBR formed π bonds with the alkyl group of Met79; ring d of BBR formed π bonds with the alkyl group of Ala256; ring e of BBR simultaneously formed π bonds with the alkyl group of Ala256 and the sulfhydryl group of Cys394; the oxygen in the 10th methoxy group of BBR formed hydrogen bonds with Val395).
3.2. Pharmacokinetics of BBR in vivo
According to the above results, it could be inferred that BBR was likely to produce demethylated metabolites by CYP51-mediated metabolism. Therefore, a BBR metabolism experiment was conducted in ICR mice, and the BBR metabolic mechanism in vivo was explored. As shown in Table 1, the AUC(0–24 h) increased by 9.8%, t1/2 increased by 17.4%, and Cmax increased by 45.7% after addition of voriconazole (Fig. 1E) compared with the values after oral administration of 200 mg/kg BRR alone (Fig. 1D). Moreover, there were double peaks in the drug-time curve. It was speculated that this might be attributed to enterohepatic circulation after oral administration.
Table 1.
The pharmacokinetic parameters of ICR mice after oral administration of berberine (BBR) simulated by a two-compartment model.
| Parameters | BBR (200 mg/kg) | BBR (200 mg/kg) + voriconazole (30 mg/kg) |
|---|---|---|
| AUC(0–24 h) (ng/mL·h) | 272.21 ± 68.42 | 298.93 ± 56.35 |
| AUC(0−∞) (ng/mL·h) | 333.61 ± 79.26 | 388.00 ± 103.32 |
| MRT(0–24 h) (h) | 7.39 ± 1.71 | 6.50 ± 1.40 |
| MRT(0−∞) (h) | 12.73 ± 5.85 | 10.43 ± 2.32 |
| t1/2 (h) | 8.44 ± 5.55 | 9.91 ± 5.26 |
| Cmax (ng/mL) | 44.26 ± 10.44 | 64.50 ± 20.12 |
Data are presented as mean ± SD. MRT: mean residence time.
Thus, it could be inferred that voriconazole reduced the metabolic capacity of BBR by inhibiting the enzyme activity and slowing the BBR metabolic process, which finally increased the t1/2, AUC and Cmax of BBR in mouse plasma.
3.3. Pharmacokinetics of M1 and M2 in vivo
The production of M1 was more than that of M2 in mice after oral administration of BBR. In addition, the M1 and M2 levels were all below the lower limit of quantitation (1 ng/mL) and could not be detected at any of the time points under the influence of voriconazole (Figs. 1F and G), which indirectly proved that voriconazole had entered the gut microbiota. It indicated that after oral administration of voriconazole, BBR could not be converted to M1 or M2 by the demethylation pathway in vivo. Therefore, combined with the results of pharmacokinetic studies, this result indicated that the demethylation process of BBR in vivo was mediated by CYP51.
3.4. Metabolism of BBR by the mice gut microbiota in vitro
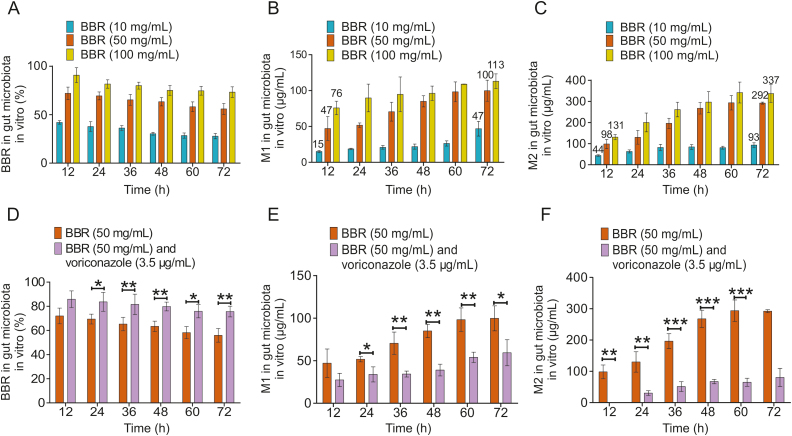
