Abstract
It has been suggested that specific molecular features could characterize the protease-resistant prion protein (PrP res) detected in animal species as well as in humans infected by the infectious agent strain that causes bovine spongiform encephalopathy (BSE). Studies of glycoform patterns in such diseases in French cattle and cheetahs, as well as in mice infected by isolates from both species, revealed this characteristic molecular signature. Similar studies of 42 French isolates of natural scrapie, from 21 different flocks in different regions of France, however, showed levels of the three glycoforms comparable to those found in BSE-linked diseases. Moreover, the apparent molecular size of the unglycosylated form was also indistinguishable among all different sheep isolates, as well as isolates from BSE in cattle. Overall results suggest that scrapie cases with features similar to those of BSE could be found more frequently in sheep than previously described.
The bovine spongiform encephalopathy (BSE) agent is assumed to have caused prion diseases not only in cattle but also in a variety of other species such as domestic cats and some exotic felines and ruminants (3). Strong evidence indicates that new-variant Creutzfeldt-Jakob disease is due to the same agent as well, which could also have infected other species in field conditions, such as sheep or goats (3). The ultimate evidence that infectious agents from different isolates are identical eventually requires transmission of the disease to mice and characterization of the lesion profiles in the brain. It has, however, been found that the qualitative and quantitative analysis of the different glycoforms of the protease-resistant prion protein (PrP res) detected by Western blotting showed a consistent and unique pattern in BSE-linked diseases in experimentally infected macaques or mice and in naturally infected domestic cats, as well as in humans developing new-variant Creutzfeldt-Jakob disease (4, 8). Such results argue for a link between molecular features of PrP res and strain variation.
Extraction and glycoform analysis of PrP res.
We studied PrP res in cattle BSE and in a cheetah with spongiform encephalopathy (SE) and in mice (C57BL/6) infected by these SE isolates, as well as in sheep with natural scrapie. PrP res was obtained by dissociation of 0.5 g of brain tissue in 4.5 ml of 5% glucose in distilled water with an Ultra-Turax grinder (IKA T25) and complete homogenization by forcing the brain suspension through a 0.4-mm-diameter needle. After addition of N-lauroyl sarcosyl (Sigma) (final concentration, 10% [vol/vol]), a 1,200-μl volume was incubated with proteinase K (Boehringer) (10 μg/100 mg of brain tissue) for 1 h at 37°C. Samples were then centrifuged at 240,000 × g for 4 h on a 10% sucrose cushion, in a Beckman TL100 ultracentrifuge. Pellets were resuspended and boiled for 5 min in denaturing buffer (4% sodium dodecyl sulfate, 2% β-mercaptoethanol, 192 mM glycine, 25 mM Tris, 5% sucrose). Appropriate dilutions of samples, between 0.5 and 5 mg of brain tissue equivalent per lane, were run in sodium dodecyl sulfate–15% polyacrylamide gels and electroblotted to nitrocellulose membranes in transfer buffer (25 mM Tris, 192 mM glycine, 10% isopropanol) at 400 mA (constant) for 1 h. The membranes were blocked for 1 h with 5% nonfat dried milk in phosphate-buffered saline–Tween 20 (0.1%) (PBST). After two washes in PBST, membranes were incubated (1 h at room temperature) with either RS1 or RB1 rabbit antisera (1/2,500 in PBST), raised against synthetic ovine (SHSQWNKPSKPKTNMK) (amino acids 98 to 113) (12) or bovine (THGQWNKPSKPKTNMK) PrP peptides, or with monoclonal antibody 3F4 (Pierce) (1/1,000 in PBST) directed to the amino acid 109 to 112 sequence of hamster PrP (2). After three washes in PBST, the membranes were incubated (30 min at room temperature) with peroxidase-labelled conjugates against rabbit or mouse immunoglobulins (Clinisciences) (1/2,500 in PBST). After three washes in PBST, bound antibodies were then detected with the ECL chemiluminescent substrate (Amersham). Chemiluminescent signals corresponding to the three glycoforms of the protein were quantified by using a Fluor S-Multimager (Bio-Rad) analysis system. Results were expressed as mean percentages of the total signal for the three glycoforms (high glycosylated [H], low glycosylated [L], and unglycosylated [U] forms), obtained from at least three separate gel runs for each individual. In order to assess if the variability of glycoform patterns between the individuals (animals) could be interpreted with regard to the reproducibility of the measurements, we analyzed the three runs for the 54 animals with a model 1 analysis of variance (variance analysis with one controlled factor, the variable individual with 54 levels [Proc GLM; SAS Institute, Inc., software]). By this analysis, the individuals were highly different (P = 0.001), thus allowing interpretation of the variability between individuals.
Glycoform ratios of PrP res in BSE cattle and BSE isolate-infected mice.
