Summary
Small molecules can be imaged in living cells using biosensors composed of RNA. However, RNA-based devices are difficult to design. Here we describe a versatile platform for designing RNA-based fluorescent small molecule sensors using naturally occurring highly stable three-way junction RNAs. We show that ligand-binding aptamers and fluorogenic aptamers can be inserted into three-way junctions, and connected in a way that enables the three-way junction to function as a small molecule-regulated fluorescent sensor in vitro and in cells. The sensors are designed so that the inter-helical stabilizing interactions in the three-way junction are only induced upon ligand binding. We use these RNA-based devices to measure the dynamics of S-adenosylmethionine levels in mammalian cells in real time. We show that this strategy is compatible with diverse metabolite-binding RNA aptamers, fluorogenic aptamers, and three-way junctions. Overall, these data demonstrate a versatile method for readily generating RNA-devices that function in living cells.
Graphical Abstract
eTOC blub
Genetically encoded RNA biosensors are an emerging tool for imaging small-molecule metabolites in cells. In this manuscript, Moon et. al. describe a versatile method for developing RNA biosensors by connecting fluorogenic RNA aptamers and metabolite-binding aptamers via reengineered, naturally occurring RNA three-way junctions.
Introduction
An emerging tool for imaging and manipulating cellular functions are genetically encoded RNA-based devices. RNA-based devices are typically short RNAs (~50-200 nt) that can fold into complex structures and perform useful cellular functions. Typically, RNA-based devices transduce one type of information into a specific measurable output. For example, riboswitches, which are naturally occurring RNA-based devices, undergo conformational changes in response to natural ligands and thereby influence translation, splicing, stability, or transcription (Nahvi et al., 2002; Winkler et al., 2002).
The discovery of naturally occurring RNA-based devices has stimulated efforts to engineer genetically encoded RNA-based devices that perform new functions. For example, RNA-based biosensors are RNAs that exhibit fluorescence upon binding specific ligands (Hallberg et al., 2017; Sun et al., 2019). Ligand binding induces a conformational change that regulates the folding of a fluorogenic aptamer, such as Spinach or Broccoli (Bose et al., 2016; Kellenberger et al., 2013, 2015; Paige et al., 2011, 2012; Song et al., 2013). These genetically encoded biosensors allow small molecules and proteins to be imaged in real-time in living cells (Chakraborty et al., 2016; Sun et al., 2019).
The initial “allosteric Spinach sensors” (Paige et al., 2012) utilize simple quasi-stable helices, termed transducer stems (Kellenberger et al., 2013; Paige et al., 2012). The transducer stem connects the ligand-binding aptamer to Spinach, in such a way that Spinach is in unfolded or misfolded conformations. Upon ligand binding, the stem becomes stabilized, which allosterically stabilizes Spinach allowing it to produce fluorescence (Paige et al., 2012). This design was based on early allosteric ribozyme devices (Robertson and Ellington, 1999; Tang and Breaker, 1997) and has now been used to design numerous RNA-based biosensors (Filonov et al., 2014; Kellenberger et al., 2013; Nakayama et al., 2012; Porter et al., 2017; Song et al., 2013). However, a major challenge is achieving high ligand-induced fluorescence activation, and low fluorescence in the absence of the ligand.
The second approach, termed “Spinach riboswitches,” (You et al., 2015) involves modifying naturally occurring riboswitches. In this approach, Spinach is inserted into a riboswitch in place of its naturally occurring ligand-regulated output domain. In many riboswitches, this domain undergoes a ligand-regulated switch from single-stranded to double-stranded RNA. In a Spinach riboswitch, the Spinach aptamer is embedded so that the ligand-regulated structural switch results in folding of Spinach (You et al., 2015). This approach is limited by the availability of naturally occurring riboswitches that bind ligands of interest.
Here, we describe a robust design strategy for constructing RNA-based sensors. The key criteria for RNA-based sensors is that they need to exhibit high signal-to-noise ratios, and they need to be widely adaptable so that diverse aptamers can be readily inserted and used for device development. We show that naturally occurring three-way junction structures can be converted into highly efficient ligand-regulated RNA devices. We show that the structural mechanisms that confer high structural stability to three-way junctions can be exploited to develop allosteric sensors that detect S-adenosylmethionine (SAM), and other metabolites. Several different three-way junctions, each using different structural mechanisms for their high stability, can be converted into RNA-based sensors. Furthermore, the sensors based on the Phi29 three-way junction function well in cells and enable imaging of dynamic changes in SAM levels in mammalian cells. Overall these studies reveal a new and versatile strategy for generating RNA-based devices based on naturally occurring three-way junctions.
