Abstract
In the current study, Chryseobacterium cucumeris strain MW-6 isolated from Arabian seawater exhibited broad-spectrum antibacterial activity against indicator bacterial pathogens. The partially extracted antibacterial metabolites with ethyl acetate revealed promising activity against Escherichia coli, Pseudomonas aeruginosa, Salmonella typhimurium, Listeria monocytogenes, and Staphylococcus aureus. The minimum inhibitory concentrations (MICs) were determined against indicator strains that ranged from 65–90 µg/ml. The genome size of C. cucumeris MW-6 is 4.81 Mbs containing 4227 coding DNA sequences, 74 tRNAs, 3 rRNAs, and 3 ncRNAs genes with 36.90% GC contents. The genome harbors nine putative biosynthetic gene clusters (BGCs) involved in the biosynthesis of lanthipeptide, NRPS-like, RiPPs-like, terpene, microviridin, T1PKS (hg1E-KS), resorcinol, and siderophore. Additionally, the strain encodes genes for sodium/proton antiporter, glutathione, superoxide dismutase, and cold shock protein to survive under stress conditions such as osmotic, oxidative, and cold shock. These putative BGCs and stress-related genes can be associated with in-vitro antibacterial activities and adaptation of this strain to the marine environment.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-03039-5.
Keywords: Chryseobacterium cucumeris, Antibacterial activity, Biosynthetic gene clusters, Adaptation, Marine water
The inappropriate use of antibiotics has driven a rapid emergence of multi-drug resistance (MDR) in pathogenic bacteria which is related to worldwide public health problems (Wang et al. 2020). The prevalence of MDR bacteria increases morbidity, mortality, healthcare costs and extends the duration of hospitalization for patients suffering from nosocomial infections (Medina and Pieper 2016). Therefore, searching for novel and efficient therapeutic agents to treat MDR infections is essential. Marine environment provides a great source of microbial diversity with great potential for the biosynthesis of diverse natural products (Choudhary et al. 2017). Additionally, whole-genome sequence analysis offers faster and cost-efficient system to explore gene clusters associated with the synthesis of bioactive metabolites (Albarano et al. 2020). The genome analysis may also help to understand the ecological function as well as the taxonomic position of bacterial isolates (Goodwin et al. 2017). Hence, genome mining of unexplored marine-derived bacterial species broadens our understanding regarding their cryptic biosynthetic potential and their use in biotechnological and pharmaceutical applications. C. cucumeris is a Gram-negative, rod-shaped, non-motile, and non-spore-forming bacteria isolated from various sources such as green lizard, wastewater, Nile tilapia fish, and plant surface-sterilized roots (Jeong et al. 2017). However, the broad-spectrum antibacterial activity of Chryseobacterium species is not yet reported except C. antibioticum sp. nov. (Dahal et al. 2021).
