Abstract
Exposure of yeast cells to increases in extracellular osmolarity activates the Hog1 mitogen-activated protein kinase (MAPK). Activation of Hog1 MAPK results in induction of a set of osmoadaptive responses, which allow cells to survive in high-osmolarity environments. Little is known about how the MAPK activation results in induction of these responses, mainly because no direct substrates for Hog1 have been reported. We conducted a two-hybrid screening using Hog1 as a bait to identify substrates for the MAPK, and the Rck2 protein kinase was identified as an interactor for Hog1. Both two-hybrid analyses and coprecipitation assays demonstrated that Hog1 binds strongly to the C-terminal region of Rck2. Upon osmotic stress, Rck2 was phosphorylated in vivo in a Hog1-dependent manner. Furthermore, purified Hog1 was able to phosphorylate Rck2 when activated both in vivo and in vitro. Rck2 phosphorylation occurred specifically at Ser519, a residue located within the C-terminal putative autoinhibitory domain. Interestingly, phosphorylation at Ser519 by Hog1 resulted in an increase of Rck2 kinase activity. Overexpression of Rck2 partially suppressed the osmosensitive phenotype of hog1Δ and pbs2Δ cells, suggesting that Rck2 is acting downstream of Hog1. Consistently, growth arrest caused by hyperactivation of the Hog1 MAPK was abolished by deletion of the RCK2 gene. Furthermore, overexpression of a catalytically impaired (presumably dominant inhibitory) Rck2 kinase resulted in a decrease of osmotolerance in wild-type cells but not in hog1Δ cells. Taken together, our data suggest that Rck2 acts downstream of Hog1, controlling a subset of the responses induced by the MAPK upon osmotic stress.
Mitogen-activated protein kinase (MAPK) cascades are common signaling modules found in both higher and lower eukaryotic cells. A typical MAPK cascade is composed of three tiers of protein kinases, a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK) (39). Yeast cells have several distinct MAPK cascades that transduce distinct extracellular stimuli (e.g., mating pheromone, high osmolarity, low osmolarity, and nitrogen starvation) (16, 14). Budding yeast (Saccharomyces cerevisiae) responds to increases in osmolarity in the extracellular environment by activating the Hog1 MAPK cascade. This cascade is essential for the survival of yeast in high-osmolarity environments (5, 6). Because a major outcome of the activation of this MAPK pathway is the elevated synthesis of glycerol, this pathway is referred to as the HOG (high-osmolarity glycerol response) pathway (1, 6).
The yeast HOG pathway is activated by two independent mechanisms. The first mechanism involves a two-component osmosensor, composed of the Sln1-Ypd1-Ssk1 proteins (29), which activate the Ssk2 and Ssk22 MAPKKKs (26). Once activated, those MAPKKKs phosphorylate the Pbs2 MAPKK. Pbs2 phosphorylation can also be achieved by a second mechanism, which involves the transmembrane protein Sho1, the MAPKKK Ste11, and the Ste11 binding protein Ste50 (20, 25, 28). Thus, both independent activation pathways converge at the level of Pbs2, and either the Ssk2, Ssk22, or Ste11 MAPKKK can activate Pbs2 by phosphorylation. Once activated, the Pbs2 MAPKK phosphorylates the Hog1 MAPK, which induces diverse stress responses.
There are a great number of genes known to be induced by the Hog1 MAPK in response to osmotic stress. Among them are genes responsible for osmotic adaptation, such as those for enzymes involved in glycerol synthesis (i.e., GPD1, GPP2, and GLO1), and genes involved in general stress responses like those for cytosolic catalase (CTT1), heat shock proteins (HSP12, HSP70, and HSP104), and a DNA damage-induced protein (DDR2) (14). The mechanism of gene regulation through activated Hog1 is still unknown, mainly because no definitive substrates for Hog1 have been reported. Several candidates, however, have been described. Those are the transcription factors Msn2 and Msn4, which bind to the promoter element defined as STRE, found in several stress-response induced genes (31), and the bZIP-type protein Sko1, which binds to CREB-like sequences (23, 30). Although these transcription factors seem to be controlled by Hog1, no direct interaction with Hog1 has been reported to date.
It is known that there exist other substrates for MAPK apart from transcription factors. In yeast, the cyclin-dependent kinase inhibitor Far1 is phosphorylated by the Fus3 MAPK in the mating pathway, which results in cell cycle arrest (24), and the tyrosine phosphatases Ptp2 and Ptp3, which are responsible for dephosphorylation and inactivation of Hog1 and Fus3, may be targets for their respective MAPKs (40, 41). In mammalian cells, several kinases involved in a number of cellular processes have been identified as substrates for MAPKs. This is the case, for example, for p90/rsk and MAPKAP kinase 2, substrates for the ERK kinases and p38 stress-activated MAPK (32, 34).
To understand the complex mechanism required for osmotic stress adaptation, it is necessary to identify substrates for the Hog1 MAPK. We conducted a two-hybrid screening to identify targets for Hog1. We found that Rck2, a protein kinase previously isolated by virtue of its sequence similarity to the yeast and mammalian calmodulin kinases (21) and as a suppressor of Schizosaccharomyces pombe checkpoint mutants (9), interacts with Hog1 and that Rck2 kinase activity is regulated by phosphorylation by the Hog1 MAPK.
MATERIALS AND METHODS
Yeast strains.
The following yeast strains were used: W303-1A (MATa leu2-3,112 ura3-1 his3-11 trp1-1a can100), EBΔR2Wa (MATa rck2::kanMX4 leu2-3,112 ura3-1 his3-11 trp1-1a can100), hog1Δ (MATa hog1::LEU2 leu2- 3,112 ura3-1 his3-11 trp1-1a can100), EBΔHR2Wa (MATa hog1::LEU2 rck2::kanMX4 leu2-3,112 ura3-1 his3-11 trp1-1a can100), DMΔpWa (MATa pbs2::kanMX4 leu2-3,112 ura3-1 his3-11 trp1-1a can100) (all strains up to this point are derivatives of W303-1A), L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ), PJ69-4a (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1:HIS3 GAL2:ADE2 met2::GAL7:lacZ), PJ69-4α (MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1:HIS3 GAL2:ADE2 met2::GAL7:lacZ), and FY1679 (MATa/α ura3-52/ura3-52 his3-Δ200/+ leu2-Δ1/+ trp1-Δ63). Genomic disruptions were made by long-flanking-homology PCR-based gene disruption (38).
Buffers and media.