Results
Intramolecular Interactions that Mutually Stabilize Two Stems in RNA Three-Way Junctions
To identify alternative designs for RNA device development, we considered RNA three-way junctions. Three-way junctions are RNAs that fold into a structure comprising three RNA helices joined at a single junction. Three-way junctions are unusually stable (Hill and Schroeder, 2017) since they contain intramolecular interactions between helical stems, or between bulge regions and helical stems, that allosterically stabilize the overall junction structure (Lescoute and Westhof, 2006). These structural features interact in different ways to form structures with unusually high stability.
We reasoned that three-way junctions could be converted into RNA-based sensors if a fluorogenic aptamer is inserted into one helical stem of a three-way junction, and a metabolite-binding aptamer is inserted into another stem. Metabolite binding can stabilize or induce folding of stem sequences of metabolite-binding aptamers. If this stem exhibits intramolecular interactions with another stem in the three-way junction, then ligand binding of an aptamer in one stem could be expected to induce folding of another stem. Thus, the three-way junction could act as a transducer that communicates information on ligand binding from a metabolite-binding aptamer to a fluorogenic aptamer.
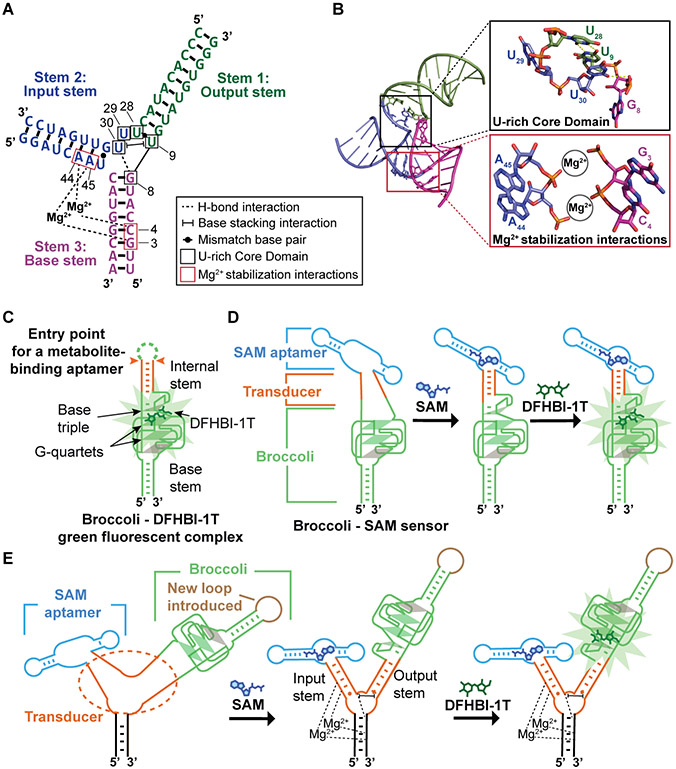
We first tested the three-way junction from the packaging RNA of bacteriophage Phi29 (Zhang et al., 2013). The Phi29 three-way junction is stabilized by two magnesium (Mg2+) ions that coordinate phosphates on two coaxially stacked helical stems (Stem 2 and Stem 3) (Figure 1A-B). The stabilized Phi29 three-way junction positions a group of four uridines (U9, U28, U29, and U30) in a nonstandard geometry at the point where Stem 1 meets the junction core (Figure 1A). We define this region as the “U-rich core domain.” Two of the U’s (U9 and U28) form a non-canonical U•U base pair that serves as the first base pair of Stem 1 from the junction core. U30 forms H-bonding and stacking interactions to stabilize the core domain and the first base pair of Stem 1 (Figure 1A-B). Notably, Stem 2 must be a stable helical duplex in order to position these four U’s into this unique geometry, which then stabilizes Stem 1. If Stem 2 does not fold into a helical form, then this U-rich core domain would not be positioned in a way that would stabilize Stem 1 (Ding et al., 2011; Zhang et al., 2013). We reasoned that we could exploit this allosteric stabilization mechanism to design new RNA-based sensors.
Figure 1. Tertiary interactions in the Phi29 three-way junction that are used in sensor design.
