Abstract
In the early 1980s, the Nobel Prize winning cellular and molecular work of Mike Brown and Joe Goldstein led to the identification of the LDL receptor gene as the first gene where mutations cause the familial hypercholesterolemia (FH) phenotype. We now know that autosomal dominant monogenic FH can be caused by pathogenic variants of three additional genes (APOB/PCSK9/APOE) and that the plasma LDL-C concentration and risk of premature coronary heart disease differs according to the specific locus and associated molecular cause. It is now possible to use next-generation sequencing to sequence all exons of all four genes, processing 96 patient samples in one sequencing run, increasing the speed of test results, and reducing costs. This has resulted in the identification of not only many novel FH-causing variants but also some variants of unknown significance, which require further evidence to classify as pathogenic or benign. The identification of the FH-causing variant in an index case can be used as an unambiguous and rapid test for other family members. An FH-causing variant can be found in 20–40% of patients with the FH phenotype, and we now appreciate that in the majority of patients without a monogenic cause, a polygenic etiology for their phenotype is highly likely. Compared with those with a monogenic cause, these patients have significantly lower risk of future coronary heart disease. The use of these molecular genetic diagnostic methods in the characterization of FH is a prime example of the utility of precision or personalized medicine.
Supplementary key words: monogenic, polygenic, LDL-C, SNP score, variants of unknown significance, clinical utility, coronary heart disease, next-generation sequencing, index case, LDLR
Abbreviations: ACGS, UK Association for Clinical Genomic Science; ACMG, American College of Medical Genetics and Genomics; CHD, coronary heart disease; CT, cascade testing; DFH, definite FH; dHPLC, denaturing HPLC; FH, familial hypercholesterolemia; LDLR, LDL receptor; LLT, lipid-lowering therapy; Lp(a), lipoprotein (a); MLPA, multiplex ligation-dependent probe amplification; NGS, next-generation sequencing; PFH, possible FH; PRS, polygenic risk score; RFLP, restriction fragment length polymorphism; SSCP, single-strand conformation polymorphism; TC, total cholesterol; VUS, variants of unknown significance; WES, whole exome sequencing; WGS, whole genome sequencing
Since the identification in 1983 of mutations in the LDL receptor (LDLR) gene as causing the familial hypercholesterolemia (FH) phenotype, pathogenic variants (mutations) in the APOB gene, which reduce binding of the LDL particle to the LDLR (1), gain-of-function variants in PCSK9 (2), and one specific amino acid deletion in APOE (3) are known to also cause autosomal dominant FH. For LDLR, over 2,300 different variants have been reported (4, 5) with some being found relatively commonly in some countries, such as Finland (6), South Africa (7), and in French Canadians (8), due to “founder effects” because of recent immigration and population expansion.
This knowledge has opened the way for molecular genetic diagnosis approaches to be developed to allow the unambiguous genetic proof that an individual with the FH phenotype is carrying an FH-causing variant and to distinguish such individuals from those with environmental and/or polygenic underlying causes of their elevated plasma cholesterol concentrations. Initial diagnostic methods used cosegregation with common genetic variants at the LDLR locus, but these were superseded by methods that enabled rapid screening of all exons of LDLR for any variant, followed by Sanger sequencing of the identified exon.
One of the most significant findings about FH emerging from next-generation sequencing (NGS) studies is the DNA-based confirmation that the prevalence of carriers of an FH-causing variant is ∼1/250 in many Caucasian populations worldwide (9). Since the textbook figure for FH is 1/500, this new knowledge has essentially doubled the number of FH patients predicted to exist and emphasizes the clinical and health economic value of testing relatives to identify individuals at a young age and offering them appropriate lifestyle and lipid-lowering therapy (LLT) to reduce their risk of future premature coronary heart disease (CHD). This process is called cascade testing (CT) and has led to the identification of many thousand previously undiagnosed individuals with FH who can then be offered appropriate lifestyle advice and LLT.
In the United Kingdom, the 2019 National Health System long-term plan pledged to find and offer treatment to 25% of the predicted number of FH patients by 2023 (https://www.longtermplan.nhs.uk/). DNA testing in index cases with a clinical diagnosis of FH and CT of their relatives for the family variant is a mainstay in such programs to find these individuals before the onset of CHD.
First diagnosis using restriction fragment length polymorphisms
Before sequencing was routine in laboratories, it was still possible to carry out the presymptomatic identification of those relatives carrying a pathogenic variant inherited from an affected relative, and this approach had already been used for many clinically important inherited diseases. The method first required finding a restriction fragment length polymorphism (RFLP) at the disease gene locus and required the index case to be heterozygous for the polymorphism. As shown in Fig. 1, in 1985 (10), we used the cloned gene probe for the LDLR gene (kindly donated by Dr D. Russell) to detect an RFLP with the enzyme PvuII using Southern blotting technology. Three different patterns can be seen. Individuals who have both LDLR alleles with the cutting site lead to the detection of only the shorter fragment of 14 kb (designated V1) and have the genotype V1V1, whereas those who have both alleles lacking the cutting site result in the detection of only the longer fragment of 16.5 kb (designated V2) and have the genotype V2V2. To be useful for following the alleles through the family, the index case must be heterozygous for the polymorphism, (i.e., having the genotype V1V2). To establish whether the FH-causing variant is segregating with the common V1 or the rare V2 allele of the RFLP, at least one affected relative is required before a DNA diagnosis on other relatives can be made. This polymorphism was used to follow the inheritance of the defective LDLR gene in two families with FH, where it could be used for unambiguous diagnosis (10). As shown in Fig. 1 in family S, the deceased proband (arrowed) must have had the genotype V1V2 since his wife had only the V1 allele and two of his children had the genotype V1V2. Both children with LDL-C concentration in the normal range had inherited the father's V2 allele, and the child with elevated LDL-C had inherited the father's V1 allele, allowing the inference that in this family, the defective LDLR gene is segregating with the V1 allele. Both the (affected) siblings of the proband also had the genotype V1V2, and thus when any of their children inherits the V1 allele, they will have FH, which was confirmed by measures of LDL-C in these subjects. Since in the UK population around 30% of individuals have the genotype V1V2, this method was only potentially in 30% of all probands. Wider application of DNA-based diagnosis required identification of the specific causal variants, and several different methods evolved to achieve this.
Fig. 1.
The first DNA diagnosis in relatives of an FH index case (data from Humphries et al. (10)). A: Southern blot using a radioactive probe for human LDLR, hybridized to a Southern blot filter of a digestion with the restriction enzyme PvuII of three samples of human DNA, showing the RFLP. The detection of an RFLP is due to a sequence change altering the restriction site and usually occurs in flanking regions or introns of the gene and is not usually itself the FH-causing variant. Identifying a potentially useful RFLP was a time-consuming and rate-limiting step in this methodology. The common allele designated V1 contains a cutting site for PvuII and is seen as a band of 14 kb. The rare allele V2 lacks the cutting site and is seen as a band of 16.5 kb. Individuals shown are homozygous for the V1 allele (genotype V1V1), homozygous for the V2 allele (genotype V2V2), and heterozygous for both alleles (genotype V1V2). B: Segregation of the PvuII RFLP in family S. The proband (deceased from an early myocardial infarction) is indicated by an arrow. It can be deduced from the genotypes of his children that this individual must have had the genotype V1V2, as do both his older sister and younger brother. From inspection of the child of the older sister, in this family, the FH causing variant is inherited with the V1 allele, since the child has inherited the V1 allele from his Fh mother and has the FH phenotype. This is confirmed in the children of the proband, where those who have inherited the V2 allele from the deceased father do not have FH, whereas the child who inherited the V1 allele has FH. In the children of the youngest brother, all have inherited the V1 allele and can all be predicted to have FH.
Early diagnostic methods—deletions, direct assays, and rapid screening of exons for any sequence variant
Southern blot analysis is a method of detecting the presence of gross deletions/insertions in a gene of interest. Because of a “founder” effect of recent immigration, a 10 kb deletion of the 5′ region and including part of exon 1 of the LDLR gene was found to be the cause of FH in individuals of French-Canadian origin (8), whereas the FH-Helsinki variant is a deletion of exons 16, 17, and a portion of exon 18 (11). Following the development of the PCR, laboratories used PCR-based methods to detect these deletions rapidly and cheaply, allowing many samples to be screened, and the characteristics of carriers of different variants compared (6).