Buffer A is 50 mM Tris-HCl (pH 7.5), 15 mM EDTA, 15 mM EGTA, 2 mM dithiothreitol (DTT), 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, and 5 μg of leupeptin per ml. Buffer B is 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2 mM DTT, 1 mM PMSF, 1 mM benzamidine, and 5 μg of leupeptin per ml. Buffer C is 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.2% Triton X-100, 0.1 mM PMSF, and 0.5 mM DTT. Kinase buffer is 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT. Phosphatase inhibitor mixture contains 10 mM NaF, 1 mM sodium vanadate, and 1 mM sodium pyrophosphate. Sodium dodecyl sulfate (SDS) loading buffer is 50 mM Tris-HCl (pH 6.8), 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol. YPD medium contains, per 1 liter, 10 g of yeast extract, 20 g of peptone, and 20 g of dextrose. Selective medium contains 1.7 g of yeast nitrogen base (Difco) per liter, 5 g of (NH4)2SO4 per liter, 20 g of dextrose per liter, and supplements (100 mg each of amino acids, uracil, or adenine per liter, as appropriate, except where indicated). All yeast growth was at 30°C. Osmotolerance in liquid cultures was evaluated by diluting fresh cultures with medium (to achieve an optical density of 0.005). Two hundred microliters of the diluted cultures in the presence of different NaCl concentrations was placed in 96-well microtiter plates. Cells were grown for 16 h with shaking at 30°C, and growth was determined by measuring the optical density of the cultures at 630 nm. Relative growth was calculated as the ratio between growth in the presence and that in the absence of added NaCl and expressed as a percentage.
Plasmids.
Plasmid pRSR2 carries a 5.4-kbp BamHI/XhoI fragment containing RCK2 in the multicopy plasmid pRS426 (Stratagene). pCMR2 was constructed by inserting the entire wild-type coding RCK2 sequence into pCM262, a derivative of pCM190 (3, 13) kindly provided by E. Herrero, by homologous recombination in yeast (22). The RCK2 fragment was PCR amplified from YEpRCK2 using Pwo DNA polymerase and hybrid primers EbtetRCK2F and EbtetRCK2R (Table 1). The resulting PCR product was cotransformed with pCM262 digested with NotI and PstI into yeast strain FY1679, followed by selection for uracil prototrophy. The desired construct was recovered by transformation of Escherichia coli DH5α to ampicillin resistance. This plasmid encodes RCK2 carboxy terminally fused to three hemagglutinin (HA) tags and one (His)6 tag, under transcriptional control of the Tet promoter. Its authenticity was verified by restriction mapping; we also verified, by Western blotting using anti-HA antibodies, that the full-length tagged protein was produced in yeast. pCMkdR2 was constructed in an analogous way, except that a Lys201→Met mutation was introduced by PCR using the mutagenic primers EbkdR2F and EbkdR2R (Table 1). The plasmid construct was finalized by cotransformation in yeast and recovery in E. coli as described above.
TABLE 1.
Oligonucleotides used
| Primer name | Homologya | Sequence (5′ to 3′)b | Use |
|---|---|---|---|
| AD8 | RCK2 −25 to −1 | GAG AGG ATC AAA CAA AAT CTC TTC G | RCK2 disruption |
| EBL2RCK2 | kanMX4 −409 to −433, RCK2 331 to 312 | GGG GAT CCG TCG ACC TGC AGC GTA CCA CTA CTT TCA TGG ACA TCG | RCK2 disruption |
| EBL3RCK2 | kanMX4 1052 to 1076, RCK2 1443 to 1462 | AAC GAG CTC GAA TTC ATC GAT GAT ACG ATT GTT TAC CAA AGG AGG | RCK2 disruption |
| EBL4RCK2 | RCK2 1749 to 1731 | CGT ATC CAG CGT TAA TTG G | RCK2 disruption |
| EBPBS2L1 | PBS2 −225 to −208 | GGA CAC AGA GAT AAC TGC | PBS2 disruption |
| EBPBS2L2 | kanMX4 −409 to −433, PBS2 22 to 3 | GGG GAT CCG TCG ACC TGC AGC GTA CGG TTA GCA AAC TTG TCT TCC | PBS2 disruption |
| EBPBS2L3 | kanMX4 1052 to 1076, PBS2 1947 to 1964 | AAC GAG CTC GAA TTC ATC GAT GAT AGG AAC GTG GTG AGA ATG G | PBS2 disruption |
| EBPBS2L4 | PBS2 2342 to 2326 | TTC GAG GAG TCG ATG GC | PBS2 disruption |
| EBHOG1F | pACTII 947 to 964, HOG1 1 to 20 | GAG GCC CCG GGG ATC CGA ATG ACC ACT AAC GAG GAA TT | HOG1-GAL4 AD fusion |
| EBHOG1R | pACTII 985 to 968, HOG1 1308 to 1288 | ATA GAT CTC TCG AGC TCG TTA CTG TTG GAA CTC ATT AGC | HOG1-GAL4 AD fusion |
| DMRCK2B-F1 | pGBT9 845 to 874, RCK2 1 to 17 | AAC AAA GGT CAA AGA CAG TTG ACT GTA TCG ATG CTT AAA ATA AAG GC | RCK2-GAL4 DB fusions |
| DMRCK2B-R1 | pGBT9 932 to 903, RCK2 507 to 488 | TAA GAA ATT CGC CCG GAA TTA GCT TGG CTG CTA GAT TTT ATT GAT CAG CTT G | RCK2-GAL4 DB fusions |
| DMRCK2B-F2 | pGBT9 845 to 874, RCK2 504 to 519 | AAC AAA GGT CAA AGA CAG TTG ACT GTA TCG ATC GGT GAA GGT GCT TT | RCK2-GAL4 DB fusions |
| DMRCK2B-R2 | pGBT9 932 to 903, RCK2 1401 to 1383 | TAA GAA ATT CGC CCG GAA TTA GCT TGG CTG CTA GTA TCT TTT AGA CGG CTC | RCK2-GAL4 DB fusions |
| DMRCK2B-F3 | pGBT9 845 to 874, RCK2 1395 to 1413 | AAC AAA GGT CAA AGA CAG TTG ACT GTA TCG AGA TAC GAC ATT GAC CAG | RCK2-GAL4 DB fusions |
| DMRCK2B-R3 | pGBT9 932 to 903, RCK2 1833 to 1816 | TAA GAA ATT CGC CCG GAA TTA GCT TGG CTG CTA TTC CCT GAT AGT GG | RCK2-GAL4 DB fusions |
| EbkdR2R | RCK2 619 to 589 (mutation at 602 and 603) | CTG CCT TTT TGA TAA CCA TAA TGG CAA CAG C | RCK2 KD mutation |
| EbkdR2F | RCK2 589 to 619 (mutation at 602 and 603) | GCT GTT GCC ATT ATG GTT ATC AAA AAG GCA G | RCK2 KD mutation |
| 5RCK2SA | RCK2 1544 to 1578 (mutation at 1558 and 1563) | CCT CGC TAC TGT TTG CAC CGG CTG CTG TTG CTA TG | RCK2 SA mutation |
| 3RCK2SA | RCK2 1544 to 1578 (mutation at 1558 and 1563) | CAT AGC AAC AGC AGC CGG TGC AAA CAG TAG CGA GG | RCK2 SA mutation |
| EbtetRCK2F | pCM262 2492 to 2522, RCK2 1 to 27 | CCG GAT CAA TTC GGG GGA TCA GTT TAA ACG CAT GCT TAA AAT AAA GGC CCT TTT CTC G | pCMR2, pCMkdR2 overexpression |
| EbtetRCK2R | pCM262 2551 to 2520, RCK2 1830 to 1811 | CCT GGT GAT CCG TCG ACC TGC AGG CGG CCG CGT TCC CTG ATA GTG GCG CTT A | pCMR2, pCMkdR2 overexpression |
For genes, nucleotide 1 is defined as the A of the start codon. For vectors, nucleotide 1 is defined according to the published sequence. For the kanMX module (38), nucleotide 1 is defined as the A of the start codon of the E. coli Kanr gene.