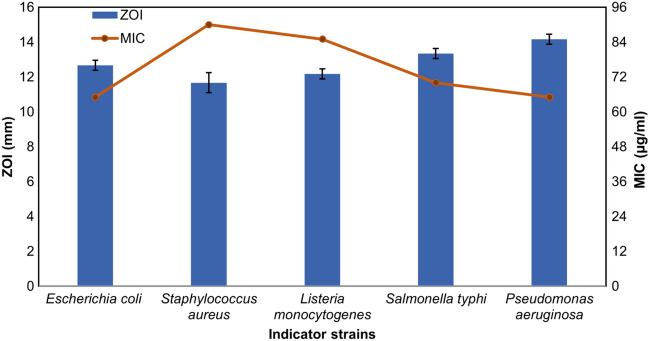
Herein, we first time report a C. cucumeris strain MW-6 from marine environment of the Arabian sea (24°47′17.1″N 67°02′37.5″E) Clifton beach Karachi, Pakistan. The sample was collected from subsurface marine water (10–12 inches) in a 500-ml sterile screw cap bottle, and transported to the laboratory at 4 °C. The temperature, pH, and electrical conductivity of the sample were recorded as 24 °C, 8.1, and 1400 µS/cm, respectively. The sample was serially diluted and 200 µl from each dilution was plated over marine agar 2216 (HiMedia, India) plates and incubated for 48 h at 30 °C. Colonies were purified by repeated subculturing. In total, 23 isolates were isolated and preliminarily screened for antibacterial activities using soft agar overlay method (Iqbal et al. 2021). The strain MW-6 showed promising antibacterial activity against all indicator strains and selected for further study. The secondary screening was performed on Muller-Hinton agar (MHA) (Oxoid, UK) using the agar well diffusion assay (Skariyachan et al. 2014). Briefly, the strain MW-6 was inoculated in marine broth 2216 (HiMedia, India) and incubated in a shaking incubator (120 rpm) for 48 h at 30 °C. Cell-free supernatant was obtained by centrifuging the culture at 4 °C and 15,000 g for 15 min. The cell-free supernatant was extracted using an equal volume (1:1) of ethyl acetate (Thermo Fisher chemical™) and the metabolites produced by strain MW-6 were concentrated in a rotary evaporator at 60 °C until ethyl acetate was completely evaporated. The dry extract was dissolved in phosphate buffer solution (one tablet was dissolved in 100 ml sterile distilled water) (Sigma-Aldrich, P4417) (100 mg/mL) and antagonistic potential was determined against a set of American type culture collection (ATCC) strains. Subsequently, minimum inhibitory concentration (MIC) of the partial extract was determined as per clinical and laboratory standard institute (CLSI) protocols (CLSI 2018). The strain MW-6 demonstrates promising antibacterial activity against Gram-positive and Gram-negative bacterial strains as measured by their zones of inhibitions (ZOI) (Figure S1). The broader ZOI was recorded against Gram-negative strains; Pseudomonas aeruginosa ATCC 27,853, Escherichia coli ATCC 25,922, and Salmonella typhimurium ATCC 14,028, while comparatively less ZOI was observed against Listeria monocytogenes ATCC 35,152 and Staphylococcus aureus ATCC 25,923 (Fig. 1).
Fig. 1.
Antibacterial activity (left-hand Y-axis zones of inhibition) and minimum inhibitory concentration (MIC) (right-hand Y-axis) of C. cucumeris MW-6 culture extract against indicator strains
The MIC values of partially purified metabolites extract against indicator strains are ranging from 65–90 µg/ml (Fig. 1). The lowest concentration (65 µg/ml) was recorded against P. aeruginosa and E. coli while the highest (90 µg/ml) was observed against S. aureus. These results are in agreement with the earlier study where partially purified metabolites from Actinomycetes kocuria inhibits the growth of P. aeruginosa and E. coli at concentrations of 40 and 60 µg/ml, respectively (Kumar and Jadeja 2018). The complete bioactive metabolites synthesis capabilities of a bacteria could be explored by the BGCs carries by its genome (diCenzo and Finan 2017). Next-generation sequencing technology and robust genome mining tools offer new prospects for discovering the biosynthetic potential of bioactive metabolites producing bacteria (Wright 2019). Therefore, to get a deeper insight into biosynthetic potential, the genome of C. cucumeris MW-6 was sequenced.
The genomic DNA of strain MW-6 was extracted from fresh marine broth (HiMedia, India) culture using a PureLink® genomic DNA extraction kit (Invitrogen). Purity and concentration of the isolated DNA was estimated through Nanodrop spectrophotometer (Titertek Berthold, Germany). Genomic DNA library was prepared using Nextera XT library prep kit (Illumina Inc., SDCA, USA) and was sequenced using Hiseq Illumina 2500 platform with paired-end reads (Illumina Inc.). The reads were trimmed using trimmomatic v 0.36 (Bolger et al. 2014) and de novo assembly was performed using SPAdes v 3.12 (Bankevich et al. 2012). Genome annotation was performed through National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) v 4.10 (Tatusova et al. 2016).