In the United Kingdom and in many populations worldwide, gross deletions/insertions explain ∼5% of the molecularly defined FH index cases (12), indicating that a PCR-based assay method is a key element of the diagnostic strategy. Multiplex ligation-dependent probe amplification (MLPA) is a commercially available kit-based method using amplification probes for all 18 exons of LDLR, alongside control probes to check for amplification efficiency. The exon-specific probes are modified to create fragments of different lengths, and the products are analyzed by capillary electrophoresis (13). Peak heights are compared, with a heterozygous deletion showing as a peak of 50% and a duplication as 150% of the expected height. MLPA has now been largely superseded by NGS copy number bioinformatics data analysis methods, which also provide data on sequencing coverage across the exons, enabling the comparison of a collective read depth per each gene exon in a single sequenced sample versus the average of the sequenced sample batch (14). While deletions and insertions have been reported in all parts of LDLR, the majority are located in introns 1–8 and 12 through the 3′ untranslated region, which corresponds to the distribution of “Alu” repeat sequences in the gene (15) and suggests that these rearrangements are due to mispairing and crossover at meiosis. Interestingly, a patient with the FH phenotype and a poor/limited response resistance to statin LLT has been found to have an entire duplication of the wild-type PCSK9 gene (16), which will clearly result in the FH phenotype because of higher plasma levels of the PCSK9 protein and greater hepatic LDLR degradation.
Since the majority of FH-causing variants are due to a single base change in the gene, methods to screen the entire gene for such changes were required. Early methods included single-strand conformation polymorphism (SSCP) analysis (17) and denaturing HPLC (dHPLC) electrophoresis (18). In both these methods, any exon harboring a sequence variant could be rapidly identified and selected for Sanger sequencing to determine if the changes were likely to be FH causing or not. While such methods have been superseded, they set the foundation and were integral in identifying the mutation spectrum in the LDLR gene in patients with the FH phenotype from many different countries.
To enable rapid triage diagnostic testing, several PCR-based approaches were developed to test for FH-causing variants common in the population under study. In the United Kingdom, this was first done using a multiplex amplification refractory mutation system assay for 12 LDLR variants and the common APOB c.10580G>A, p.(Arg3527Gln) variant (19). This method was rapid and cheap, and if a patient sample gave a positive result, this could be reported quickly, with negative patients entering a lengthy pathway of SSCP/dHPLC testing followed by Sanger sequencing and MLPA, often taking several months to complete. As sequencing became faster and cheaper, SSCP and dHPLC methods were dropped, and Sanger sequencing of all exons of LDLR, but only for APOB and PCSK9 hotspots, plus MLPA testing was adopted as the diagnostic protocol.
Using these available tools, DNA testing laboratories were established in a number of countries including in Holland (20), Spain (21), and Norway (22). In the United Kingdom, the FH molecular diagnostic service was established in 1997 in the Clinical Molecular Genetics Laboratory at Great Ormond Street Hospital for Children (23). Over a 4-year period, the laboratory analyzed 227 probands and 141 family members, from lipid clinics. Pathogenic variants were found in 76 probands, 67 in LDLR (23 previously undescribed) and nine carriers of the common APOB mutation. The mutation detection rate was 53% in pediatric probands, 32% in adults with tendon xanthoma-positive definite FH (DFH) and 14% in adults with a tendon xanthoma-negative possible FH (PFH). By 2010, using similar methods but with improved sensitivity and speed, 635 probands were analyzed in 2 years using the Amplification Refractory Mutation kit for 18 different LDLR mutations, one APOB and one common PCSK9 variant c.1120G>T, p.(Asp374Tyr), followed by SSCP/dHPLC and finally MLPA. The detection rate in DFH adult patients had improved to 56% and in PFH to 25% (24).
FH-mutation databases
As more and more countries have established molecular diagnostic laboratories for FH testing, and as commercial FH testing has developed (e.g., (25)), the number of published reports of FH-causing variants has increased significantly. University College London established an LDLR mutation database in 1998 (26) with regular updates since then. Analysis of worldwide-reported variants identified that LDLR exon 4 appears to be a “hot spot” for pathogenic changes, with significantly more variants reported here than in any other single exon (5, 27). However, exon 4 is also one of the largest in the gene, and when corrected for size, this hot spot appears less impressive. This exon codes for three of the crucial 7-finger repeat motifs involved in LDL binding (28), which suggests that any missense mutation that alters the structure of this part of the protein may be pathogenic.
One of the major problems in curating such a database is to have agreement as to the correct transcript and nomenclature to report a DNA or predicted protein change. Another problem is that simply finding a DNA change in a patient with FH does not prove that the change is actually FH causing. In 2015, the American College of Medical Genetics and Genomics (ACMG) published guidelines for the classification of variants (29), with five categories for variant classification: benign, likely benign, variants of unknown significance (VUS), likely pathogenic, and pathogenic. The UK Association for Clinical Genomic Science (ACGS) has published annual best practice guidelines for variant classification and reporting for UK clinical genomic laboratories based on the implementation of ACMG guidelines. The recently updated LDLR variant database with variants classified according to these guidelines may be accessed via http://databases.lovd.nl/shared/genes/LDLR. Although 93% of LDLR variants in the current upgrade of the database have been assigned to an ACGS pathogenicity category, ∼7% could not be classified with available data and remain as VUS. Recently, the ClinVar consortium has published LDLR-specific criteria (30), which will be extremely useful in helping laboratories make uniformly consistent decisions regarding whether a novel variant is pathogenic.
As an illustration of the immense genetic heterogeneity of molecular causes of FH, a recent study (31) of over 2,500 children with molecularly defined FH from just eight countries in Europe (Norway, Holland, France, the United Kingdom, Portugal, Czech Republic, Austria, and Greece) found 297 different LDLR pathogenic variants, with the highest degree of heterogeneity seen in the Czech Republic and the United Kingdom, with Greece showing the lowest (81, 67, and 16 different variants, respectively). Across countries, the three most common variants in each showed no overlap except for an intron 3 c.313+1G>A mutation, which occurred commonly in Norway and the Netherlands, and c.131G>A, p.(Trp44∗) in the Netherlands and Czech Republic, demonstrating the extreme heterogeneity of LDLR pathogenic variation across these eight countries. Interestingly, the prevalence of the APOB p.(Arg3527Gln) variant varied significantly (ranging from 0% in Greece to 39% in Czech Republic).
NGS methods
The technical advances realized during the Human Genome project has enabled significant increases in the speed of return for diagnostic test results and significant reductions in the costs. DNA diagnostic laboratories have now developed NGS protocols whereby all the protein coding regions for all the four known autosomal dominant FH genes as well as LDLRAP1, a gene showing autosomal recessive inheritance (two pathogenic variants required to cause the condition) (32), can be captured and sequenced together (33, 34, 35, 36). With the addition of small “barcoding” sequence identifiers into primers used for PCR amplification, it is also possible to batch samples from up to 96 individuals and analyze them in one run with high accuracy (34). This economy of scale has reduced costs so that now a full FH diagnostic screen including copy number assessment (deletions and duplications found in 5% of cases) can be completed for under £300. While laboratories routinely confirm the identified variant by Sanger sequencing, the Bristol laboratory has carried out a validation exercise that negates the need for Sanger confirmation of an NGS variant if specific quality parameters are met (QUAL score and allele read split), reducing the number of variants that require confirmation and reducing reporting time by around 2 weeks.
Clinical utility of a molecular diagnosis of monogenic FH
When an individual with a clinical diagnosis of FH is found to carry an FH-causing variant, this creates a definitive DNA-based diagnosis of monogenic FH (37). All recent guidelines on the management of FH recognize the utility of a DNA confirmed diagnosis. In those where a pathogenic variant is found, CT using the family variant for unambiguous identification of FH relatives is recommended by all recent published guidelines (e.g., (37, 38, 39, 40, 41)), with identified subjects treated with high-intensity LLTs to reduce their very considerable risk of early CHD. CT has been shown to be a feasible and highly cost-effective strategy in many countries (e.g., (42, 43)) and particularly in Holland (44).