For mutagenic primers, mutated positions are in boldface.
The bacterial expression plasmids pET-16b and pRSETB (Stratagene) allow the expression of His-tagged proteins in E. coli. Full-length wild-type and several mutant RCK2 alleles were cloned into the pET-16b and pRSETB plasmids. Mutations in Ser519→Ala and Lys201→Met were made by PCR and verified by either DNA sequencing or digestion with specific restriction enzymes. The yeast expression vector YCpIF16 (PGAL1-HA TRP1 CEN) allowed the expression of HA fusion proteins using the GAL1 promoter (12). HOG1 was cloned into the BamHI site of YCpIF16 for expression of HA-tagged HOG1. HOG1 was cloned into the BamHI site of the pBTM116 plasmid (37) to fuse it to the LexA binding domain. The LexA-HOG1 fusion protein fully complemented the osmosensitivity observed for a hog1Δ strain. pACTII-RCK2 was obtained by fusion of the full-length RCK2 with the GAL4 activation domain (AD) in pACTII (18). A fusion of full-length HOG1 with the GAL4 AD was made by cotransformation of yeast strain PJ69-4a with pACTII cut with BamHI and SacI and full-length HOG1 PCR product from the hybrid primers EBHOG1F and EBHOG1R (Table 1). Similarly, various fragments of RCK2 were fused with the GAL4 DNA binding domain (DB) by cotransformation of PJ69-4α with pGBT9 (2) cut with EcoRI and BamHI and PCR products from hybrid primers DMRCK2B-F1 through DMRCK2B-R3 (Table 1). Yeast expression plasmid pGal-PBS2DD has been described previously (40).
Two-hybrid analysis.
The two-hybrid analysis was carried out essentially according to the method of Durfee et al. (11), using pACTII (18) and pBTM116 (37), as the AD plasmid and the LexA DB plasmid, respectively. A yeast cDNA library in the pACTII plasmid (ATCC 87293) was screened for proteins that interact with LexA-HOG1 using the L40 reporter strain. One million clones were analyzed by growth in histidine-deficient plates containing 40 mM 3-aminotriazole (3-AT) (Sigma). 3-AT was included in the screening plates to abolish any transactivator activity of the LexA-HOG1 bait. Positive clones were selected and further tested for β-galactosidase activity as follows. Cells (∼5 × 106) were spotted onto YPD plates and incubated for 5 h at 30°C, and the cells were replicated onto nitrocellulose membranes. β-Galactosidase activity was visualized in situ using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described previously (37). For analysis of interactions between various fragments of RCK2, cloned in a DB vector, and HOG1, cloned in an AD vector, constructs were introduced into the same diploid cell by mating (4). Taking advantage of the GAL1:HIS3 and GAL2:ADE2 reporters of the host PJ69-4a/α strains (17), the strength of interactions was then assayed on selective medium lacking histidine and containing 3 mM 3-AT and 2 μg of adenine per ml.
In vivo coprecipitation assay.
Cells in mid-log phase (10 ml) were collected by brief centrifugation at 4°C. The pellet was washed in 1 ml of buffer C containing 1 tablet of Complete protease inhibitor mix (Roche) per 25 ml of buffer and resuspended in 80 μl of the same buffer. Five hundred microliters of glass beads was added, and cells were disrupted in a FastPrep 120 apparatus (Bio 101) at speed 4 for 15 s. One milliliter of buffer C containing 0.25% Nonidet P-40 was added, followed by a 10-min centrifugation at 10,000 ×g in a chilled microcentrifuge. The extract was precleared by addition of 20 μl of Pansorbin (formalin-fixed Staphylococcus aureus cells) followed by head-over-head incubation for 2 h at 4°C. After 2 min of centrifugation at 10,000 × g, 4 μl of the precipitating antibody was added followed by a 4-h head-over-head incubation. After addition of 10 μl of Pansorbin, incubation was continued overnight. The extract was then centrifuged for 2 min at 10,000 × g, and the pellet was washed twice in 500 μl of buffer C containing 0.25% NP-40 and finally resuspended in 500 μl of SDS loading buffer prior to electrophoresis.
Expression and purification of epitope-tagged proteins.
His-tagged wild-type and mutant RCK2 alleles were constructed using pET-16b, expressed in E. coli BL21(DE3) cells (33), purified using TALON metal affinity resin (Clontech), and eluted using imidazole buffer, according to the manufacturer's instructions. RCK2 (434–610) was constructed using pRSETB (Invitrogen) and expressed as described above. Glutathione S-transferase (GST) fusion proteins encoding PBS2(EE) and HOG1 were constructed using pGEX-4T (Pharmacia), expressed in E. coli DH5, and purified using glutathione-Sepharose beads (Pharmacia) in buffer B as described previously (29). HA-tagged HOG1 was expressed in yeast, and purification was carried out by immunoprecipitation with anti-HA monoclonal antibody 12CA5 and protein A-Sepharose beads (Roche). Beads were washed extensively with buffer A plus 150 mM NaCl and resuspended in kinase buffer.
In vivo RCK2 phosphorylation assays.
Wild-type, pbs2Δ, and hog1Δ cells were grown in synthetic complete (sc) medium and subjected or not to a brief osmotic shock (0.4 M NaCl, 5 to 10 min). Cell extracts were prepared as described above but in the presence of buffer A without EGTA and EDTA. Extracts were treated with or without λ phosphatase (300 U for 60 min) in the presence or absence of phosphatase inhibitors. Rck2 was detected by immunoblotting using partially purified polyclonal antibodies against Rck2.
In vitro kinase assays.
HA-HOG1 was purified as described above from untreated yeast cells or cells treated with a brief osmotic shock (0.4 M NaCl for 10 min). One microgram of recombinant GST-HOG1 from E. coli was activated by phosphorylation using 0.5 μg of GST-PBS2(EE) in the presence of kinase buffer and ATP. After 15 min at 30°C, 5 μg of His-tagged Rck2 proteins, purified from E. coli, was added to the previous mixture together with [γ-32P]ATP (0.2 μCi/μl). The mixture was then incubated for 5 min at 30°C, and the reactions were terminated by the addition of 2× SDS loading buffer.