Different concentrations of BBR (10, 50, and 100 mg/mL) and mouse colon contents were mixed and incubated for 72 h, and the contents of BBR, M1 and M2 were determined. After 12 h of incubation, the production of M1 was 15, 47, and 76 μg/mL while the production of M2 was 44, 98, and 131 μg/mL, respectively. After 72 h of incubation, the production of M1 was 47, 100, and 113 μg/mL while the production of M2 was 93, 292, and 337 μg/mL. From the above results, it could be known that the production of M2 (Fig. 2C) was nearly 3 times (the range was 1.73–2.98) higher that of M1 (Fig. 2B) when BBR was metabolized by gut microbiota in vitro. After 72 h of incubation, BBR could not be completely metabolized (Fig. 2A). At the same time, the percentage of BBR metabolized at each concentration was different, and the percentage of BBR metabolized at 10, 50, and 100 mg/mL concentrations decreased sequentially. It was speculated that the concentration of intestinal bacteria and the activity of CYP51 in each system were constant, so the metabolic capacity of BBR was certain, which further resulted in the difference in the percentage of BBR metabolism at each concentration. In addition, BBR could be metabolized in the mouse intestinal microsomes and M1 and M2 were produced. However, the production of M1 and M2 was very low (Figs. S1A and B). The production of M1 was decreased by 21%, 25%, and 20% at 30, 60, and 120 min after adding voriconazole (Fig. S1A) and the production of M2 was decreased by 28%, 26%, and 30% at 30, 60, and 120 min, respectively (Fig. S1B). This showed that the demethylation of BBR in the intestine was mainly mediated by gut microbiota and CYP51 played a key role in the metabolism of BBR.
Fig. 2.
Metabolism of BBR by the mice gut microbiota in vitro. (A) Percentage of residual BBR (10, 50, and 100 mg/mL) after intestinal metabolism in mice in vitro. (B) Level of M1 produced from BBR (10, 50, and 100 mg/mL) after intestinal metabolism in mice in vitro. (C) Level of M2 produced from BBR (10, 50, and 100 mg/mL) after intestinal metabolism in mice in vitro. (D) Comparison of the percentage of residual BBR (50 mg/mL) metabolized in the gut microbiota under the influence of an inhibitor (voriconazole, 3.5 μg/mL). ∗ P < 0.05; ∗∗ P < 0.01. (E) Comparison of the M1 content produced by BBR (50 mg/mL) metabolism in the gut microbiota under the influence of an inhibitor (voriconazole, 3.5 μg/mL). ∗∗ P < 0.01; ∗∗∗ P < 0.001. (F) Comparison of the M2 content produced by BBR (50 mg/mL) metabolism in the gut microbiota under the influence of an inhibitor (voriconazole, 3.5 μg/mL). ∗ P < 0.05; ∗∗ P < 0.01 .
However, the percentage of residual BBR was significantly increased (Fig. 2D) after the addition of voriconazole (3.5 μg/mL). Furthermore, the production of M1 (Fig. 2E) and M2 (Fig. 2F) was significantly decreased sequentially compared with that in the BBR group. This further demonstrated that voriconazole could specifically inhibit the activity of CYP51, thereby reducing the ability of intestinal bacteria to metabolize BBR and produce M1 and M2.