Knowing the specific gene (and specific variant) may also help with clinical management, but this is mainly because of the direct or indirect effect of the genetic variant on LDLR activity and thus the impact of the variant on the individual's untreated LDL-C concentrations (45). Mutations in LDLR, which completely destroy receptor function (such as large deletions or the introduction of a premature stop codon), are often associated with higher LDL-C than, for example, some missense mutations or mutations influencing correct intron-exon splicing, which may result in some degree of residual receptor function. In the recent analysis of the 2,500 Pan-European children discussed previously (31), the highest mean concentrations were observed in the large insertion/deletion mutation carriers, which were similar to nonsense mutation carriers, but significantly higher than in promoter, splicing, and missense mutation carriers. When examining the untreated mean LDL-C concentrations between the 22 most common mutations, a 1.6-fold difference was seen, but with each group showing a wide variation, probably reflecting the influence of both polygenetic and environmental influences. While the LDL-C concentration varies considerably within a group of individuals who all carry the same LDLR variant, it is well documented that compared with those with a variant in LDLR, those with the common APOB variant tend to have lower mean untreated plasma total and LDL-C concentrations, whereas in those with a gain-of-function PCSK9 variant, mean concentrations tend to be higher, particularly for the variant observed in the UK p.(Asp374Tyr) (46). This is illustrated in Fig. 2, using data from the Simon Broome Register, where 410 DFH (tendon xanthoma positive) patients were examined, 41% of whom had documented CHD (46). Compared with those where no pathogenic variant could be found (using pre-NGS methods), the median pretreatment total cholesterol (TC) was 33% higher in the PCSK9 group, 5% higher in the LDLR group, and similar in the APOB group. All four groups showed similar and clinically useful reduction in TC concentrations upon statin treatment, but because pretreatment concentrations were so high in the PCSK9 patients, their median on-treatment concentrations were still higher than recommended (9.3 mmol/l). Compared with those with no mutation, the odds ratio for CHD in the LDLR group was 84% higher and in the PCSK9 group was 20-fold higher, estimates that were only modestly diminished by adjusting for pretreatment TC. In this selected sample of DFH patients, those carrying the APOB mutation had an intermediate but nonsignificant odds ratio for CHD.
Fig. 2.
Box and whisker plot of untreated TC before and after statin treatment and odds ratio for CHD by FH-causing gene (data from Huijgen et al. (46)). Boxes show median and interquartile range, with 95% range shown by bars. Outliers (more than 1.5 times the interquartile range from the edge of the box) are shown as dots. ∗ indicate P < 0.005 versus none, values from two-tailed unpaired Student's t-tests.
The Copenhagen General population study of over 98,000 participants has also examined this, with 111 carrying an LDLR mutation and 63 the common APOB mutation. Both groups had an earlier median age of myocardial infarction than the general population (14 and 10 years, respectively), with the odds ratio for myocardial infarction being 5.3 (2.4–12) and 1.8 (0.7–4.6), respectively (47). The specific mutation in the LDLR is also known to be associated with pretreatment TC concentrations and with risk of early CHD, with for example, “null mutations,” which lead to no functional receptors being produced being more severe than missense mutations (48) where some functional receptors may still be formed.
VUS
The full gene screening approaches available through NGS have increased the number of diagnoses, but in also the number of occasions whereby a VUS is identified, creating a diagnostic conundrum. These variants are more often novel with no or little published evidence to either support refute classification. The proportion of UK tested samples showing a VUS is small but is not trivial. The UK FH PASS database reports that as June 2019 there were 19,742 index cases tested with a clinical diagnosis of Simon Broome DFH or PFH, which resulted in 4,145 mutation-positive cases (21%) and 670 patients (3.3%) where a VUS was reported (49). ACGS best practice variant interpretation and reporting guidelines in 2020 now advocate only reporting a VUS where there is significant evidence supporting the VUS status, such that segregation or functional testing will enable classification. This should further reduce the number of VUS reported.
It is clearly of importance to be able to assess the probability of whether a variant identified in a clinical setting or as an incidental finding in genomics projects, is pathogenic or not. Predicting this is not always straightforward, especially for synonymous and missense variants. For LDLR, definitive proof that a variant is pathogenic requires either in vitro molecular assays to examine impact on transcription (50) or correct splicing (51) or LDLR expression (52), and although such studies have been reported for some variants, for the majority of LDLR variants such data are lacking.
The ApoB protein is highly polymorphic, with many common and rare variants that do not cause FH (e.g., (53)). However, as well as the two common causes of the FH phenotype c.10580G>A, p.(Arg3527Gln) (46) and c.10579C>T, p.(Arg3527Trp) (54), several novel variants in the APOB gene have been shown to be FH causing using in vitro assays (55, 56, 57). For PCSK9, the situation is more complex, since in silico prediction algorithms may predict that a missense change is likely to affect function but cannot distinguish between a gain of function, possibly FH-causing variant, and a loss of function, low LDL-C variant. For LDLRAP1, pathogenic variants cause a premature truncation and loss of function (32); however, where an individual is heterozygous for two different variants, determining phase and proof of recessive inheritance requires family studies.
In 2018, ClinVar published an update of all reported FH causing variants (4), and as shown in Fig. 3, this included 2,314 LDLR variants of which 1,620 were predicted to be pathogenic, 353 in APOB of which 35 were designated as pathogenic, and 216 in PCSK9, of which 28 were designated as pathogenic. Figure 3 shows that while only 8% of reported LDLR variants are VUS, 58% of APOB and 46% of PCSK9 variants are currently designated as VUS using available evidence.
Fig. 3.
Distribution of ClinVar benign, VUS, pathogenic, and unclear variants in LDLR, APOB, and PCSK9 (data from Iacocca et al. (4)).
The gold standard for proof of a variant being pathogenic requires family studies to see if other relatives who have inherited this variant have also shown high LDL-C levels, while the relatives without the inherited variant have normal levels of LDL-C. Such studies are time consuming and resource costly, and a method to triage the VUS would be of considerable utility.
The LDL-C polygenic risk score
The proportion of those with a clinical diagnosis of FH who are found to carry an FH-causing variant is of course strongly dependent on the precise selection criteria being used. While in those with the highest clinical suspicion of FH (Simon Broome DFH or a Dutch Lipid Clinic Network score >8) between 40 and 80% have a monogenic cause, in those with a lower clinical suspicion, the detection rate is usually 20–30% (24, 58, 59). A modification of the Dutch Lipid Clinic Network score has been proposed (60), which allocates “negative” points to the score depending on fasting plasma triglyceride concentrations, on the grounds that these would lower the probability of an individual having FH. However, with the reduction in costs of NGS, it is becoming less important to only select individuals with the highest probability of carrying an FH-causing variant, since identifying the monogenic cause in an individual with a low score is clinically useful for managing treatment and for testing relatives.
In the patients with clinical FH but where no FH-causing variant has been found, a polygenic etiology should be considered because of the coinheritance of a greater-than-average number of common LDL-C raising genetic variants (SNPs). This can be examined using a validated 12-SNP “LDL-SNP score,” which in combination gives a “polygenic risk score” (PRS) (61). Data from UK patients (and in international collaborations) suggest that in more than 80% of those with a clinical diagnosis of FH but with no detectable monogenic cause, the polygenic explanation is the most likely cause of their hypercholesterolemia (62).
The clinical use of the PRS in the management of FH is shown in Fig. 2. In the reports that have been returned to clinicians from several UK Diagnostic Laboratories for the last 3 years, the PRS is included by reporting the decile in which the individual scores. Deciles have been calculated using over 400,000 healthy UK White Caucasians from the UK BioBank study (63). Although using smaller sample numbers, the data suggest that these decile cutoffs are also appropriate for individuals from the Indian subcontinent, but not in those of Afro-Caribbean origin, and different values will need to be used in reporting for this group. In those without a monogenic cause, the report states that that there is a “high” likelihood of an individual in the 6th–10th decile of the PRS having a genetic cause for their hyperlipidemia, those in the 4th–6th decile an “intermediate,” and those in the 1st–3rd decile as having a “low” likelihood. In individuals where no monogenic cause has been identified, the PRS allows a genetic cause to be reported as highly likely in an additional 40% (50% of 80%) and intermediate likelihood in an additional 16% (20% of 80%), thus increasing the overall diagnostic yield of FH testing from ∼20–30% to >75%. The PRS also has diagnostic utility in triaging identified VUSs for family or in vitro studies. Finding a VUS in an individual with a low PRS score in an individual with a VUS would suggest that the variant is more likely to be pathogenic. Such families, which would represent only 20–30% of the VUS subjects (i.e., in the lowest three score deciles) would then allow these families to be prioritized for cosegregation or functional studies to confirm or refute pathogenicity.