The induction of Rck2 activity by Hog1 phosphorylation was monitored by the following in vitro kinase assay. One microgram of recombinant GST-HOG1 or GST-HOG1(KN) from E. coli was phosphorylated by using 0.5 μg of GST-PBS2(EE) in the presence of kinase buffer and ATP. After 15 min at 30°C, 5 μg of wild-type or mutant versions of Rck2 were added to the mixture and incubation was maintained at 30°C for 15 min. GST-HOG1 and GST-HOG1(KN) were removed from their mixture by affinity chromatography using glutathione-Sepharose beads, and Rck2 proteins were further incubated for 15 min in the presence of kinase buffer and radioactive ATP.
Labeled proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography using dried gels or after transfer of the proteins to Immobilin P membranes (Millipore). His-tagged proteins were probed by immunoblotting with the anti-His monoclonal antibody BMG-His-1 (Roche).
RESULTS
The Hog1 MAPK binds to the C-terminal regulatory domain of the Rck2 kinase.
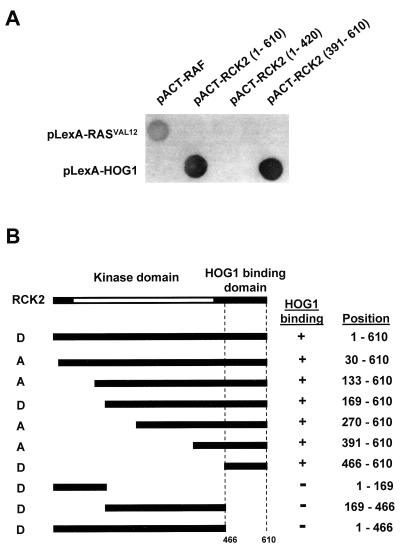
Osmotic stress induces activation of the Hog1 MAPK, which mediates induction of a set of osmoadaptive responses. Full understanding of Hog1-mediated responses requires the identification of direct substrates for Hog1, a goal, however, not achieved so far. To identify substrates for this MAPK, a two-hybrid screening was conducted. A yeast library constructed with a GAL4 activator domain vector was screened using as bait a fusion construct between the LexA DB and the full-length HOG1. The LexA-HOG1 fusion protein used in this screening fully complemented the osmosensitive phenotype of a hog1-deficient strain and showed the same time-dependent phosphorylation pattern after osmotic stress as that of the wild-type protein (data not shown). Fourteen yeast cDNA clones that interacted with Hog1 were isolated from the screening. Of these clones, six were overlapping clones encoding Rck2, a 610-residue protein which resembles the yeast and mammalian calmodulin kinases as well as S. cerevisiae protein kinase Dun1 (10, 21). In addition to the catalytic domain, Rck2 contains a 132-residue C-terminal extension speculated to play an autoinhibitory role (21). Interestingly, the clones obtained from the two-hybrid screening allowed us to approximately map the domain of interaction of Rck2 with Hog1 (Fig. 1). A partial cDNA clone encoding amino acid residues 391 to 610 was able to interact with Hog1 as efficiently as did the full-length clone (Fig. 1A). To extend these analyses and further confirm the specific domain in Rck2 that is essential for binding to Hog1, various segments of Rck2 were fused to the GAL4 activator domain, and their interaction with the full-length Hog1 protein was tested. Figure 1A shows typical results, and Fig. 1B summarizes the interaction between several Rck2 segments and Hog1. The results obtained with both orientations of AD and DB fusions were completely consistent, indicating that Rck2 residues 466 to 610 are sufficient for binding to Hog1. We thus designated this segment the HOG1 binding domain.
FIG. 1.
Identification by two-hybrid analysis of the HOG1 binding domain in Rck2. (A) Interactions of various Rck2 fragments fused to the GAL4 activator domain with the full-length Hog1 fused to the LexA DB were examined. Representative filter β-galactosidase assays demonstrating interactions between Hog1 and Rck2 are shown. Positions of the Rck2 fragments included in the constructs are indicated in parentheses. Proteins encoded by the control plasmids pLexA-RASV12 and pACT-RAF, which are known to interact with each other (37), are shown for comparison. (B) Summary of the two-hybrid interaction analysis between Rck2 and Hog1. The Rck2 segments included in the AD (denoted as A) or DB (denoted as D) constructs are schematically shown on the left, with their precise amino acid positions indicated on the right. The presence or absence of interaction is depicted by a plus or minus sign, respectively.
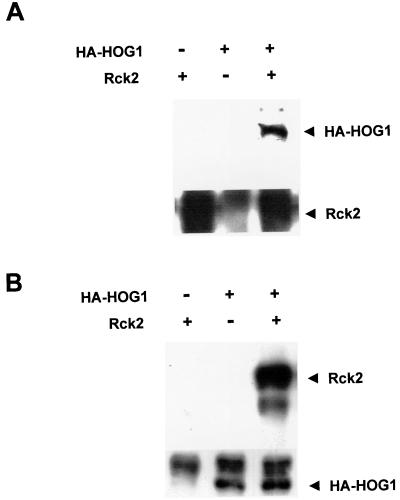
The interaction between Hog1 and Rck2, concluded on the basis of the two-hybrid data, was confirmed by in vivo immunoprecipitation experiments. Yeast rck2Δ mutant cells were cotransformed with plasmids that express HA-tagged HOG1 and a multicopy plasmid carrying wild-type Rck2. Rck2 was immunoprecipitated using specific polyclonal antibodies against this protein, and the presence of HA-HOG1 in the precipitates was determined with an anti-HA monoclonal antibody. As shown in Fig. 2A, Rck2 was able to coprecipitate Hog1. Conversely, when HA-HOG1 was immunoprecipitated using monoclonal antibodies against HA, and the presence of Rck2 in the precipitates was determined with specific antibodies against Rck2, Hog1 was able to coprecipitate Rck2 (Fig. 2B). Thus, these in vivo binding assays confirmed the conclusion of the two-hybrid analyses indicating that Hog1 interacts with Rck2.
FIG. 2.
In vivo association of Hog1 and Rck2 proteins. rck2Δ yeast strain EBΔR2Wa was transformed with a plasmid expressing HA-HOG1 and a multicopy plasmid carrying RCK2. (A) Hog1 coprecipitates with Rck2. Rck2 was immunoprecipitated using specific antibodies against this kinase (lower panel), and the presence of HA-HOG1 in the precipitates was determined with an anti-HA antibody (upper panel) (B) Rck2 coprecipitates with Hog1. HA-HOG1 was immunoprecipitated using anti-HA antibody (lower panel), and the presence of Rck2 was determined with specific antibodies against Rck2 (upper panel).
Rck2 is phosphorylated after osmotic stress in a Hog1-dependent manner.