3.5. Metabolism of BBR in the liver and kidney system in vitro
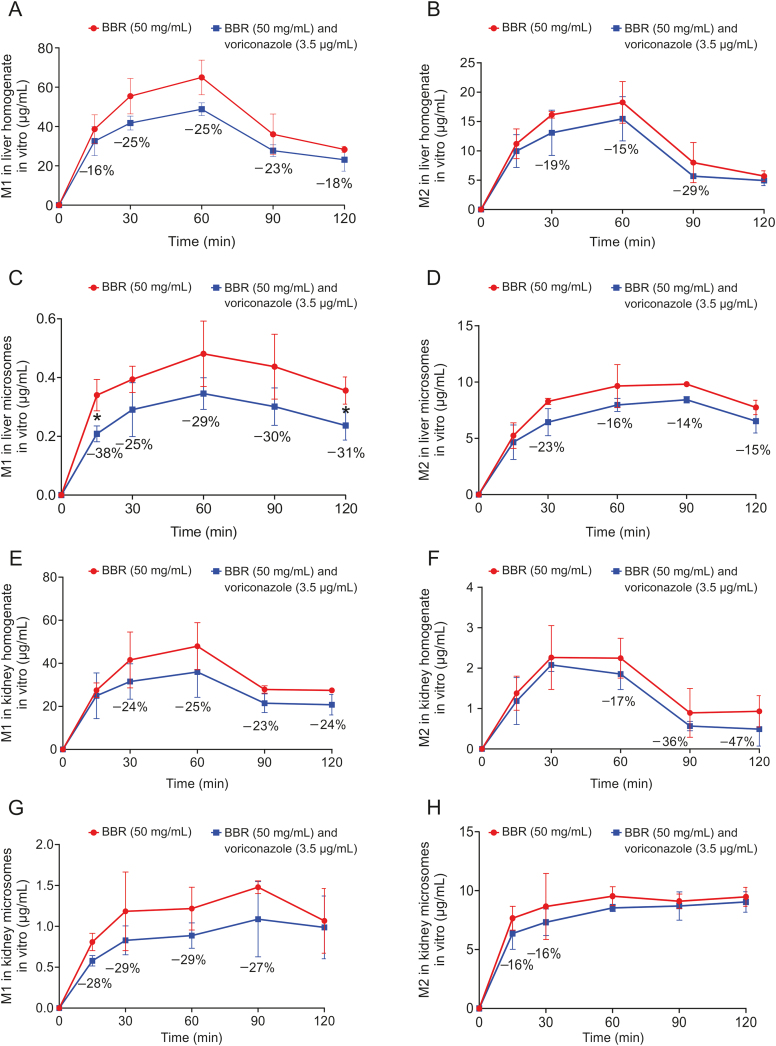
The mouse liver homogenates and liver microsomes were mixed and incubated with BBR (50 mg/mL) and voriconazole (3.5 μg/mL), respectively. Then, the levels of M1 and M2 in the mixture were determined at different time points. After the addition of voriconazole (3.5 μg/mL), BBR could still be metabolized in the liver homogenate, and M1 and M2 were produced, but the production was partially reduced (Figs. 3A and B). Similar results were also observed in the liver microsomes (Figs. 3C and D).
Fig. 3.
Metabolism of BBR in the liver and kidney system in vitro. (A and B) Comparison of M1 and M2 levels produced by BBR (50 mg/mL) metabolism in liver homogenate under the influence of an inhibitor (voriconazole, 3.5 μg/mL). (C and D) Comparison of M1 and M2 levels produced by BBR (50 mg/mL) metabolism in kidney microsomes under the influence of an inhibitor (voriconazole, 3.5 μg/mL). (E and F) Comparison of M1 and M2 levels produced by BBR (50 mg/mL) metabolism in kidney homogenate under the influence of an inhibitor (voriconazole, 3.5 μg/mL). (G and H) Comparison of M1 and M2 levels produced by BBR (50 mg/mL) metabolism in kidney microsomes under the influence of an inhibitor (voriconazole, 3.5 μg/mL).
We also performed a comparison with the metabolic process of BBR in the renal system in vitro. The mouse kidney homogenates and kidney microsomes were treated as described above. After the addition of voriconazole (3.5 μg/mL), BBR could still be metabolized in the kidney homogenate, and M1 and M2 were produced, although also at reduced levels (Figs. 3E and F). In addition, similar results were observed in kidney microsomes (Figs. 3G and H).