There are several lines of evidence to suggest that the extent of atherosclerosis is higher in monogenic compared with polygenic FH patients. We have recently demonstrated (64) that the degree of thickening in the carotid artery, as measured by ultrasound, is considerably greater in a group of monogenic FH patients compared with a group of patients with a polygenic etiology, even though total and LDL-C levels were similar. In addition, coronary calcium score was significantly higher in monogenic versus polygenic patients. Many articles report that the prevalence of CHD is higher in groups of FH patients where a mutation can be found compared with those with a clinical diagnosis of FH but where no mutation can be found (46, 65, 66). Using NGS for the known FH genes among 20,485 CHD-free individuals, 1,386 (6.7%) had LDL-C >4.9 mmol/l, and of these, 24 (1.7%) carried a known FH mutation. As expected, there was a clear and continuous gradient of increasing CVD risk in individuals with increasing quintiles of LDL-C concentration, but in each quintile, those carrying an FH-causing variant had markedly higher CVD risk than noncarriers. Compared with individuals with LDL-C <3.7 mmol/l and no mutation, those with LDL-C >4.9 mmol/l and no FH mutation had a 6-fold higher risk for CHD, but those with both LDL-C >4.9 mmol/l and an FH mutation had a 22-fold higher risk. This elevated risk for CHD in individuals carrying an FH-causing variant has also been convincingly confirmed in a population-based analysis (66). This higher risk is likely explained by the substantially higher accumulated “LDL-C burden” in monogenic FH subjects, since these individuals will have had genetically determined lifelong high LDL-C.
There is now mounting evidence of clinical utility of including the PRS in a diagnostic report of those where no FH-causing variant is found. For the clinician, patients who have a PRS greater than the 8th decile, it is appropriate to consider that, because of their high genetic burden, and therefore life-long exposure to elevated LDL-C, their risk of developing CVD is high and that they should be considered for more intensive LLT. In addition, for all patients with a PRS greater than the 3rd decile, this establishes a probable genetic cause for their hyperlipidemia, which should help the physician (and patient) from embarking on a search for other esoteric causes of their dyslipidemia. It also gives more confidence about recommending treatment for primary prevention to younger patients with a high PRS, in the knowledge that, as they have a genetic cause, they will be more likely to benefit. It is relevant to note that the low PRS score clinical FH patients who have a dominant family history of high cholesterol may have a novel monogenic cause, and this information could be helpful in selecting patients for recruitment for novel gene discovery research studies.
For the patient, the PRS is also helpful. Clinicians who have been using this PRS for some time report that patients find the information gives them clarity about their diagnosis and less guilt that they have a high cholesterol because of their lifestyle. They also report that patients are more likely to accept statin treatment and less likely to pursue long and potentially futile attempts to treat their dyslipidemia with diet and lifestyle measures alone. The PRS offers a likely explanation for their hypercholesterolemia, and its genetic cause may promote adherence with lifestyle and treatment. While there are limited data in this field, one randomized control study of reporting CVD risk using conventional risk factors plus or minus a 22 CVD risk SNP score found that significantly more subjects receiving the score started LLT (and had a greater reduction in LDL-C) than those not told their genetic risk (67). This supports the clinical view that the LDL-SNP PRS will be motivational for statin use and adherence.
Finally, for those individuals with clinical FH and both a monogenic cause and a high polygenic score, there is evidence for having a higher CVD risk than those with monogenic FH and a low score (66, 68). In a meta-analysis of over 1,000 FH mutation positive individuals from three different cohorts (including UK BioBank), those with an LDL-SNP PRS above the 80th percentile had a 48% higher heart rate for CVD (68). This risk was in part, but not fully, explained by their higher LDL-C. Based on these data, it is appropriate to consider using more intense LLT and even lower LDL-C on treatment targets for those with a PRS >8th decile, for example, using PSCK9i agents (http://www.nice.org.uk/guidance/TA394).
Future prospects
CT
Although CT has been used very effectively in Holland (44) and has been shown to be highly cost effective using economic modeling (42), this is still not widely available in many countries (37). Apart from hesitation from family members to accept DNA testing (69), the main barriers to this being carried out are the lack of trained health care professionals (i.e., an FH nurse) to perform the necessary pedigree construction, plus the logistics of contacting relatives (especially those living at a distance) to obtain informed consent for genetic testing and a blood sample (particularly in the time of coronavirus disease pandemic). Obtaining funding for this work from health service providers requires a co-ordinated effort from local clinical champions, with support from patient groups. This will both raise public and professional awareness of genomic and precision medicine and be essential to persuade funders that the short-term investment for this process is cost saving in the long term, with regard to CVD events prevented.
Other PRSs
Although we have focused here on the use of the LDL-C SNP PRS, it is also of potential clinical utility to consider other PRS in the management of individuals with FH. For example, a PRS based on more than 1.6 million SNPs obtained from a genome-wide SNP chip has been shown to predict future risk of CHD in healthy individuals in the UK BioBank study (70). The magnitude of this effect was as large as that of the convention that CHD risk factors were combined and added significantly to overall risk prediction in a model including all classical risk factors. A particular genetic risk factor that is known to influence CHD risk in FH patients is their plasma concentration of the lipoprotein (a) Lp(a) (71). Since concentrations of Lp(a) are almost entirely genetically determined, a Lp(a) PRS, such as the one reported (72), may have clinical utility in patients with FH. Although neither of these PRS have yet been examined in patients with FH, it is likely that their use could identify those at highest risk of future CHD (based on non-LDL-C causes), and that such individuals would benefit from more intensive risk-factor management (e.g., even more intensive LDL-C lowering).
Testing children
Would it be feasible, affordable, and ethically acceptable to use whole exome sequencing (WES)/whole genome sequencing (WGS) of samples from babies at birth to identify those carrying an FH-causing variant? Currently, many countries already use biochemical and molecular approaches to test for a range of inherited metabolic diseases, usually using heel-prick blood samples taken within a few days of birth. It is certainly feasible to use this material for WES/WGS, and as costs fall, it is likely to become affordable in the next few years. Since an individual's genome information can be used over a lifetime, the cost effectiveness could be high. However, the ethical acceptability of such an approach is still open to debate, and this is beyond the scope of the current review. It is worth pointing out that the metabolic diseases currently tested for at birth, for example, phenylketonuria, have a major consequence on the health and development of the infant if undetected, and treatment has a huge impact on the child's future health. For FH, the most relevant immediate consequence of a diagnostic finding is for the parents, one of whom will have FH, of which many are likely to be as yet undiagnosed. The subsequent LLT of the affected parent and other siblings and relatives will improve their health and life expectancy.
A related approach has been proposed of first measuring cholesterol concentrations in infants at the time of their routine 12 month vaccination visit, and then using targeted NGS in those with elevated cholesterol to identify FH-variant carriers (73), an approach designated “child-parent screening.” In the United Kingdom, this approach has been shown to be feasible and acceptable in a pilot project, which confirmed the prevalence of FH-causing variant carriers of ∼1/270 (74). As a source of new FH index cases, if implemented, this would make a significant contribution to achieving the UK National Health System target of finding 25% of the predicted FH cases in the next 5 years (75). Health economic modeling has shown that this approach is cost effective (76), but the UK National Screening Committee has not yet endorsed this approach (https://legacyscreening.phe.org.uk/familialhypercholesterolaemia-child) and requested additional information about appropriate cholesterol thresholds, long-term benefits, and acceptability, which is being obtained through a larger pilot scheme. Measuring the lipid profile in children, for example, at school age and genetic screening of those above a threshold is already being run successfully in some countries (77).
Incidental findings
An additional way in which an individual carrying an FH-causing variant may be picked up is through having an “incidental finding” as part of a WES/WGS activity for an unrelated disorder. The ACMG has published a list of 57 genes involved in 27 disorders, where reporting such incidental findings has clinical utility (78), and this list includes FH. As part of the UK 100,000 Genomes Project (https://www.genomicsengland.co.uk/the-100000-genomes-project/), more than 70,000 individuals have had WGS carried out, and based on the prevalence of 1/250, between 200 and 300 FH patients are likely to be identified. Results will be fed back under the additional findings arm of the project and evaluation of the acceptability of receiving an FH diagnosis by this route, and the extent to which this leads to appropriate LLT and CT will be carried out. Several similar large-scale WES/WGS projects are ongoing around the world, both as part of research protocols and as part of routine clinical practice, and follow-up of the identified individuals will be important.
One of the aims of such WES/WGS projects is to identify novel genes where variants cause FH. To date, while potential candidates have been identified (79), these approaches have not yielded definitive results, and every candidate must be examined in detail before adding to diagnostic FH panel. As an example, the gene STAP1 was suggested to be a novel FH gene by linkage analysis and NGS in a large family from Holland (80). Although these findings appeared robust, functional analysis (81) and failure in cosegregation analysis (82) has definitively ruled out STAP1 as being an FH gene, demonstrating the degree to which caution must be used and a high threshold set for such candidates. Finally, WGS would also allow the identification of potential FH-causing variants in regions of the known FH genes not usually examined, such as in distant flanking transcriptional control elements or deep introns (83), which would then require function studies to verify the effect.