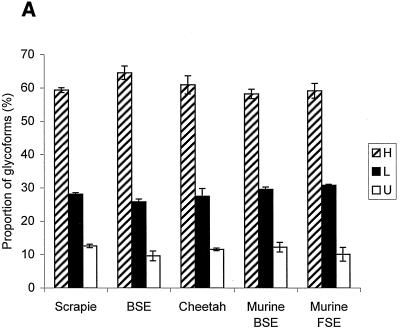
PrP res from mice that developed the disease after infection with an isolate from French BSE cattle, detected by RS1 antibody, presented comparable ratios of glycoforms in all animals (H, minimum = 56.2 and maximum = 61.0; L, minimum = 28.8 and maximum = 31.0; U, minimum = 10.1 and maximum = 15.0) (Fig. 1), similar to those previously reported for mice inoculated with isolates from British cattle (4). In the same way, PrP res from cattle was identified by using rabbit antiserum RB1, since RS1 failed to label bovine PrP, and electrophoresis also showed comparable patterns in the four animals studied from four different outbreaks with very high levels of diglycosylated protein (H, minimum = 60.4 and maximum = 68.7; L, minimum = 24.6 and maximum = 28.4; U, minimum = 6.7 and maximum = 12.9) (Fig. 1). These results are consistent with a common origin for the BSE cases.
FIG. 1.
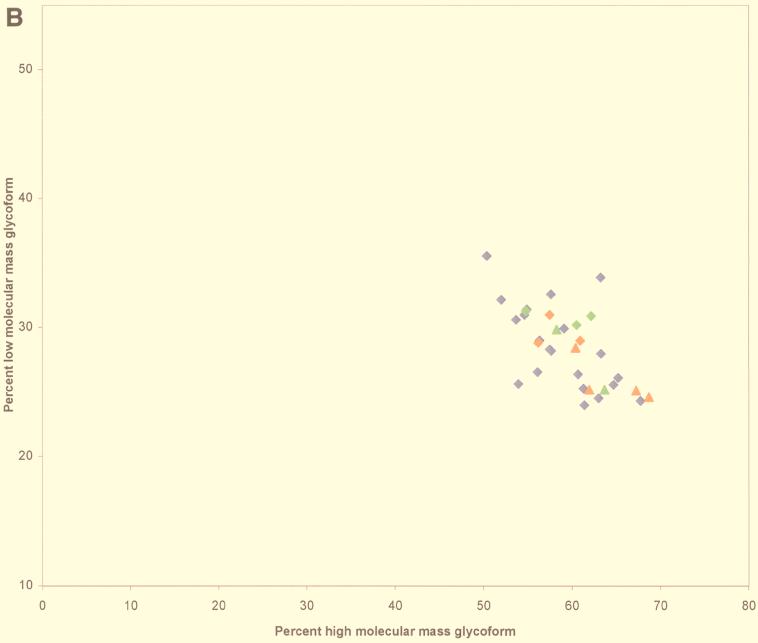
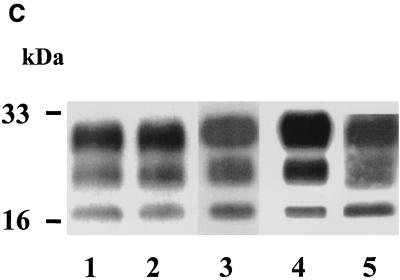
(A) Proportions of PrP res glycoforms in transmissible SE from sheep (n = 42), cattle (n = 4), and cheetahs (n = 2) and from mice infected with BSE (n = 3) or transmissible SE from cheetahs (n = 3). Percentages are indicated as means ± standard errors of the means of the PrP res bands, H, L, and U. FSE, feline SE. (B) Scattergraph of proportions of H and L glycoforms of PrP res. Red diamonds, cattle BSE isolate-infected mice; green diamonds, cheetah SE isolate-infected mice; blue diamonds, scrapie sheep; red triangles, cattle BSE; green triangles, cheetah SE. (C) Western blot of PrP res. Lane 1, cattle BSE isolate-infected mice; lane 2, cheetah SE isolate-infected mice; lane 3, scrapie sheep; lane 4, cattle BSE; lane 5, cheetah SE.
Molecular analysis of PrP res in cheetahs with SEs.
PrP res from two cheetahs imported from Great Britain that developed SEs in France was examined (1). Such cases are assumed to be caused by the BSE agent, as in other captive exotic felines or ruminants and in domestic cats (3). Western blot studies with the 3F4 monoclonal antibody showed for both cheetahs a pattern identical to that previously described for a cat with feline SE (H, minimum = 58.2 and maximum = 63.7; L, minimum = 25.2 and maximum = 29.8; U, minimum = 11.2 and maximum = 12.0) (Fig. 1) (4). A similar pattern was also found, with RS1 antibody, for mice inoculated with one cheetah isolate (H, minimum = 54.8 and maximum = 62.1; L, minimum = 30.2 and maximum = 31.2; U, minimum = 7.0 and maximum = 14.0) (Fig. 1).