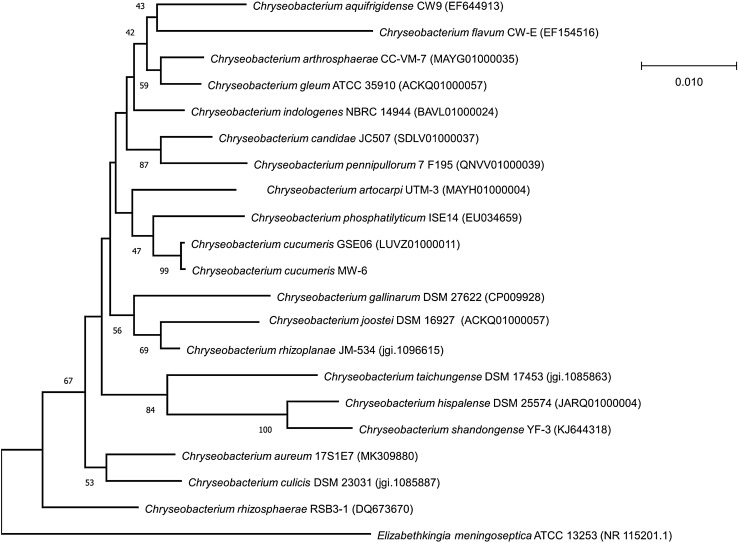
The 16S rRNA gene sequence was retrieved from MW-6 genome and closely related bacterial strains were determined by global alignment using EzBioCloud (https://www.ezbiocloud.net/) online server. The 16S rRNA genes of closely related strains were retrieved and phylogenetic tree was constructed using MEGA-X package (Kumar et al. 2018).
The BLASTn results of the 16S rRNA gene sequence of strain MW-6 exhibit maximum similarity (99.93%) with Chryseobacterium cucumeris strain GSE06T. The 16S rRNA gene similarity of strain MW-6 with C. cucumeris GSE06 is higher than the cutoff value (98.65%) thus indicating strong affiliation with the type strain C. cucumeris (Kim et al. 2014). Phylogenetic tree also showed that the strain MW-6 is closely related to C. cucumeris GSE06T (Fig. 2).
Fig. 2.
The phylogenetic tree was constructed using MEGA-X. The evolutionary history was inferred using the Neighbor-Joining method and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Elizabethkingia meningoseptica ATCC 13253 was used as an outgroup
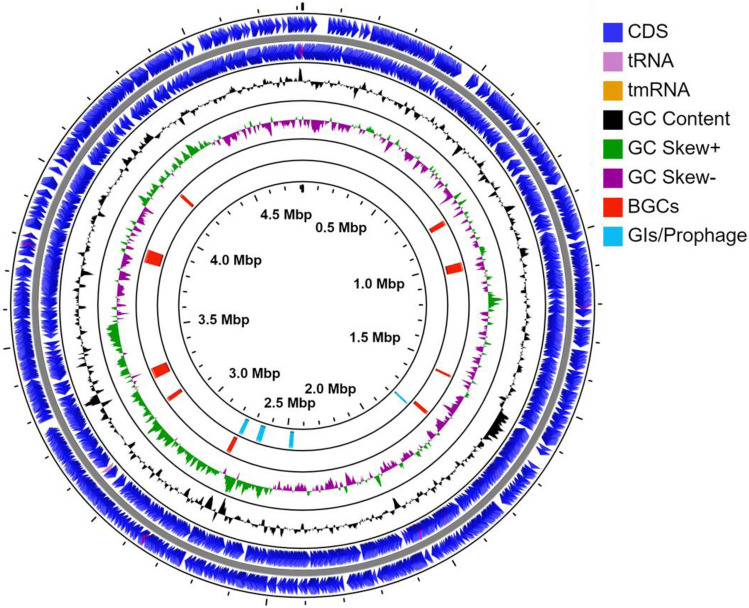
The draft genome of strain MW-6 is 4,813,464 bp in size with 36.90% G + C contents and contains 4,227 coding sequences, 74 tRNAs, 3 rRNAs, 3 ncRNA genes, and 17 pseudogenes (Table 1). The whole-genome sequence and prominent features of strain MW-6 are presented in a circular map using a CGveiw (http://cgview.ca/) server (Fig. 3).