Conclusions
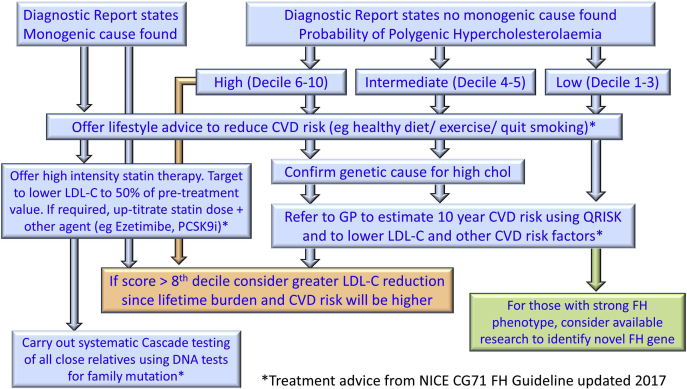
Since the identification of the LDLR gene in 1983 and demonstration that pathogenic variants in this gene cause FH, enormous progress has been made in developing rapid, affordable, and sensitive methods for the identification of the underlying molecular cause of the FH phenotype in an individual. The estimated prevalence of FH in Caucasian populations is 1/250 individuals, and although we yet have limited data to substantiate this, if the prevalence is similar in all ethnic groups, there may be up to 31 million individuals carrying an FH-variant worldwide. Although there is currently no global database of the number of individuals with a genetic confirmation of their diagnosis, published data suggest that, for example, 40% of the predicted number in Holland, 20% in Spain, and ∼10% in Canada and the United Kingdom have been identified, with the vast majority of countries having fewer than 1% identified to date (84, 85). While all patients with a clinical diagnosis of FH need cholesterol and CHD risk factor management, the demonstration of higher levels of atherosclerotic burden in those with an identified monogenic cause supports recommendations that they warrant intensive LDL-C lowering, and that this should be performed under the management of a lipid specialist. In some patients, this may include treatment with PCSK9 inhibitors in order to achieve LDL-C lowering target. By contrast, in those who do not have a monogenic cause for their lipid phenotype, estimation of their CHD risk using risk algorithms is appropriate, and they may be able to be managed in general practice. This use of genetic information to risk stratify patients with a clinical diagnosis of FH is a paradigm example of the utility of genetic in precision medicine (86) (Fig. 4).
Fig. 4.
Flowchart showing the risk stratification clinical management for adding the 12-SNP polygenic risk score to the NGS strategy used in Diagnostic Laboratories.
Conflict of interest
M. F. reports speakers' fees from Sanofi, and S. E. H. reports receiving fees for Advisory Boards of Novartis and Amryt. S. E. H. is the Medical Director of a University College London spin-out company StoreGene that offers to clinicians genetic testing for patients with FH. S. E. H. directs the UK Children's FH Register, which has been supported by a grant from Pfizer (grant no.: 24052829) given by the International Atherosclerosis Society. M. W. reported speaker fees from Amgen and Akcea. A. T.-B. declares no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
S. E. H. writing–original draft; M. F., M. W., and A. T.-B. writing–review and editing.
Funding and additional information
S. E. H. and M. F. were supported by a grant from the British Heart Foundation (grant no.: PG 08/008) and by funding from the Department of Health's National Institute for Health Research Biomedical Research Centers funding scheme. M. F. is supported by the Fondation Leducq Transatlantic Networks of Excellence Program grant (grant no.: 14 CVD03).
References
- 1.Boren J., Ekstrom U., Agren B., Nilsson-Ehle P., Innerarity T.L. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J. Biol. Chem. 2001;276:9214–9218. doi: 10.1074/jbc.M008890200. [DOI] [PubMed] [Google Scholar]
- 2.Abifadel M., Varret M., Rabes J.P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derre A., Villeger L., Farnier M., Beucler I., Bruckert E. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 3.Marduel M., Ouguerram K., Serre V., Bonnefont-Rousselot D., Marques-Pinheiro A., Erik Berge K., Devillers M., Luc G., Lecerf J.M., Tosolini L., Erlich D., Peloso G.M., Stitziel N., Nitchke P., Jais J.P. Description of a large family with autosomal dominant hypercholesterolemia associated with the APOE p.Leu167del mutation. Hum. Mutat. 2013;34:83–87. doi: 10.1002/humu.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iacocca M.A., Chora J.R., Carrie A., Freiberger T., Leigh S.E., Defesche J.C., Kurtz C.L., DiStefano M.T., Santos R.D., Humphries S.E., Mata P., Jannes C.E., Hooper A.J., Wilemon K.A., Benlian P. ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum. Mutat. 2018;39:1631–1640. doi: 10.1002/humu.23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leigh S., Futema M., Whittall R., Taylor-Beadling A., Williams M., den Dunnen J.T., Humphries S.E. The UCL low-density lipoprotein receptor gene variant database: pathogenicity update. J. Med. Genet. 2017;54:217–223. doi: 10.1136/jmedgenet-2016-104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuorio A.F., Turtola H., Piilahti K.M., Repo P., Kanninen T., Kontula K. Familial hypercholesterolemia in the Finnish north Karelia. A molecular, clinical, and genealogical study. Arterioscler. Thromb. Vasc. Biol. 1997;17:3127–3138. doi: 10.1161/01.atv.17.11.3127. [DOI] [PubMed] [Google Scholar]
- 7.Leitersdorf E., Van der Westhuyzen D.R., Coetzee G.A., Hobbs H.H. Two common low density lipoprotein receptor gene mutations cause familial hypercholesterolemia in Afrikaners. J. Clin. Invest. 1989;84:954–961. doi: 10.1172/JCI114258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobbs H.H., Brown M.S., Russell D.W., Davignon J., Goldstein J.L. Deletion in the gene for the low-density-lipoprotein receptor in a majority of French Canadians with familial hypercholesterolemia. N. Engl. J. Med. 1987;317:734–737. doi: 10.1056/NEJM198709173171204. [DOI] [PubMed] [Google Scholar]
- 9.Akioyamen L.E., Genest J., Shan S.D., Reel R.L., Albaum J.M., Chu A., Tu J.V. Estimating the prevalence of heterozygous familial hypercholesterolaemia: a systematic review and meta-analysis. BMJ Open. 2017;7 doi: 10.1136/bmjopen-2017-016461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphries S.E., Kessling A.M., Horsthemke B., Donald J.A., Seed M., Jowett N., Holm M., Galton D.J., Wynn V., Williamson R. A common DNA polymorphism of the low-density lipoprotein (LDL) receptor gene and its use in diagnosis. Lancet. 1985;1:1003–1005. doi: 10.1016/s0140-6736(85)91611-3. [DOI] [PubMed] [Google Scholar]
- 11.Aalto-Setala K., Helve E., Kovanen P.T., Kontula K. Finnish type of low density lipoprotein receptor gene mutation (FH-Helsinki) deletes exons encoding the carboxy-terminal part of the receptor and creates an internalization-defective phenotype. J. Clin. Invest. 1989;84:499–505. doi: 10.1172/JCI114192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosi I., Toledo-Leiva P., Neuwirth C., Naoumova R.P., Soutar A.K. Genetic defects causing familial hypercholesterolaemia: identification of deletions and duplications in the LDL-receptor gene and summary of all mutations found in patients attending the Hammersmith Hospital Lipid Clinic. Atherosclerosis. 2007;194:102–111. doi: 10.1016/j.atherosclerosis.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Taylor A., Martin B., Wang D., Patel K., Humphries S.E., Norbury G. Multiplex ligation-dependent probe amplification analysis to screen for deletions and duplications of the LDLR gene in patients with familial hypercholesterolaemia. Clin. Genet. 2009;76:69–75. doi: 10.1111/j.1399-0004.2009.01168.x. [DOI] [PubMed] [Google Scholar]
- 14.Futema M., Plagnol V., Whittall R.A., Neil H.A., Humphries S.E. Use of targeted exome sequencing as a diagnostic tool for Familial Hypercholesterolaemia. J. Med. Genet. 2012;49:644–649. doi: 10.1136/jmedgenet-2012-101189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leigh S.E., Foster A.H., Whittall R.A., Hubbart C.S., Humphries S.E. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann. Hum. Genet. 2008;72:485–498. doi: 10.1111/j.1469-1809.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 16.Iacocca M.A., Wang J., Sarkar S., Dron J.S., Lagace T., McIntyre A.D., Lau P., Robinson J.F., Yang P., Knoll J.H., Cao H., McPherson R., Hegele R.A. Whole-gene duplication of PCSK9 as a novel genetic mechanism for severe familial hypercholesterolemia. Can. J. Cardiol. 2018;34:1316–1324. doi: 10.1016/j.cjca.2018.07.479. [DOI] [PubMed] [Google Scholar]