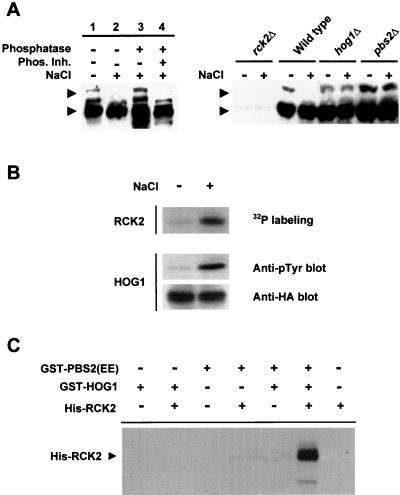
When yeast cells are exposed to osmotic stress, Hog1 MAPK is rapidly phosphorylated and activated (20). We decided to test whether Rck2 was also phosphorylated after osmotic stress. Wild-type, hog1Δ, and pbs2Δ strains were subjected to a brief osmotic shock, and wild-type Rck2 protein was monitored by Western blotting using anti-Rck2 antibodies. As shown in Fig. 3A, Rck2 was detected as multiple bands under nonstress conditions (lane 1). When subjected to osmotic stress, the mobility pattern of Rck2 was altered (Fig. 3A, lane 2). Mainly, the slower band shifted to a faster-migrating form. The exact nature of the upper band is currently unknown. However, we believe that the band represents a phosphorylated form of Rck2 and that its disappearance is caused by additional phosphorylation events as suggested by the following results. The mobility change of RCK2 was induced by phosphorylation because, when extracts from osmotically stressed cells were treated with λ phosphatase, the mobility pattern could be reversed (Fig. 3A, lane 3). In control experiments using phosphatase inhibitors, Rck2 mobility was not affected by λ phosphatase treatment (lane 4). Thus, after osmotic stress Rck2 is phosphorylated. Interestingly, when osmotic stress Rck2 phosphorylation was studied with mutant cells deficient in the HOG pathway (pbs2Δ and hog1Δ strains) no differences in Rck2 mobility were observed after osmotic stress compared with the wild-type strain (Fig. 3A, right panel). Therefore, Rck2 is phosphorylated after osmotic stress in a HOG-dependent manner.
FIG. 3.
In vivo and in vitro phosphorylation of Rck2 by Hog1. (A) Dependence of the HOG pathway for the in vivo osmotic stress-induced phosphorylation of Rck2. Wild-type cells were grown in sc medium and subjected (+) or not (−) to a brief osmotic shock (0.4 M NaCl, 10 min). Cell extracts were prepared and treated with (+) or without (−) λ phosphatase in the presence (+) or absence (−) of a mixture of phosphatase inhibitors as described in Materials and Methods (left panel). Rck2 was detected by immunoblotting using anti-Rck2 antibodies (arrowheads), and its phosphorylation state was monitored by noting changes in the electrophoretic mobility of the protein. Results from several strains subjected (+) or not (−) to brief osmotic stress (0.4 M NaCl, 5 min) are shown in the right panel. Relevant genotypes are depicted. Extracts from rck2Δ cells are included in order to monitor the specificity of the anti-Rck2 antibodies. (B) In vivo-activated Hog1 phosphorylates Rck2. HA-HOG1 was immunoprecipitated by using anti-HA monoclonal antibody from yeast cells before (−) or after (+) the addition of NaCl to a final concentration of 0.4 M. The presence of HA-HOG1 in the precipitates was detected with an anti-HA antibody, and Hog1 activation was monitored by immunoblot analysis using a monoclonal antibody specific to phosphotyrosine (4G10) (middle panel). After immunoprecipitation, HA-HOG1 was incubated with purified His-tagged RCK2(KD) in the presence of kinase buffer and radioactive ATP. Phosphorylated proteins were separated by SDS-PAGE and detected by autoradiography. (C) In vitro phosphorylation of Rck2 by Hog1. Recombinant tagged proteins were purified from E. coli as described in Materials and Methods. Hog1 and the constitutively activated Pbs2 allele [PBS2(EE)] were incubated in the presence of kinase buffer and ATP. Rck2 was then added (when indicated) in the presence of radioactive ATP. Phosphorylated proteins were resolved by SDS-PAGE and detected by autoradiography. The position of tagged Rck2 is indicated on the left.
Both in vivo- and in vitro-activated Hog1 phosphorylates Rck2.
We then tested whether activated Hog1 was able to phosphorylate Rck2. For this purpose, yeast cells expressing HA-tagged Hog1 were treated with a brief osmotic shock. HA-HOG1 was then immunoprecipitated by using monoclonal anti-HA antibodies and protein A-Sepharose beads. The activation of HA-HOG1 was assessed by Western blotting using the monoclonal antibody 4G10 against phosphorylated tyrosine residues. Immunoprecipitated Hog1 was incubated together with a catalytically inactive, His-tagged Rck2 in the presence of [γ-32P]ATP. The use of a kinase-deficient Rck2 protein, termed RCK2(KD), which contains a Lys201→Met mutation, was necessary because Rck2 has a background kinase activity that results in autophosphorylation. As shown in Fig. 3B, RCK2(KD) phosphorylation significantly increased when the protein was incubated with activated Hog1 from osmotically stressed cells.
We then tested whether Rck2 phosphorylation was carried out directly by the Hog1 MAPK, using purified proteins in an in vitro kinase assay. For this purpose, Hog1 and a constitutively activated version of Pbs2 [PBS2(EE)] (EE denotes Ser514→Glu and Thr518→Glu mutations in the activation loop of the Pbs2 MAPKK) were purified as GST fusion proteins from E. coli. In the first step of the reaction, Hog1 was activated by phosphorylation in the presence of PBS2(EE) and ATP. Then, RCK2(KD), purified from E. coli as a His-tagged protein, and [γ-32P]ATP were added to the reaction. As shown in Fig. 3C, only when RCK2(KD) was incubated with preactivated Hog1 did phosphorylation of Rck2 result. Taken together, these results indicate that Rck2 is a direct substrate for the MAPK Hog1.
Hog1 phosphorylates Ser519 at the autoinhibitory domain of Rck2.
To map the phosphorylation site for Hog1 in Rck2, we created several truncated RCK2 alleles and expressed them as His-tagged proteins in E. coli. It was found, however, that removal of the C-terminal region of Rck2 results in proteins with a very high in vitro kinase activity (data not shown), which is consistent with the putative autoinhibitory role of this region. For this reason, the Lys201→Met mutation (referred to as KD) was used to create catalytically deficient enzymes. Truncated forms of RCK2(KD) expressed and purified from E. coli were subjected to in vitro phosphorylation by activated Hog1 (as described above). A 63-residue C-terminal deletion had no effect on Rck2 phosphorylation compared to the full-length protein (Fig. 4A, lanes 1 and 2). However, a 200-residue C-terminal deletion completely abolished Rck2 phosphorylation (Fig. 4A, lane 3). Furthermore, when a C-terminal polypeptide (amino acids 434 to 610) was tested, it was phosphorylated by Hog1, suggesting that the carboxy-terminal regulatory domain and not the kinase domain of Rck2 was the target for Hog1 phosphorylation. Thus, phosphorylation of Rck2 by Hog1 occurs in a region which is coincident with the HOG1 binding domain.
FIG. 4.