This indicated that the addition of voriconazole to the liver and kidney homogenates could inhibit the activity of CYP51, thereby reducing the amount of M1 and M2 produced by BBR metabolism, but the reduction ratio was much lower than that in the gut microbiota. Therefore, it was hypothesized that the key enzyme in the demethylation pathway in the gut was CYP51. In addition, the production of M1 and M2 in kidney homogenate was the lowest (Figs. S1C and D). Compared with the group of kidney homogenate, the production of M1 in liver homogenate was increased by 27%, 38%, and 4% at 30, 60, and 120 min (Fig. S1C); the production of M2 in liver homogenate increased by 657%, 724%, and 315% at 30, 60, and 120 min, respectively (Fig. S1D). Nevertheless, it could still be seen that the production of M1 and M2 in liver homogenate was much lower than that in gut microbiota (Figs. S1E and F). The production of M1 in gut microbiota was 52.59, 69.31, and 84.16 μg/mL at 30, 60, and 120 min (Fig. S1E); the production of M2 in gut microbiota was 132.34, 212.45, and 267.40 μg/mL at 30, 60, and 120 min, respectively (Fig. S1F). Therefore, it was hypothesized that the gut microbiota was more important to the demethylation of BBR and the key enzyme in the demethylation pathway in the gut was CYP51. In view of the fact that enzymes in the liver and kidneys were likely to participate in the metabolism of BBR in the body, and under the action of voriconazole, BBR still had metabolic processes in the liver and kidneys. Therefore, other metabolic enzymes in the liver and kidneys including drug metabolism P450 enzymes might be involved in the metabolism of BBR. Due to the low absorption rate of BBR, less than 10% of BBR could enter the blood [25]. Therefore, although the liver and kidneys had a certain influence on the metabolism of BBR, the demethylation metabolism of BBR in the intestine was more important.
3.6. Metabolism of BBR in standard bacterial strains in vitro
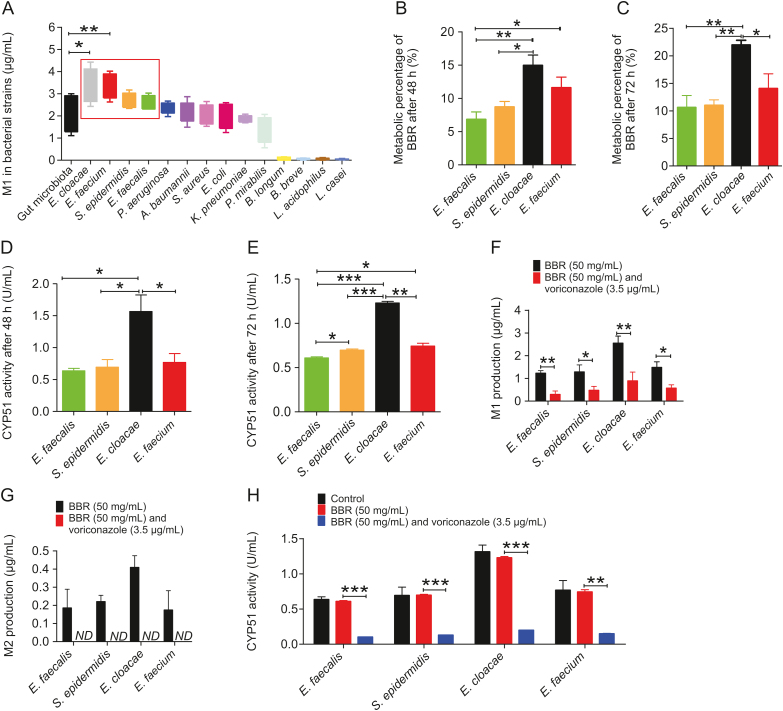
Fourteen standard bacterial strains present in the gut microbiota were individually used to investigate the ability of BBR metabolism in vitro, and four standard bacterial strains were selected due to their high production of M1 (Fig. 4A), namely, E. faecalis, S. epidermidis, E. cloacae, and E. faecium.
Fig. 4.
Metabolism of BBR in standard bacterial strains in vitro. (A) Comparison of M1 content produced by BBR metabolism in 14 standard bacterial strains. ∗ P < 0.05; ∗∗ P < 0.01. (B and C) Percentage of BBR metabolized by 4 standard bacterial strains after 48 h and 72 h. ∗ P < 0.05; ∗∗ P < 0.01. (D and E) Activity of CYP51 in 4 standard bacterial strains after metabolism of BBR for 48 h and 72 h. ∗ P < 0.05; ∗∗ P < 0.01; ∗∗∗ P < 0.001. (F and G) Comparison of M1 and M2 levels produced by BBR (50 mg/mL) metabolism in 4 standard bacterial strains for 72 h under the influence of an inhibitor (voriconazole, 3.5 μg/mL). ∗ P < 0.05; ∗∗ P < 0.01; ND: not detected. (H) Comparison of CYP51 activity in 4 standard bacterial strains after metabolism of BBR (50 mg/mL) for 72 h under the influence of an inhibitor (voriconazole, 3.5 μg/mL) ∗∗ P < 0.01; ∗∗∗ P < 0.001.