- 17.Jensen H.K., Jensen L.G., Hansen P.S., Faergeman O., Gregersen N. High sensitivity of the single-strand conformation polymorphism method for detecting sequence variations in the low-density lipoprotein receptor gene validated by DNA sequencing. Clin. Chem. 1996;42:1140–1146. [PubMed] [Google Scholar]
- 18.Bodamer O.A., Bercovich D., Schlabach M., Ballantyne C., Zoch D., Beaudet A.L. Use of denaturing HPLC to provide efficient detection of mutations causing familial hypercholesterolemia. Clin. Chem. 2002;48:1913–1918. [PubMed] [Google Scholar]
- 19.Taylor A., Tabrah S., Wang D., Sozen M., Duxbury N., Whittall R., Humphries S.E., Norbury G. Multiplex ARMS analysis to detect 13 common mutations in familial hypercholesterolaemia. Clin. Genet. 2007;71:561–568. doi: 10.1111/j.1399-0004.2007.00807.x. [DOI] [PubMed] [Google Scholar]
- 20.Lombardi M.P., Redeker E.J., Defesche J.C., Kamerling S.W., Trip M.D., Mannens M.M., Havekes L.M., Kastelein J.J. Molecular genetic testing for familial hypercholesterolemia: spectrum of LDL receptor gene mutations in The Netherlands. Clin. Genet. 2000;57:116–124. doi: 10.1034/j.1399-0004.2000.570205.x. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Garcia A.B., Real J.T., Puig O., Cebolla E., Marin-Garcia P., Martinez Ferrandis J.I., Garcia-Sogo M., Civera M., Ascaso J.F., Carmena R., Armengod M.E., Chaves F.J. Molecular genetics of familial hypercholesterolemia in Spain: Ten novel LDLR mutations and population analysis. Hum. Mutat. 2001;18:458–459. doi: 10.1002/humu.1218. [DOI] [PubMed] [Google Scholar]
- 22.Leren T.P., Tonstad S., Gundersen K.E., Bakken K.S., Rodningen O.K., Sundvold H., Ose L., Berg K. Molecular genetics of familial hypercholesterolaemia in Norway. J. Intern. Med. 1997;241:185–194. doi: 10.1046/j.1365-2796.1997.78119000.x. [DOI] [PubMed] [Google Scholar]
- 23.Heath K.E., Humphries S.E., Middleton-Price H., Boxer M. A molecular genetic service for diagnosing individuals with familial hypercholesterolaemia (FH) in the United Kingdom. Eur. J. Hum. Genet. 2001;9:244–252. doi: 10.1038/sj.ejhg.5200633. [DOI] [PubMed] [Google Scholar]
- 24.Taylor A., Wang D., Patel K., Whittall R., Wood G., Farrer M., Neely R.D., Fairgrieve S., Nair D., Barbir M., Jones J.L., Egan S., Everdale R., Lolin Y., Hughes E. Mutation detection rate and spectrum in familial hypercholesterolaemia patients in the UK pilot cascade project. Clin. Genet. 2010;77:572–580. doi: 10.1111/j.1399-0004.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- 25.Berger M.J., Williams H.E., Barrett R., Zimmer A.D., McKennon W., Hong H., Ginsberg J., Zhou A.Y., Neben C.L. Color Data v2: a user-friendly, open-access database with hereditary cancer and hereditary cardiovascular conditions datasets. Database (Oxford) 2020;2020 doi: 10.1093/database/baaa083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson D.J., Gahan M., Haddad L., Heath K., Whittall R.A., Williams R.R., Humphries S.E., Day I.N. A World Wide Web site for low-density lipoprotein receptor gene mutations in familial hypercholesterolemia: sequence-based, tabular, and direct submission data handling. Am. J. Cardiol. 1998;81:1509–1511. doi: 10.1016/s0002-9149(98)00215-x. [DOI] [PubMed] [Google Scholar]
- 27.Esser V., Limbird L.E., Brown M.S., Goldstein J.L., Russell D.W. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 1988;263:13282–13290. [PubMed] [Google Scholar]
- 28.Hobbs H.H., Brown M.S., Goldstein J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 29.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chora J.R., Iacocca M.A., Tichy L., Wand H., Kurtz C.L., Zimmermann H., Leon A., Williams M., Humphries S.E., Hooper A.J., Trinder M., Brunham L.R., Costa Pereira A., Jannes C.E., Chen M. The Clinical Genome Resource (ClinGen) Familial Hypercholesterolemia Variant Curation Expert Panel consensus guidelines for LDLR variant classification. medRxiv. 2021 doi: 10.1101/2021.03.17.21252755. [DOI] [PubMed] [Google Scholar]
- 31.Futema M., Ramaswami U., Tichy L., Bogsrud M.P., Holven K.B., Roeters van Lennep J., Wiegman A., Descamps O.S., De Leener A., Fastre E., Vrablik M., Freiberger T., Esterbauer H., Dieplinger H., Greber-Platzer S. Comparison of the mutation spectrum and association with pre and post treatment lipid measures of children with heterozygous familial hypercholesterolaemia (FH) from eight European countries. Atherosclerosis. 2021;319:108–117. doi: 10.1016/j.atherosclerosis.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Cohen J.C., Kimmel M., Polanski A., Hobbs H.H. Molecular mechanisms of autosomal recessive hypercholesterolemia. Curr. Opin. Lipidol. 2003;14:121–127. doi: 10.1097/00041433-200304000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Hinchcliffe M., Le H., Fimmel A., Molloy L., Freeman L., Sullivan D., Trent R.J. Diagnostic validation of a familial hypercholesterolaemia cohort provides a model for using targeted next generation DNA sequencing in the clinical setting. Pathology. 2014;46:60–68. doi: 10.1097/PAT.0000000000000026. [DOI] [PubMed] [Google Scholar]
- 34.Yarram-Smith L., Dean P., O'Shea S., Dennis G., Bayly G., Taylor A., Day A., Watson M., Giles P., Ayling R., Haralambos K., Whatley S., McDowell I., Williams M. The impact of routine next generation sequencing testing for familial hypercholesterolaemia - 5 months service experience. Atherosclerosis. 2014;236:3304. [Google Scholar]
- 35.Norsworthy P.J., Vandrovcova J., Thomas E.R., Campbell A., Kerr S.M., Biggs J., Game L., Soutar A.K., Smith B.H., Dominiczak A.F., Porteous D.J., Morris A.D., Scotland G., Aitman T.J. Targeted genetic testing for familial hypercholesterolaemia using next generation sequencing: a population-based study. BMC Med. Genet. 2014;15:70. doi: 10.1186/1471-2350-15-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maglio C., Mancina R.M., Motta B.M., Stef M., Pirazzi C., Palacios L., Askaryar N., Boren J., Wiklund O., Romeo S. Genetic diagnosis of familial hypercholesterolaemia by targeted next-generation sequencing. J. Intern. Med. 2014;276:396–403. doi: 10.1111/joim.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturm A.C., Knowles J.W., Gidding S.S., Ahmad Z.S., Ahmed C.D., Ballantyne C.M., Baum S.J., Bourbon M., Carrie A., Cuchel M., de Ferranti S.D., Defesche J.C., Freiberger T., Hershberger R.E., Hovingh G.K. Convened by the familial hypercholesterolemia, F. clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J. Am. Coll. Cardiol. 2018;72:662–680. doi: 10.1016/j.jacc.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 38.Watts G.F., Gidding S., Wierzbicki A.S., Toth P.P., Alonso R., Brown W.V., Bruckert E., Defesche J., Lin K.K., Livingston M., Mata P., Parhofer K.G., Raal F.J., Santos R.D., Sijbrands E.J. Integrated guidance on the care of familial hypercholesterolemia from the International FH Foundation. J. Clin. Lipidol. 2014;8:148–172. doi: 10.1016/j.jacl.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Familial hypercholesterolaemia: identification and management. National Institute for Health and Care Excellence: Clinical Guidelines; London, UK: 2019. [PubMed] [Google Scholar]