Hog1 phosphorylates Ser519 at the regulatory domain of Rck2. (A) Phosphorylation of different fragments of Rck2 by Hog1. Various Rck2 fragments were tested for their ability to be phosphorylated by an in vitro-activated Hog1 (as described in Materials and Methods). After the in vitro kinase assay, phosphorylated proteins were resolved by SDS-PAGE and detected by autoradiography. Positions of the Rck2 fragments included in the constructs are indicated in parentheses. Proteins were His tagged and contained a Lys201→Met mutation which results in catalytically inactive enzymes, to avoid autophosphorylation. (B) Mutation of Rck2 Ser519→Ala abolishes Hog1 phosphorylation. The full-length RCK2(KD) and its Ser519 mutant form were tested for Hog1 phosphorylation as described for panel A. After phosphorylation, proteins were resolved by SDS-PAGE and transferred to a nylon membrane. Phosphorylated proteins were detected by autoradiography (upper panel). His-tagged Rck2 proteins were detected by immunoblotting by using the anti-His monoclonal antibody BMG-His-1 (lower panel).
A sequence corresponding to the consensus phosphorylation site for MAPK (Ser-Pro) is present in the region between residues 434 and 546 (corresponding to the protein sequence LLFSP). We created a point mutant version to replace Ser519 with Ala (RCK2-Ser519A) and tested it for phosphorylation by Hog1. As shown in Fig. 4B, phosphorylation of Rck2 was completely abolished in the mutant version, indicating that Ser519 at the regulatory domain of Rck2 is the unique phosphorylation site for Hog1.
Phosphorylation of Rck2 by Hog1 results in induction of Rck2 activity.
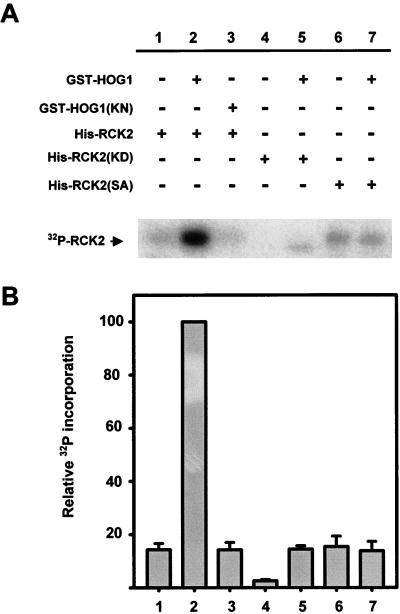
To test if Rck2 kinase activity can be regulated by phosphorylation, we developed the following in vitro kinase assay. Because no substrates are known for this kinase, we based the in vitro kinase assay on the fact that Rck2 is able to autophosphorylate (reference 21 and our results). Therefore, we assayed Rck2 activity by measuring its level of autophosphorylation. To test the role of Hog1 phosphorylation in Rck2 kinase activity, we incubated wild-type Rck2, the catalytically impaired RCK2(KD), or the unphosphorylatable RCK2(SA) mutant with activated GST-HOG1 in the presence of nonradioactive ATP. After incubation, Hog1 was removed from the mixture by affinity chromatography and Rck2 autophosphorylation was assayed in the presence of kinase buffer and radioactive ATP. As expected, the catalytically impaired Rck2 was unable to autophosphorylate (Fig. 5, lane 4) and increased its level of phosphorylation slightly when incubated with Hog1, as a result of Hog1 activity (Fig. 5, lane 5). Interestingly, when the wild-type Rck2 was incubated with Hog1, a very marked increase in phosphorylation was observed which could not be accounted for by Hog1 phosphorylation alone (Fig. 5, lane 2). When the catalytically impaired HOG1(KN) mutant was used, Rck2 phosphorylation was not induced (Fig. 5, lane 3). Similarly, when wild-type Hog1 was incubated with Rck2 but no ATP was present in the reaction mixture, Rck2 autophosphorylation was not increased (data not shown). This clearly indicates that Hog1 phosphorylation is required for the induction of Rck2 activity. The Ser519 mutant of Rck2, which cannot be phosphorylated by Hog1, still has some autophosphorylation activity, but it did not show any increase in phosphorylation when incubated with Hog1 (Fig. 5, lanes 6 and 7). Taken together, our results suggest that, after incubation with active Hog1, Rck2 is phosphorylated at Ser519 in the autoinhibitory domain, and this phosphorylation markedly increases Rck2 kinase activity.
FIG. 5.
Induction of Rck2 activity by Hog1 phosphorylation. (A) Purified wild-type Rck2 protein or the indicated mutant forms of Rck2 were incubated with (+) or without (−) wild-type GST-HOG1 or the catalytically inactive mutant version GST-HOG1(KN), in the presence of kinase buffer and cold ATP. After 15 min at 30°C, HOG1 was removed by affinity chromatography and Rck2 was further incubated in the presence of kinase buffer and radioactive ATP. Phosphorylated proteins were separated by SDS-PAGE and detected by autoradiography. The position of Rck2 is indicated on the left. A representative experiment is shown. (B) The intensity of autophosphorylation was quantified with a phosphorimager (Fuji BAS1000). Samples from three independent experiments were measured, and the intensity of each band was normalized to that of lane 2. Error bars indicate standard errors of the means.
Overexpression of a dominant negative RCK2 allele results in osmosensitivity.
It was reported previously that RCK2 disruption does not cause osmosensitivity (9, 21). Prompted by our current findings, we have reexamined osmotic stress-related phenotypes of rck1, rck2, and rck1 rck2 mutants in different strain backgrounds but again found no difference compared to the wild type (data not shown). However, the absence of an evident osmosensitive phenotype can still be compatible with the notion of Rck2 being involved in the HOG pathway. Thus, we tested whether overexpression of a catalytically impaired RCK2 mutant [RCK2(KD)] was able to alter activation of the osmotic stress responses driven by Hog1. Because RCK2(KD) does not have any detectable kinase activity (as shown in Fig. 5), overexpression of RCK2(KD) might inhibit the Hog1 signal transduction pathway by causing RCK2(KD) to abortively interact with Hog1 and/or downstream elements.
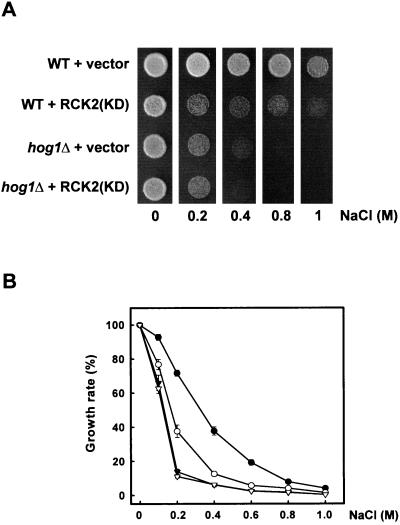
Yeast cells overexpressing RCK2(KD) were cultured on plates in the absence or presence of various NaCl concentrations. As shown in Fig. 6A, wild-type cells expressing the RCK2(KD) allele grew slower than cells carrying an empty vector in the presence of virtually any NaCl concentration tested. A similar effect was observed when sorbitol was used instead of NaCl (data not shown). This behavior was consistent for different plasmid isolates tested and reproducible in different genetic backgrounds. A nonspecific toxic effect of overexpression of the RCK2(KD) allele was ruled out, as, on normal-osmolarity medium, the growth rate was similar (Fig. 6A). Interestingly, when RCK2(KD) was overexpressed in hog1Δ cells, no effect on osmosensitivity was observed compared to the hog1Δ cells transformed with an empty vector.