These results all directly confirmed the presence of CYP51 in the gut microbiota after each strain was incubated with BBR for 48 h and 72 h. The metabolism of BBR differed among the four strains (Figs. 4B and C). Moreover, the activity of CYP51 differed among different bacterial strains, and the ability of the strains to demethylate BBR was directly proportional to the activity of CYP51 (Figs. 4D and E). It was verified that CYP51 was indeed present in the gut microbiota and participated in demethylation-related metabolism.
However, the level of M1 produced by each strain was significantly reduced (Fig. 4F), while the level of M2 produced by each strain was below the lower limit of quantitation and could not be detected (Fig. 4G) after the addition of voriconazole (3.5 μg/mL) for 72 h. In addition, the activity of CYP51 in the four standard bacterial strains incubated with 50 mg/mL BBR for 72 h was not significantly different from that in the strains alone. After the addition of voriconazole (3.5 μg/mL), CYP51 activity was reduced (Fig. 4H).
This result indicated that voriconazole, a specific inhibitor of CYP51, inhibited most of the activity of CYP51 in bacterial strains, significantly reduced the ability of CYP51 to demethylate BBR for metabolism, and prevented the production of M1 and M2. It was further verified that CYP51 was present in the gut microbiota and is the key enzyme used by the gut microbiota to metabolize BBR to produce a demethylated metabolite.
3.7. 16S rRNA analysis of the feces of ICR mice after oral administration of BBR
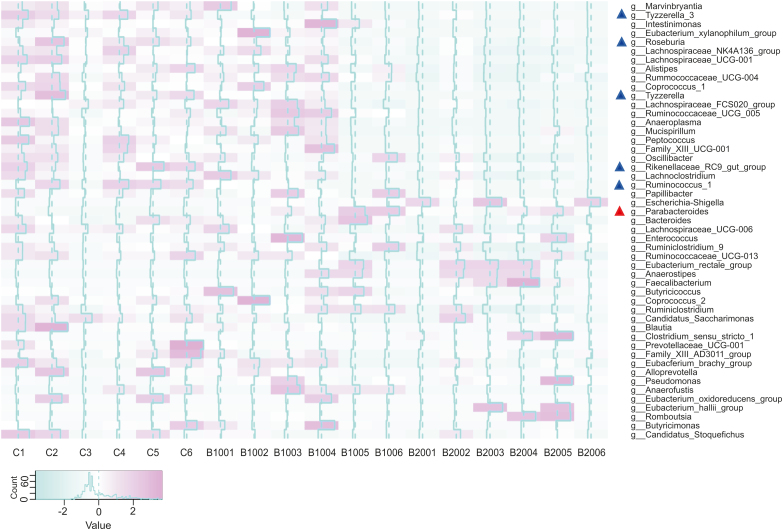
To explore how BBR affects the gut microbiota, ICR mice were orally administered BBR. Then, the feces of the ICR mice were collected 24 h later and subjected to 16S rRNA sequencing and data analysis, as shown in Fig. 5. Compared with the control group, the BBR-treated group exhibited a significant increase in the abundance of one genus (Parabacteroides) (P < 0.05, marked in a red “Δ”) and a significant decrease in the abundances of five genera (Tyzzerella 3, Rikenellaceae RC9 gut group, Roseburia, Tyzzerella, Ruminococcus 1) (P < 0.05, marked in a blue “Δ”). Parabacteroides is a potential probiotic genus. Recent studies have found that Parabacteroides species could produce succinate and secondary bile acids to resist body obesity and metabolic dysfunction [26,27]; the reduced abundance of Ruminococcus 1 validated the role of BBR. Zhang et al. [28] found that the abundance of Ruminococcus 1 was significantly increased in rats with high-fat-induced obesity but significantly decreased in rats after BBR administration; Rikenellaceae RC9 gut group has also been shown to have important effects on carbohydrates and carbohydrate and lipid metabolism [29]. Tyzzerella has been reported to be greatly enriched in individuals with a high risk of cardiovascular disease [30]. In addition, studies have reported a positive correlation between Tyzzerella 3 and inflammation and hypertension [31,32]. Furthermore, the abundance of one taxon (Porphyromonadaceae) exhibited a significant increase (Fig. S2) at the family level (P < 0.05), which also confirmed the regulatory effect of BBR on the gut microbiota [33]. The above results proved that BBR changed the gut microbiota after oral administration and altered its own metabolism by the gut microbiota. To investigate whether voriconazole affected the gut microbiota, ICR mice were orally administered BBR and voriconazole. The abundances of Parabacteroides, Tyzzerella 3, Rikenellaceae RC9 gut group, Roseburia, Tyzzerella, Ruminococcus 1 and Porphyromonadaceae, which were originally changed, did not change significantly after the addition of voriconazole (Fig. S3).