- 40.Brunham L.R., Ruel I., Aljenedil S., Riviere J.B., Baass A., Tu J.V., Mancini G.B.J., Raggi P., Gupta M., Couture P., Pearson G.J., Bergeron J., Francis G.A., McCrindle B.W., Morrison K. Canadian Cardiovascular Society position statement on familial hypercholesterolemia: Update 2018. Can. J. Cardiol. 2018;34:1553–1563. doi: 10.1016/j.cjca.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Mach F., Baigent C., Catapano A.L., Koskinas K.C., Casula M., Badimon L., Chapman M.J., De Backer G.G., Delgado V., Ference B.A., Graham I.M., Halliday A., Landmesser U., Mihaylova B., Pedersen T.R. 2019 ESC/EAS guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Atherosclerosis. 2019;290:140–205. doi: 10.1016/j.atherosclerosis.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Kerr M., Pears R., Miedzybrodzka Z., Haralambos K., Cather M., Watson M., Humphries S.E. Cost effectiveness of cascade testing for familial hypercholesterolaemia, based on data from familial hypercholesterolaemia services in the UK. Eur. Heart J. 2017;38:1832–1839. doi: 10.1093/eurheartj/ehx111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazaro P., Perez de Isla L., Watts G.F., Alonso R., Norman R., Muniz O., Fuentes F., Mata N., Lopez-Miranda J., Gonzalez-Juanatey J.R., Diaz-Diaz J.L., Blasco A.J., Mata P. Cost-effectiveness of a cascade screening program for the early detection of familial hypercholesterolemia. J. Clin. Lipidol. 2017;11:260–271. doi: 10.1016/j.jacl.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Louter L., Defesche J., Roeters van Lennep J. Cascade screening for familial hypercholesterolemia: Practical consequences. Atheroscler. Suppl. 2017;30:77–85. doi: 10.1016/j.atherosclerosissup.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 45.Huijgen R., Kindt I., Defesche J.C., Kastelein J.J. Cardiovascular risk in relation to functionality of sequence variants in the gene coding for the low-density lipoprotein receptor: a study among 29,365 individuals tested for 64 specific low-density lipoprotein-receptor sequence variants. Eur. Heart J. 2012;33:2325–2330. doi: 10.1093/eurheartj/ehs038. [DOI] [PubMed] [Google Scholar]
- 46.Humphries S.E., Whittall R.A., Hubbart C.S., Maplebeck S., Cooper J.A., Soutar A.K., Naoumova R., Thompson G.R., Seed M., Durrington P.N., Miller J.P., Betteridge D.J., Neil H.A. Genetic causes of familial hypercholesterolaemia in patients in the UK: relation to plasma lipid levels and coronary heart disease risk. J. Med. Genet. 2006;43:943–949. doi: 10.1136/jmg.2006.038356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benn M., Watts G.F., Tybjaerg-Hansen A., Nordestgaard B.G. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur. Heart J. 2016;37:1384–1394. doi: 10.1093/eurheartj/ehw028. [DOI] [PubMed] [Google Scholar]
- 48.Lahtinen A.M., Havulinna A.S., Jula A., Salomaa V., Kontula K. Prevalence and clinical correlates of familial hypercholesterolemia founder mutations in the general population. Atherosclerosis. 2015;238:64–69. doi: 10.1016/j.atherosclerosis.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Haralambos K., Humphries S.E., Whitmore J., Datta D., Cather M., Miedzybrodzka Z., Breen J., Gritzmacher L., Hamlen A., Potter A., Cazeaux A., Sherman C., Moore A., Tyler M., Burton C. Familial hypercholesterolaemia (fh) genetic testing in the UK. Atheroscler. Suppl. 2018;34:e1–e9. [Google Scholar]
- 50.Khamis A., Palmen J., Lench N., Taylor A., Badmus E., Leigh S., Humphries S.E. Functional analysis of four LDLR 5'UTR and promoter variants in patients with familial hypercholesterolaemia. Eur. J. Hum. Genet. 2015;23:790–795. doi: 10.1038/ejhg.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb J.C., Patel D.D., Shoulders C.C., Knight B.L., Soutar A.K. Genetic variation at a splicing branch point in intron 9 of the low density lipoprotein (LDL)-receptor gene: a rare mutation that disrupts mRNA splicing in a patient with familial hypercholesterolaemia and a common polymorphism. Hum. Mol. Genet. 1996;5:1325–1331. doi: 10.1093/hmg/5.9.1325. [DOI] [PubMed] [Google Scholar]
- 52.Duskova L., Nohelova L., Loja T., Fialova J., Zapletalova P., Reblova K., Tichy L., Freiberger T., Fajkusova L. Low density lipoprotein receptor variants in the beta-propeller subdomain and their functional impact. Front. Genet. 2020;11:691. doi: 10.3389/fgene.2020.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabes J.P., Varret M., Devillers M., Aegerter P., Villeger L., Krempf M., Junien C., Boileau C. R3531C mutation in the apolipoprotein B gene is not sufficient to cause hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2000;20:E76–82. doi: 10.1161/01.atv.20.10.e76. [DOI] [PubMed] [Google Scholar]
- 54.Gaffney D., Reid J.M., Cameron I.M., Vass K., Caslake M.J., Shepherd J., Packard C.J. Independent mutations at codon 3500 of the apolipoprotein B gene are associated with hyperlipidemia. Arterioscler. Thromb. Vasc. Biol. 1995;15:1025–1029. doi: 10.1161/01.atv.15.8.1025. [DOI] [PubMed] [Google Scholar]
- 55.Thomas E.R., Atanur S.S., Norsworthy P.J., Encheva V., Snijders A.P., Game L., Vandrovcova J., Siddiq A., Seed M., Soutar A.K., Aitman T.J. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol. Genet. Genomic Med. 2013;1:155–161. doi: 10.1002/mgg3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alves A.C., Etxebarria A., Soutar A.K., Martin C., Bourbon M. Novel functional APOB mutations outside LDL-binding region causing familial hypercholesterolaemia. Hum. Mol. Genet. 2014;23:1817–1828. doi: 10.1093/hmg/ddt573. [DOI] [PubMed] [Google Scholar]
- 57.Alves A.C., Benito-Vicente A., Medeiros A.M., Reeves K., Martin C., Bourbon M. Further evidence of novel APOB mutations as a cause of familial hypercholesterolaemia. Atherosclerosis. 2018;277:448–456. doi: 10.1016/j.atherosclerosis.2018.06.819. [DOI] [PubMed] [Google Scholar]
- 58.Futema M., Whittall R.A., Kiley A., Steel L.K., Cooper J.A., Badmus E., Leigh S.E., Karpe F., Neil H.A., Humphries S.E. Analysis of the frequency and spectrum of mutations recognised to cause familial hypercholesterolaemia in routine clinical practice in a UK specialist hospital lipid clinic. Atherosclerosis. 2013;229:161–168. doi: 10.1016/j.atherosclerosis.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graham C.A., McIlhatton B.P., Kirk C.W., Beattie E.D., Lyttle K., Hart P., Neely R.D., Young I.S., Nicholls D.P. Genetic screening protocol for familial hypercholesterolemia which includes splicing defects gives an improved mutation detection rate. Atherosclerosis. 2005;182:331–340. doi: 10.1016/j.atherosclerosis.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 60.Haralambos K., Whatley S.D., Edwards R., Gingell R., Townsend D., Ashfield-Watt P., Lansberg P., Datta D.B., McDowell I.F. Clinical experience of scoring criteria for Familial Hypercholesterolaemia (FH) genetic testing in Wales. Atherosclerosis. 2015;240:190–196. doi: 10.1016/j.atherosclerosis.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Talmud P.J., Shah S., Whittall R., Futema M., Howard P., Cooper J.A., Harrison S.C., Li K., Drenos F., Karpe F., Neil H.A., Descamps O.S., Langenberg C., Lench N., Kivimaki M. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet. 2013;381:1293–1301. doi: 10.1016/S0140-6736(12)62127-8. [DOI] [PubMed] [Google Scholar]
- 62.Futema M., Shah S., Cooper J.A., Li K., Whittall R.A., Sharifi M., Goldberg O., Drogari E., Mollaki V., Wiegman A., Defesche J., D'Agostino M.N., D'Angelo A., Rubba P., Fortunato G. Refinement of variant selection for the LDL cholesterol genetic risk score in the diagnosis of the polygenic form of clinical familial hypercholesterolemia and replication in samples from 6 countries. Clin. Chem. 2015;61:231–238. doi: 10.1373/clinchem.2014.231365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J., Cortes A., Welsh S., Young A., Effingham M., McVean G. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharifi M., Higginson E., Bos S., Gallivan A., Harvey D., Li K.W., Abeysekera A., Haddon A., Ashby H., Shipman K.E., Cooper J.A., Futema M., Roeters van Lennep J.E., Sijbrands E.J.G., Labib M. Greater preclinical atherosclerosis in treated monogenic familial hypercholesterolemia vs. polygenic hypercholesterolemia. Atherosclerosis. 2017;263:405–411. doi: 10.1016/j.atherosclerosis.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khera A.V., Won H.H., Peloso G.M., Lawson K.S., Bartz T.M., Deng X., van Leeuwen E.M., Natarajan P., Emdin C.A., Bick A.G., Morrison A.C., Brody J.A., Gupta N., Nomura A., Kessler T. Diagnostic yield of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J. Am. Coll. Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trinder M., Li X., DeCastro M.L., Cermakova L., Sadananda S., Jackson L.M., Azizi H., Mancini G.B.J., Francis G.A., Frohlich J., Brunham L.R. Risk of premature atherosclerotic disease in patients with monogenic versus polygenic familial hypercholesterolemia. J. Am. Coll. Cardiol. 2019;74:512–522. doi: 10.1016/j.jacc.2019.05.043. [DOI] [PubMed] [Google Scholar]