Similar signatures of PrP res in natural scrapie and in BSE.
Scrapie in sheep may be the origin of BSE, but the BSE agent could also have secondarily infected sheep through feeding with contaminated meat and bone meal. So, we studied PrP res, with RS1 antibody, from 42 natural sheep scrapie isolates found in 21 French flocks (1 sheep each in 18 flocks and 3, 7, and 14 in the others). PrP res was easily detected in all 42 cases, allowing the evaluation of the proportions of each glycoform of the protein and the size of the U form. Variable ratios of glycoforms were found (H, minimum = 49.7 and maximum = 69.7; L, minimum = 22.5 and maximum = 35.5; U, minimum = 2.9 and maximum = 20.5); however, these ratios were not significantly different for the three isoforms between natural scrapie sheep and BSE-linked diseases P = 0.11, 0.38, and 0.17 for H, L, and U values, respectively) (Fig. 1). These results demonstrate that, in our series, natural scrapie cases cannot be distinguished from BSE by means of studies of the glycoform ratios.
Relevance of PrP res extraction methods to the glycoform analysis.
The purification method used in this study, involving PrP harvesting by ultracentrifugation of the sample, differs from the direct extraction of PrP used in initial molecular typing studies of the human protein (4). However, this method has since been used for the characterization of Creutzfeldt-Jakob disease in humans and similarly was able to recognize the four described molecular types of the protein (5). In our studies, PrP res obtained by this purification method from BSE-infected mice and from cheetahs also showed profiles identical to those described for brain homogenates of BSE-infected mice and cats, respectively (4). Furthermore, whereas the three glycoforms of PrP res could not be detected in a number of scrapie cases following direct identification of the protein in proteinase K-treated brain homogenates (9, 10), the purification method used in this study allowed the consistent detection of PrP res and the analysis of the three glycoforms in each case.
Studies of apparent molecular masses of the unglycosylated PrP res.
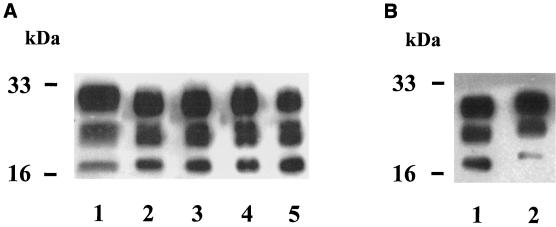
Whereas PrP res from mice infected by a passaged scrapie strain (C506 M3) (11) or by BSE isolates from cattle clearly had distinguishable sizes of the unglycosylated form (Fig. 2B), we did not detect significant differences among PrP res proteins from sheep, all of them displaying an unglycosylated form with a size similar to that found in cattle BSE (Fig. 2A). Previous studies of British scrapie cases claimed that natural scrapie cases could be distinguished from BSE-inoculated sheep, by either a higher (9, 10) or in some cases a lower (10) size of the unglycosylated form. In these last studies, electrophoretic profiles were, however, found to be similar between BSE-infected and experimentally CH1641-infected sheep, leading to the hypothesis that BSE could have originated from a scrapie strain such as CH1641 (10). Our results now show that natural scrapie cases with features indistinguishable from those of BSE can be found more frequently under field conditions than previously recognized.
FIG. 2.
Western blot of PrP res. (A) Lane 1, cattle BSE; lanes 2 to 5, natural sheep scrapie cases. (B) Lane 1, cattle BSE isolate-infected mouse; lane 2, scrapie-infected mouse (C506 M3 strain).
Relationship of PrP res molecular analysis with geographic distribution, breeds, and PrP genotypes in natural sheep scrapie.
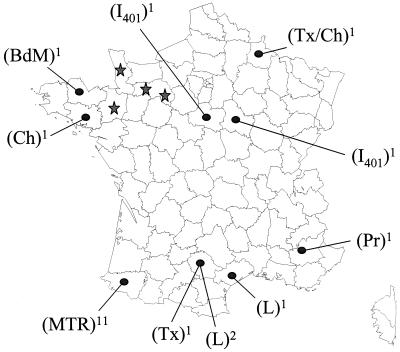
Our series of scrapie cases includes 42 individual sheep from 21 different outbreaks of the disease, in different regions of France and in different breeds (Fig. 3). These scrapie cases were identified in these different herds between October 1996 and August 1997 by the French National Surveillance Scheme. Eleven outbreaks originated from the Pyrénées Atlantiques department (southwest France, Manech tête rousse breed), where 70% of the scrapie outbreaks until now have been identified. The other 10 outbreaks were distributed throughout France (eight different departments) in six different breeds (dairy Lacaune, Bleu du Maine, Préalpes, Texel, Charollais, and INRA 401). These data show that the homogeneous glycoform pattern observed in this study is probably not the result of a possible transmission by a single strain between nearby flocks.