Fig. 5.
16S rRNA analysis of the feces of ICR mice after oral administration of BBR. (△Bacteria with abundance increased (red) and decreased (blue) after oral administration of BBR).
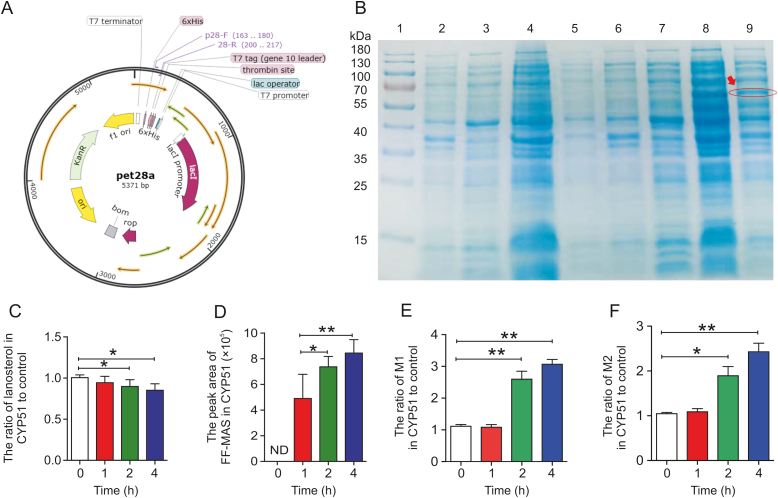
3.8. Metabolism of BBR in CYP51 enzyme lysate
Recombinant plasmid containing the CYP51 gene was constructed (Figs. 6A and S4A), and transformed into E. coli BL21(DE3) cells. The CYP51 protein was expressed in E. coli as accessed by SDS-PAGE analysis (Fig. 6B). Lanosterol, the classic substrate of CYP51, was selected to verify the function of the CYP51 enzyme lysate. As shown in Fig. 6C, the lanosterol level decreased significantly under the action of CYP51 enzyme lysate compared with the control. The production ratio of its demethylated metabolite, FF-MAS, increased significantly (Fig. 6D), while the FF-MAS level was below the lower limit of quantitation after the reaction of lanosterol with the control enzyme lysate (Fig. S4B). BBR was added to the CYP51 enzyme lysate system, and the production ratios of M1 and M2 were significantly increased compared with that of the control (Figs. 6E and F), which verified that CYP51 was the key enzyme of the gut microbiota that promoted the metabolism of BBR to produce demethylated metabolites.
Fig. 6.