Table 1.
Features and minimum information about a genome sequence (MIGS) of C. cucumeris strain MW-6
| MIGS data | |
|---|---|
| Investigation type | Bacteria |
| BioProject accession | PRJNA594265 |
| BioSample accession | SAMN17526039 |
| Geographical location | Arabian sea, Pakistan |
| Longitude and latitude | 24°47′17.1″N 67°02′37.5″E |
| Sampling date | April 2018 |
| Sequencing method | HiSeq Illumina 2500 |
| Assembly | SPAdes v 3.12 |
| Coverage | 30.0× |
| Finishing strategy | High-quality draft genome |
| Genome features | |
| Genome size | 4,813,464 bps |
| G + C content | 36.90% |
| CDS | 4227 |
| Pseudogenes | 17 |
| tRNAs | 74 |
| rRNAs | 3 |
| ncRNAs | 3 |
| Accession number | CP068760.1 |
Fig. 3.
Circular map of C. cucumeris strain MW-6 genome. The outermost two circles represent coding DNA sequence (CDS) on forward and reverse strand, 3rd circle (from outer to inner) represents GC contents, 4th circle represents GC skew deviation from the average, 5th circle showing the position of biosynthetic gene clusters (BGCs) and the innermost circle shows the genomic position of GIs/prophage in MW-6 genome
The bacterial strains with antibacterial properties compete with other microbes by secreting bioactive secondary metabolites such as siderophore and lanthipeptides (Repka et al. 2017; Chen et al. 2019). In the strain MW-6 genome, nine secondary metabolites BGCs were identified using AntiSMASH (https://antismash.secondarymetabolites.org/) with enabling all extended parameters. These BGCs are involved in the biosynthesis of lanthipeptide, NRPS-like, RiPPs-like, terpene, microviridin, T1PKS (hg1E-KS), resorcinol (arylpolyene), and siderophore (Figure S2). The biosynthetic cluster of lanthipeptide, NRPS-like, RiPP, terpene, microviridin, and T1PKS showed no similarity with known clusters; while, resorcinol, terpene, and siderophore showed 75, 28, and 20% similarity with known clusters, respectively (Table 2). The broad-spectrum antimicrobial activities of lanthipeptides, NRPs, and RiPPs were reported earlier (Zhao and van der Donk 2016; Agrawal et al. 2017; Zhang et al. 2018). Biosynthesis of resorcinol remains unexplored, however, there is an evidence showing that it is involved in antioxidative activities (Schöner et al. 2016). The BGC in MW-6 genome coding resorcinol exhibits 75% similarity with flexirubin (BGC0000838) of Flavobacterium johnsoniae (McBride et al. 2009). Flexirubins are unique bacterial pigments that can be applied as a therapeutic agent for gastric ulcers, skin diseases, and eczema (Venkatesan and Kim 2013). To confirm flexirubin synthesis, in vitro flexirubin assay was performed directly on the MW-6 colonies placed on a glass slide and 200 µl of 20% potassium hydroxide (KOH) was poured down on the colonies. The color shift from yellow (colony color) to reddish was observed (Figure S3) which indicates flexirubin production (Venil et al. 2014). The BGC coding terpene and siderophore exhibited 28 and 20% gene similarity with carotenoid (BGC0000650) of Algoriphagus sp. KK10202C and putrebactin/avaroferrin (BGC0001870) of Xenorhabdus budapestensis, respectively. Terpenes are the largest class of natural products with diverse biological activities such as anticancer and antimalarial activities (Helfrich et al. 2019). Siderophores are produced by several bacterial species to gain essential Fe (III) from the environment in response to low intracellular iron which is pertinent in understanding the metal ligand-speciation in seawater (Codd 2008; Schalk et al. 2011). Siderophores are also associated with plant-growth-promoting and antibacterial activities. Therefore, chrome azurol sulphonate (CAS) assay was performed to assess siderophore production by strain MW-6. The CAS assay was conducted as described earlier with little modification (Alexander and Zuberer 1991). The cell-free supernatant of strain MW-6 culture was mixed with CAS reagent and after 15-min optical density (O.D) was taken at 630 nm. The supernatant of uninoculated marine broth mixed with CAS was used as a negative control. The higher O.D (0.21 ± 0.015) for MW-6 supernatant was observed which indicates siderophore production as compared to the negative control (0.004 ± 0.001). The experiment was performed in triplicates.