FIG. 6.
Cellular osmosensitivity induced by overexpression of a catalytically inactive RCK2(KD) allele. (A) Wild-type (WT) and hog1Δ cells transformed with an empty vector (pCM262) or a plasmid expressing the catalytically inactive RCK2(KD) (pCMkdR2) were grown on YPD or YPD containing NaCl at different concentrations, in the absence of doxycycline (allowing full expression from the Tet promoter). Growth in plates was scored after 3 days at 30°C. (B) Wild-type cells transformed with pCM262 (●) or pCMkdR2 (○) and hog1Δ cells transformed with pCM262 (▾) or pCMkdR2 (▿) were grown in liquid medium in the presence of different concentrations of NaCl, and the effect of stress on cell growth was determined as described in Materials and Methods. Results are means ± standard errors of the means from three independent experiments.
To further characterize this phenomenon and to obtain a more quantitative estimate of the effect of the RCK2(KD) allele on cellular osmosensitivity, cell growth in the presence of various NaCl concentrations was measured in liquid medium. Wild-type cells expressing RCK2(KD) were much more sensitive to the presence of NaCl than were cells carrying an empty vector (Fig. 6B, open circles versus solid circles). For instance, at 0.4 M NaCl, the growth rate of cells carrying the RCK2(KD) allele was reduced to less than 50% compared to the same cells carrying an empty vector. Again, no alterations in osmosensitivity were observed in hog1Δ cells expressing the RCK2(KD) allele (Fig. 6B, open triangles versus solid triangles). Thus, the dominant inhibitory effect of RCK2(KD) appears specific to the stress-induced activation of the HOG pathway.
Overexpression of RCK2 suppresses the osmotic sensitivity of hog1 and pbs2 mutants.
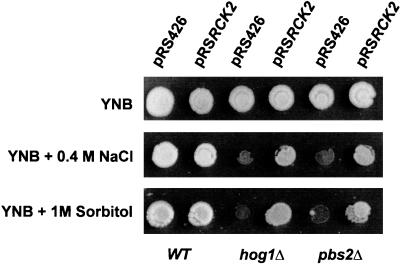
A possible explanation for the osmosensitivity observed with overexpression of the dominant negative RCK2(KD) is the involvement of Rck2 in the Hog1 osmotic stress signal transduction. To confirm this possibility, we tested whether overexpression of wild-type RCK2 was able to suppress the hog1Δ and pbs2Δ mutant osmosensitive phenotype. Overexpression of RCK2 from a multicopy vector in hog1Δ mutants resulted in an over 100-fold increase in the survival rate on high-NaCl-containing medium. Moreover, a comparable effect was seen for sorbitol-containing plates (Fig. 7), indicating general suppression of the osmotic sensitivity of this mutant. For a strain disrupted in the PBS2 gene, which encodes the Hog1-activating MAPKK, the outcome was similar (Fig. 7). By contrast, overexpression of the RCK2-related gene RCK1 (10) did not affect osmosensitivity of these mutants (data not shown). Therefore, these data also support the notion of a link between Hog1 and Rck2 kinases to control osmotic stress responses in yeast.
FIG. 7.
Suppression of hog1Δ and pbs2Δ cell osmosensitivity by RCK2 overexpression. Yeast cells deficient in the HOG1 or PBS2 gene were transformed with either an empty vector (pRS426) or a multicopy plasmid carrying wild-type (WT) RCK2 (pRSRCK2). Cells were grown in YPD, YPD containing NaCl at a concentration of 0.4 M, or sorbitol at 1 M. Growth was scored after 3 days at 30°C.
Deletion of RCK2 suppresses cell lethality caused by hyperactivation of the HOG pathway.
To further confirm the genetic data on the involvement of Rck2 in HOG signaling, we decided to test whether deletion of RCK2 alters HOG signaling. It has been reported previously that hyperactivation of the HOG pathway results in cell lethality (20, 29). Hyperactivation of Hog1 can be achieved by deletion of the SLN1 osmosensor (a negative regulator of the pathway) or by overexpression of constitutive alleles of the genes encoding MAPKKK Ssk2 and Ssk22 or Pbs2 MAPKK (20). This lethality can be prevented by overexpression of protein phosphatases Ptp2 and Ptc1 or by deletion of downstream elements such as the Hog1 MAPK. Based upon this observation, we have tested whether cell lethality caused by the constitutive allele of the gene for Pbs2 (PBS2DD) could be suppressed by deletion of RCK2. As shown in Fig. 8, deletion of the RCK2 gene suppressed cell lethality caused by hyperactivation of the Hog1 MAPK by PBS2DD, providing additional evidence that Rck2 is a new element of the HOG pathway, acting downstream of the Hog1 MAPK.
FIG. 8.
Deletion of the RCK2 gene suppresses the lethality of the PBS2DD mutation in yeast. The wild-type and rck2Δ yeast strains were transformed with either pGal control vector or pGal:PBS2DD. Yeast cells were grown at 30°C on sc plates containing glucose or galactose. Growth was scored after 4 days.
DISCUSSION
Yeast cells respond to increases in osmolarity in the extracellular environment by activating the HOG pathway. While the sensing and signal transduction mechanism required for activation of the Hog1 MAPK is well established (27), the proteins that are immediate targets of the Hog1 MAPK, necessary for the generation of osmostress responses, are not known. To identify substrates for the Hog1 MAPK, we undertook a two-hybrid screening using Hog1 as bait. In this report, we report that the Rck2 kinase interacts with Hog1 and that its kinase activity is regulated by the Hog1 MAPK, suggesting that Rck2 is one such substrate, the first one for the Hog1 MAPK identified so far.
The results of this study demonstrate by two-hybrid and in vivo coprecipitation experiments that Hog1 binds Rck2. Furthermore, Hog1 phosphorylates a specific serine residue at the noncatalytic C terminus of Rck2. These results, together with earlier findings, allow us to speculate on the molecular mechanism of Rck2 activation by Hog1. It was proposed previously that the C-terminal extension of the Rck2 kinase could play an inhibitory role, based on the observation that overexpression of a C-terminally truncated form of Rck2 is more toxic to the cell than is the wild-type allele (21). Consistent with these results, we observed that deletion of the C-terminal 63 residues results in an extremely active Rck2 enzyme (data not shown). Because osmotic stress-activated Hog1 binds and phosphorylates the C-terminal regulatory domain of Rck2, it can be proposed that Hog1 phosphorylation results in Rck2 activation most probably as a result of a release from the constraint imposed by the autoinhibitory domain. Once active, Rck2 autophosphorylates as well as phosphorylating its substrates, thus allowing the control of a subset of the osmotic stress responses.