FIG. 3.
Geographic distribution (spots), sheep breeds, and numbers of outbreaks studied (superscript numbers) of natural scrapie (n = 21). Comparative locations of BSE cases in cattle (n = 4) are indicated (stars). Sheep breeds were Manech tête rousse (MTR), Lacaune (L), Bleu du Maine (BdM), Préalpes (Pr), Texel (Tx), Charollais (Ch), and INRA 401 (I401).
Reported studies of experimentally BSE-infected sheep detected PrP res in only two of five sheep, both with PrP genotype AA136 RR154 RQ171, from which the disease has not been experimentally transmitted to mice (10). To the best of our knowledge, molecular analysis of PrP res in BSE-inoculated sheep from which the BSE strain could be reisolated after transmission to mice (6, 7) has not been performed yet.
Conclusion.
Finally, whereas the BSE-specific pattern of PrP res in BSE-infected sheep has to be more accurately established, our studies mainly point out that a number of field isolates from sheep scrapie could have patterns indistinguishable from those already known in BSE-linked diseases.
Acknowledgments
We gratefully thank S. Philippe for statistical analysis of the data, as well as L. Maitrias and A. Paoli for their technical help.
This work was partly funded by grants from the European Commission (proposal PL 97 3305).
REFERENCES
- 1.Baron T, Belli P, Madec J-Y, Moutou F, Vitaud C, Savey M. Spongiform encephalopathy in an imported cheetah in France. Vet Rec. 1997;141:270–271. doi: 10.1136/vr.141.11.270. [DOI] [PubMed] [Google Scholar]
- 2.Bolton D, Seligman S, Bablanian G, Windsor D, Scala L J, Kim K S, Chen C J, Kascsak R J, Bendheim P E. Molecular location of a species-specific epitope on the hamster scrapie agent protein. J Virol. 1991;65:3667–3675. doi: 10.1128/jvi.65.7.3667-3675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce M E, Will R G, Ironside J W, McConnell I, Drummond D, Suttle A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock C J. Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J, Sidle K C L, Meads J, Ironside J, Hill A F. Molecular analysis of prion strain variation and the aetiology of 'new variant' CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 5.Deslys J-P, Lasmézas C, Streichenberger N, Hill A, Collinge J, Dormont D, Kopp N. New variant Creutzfeldt-Jakob disease in France. Lancet. 1997;349:30–31. doi: 10.1016/s0140-6736(05)62163-0. [DOI] [PubMed] [Google Scholar]
- 6.Foster J D, Hope J, Fraser H. Transmission of bovine spongiform encephalopathy to sheep and goats. Vet Rec. 1993;133:339–341. doi: 10.1136/vr.133.14.339. [DOI] [PubMed] [Google Scholar]
- 7.Foster J D, Bruce M, McConnell I, Chree A, Fraser H. Detection of BSE infectivity in brain and spleen of experimentally infected sheep. Vet Rec. 1996;138:546–548. doi: 10.1136/vr.138.22.546. [DOI] [PubMed] [Google Scholar]
- 8.Hill A, Desbruslais M, Joiner S, Sidle K C L, Gowland I, Collinge J. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 9.Hill A F, Sidle K C L, Joiner S, Keyes P, Martin T C, Dawson M, Collinge J. Molecular screening of sheep for bovine spongiform encephalopathy. Neurosci Lett. 1998;255:159–162. doi: 10.1016/s0304-3940(98)00736-8. [DOI] [PubMed] [Google Scholar]
- 10.Hope J, Wood S C E R, Birkett C R, Chong A, Bruce M E, Cairns D, Goldmann W, Hunter N, Bostock C J. Molecular analysis of ovine prion protein identifies similarities between BSE and an experimental isolate of natural scrapie, CH1641. J Gen Virol. 1999;80:1–4. doi: 10.1099/0022-1317-80-1-1. [DOI] [PubMed] [Google Scholar]
- 11.Lasmézas C I, Deslys J-P, Demaimay R, Adjou K T, Hauw J-J, Dormont D. Strain specific and common pathogenic events in murine models of scrapie and bovine spongiform encephalopathy. J Gen Virol. 1996;77:1601–1609. doi: 10.1099/0022-1317-77-7-1601. [DOI] [PubMed] [Google Scholar]
- 12.Madec J-Y, Vanier A, Dorier A, Bernillon J, Belli P, Baron T. Biochemical properties of protease resistant prion protein PrP Sc in natural sheep scrapie. Arch Virol. 1997;142:1603–1612. doi: 10.1007/s007050050183. [DOI] [PubMed] [Google Scholar]