Table 2.
Biosynthetic gene clusters (BGCs) of secondary metabolites identified in C. cucumeris strain MW-6 genome
| S. No | Type | Location from–to | Size (bp) | Most similar cluster | Similarity | Compound MiBG BGC number |
|---|---|---|---|---|---|---|
| 1 | Lanthipeptide-class-i | 787,508–811,826 | 24,318 | – | – | – |
| 2 | NRPS-like | 999,485–1,042,592 | 43,107 | – | – | – |
| 3 | RiPP-like | 1,541,090–1,552,175 | 11,085 | – | – | – |
| 4 | Terpene | 1,741,184–1,758,768 | 17,584 | Terpene | 28% | BGC0000650 |
| 5 | Terpene | 2,755,908–2,776,921 | 21,013 | – | – | – |
| 6 | Microviridin | 3,132,911–3,153,421 | 20,510 | – | – | – |
| 7 | T1PKS (hglE-KS) | 3,257,557–3,308,852 | 51,295 | – | – | – |
| 8 | Resorcinol (arylpolyene) | 3,813,772–3,876,807 | 63,035 | Flexirubin | 75% | BGC0000838 |
| 9 | Siderophore | 4,166,578–4,182,113 | 15,535 | Putrebactin/avaroferrin | 20% | BGC0001870 |
IslandVeiwer 4 (http://www.pathogenomics.sfu.ca/islandviewer/) and PHASTER (https://phaster.ca/) were used to identify Genomic Islands (GIs) and prophage regions, respectively in MW-6 genome. A total of three GIs and one prophage region were identified (Fig. 3). The GI one is 24 kb in size and comprises several biotechnologically important genes such as DEAD/DEAH cox helicase and cytochrome C oxidase coding genes. While genomic island 2 (31 kb) and 3 (23 kb) include several biotechnological important genes encoding for DNA ligase D and conjugative transposons (traN, traM, traK, traD, and traJ), peptidoglycan binding proteins (lysM) and transposase, etc. The incomplete prophage region is 11 kb in size with a GC content of 37.28%. This prophage region comprises 16 coding genes of which 12 are phage hit proteins. GIs play important role in genome plasticity, environmental adaptation, and speciation (Juhas et al. 2009). Recently li et al. analyzed Enterococcus genomes and found several GIs associated with environmental adaptation and had numerous mobile genetic elements such as prophages, transposons, and integrons (Li and Wang 2021).
Proteome comparison of cluster orthologous groups (COGs) with closely related strains was performed using the web-based tool orthoVenn2 (Xu et al. 2019). The comparative proteome of MW-6 and closely related strains makes 4932 orthologous clusters (at least contain 2 strains) and 1462 singleton clusters. The strain MW-6 genome contains 3932 orthologous clusters and 255 singleton clusters (Figure S4). Rapid Annotation Subsystem Technology (RAST) server was employed to predict mapping of genes to subsystem and metabolic reconstruction (Aziz et al. 2008). The RAST subsystem analysis revealed 80% subsystem coverage with the highest feature counts 290 for amino acids and derivatives, followed by 141 for carbohydrate, 129 for protein metabolism, and 127 for cofactors, vitamins, prosthetic groups, and pigments (Figure S5). Additionally, the MW-6 genome contains 53 genes associated with stress response such as sodium/proton antiporter, glutathione, and cold shock protein-coding genes (Table S1), which were earlier reported to be associated with osmotic, oxidative, and cold shock stress, respectively (Kempf and Bremer 1998; Mishra and Imlay 2012; Haslbeck and Vierling 2015). The highest feature in the amino acids and derivatives (290) and carbohydrates (141) sub-system category indicates the strain MW-6 can utilize various types of carbon and nitrogen sources. The strain MW-6 genome also harbors genes such as dnaKJ, hemW, htpG and groEL encoding for various chaperon proteins which are capable to refold the misfolded polypeptides under stress conditions. The chaperonin groEL was recently reported to be a versatile tool in modern applied biotechnology (O’Neil et al. 2018).