Effect of the CYP51 enzyme lysate on demethylation of BBR. (A) CYP51 plasmid sequence. (B) CYP51 was present in the enzyme lysate (1: marker; 2: the protein contained in the precipitate of E. coli BL21 without pet28a-cyp51 before induction; 3: the protein contained in the supernatant of E. coli BL21 without pet28a-cyp51 before induction; 4: the protein contained in the precipitate of E. coli BL21 without pet28a-cyp51 after induction; 5: the protein contained in the supernatant of E. coli BL21 without pet28a-cyp51 after induction; 6: the protein contained in the precipitate of E. coli BL21 with pet28a-cyp51 before induction; 7: the protein contained in the supernatant of E. coli BL21 with pet28a-cyp51 before induction; 8: the protein contained in the precipitate of E. coli BL21 with pet28a-cyp51 after induction; 9: the protein contained in the supernatant of E. coli BL21 with pet28a-cyp51 after induction). (C) Relative proportions of lanosterol after incubation for 0, 1, 2, and 4 h in the enzymatic reaction system of the CYP51 enzyme lysate compared with the control enzyme lysate. ∗ P < 0.05. (D) Relative peak area of 14-demethyl-14-dehydrolanosterol (FF-MAS) after 0, 1, 2, and 4 h of incubation in the enzymatic reaction system of the CYP51 enzyme lysate. ∗ P < 0.05; ∗∗ P < 0.01; ND: not detected. (E and F) Relative proportions of M1 and M2 after 0, 1, 2, and 4 h of incubation in the CYP51 enzyme lysate compared with the control enzyme lysate. ∗ P < 0.05; ∗∗ P < 0.01.
4. Conclusion
BBR is a safe drug with multiple therapeutic effects in the clinic. Due to the low absorption rate of BBR in the gut microbiota, the activity of its metabolites is particularly important. In this research, it was first suggested that CYP51 was present in the gut microbiota, and BBR was metabolized by CYP51 in the intestine to produce demethylated metabolites M1 and M2, which provided a basis for the study of metabolic conversion mechanisms of various isoquinoline alkaloids in vivo and expanded our understanding of the role of the gut microbiota in drug metabolism with demethylation reaction.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The project was supported by CAMS Innovation Fund for Medical Sciences (CIFMS, Grant No.: 2016-I2M-3-011, China), the National Natural Science Foundation of China (Grant Nos.: 81803613 and 81973290), Beijing Key Laboratory of Non-Clinical Drug Metabolism and PK/PD study (Grant No.: Z141102004414062, China), Beijing Natural Sciences Fund Key Projects (Grant No.: 7181007) and the National Megaproject for Innovative Drugs (Grant No.: 2018ZX09711001-002-002). We would like to thank Shimadzu (China) Co., Ltd. for technological support.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.10.001.
Contributor Information
Yan Wang, Email: wangyan@imm.ac.cn.
Jian-Dong Jiang, Email: jiang.jdong@163.com.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Gareau M.G., Sherman P.M., Walker W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010;7:503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu B., Wang X., Li L. Human gut microbiome: the second genome of human body. Protein Cell. 2010;1:718–725. doi: 10.1007/s13238-010-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley R.E., Bäckhed F., Turnbaugh P. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellarin M., Warren R.L., Freeman J.D. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostic A.D., Gevers D., Pedamallu C.S. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett W.S., Gallini C.A., Yatsunenko T. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tana C., Umesaki Y., Imaoka A. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol. Motil. 2010;22:512–519. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Klipfell E., Bennett B.J. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsythe P., Sudo N., Dinan T. Mood and gut feelings. Brain Behav. Immun. 2010;24:9–16. doi: 10.1016/j.bbi.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 10.Tao J.-H., Di L.-Q., Shan J.-J. Interaction of intestinal microecology and internal metabolism of effective ingredients from Chinese materiamedica. Chin. Tradit. Herb. Drugs. 2008;39:1902–1904. [Google Scholar]
- 11.Wang Y., Tong Q., Shou J.-W. Gut microbiota-mediated personalized treatment of hyperlipidemia using berberine. Theranostics. 2017;7:2443–2451. doi: 10.7150/thno.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson I.D., Nicholson J.K. The role of gut microbiota in drug response. Curr. Pharmaceut. Des. 2009;15:1519–1523. doi: 10.2174/138161209788168173. [DOI] [PubMed] [Google Scholar]