- 67.Kullo I.J., Jouni H., Austin E.E., Brown S.A., Kruisselbrink T.M., Isseh I.N., Haddad R.A., Marroush T.S., Shameer K., Olson J.E., Broeckel U., Green R.C., Schaid D.J., Montori V.M., Bailey K.R. Incorporating a genetic risk score into coronary heart disease risk estimates: Effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial) Circulation. 2016;133:1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trinder M., Paquette M., Cermakova L., Ban M.R., Hegele R.A., Baass A., Brunham L.R. Polygenic contribution to low-density lipoprotein cholesterol levels and cardiovascular risk in monogenic familial hypercholesterolemia. Circ. Genom. Precis Med. 2020;13:515–523. doi: 10.1161/CIRCGEN.120.002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gidding S.S., Sheldon A., Neben C.L., Williams H.E., Law S., Zhou A.Y., Wilemon K., Ahmed C.D., Kindt I. Patient acceptance of genetic testing for familial hypercholesterolemia in the CASCADE FH Registry. J. Clin. Lipidol. 2020;14:218–223 e2. doi: 10.1016/j.jacl.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Inouye M., Abraham G., Nelson C.P., Wood A.M., Sweeting M.J., Dudbridge F., Lai F.Y., Kaptoge S., Brozynska M., Wang T., Ye S., Webb T.R., Rutter M.K., Tzoulaki I., Patel R.S. Genomic risk prediction of coronary artery disease in 480,000 adults: Implications for primary prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bogsrud M.P., Graesdal A., Johansen D., Langslet G., Hovland A., Arnesen K.E., Mundal L.J., Retterstol K., Wium C., Holven K.B. LDL-cholesterol goal achievement, cardiovascular disease, and attributed risk of Lp(a) in a large cohort of predominantly genetically verified familial hypercholesterolemia. J. Clin. Lipidol. 2019;13:279–286. doi: 10.1016/j.jacl.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 72.Trinder M., Uddin M.M., Finneran P., Aragam K.G., Natarajan P. Clinical utility of lipoprotein(a) and LPA genetic risk score in risk prediction of incident atherosclerotic cardiovascular disease. JAMA Cardiol. 2020;6:1–9. doi: 10.1001/jamacardio.2020.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wald D.S., Bestwick J.P., Morris J.K., Whyte K., Jenkins L., Wald N.J. Child-parent familial hypercholesterolemia screening in primary care. N. Engl. J. Med. 2016;375:1628–1637. doi: 10.1056/NEJMoa1602777. [DOI] [PubMed] [Google Scholar]
- 74.Futema M., Cooper J.A., Charakida M., Boustred C., Sattar N., Deanfield J., Lawlor D.A., Timpson N.J., Consortium U.K., Humphries S.E., Hingorani A.D. Screening for familial hypercholesterolaemia in childhood: Avon Longitudinal Study of Parents and Children (ALSPAC) Atherosclerosis. 2017;260:47–55. doi: 10.1016/j.atherosclerosis.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wald D.S., Bestwick J.P. Reaching detection targets in familial hypercholesterolaemia: Comparison of identification strategies. Atherosclerosis. 2020;293:57–61. doi: 10.1016/j.atherosclerosis.2019.11.028. [DOI] [PubMed] [Google Scholar]
- 76.McKay A.J., Hogan H., Humphries S.E., Marks D., Ray K.K., Miners A. Universal screening at age 1-2 years as an adjunct to cascade testing for familial hypercholesterolaemia in the UK: A cost-utility analysis. Atherosclerosis. 2018;275:434–443. doi: 10.1016/j.atherosclerosis.2018.05.047. [DOI] [PubMed] [Google Scholar]
- 77.Groselj U., Kovac J., Sustar U., Mlinaric M., Fras Z., Podkrajsek K.T., Battelino T. Universal screening for familial hypercholesterolemia in children: The Slovenian model and literature review. Atherosclerosis. 2018;277:383–391. doi: 10.1016/j.atherosclerosis.2018.06.858. [DOI] [PubMed] [Google Scholar]
- 78.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O'Daniel J.M., Ormond K.E., Rehm H.L., Watson M.S., Williams M.S., Biesecker L.G., American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Futema M., Plagnol V., Li K., Whittall R.A., Neil H.A., Seed M., Bertolini S., Calandra S., Descamps O.S., Graham C.A., Hegele R.A., Karpe F., Durst R., Leitersdorf E., Lench N. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J. Med. Genet. 2014;51:537–544. doi: 10.1136/jmedgenet-2014-102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fouchier S.W., Dallinga-Thie G.M., Meijers J.C., Zelcer N., Kastelein J.J., Defesche J.C., Hovingh G.K. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ. Res. 2014;115:552–555. doi: 10.1161/CIRCRESAHA.115.304660. [DOI] [PubMed] [Google Scholar]
- 81.Loaiza N., Hartgers M.L., Reeskamp L.F., Balder J.W., Rimbert A., Bazioti V., Wolters J.C., Winkelmeijer M., Jansen H.P.G., Dallinga-Thie G.M., Volta A., Huijkman N., Smit M., Kloosterhuis N., Koster M. Taking one step back in familial hypercholesterolemia: stap1 does not alter plasma ldl (low-density lipoprotein) cholesterol in mice and humans. Arterioscler. Thromb. Vasc. Biol. 2020;40:973–985. doi: 10.1161/ATVBAHA.119.313470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamiquiz-Moneo I., Restrepo-Cordoba M.A., Mateo-Gallego R., Bea A.M., Del Pino Alberiche-Ruano M., Garcia-Pavia P., Cenarro A., Martin C., Civeira F., Sanchez-Hernandez R.M. Predicted pathogenic mutations in STAP1 are not associated with clinically defined familial hypercholesterolemia. Atherosclerosis. 2020;292:143–151. doi: 10.1016/j.atherosclerosis.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 83.Kulseth M.A., Berge K.E., Bogsrud M.P., Leren T.P. Analysis of LDLR mRNA in patients with familial hypercholesterolemia revealed a novel mutation in intron 14, which activates a cryptic splice site. J. Hum. Genet. 2010;55:676–680. doi: 10.1038/jhg.2010.87. [DOI] [PubMed] [Google Scholar]
- 84.Nordestgaard B.G., Chapman M.J., Humphries S.E., Ginsberg H.N., Masana L., Descamps O.S., Wiklund O., Hegele R.A., Raal F.J., Defesche J.C., Wiegman A., Santos R.D., Watts G.F., Parhofer K.G., Hovingh G.K. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur. Heart J. 2013;34:3478–3490a. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.EAS Familial Hypercholesterolaemia Studies Collaboration. Vallejo-Vaz A.J., De Marco M., Stevens C.A.T., Akram A., Freiberger T., Hovingh G.K., Kastelein J.J.P., Mata P., Raal F.J., Santos R.D., Soran H., Watts G.F., Abifadel M., Aguilar-Salinas C.A., Al-Khnifsawi M. Overview of the current status of familial hypercholesterolaemia care in over 60 countries - The EAS Familial Hypercholesterolaemia Studies Collaboration (FHSC) Atherosclerosis. 2018;277:234–255. doi: 10.1016/j.atherosclerosis.2018.08.051. [DOI] [PubMed] [Google Scholar]
- 86.Representatives of the Global Familial Hypercholesterolemia Community. Wilemon K.A., Patel J., Aguilar-Salinas C., Ahmed C.D., Alkhnifsawi M., Almahmeed W., Alonso R., Al-Rasadi K., Badimon L., Bernal L.M., Bogsrud M.P., Braun L.T., Brunham L., Catapano A.L., Cillikova K. Reducing the clinical and public health burden of familial hypercholesterolemia: A global call to action. JAMA Cardiol. 2020;5:217–229. doi: 10.1001/jamacardio.2019.5173. [DOI] [PubMed] [Google Scholar]