There are a number of independent lines of evidence for an involvement of Rck2 in yeast osmotic stress responses. First, Rck2 is phosphorylated in vivo upon osmotic stress, and its phosphorylation is HOG1 dependent. It is worth noting that phosphorylation of Rck2 results in its activation as shown by in vitro kinase assays. Second, multicopy expression of RCK2 partially suppresses the osmosensitive phenotype of mutant cells defective in the HOG MAPK cascade, such as hog1Δ or pbs2Δ cells. Third, although the exact mechanism of the dominant negative effect of the RCK2(KD) allele is unknown at the moment, a high-level expression of the kinase-impaired RCK2(KD) causes osmosensitivity in a wild-type background but not in a hog1Δ background. However, the formal possibility that some of the effects of Rck2 in osmoregulation could be controlled by a distinct pathway that does not involve Hog1 cannot be excluded. Furthermore, cell lethality caused by hyperactivation of the HOG pathway can be suppressed by deletion of RCK2. In the light of these findings, it is noteworthy that the rck2 disruption does not cause osmosensitivity even if deleted in a background lacking the related Rck1 kinase (9, 21). A model that accommodates these observations could involve a situation for Rck2 analogous to that found for the MAPK Kss1, whose role in filamentous growth was long overlooked, since the kss1Δ mutant was not affected in this function. More detailed analysis (8, 19) showed, however, that the Kss1 kinase has two opposite functions in the filamentous growth pathway: activation in its phosphorylated form and inhibition in its unphosphorylated form. In the kss1Δ null mutant, these two effects cancel each other out. Other models are also compatible with the data, such as a model that invokes the presence of multiple redundant downstream targets for the Hog1 MAPK. This would explain the dominant inhibitory effect observed with overexpression of the RCK2(KD) protein, whereas no effect in osmosensitivity is observed for the rck2 deletion.
A scenario in which a protein kinase is under the control of a stress-activated MAPK is not unique. This is the case, for example, for the p38 pathway in mammals. The p38 pathway is the signal transduction pathway homologous to the HOG pathway. Both pathways are activated by osmotic stress, and not only do they have very similar components, but also the mammalian proteins can functionally complement the osmosensitive phenotype observed with the corresponding yeast mutants. This is the case for the p38 MAPK, which complements osmosensitivity of a hog1Δ strain (15), and the human MTK1 MAPKKK, which complements an ssk2Δ ssk22Δ deficient strain (35). Activation of the p38 pathway results in induction of a set of response genes by direct phosphorylation of several transcription factors by the MAPK (i.e., CHOP, MEF2C, ATF-2, and ELK-1) (7, 36). However, some of the responses induced by the p38 pathway are mediated by kinases lying downstream of and directly regulated by the p38 MAPK. These include kinases such as MNK, MAPKAP-K2/3, PRAK, and MSK (7, 36). Activation of those kinases results in a number of responses such as regulation of transcription factors (i.e., CREB) and essential proteins such as HSP27 and eIF4e (36). Those proteins mediate a subset of stress responses of the p38 MAPK. A similar scenario could be imagined for yeast, where Hog1 would be regulating transcription factors and kinases (i.e., Rck2) to complete the whole network of the osmotic stress-induced responses.
In summary, Rck2 protein kinase is a mediator of the osmotic stress signal transduction pathway, which is under the direct control of the MAPK Hog1. Discovery of other proteins under the control of Hog1 and Rck2 will result in a better understanding of osmotic stress responses in both yeast and mammalian cells.
ACKNOWLEDGMENTS
We thank Despina Alexandraki and Jean-Claude Jauniaux for valuable advice regarding two-hybrid technology and Mireia Zaguirre and Anna Vilalta for their technical assistance.
This work was supported by grants from the Swedish Cancer Fund (2163-B97-08XAC) and the Swedish Radiation Protection Institute (1092.98) and by grants from AstraZeneca to P.S., grants PB95-0663 and PB98-0565-C4-02 from the Dirección General de Investigación Científica y Técnica (Ministry of Education, Spain) to J.A., grant GM50909 from the National Institutes of Health to H.S., and grant PM99-0028 from the Dirección General de Investigación Científica y Técnica (Ministry of Education, Spain) to F.P. F.P. was the recipient of a postdoctoral research contract from the MEC, Spain.
REFERENCES
- 1.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel P L, Chien C, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 3.Bellí G, Garí E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendixen C, Gangloff S, Rothstein R. A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 1994;22:1778–1779. doi: 10.1093/nar/22.9.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boguslawski G. PBS2, a yeast gene encoding a putative protein kinase, interacts with the RAS2 pathway and affects osmotic sensitivity of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2425–2432. doi: 10.1099/00221287-138-11-2425. [DOI] [PubMed] [Google Scholar]
- 6.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 7.Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–361. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- 8.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 9.Dahlkvist A, Kanter-Smoler G, Sunnerhagen P. The RCK1 and RCK2 protein kinase genes from Saccharomyces cerevisiae suppress cell cycle checkpoint mutations in Schizosaccharomyces pombe. Mol Gen Genet. 1995;246:316–326. doi: 10.1007/BF00288604. [DOI] [PubMed] [Google Scholar]
- 10.Dahlkvist A, Sunnerhagen P. Two novel deduced serine/threonine protein kinases from Saccharomyces cerevisiae. Gene. 1994;139:27–33. doi: 10.1016/0378-1119(94)90519-3. [DOI] [PubMed] [Google Scholar]
- 11.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 12.Foreman P K, Davis R W. Cloning vectors for the synthesis of epitope-tagged, truncated and chimeric proteins in Saccharomyces cerevisiae. Gene. 1994;144:63–68. doi: 10.1016/0378-1119(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 13.Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Lee J-D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 16.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 17.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Elledge S J, Peterson C A, Bales E S, Legerski R J. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 20.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 21.Melcher M L, Thorner J. Identification and characterization of the CLK1 gene product, a novel CaM kinase-like protein kinase from the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271:29958–29968. doi: 10.1074/jbc.271.47.29958. [DOI] [PubMed] [Google Scholar]
- 22.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 23.Nehlin J O, Carlberg M, Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992;20:5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 25.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 26.Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- 28.Posas F, Witten E A, Saito H. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 30.Proft M, Serrano R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stokoe D, Campbell D G, Nakielny S, Hidaka H, Leevers S J, Marshall C, Cohen P. MAPKAP kinase-2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 1992;11:3985–3994. doi: 10.1002/j.1460-2075.1992.tb05492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Sturgill T W, Ray L B, Erikson E, Maller J L. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 35.Takekawa M, Posas F, Saito H. A human homolog of the yeast Ssk2/Ssk22 MAP kinase kinase kinases, MTK1, mediates stress-induced activation of the p38 and JNK pathways. EMBO J. 1997;16:4973–4982. doi: 10.1093/emboj/16.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson S, Mahadevan L C, Clayton A L. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin Cell Dev Biol. 1999;10:205–214. doi: 10.1006/scdb.1999.0302. [DOI] [PubMed] [Google Scholar]
- 37.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 38.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 39.Waskiewicz A J, Cooper J A. Mitogen and stress response pathways MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 40.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan X L, Deschenes R J, Guan K L. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]