In conclusion, the presence of low similarity and unique BGCs in C. cucumeris strain MW-6 genome suggests the synthesis of new bioactive compounds which may find their application in healthcare biotechnology to combat growing antibiotics resistance. Furthermore, the genes associated with siderophore and stress can be employed in agriculture biotechnology by considering a heterologous expression of these genes in the development of stress-resistant crops.
Genome sequence accession numbers
The whole-genome sequence of C. cucumeris strain MW-6 has been submitted to NCBI GenBank under accession number CP068760.1 and BioProject accession number PRJNA594265.
Supplementary Information
Below is the link to the electronic supplementary material.
Author’s contributions
SI and HAJ conceptualize and design the study, SI and MSV performed the experiments, SI and HAJ wrote the manuscript and analyze the data.
Funding
The current study is a part of SI PhD research project which is supported by National University of Sciences and Technology (NUST) student research funds.
Declarations
Conflict of interest
All authors declared that they have no conflict of interest.
References
- Agrawal S, Acharya D, Adholeya A, et al. Nonribosomal Peptides from marine microbes and their antimicrobial and anticancer potential. Front Pharmacol. 2017;8:828. doi: 10.3389/fphar.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarano L, Esposito R, Ruocco N, Costantini M. Genome mining as new challenge in natural products discovery. Mar Drugs. 2020;18:199. doi: 10.3390/md18040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DB, Zuberer DA. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol Fertil Soils. 1991;12:39–45. doi: 10.1007/BF00369386. [DOI] [Google Scholar]
- Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Guo Y, Lu Y, et al. Chemistry and biology of siderophores from marine microbes. Mar Drugs. 2019;17:562. doi: 10.3390/md17100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary A, Naughton LM, Montánchez I, et al. Current status and future prospects of Marine Natural Products (MNPs) as antimicrobials. Mar Drugs. 2017;15:272. doi: 10.3390/md15090272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI C and LSI (2018) M07: methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th Edn. Clinical and Laboratory Standard Institute 950 west valley road, suit 2500 Wayne, 19087 USA
- Codd R. Traversing the coordination chemistry and chemical biology of hydroxamic acids. Coord Chem Rev. 2008;252:1387–1408. doi: 10.1016/j.ccr.2007.08.001. [DOI] [Google Scholar]
- Dahal RH, Chaudhary DK, Kim D-U, et al. Chryseobacterium antibioticum sp. nov. with antimicrobial activity against Gram-negative bacteria, isolated from Arctic soil. J Antibiot (tokyo) 2021;74:115–123. doi: 10.1038/s41429-020-00367-1. [DOI] [PubMed] [Google Scholar]
- diCenzo GC, Finan TM. The divided bacterial genome: structure, function, and evolution. Microbiol Mol Biol Rev. 2017;81:e00019. doi: 10.1128/MMBR.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin KD, Thompson LR, Duarte B, et al. DNA sequencing as a tool to monitor marine ecological status. Front Mar Sci. 2017;4:107. doi: 10.3389/fmars.2017.00107. [DOI] [Google Scholar]
- Haslbeck M, Vierling E. a first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol. 2015;427:1537–1548. doi: 10.1016/j.jmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich EJN, Lin GM, Voigt CA, Clardy J. Bacterial terpene biosynthesis: challenges and opportunities for pathway engineering. Beilstein J Org Chem. 2019;15:2889–2906. doi: 10.3762/bjoc.15.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S, Vollmers J, Janjua HA (2021) Genome mining and comparative genome analysis revealed niche-specific genome expansion in antibacterial bacillus pumilus strain SF-4. Genes 12 [DOI] [PMC free article] [PubMed]