- 13.Cani P.D., Bibiloni R., Knauf C. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 14.Membrez M., Blancher F., Jaquet M. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. Faseb. J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 15.Caesar R., Reigstad C.S., Bäckhed H.K. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012;61:1701–1707. doi: 10.1136/gutjnl-2011-301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida Y., Aoyama Y., Noshiro M. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem. Biophys. Res. Commun. 2000;273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
- 17.Lepesheva G.I., Waterman M.R. Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta. 2007;1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterman M.R., Lepesheva G.I. Sterol 14α-demethylase, an abundant and essential mixed-function oxidase. Biochem. Biophys. Res. Commun. 2005;338:418–422. doi: 10.1016/j.bbrc.2005.08.118. [DOI] [PubMed] [Google Scholar]
- 19.Lepesheva G.I., Zaitseva N.G., Nes W.D. Cyp51 from trypanosoma cruzi: a phyla-specific residuein the B’ helix defines substrate preferences of sterol 14 alpha-demethylase. J. Biol. Chem. 2006;281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 20.He C.-Y., Fu J., Shou J.-W. In vitro study of the metabolic characteristics of eight isoquinoline alkaloids from natural plants in rat gut microbiota. Molecules. 2017;22:932. doi: 10.3390/molecules22060932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Čerňáková M., Košťálová D. Antimicrobial activity of berberine—a constituent ofMahoniaaquifolium. Folia Microbiol. 2002;47:375–378. doi: 10.1007/BF02818693. [DOI] [PubMed] [Google Scholar]
- 22.Kong W., Wei J., Abidi P. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat. Med. 2004;10:1344–1351. doi: 10.1038/nm1135. [DOI] [PubMed] [Google Scholar]
- 23.Ma J.-Y., Feng R., Tan X.-S. Excretion of berberine and its metabolites in oral administration in rats. J. Pharmacol. Sci. 2013;102:4181–4192. doi: 10.1002/jps.23718. [DOI] [PubMed] [Google Scholar]
- 24.Feng R., Shou J.-W., Zhao Z.-X. Transforming berberine into its intestine absorbable form by gut microbiota. Sci. Rep. 2015;5:12155. doi: 10.1038/srep12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua W., Ding L., Chen Y. Determination of berberine in human plasma by liquid chromatography-electrospray ionization-mass spectrometry. J. Pharmaceut. Biomed. Anal. 2007;44:931–937. doi: 10.1016/j.jpba.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 26.Wang K., Liao M., Zhou N. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26:222–235.e1-e5. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 27.Wu T.-R., Lin C.-S., Chang C.-J. Gut commensal parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut. 2019;68:248–262. doi: 10.1136/gutjnl-2017-315458. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F., Ma T., Cui P. Diversity of the gut microbiota in dihydrotestosterone-induced PCOS rats and the pharmacologic effects of diane-35, probiotics, and berberine. Front. Microbiol. 2019;10:175. doi: 10.3389/fmicb.2019.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P., Zhao J., Guo P. Dietary corn bran fermented by bacillus subtilis MA139 decreased gut cellulolytic bacteria and microbiota diversity in finishing pigs. Front. Cell Infect. Microbiol. 2017;7:526. doi: 10.3389/fcimb.2017.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ascher S., Reinhardt C. The gut microbiota: an emerging risk factor for cardiovascular and cerebrovascular disease. Eur. J. Immunol. 2018;48:564–575. doi: 10.1002/eji.201646879. [DOI] [PubMed] [Google Scholar]
- 31.Xu J., Chen N., Wu Z. 5-Aminosalicylic acid alters the gut bacterial microbiota in patients with ulcerative colitis. Front. Microbiol. 2018;9:1274. doi: 10.3389/fmicb.2018.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Liu B., Song J. Characteristics of gut microbiota in patients with hypertension and/or hyperlipidemia: a cross-sectional study on rural residents in Xinxiang County. Henan Province, Microorganisms. 2019;7 doi: 10.3390/microorganisms7100399. E399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue S.-J., Liu J., Wang W.-X. Berberine treatment-emergent mild diarrhea associated with gut microbiota dysbiosis. Biomed. Pharmacother. 2019;116:109002. doi: 10.1016/j.biopha.2019.109002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.