- Jeong JJ, Lee DW, Park B, et al. Chryseobacterium cucumeris sp. nov., an endophyte isolated from cucumber (Cucumis sativus L.) root, and emended description of Chryseobacterium arthrosphaerae. Int J Syst Evol Microbiol. 2017;67:610–616. doi: 10.1099/ijsem.0.001670. [DOI] [PubMed] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M, et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RR, Jadeja VJ (2018) Characterization and partial purification of an antibacterial agent from halophilic actinomycetes Kocuria sp. strain rsk4. Bioimpacts. 8: 253-261. 10.15171/bi.2018.28 [DOI] [PMC free article] [PubMed]
- Li W, Wang A. Genomic islands mediate environmental adaptation and the spread of antibiotic resistance in multiresistant Enterococci—evidence from genomic sequences. BMC Microbiol. 2021;21:55. doi: 10.1186/s12866-021-02114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Xie G, Martens EC, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina E, Pieper DH. Tackling threats and future problems of multidrug-resistant bacteria BT—how to overcome the antibiotic crisis : facts, challenges, technologies and future perspectives. In: Dersch P, editor. Stadler M. Cham: Springer International Publishing; 2016. pp. 3–33. [Google Scholar]
- Mishra S, Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 2012;525:145–160. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil PT, Machen AJ, Deatherage BC, et al. The Chaperonin GroEL: a versatile tool for applied biotechnology platforms. Front Mol Biosci. 2018;5:46. doi: 10.3389/fmolb.2018.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repka LM, Chekan JR, Nair SK, van der Donk WA. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev. 2017;117:5457–5520. doi: 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 2011;13:2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- Schöner TA, Gassel S, Osawa A, et al. Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. ChemBioChem. 2016;17:247–253. doi: 10.1002/cbic.201500474. [DOI] [PubMed] [Google Scholar]
- Skariyachan S, Rao AG, Patil MR, et al. Antimicrobial potential of metabolites extracted from bacterial symbionts associated with marine sponges in coastal area of Gulf of Mannar Biosphere, India. Lett Appl Microbiol. 2014;58:231–241. doi: 10.1111/lam.12178. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Dicuccio M, Badretdin A, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venil CK, Zakaria ZA, Usha R, Ahmad WA. Isolation and characterization of flexirubin type pigment from Chryseobacterium sp UTM-3T. Biocatal Agric Biotechnol. 2014;3:103–107. doi: 10.1016/j.bcab.2014.02.006. [DOI] [Google Scholar]
- Venkatesan J, Kim SK. Marine biomaterials: characterization, isolation and applications. CRC Press Taylor Fr; 2013. pp. 1195–1215. [Google Scholar]
- Wang H-C, Liao C-C, Chu S-M, et al. Impacts of multidrug-resistant pathogens and inappropriate initial antibiotic therapy on the outcomes of neonates with ventilator-associated pneumonia. Antibiotics. 2020;9:760. doi: 10.3390/antibiotics9110760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GD. Unlocking the potential of natural products in drug discovery. Microb Biotechnol. 2019;12:55–57. doi: 10.1111/1751-7915.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Dong Z, Fang L, et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen M, Bruner SD, Ding Y. Heterologous production of microbial ribosomally synthesized and post-translationally modified peptides. Front Microbiol. 2018;9:1801. doi: 10.3389/fmicb.2018.01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, van der Donk WA. Structural characterization and bioactivity analysis of the two-component lantibiotic flv system from a ruminant bacterium. Cell Chem Biol. 2016;23:246–256. doi: 10.1016/j.chembiol.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.