(A) Secondary structure of the Phi29 three-way junction. Stem 2 and Stem 3 are connected by two Mg2+ ions. The phosphates of nucleotides A44 and C4 interact with one Mg2+. A45 and G3 phosphates interact with the other Mg2+. The Phi29 three-way junction core contains a U-rich domain comprising U9, U28, U29, and U30 (black boxes). U9 and U28 form a mismatch base pair that serves as the closest base pair of Stem 1 (green) to the junction. Shown is the H-bond between the U30 carbonyl and the G8 phosphate. Shown is the base-stacking interaction between U30 and U9. Tertiary interaction notation is based on Leontis and Westhof (Leontis and Westhof, 2001). The input and output stems used for the initial sensor design are indicated. Structures from PDB 4KZ2 (Zhang et al., 2013). (B) Helical stems in the Phi29 three-way junction. Top, detailed view of the U-rich core domain that separates Stem 2 and Stem 3 from Stem 1. Bottom, stabilizing interactions of two Mg2+ ions between phosphates of Stem 1 nucleotides A44 and A45 and Stem 3 phosphates of C4 and G3 (magenta). (C) Schematic representation of Broccoli shows the entry point for a metabolite-binding aptamer (orange) and the stem at its base (green). DFHBI-1T binds between the topmost G-quartet (green parallelogram) and a base triple (grey triangle). The internal stem serves as an entry point for metabolite-binding aptamers since the folding of this stem directly influences the stability of the base triple. The transparent grey parallelogram indicates a mixed-sequence tetrad. (D) SAM binding to the SAM aptamer induces folding of the transducer domain. The folding of this transducer domain enables Broccoli to fold and bind DFHBI-1T, which leads to DFHBI-1T fluorescence. (E) The internal stem of Broccoli is attached to the F30 RNA three-way junction, and thus constitutes the output stem. A loop (brown) connects the previous 3’ and 5’ ends of the Broccoli aptamer. The SAM-binding aptamer is fused to the three-way junction via a stem that is designated the input stem. Before SAM binding, the input and output stems of F30 are unfolded. SAM binding to the SAM aptamer induces folding of the input stem of F30 into a stable duplex. This stable duplex facilitates the folding of the U-rich core domain (shown are the H bonds and the base stacking interactions of the core, with a dotted line and a bracketed line) and the output stem. The folding of the output stem enables Broccoli folding.
Optimization of the F30 Three-Way Junction as a Transducer
We first asked if the folding of an aptamer in one stem of the Phi29 three-way junction could be regulated by the folding of another stem. Instead of Phi29, we used F30, which is a modified version Phi29 that is optimized for expression in mammalian cells by removing a mammalian Pol III transcription termination sequence (Filonov et al., 2015). F30 was previously used as a folding scaffold for aptamers—aptamers inserted into either stem of F30 exhibited improved folding, presumably due to the highly efficient folding of F30 (Filonov et al., 2015). Therefore, we asked whether F30 could be converted into a ligand-regulated RNA device.
In F30, one stem will contain the input metabolite-binding aptamer, a second stem will contain the output effector aptamer, and third stem will serve as the base and contains the 5’ and 3’ ends. Since there are too many different combinations in which the aptamers could be inserted into F30, we chose to first test Stem 1 as the output stem and Stem 2 as the input stem (Stem 1 and Stem 2 are indicated in Figure 1A) for two reasons: First, RNA aptamers expressed in each of these stems have previously been expressed in cells (Filonov et al., 2015). Second, proper folding of Stem 2 as a duplex has been shown to stabilize the nonstandard geometry of the U-rich core domain including the U•U base pair of Stem 1 (Zhang et al., 2013). Therefore, the folding of the input stem could potentially allosterically regulate the output stem.
To test if an aptamer on one stem of F30 can regulate an output aptamer located in another stem, we used the fluorogenic Broccoli aptamer as the output aptamer (Filonov et al., 2014). Broccoli contains an internal stem that is required for its fluorogenic function since this stem stabilizes an U•A•U base-triple that forms one face of the fluorophore-binding pocket (Warner et al., 2014). This stem has previously been used as a transducer stem connecting metabolite-binding aptamers to Broccoli for the design of previous allosteric RNA-based sensors (Figure 1C-D) (Litke and Jaffrey, 2019; Paige et al., 2012). Therefore, we fused this Broccoli stem into Stem 1 of F30 (Figure 1 A and E). In this Broccoli-F30 fusion RNA, Broccoli will be fluorescent if its stem is in a base-paired duplex (Figure 1E).
After attaching Broccoli to F30, the 5’ and 3’ ends of Broccoli were fused with a loop (Figure 1E). The 5’ and 3’ ends of this Broccoli-F30 fusion remained at the base of F30.
We next asked if the folding of Broccoli in the F30 output stem can be controlled by the input stem. Since this is the core concept for converting F30 into an allosteric sensor, we sought to develop an approach to rapidly test this concept. However, in order to determine if the input stem can regulate Broccoli in the output stem, the output stem must be able to regulate Broccoli stability. If the output stem is too unstable, and thus completely unable to form a stable duplex, then it will never be able to fold, regardless of the input stem. If, on the other hand, the output stem is a highly stable helix, then Broccoli will already be fluorescent, and the input stem cannot further stabilize Broccoli. We therefore needed to find an output stem that is “metastable,” i.e., in a range of stability where it could potentially be influenced by the input stem.
To identify metastable outputs stems to regulate Broccoli, we tested several variations of the length and G/C composition of the output stem. The input stem was converted to a large unstructured loop to prevent it from providing any possible stabilization effect, including its normal ability to position the U-rich core domain (Figure 2A).
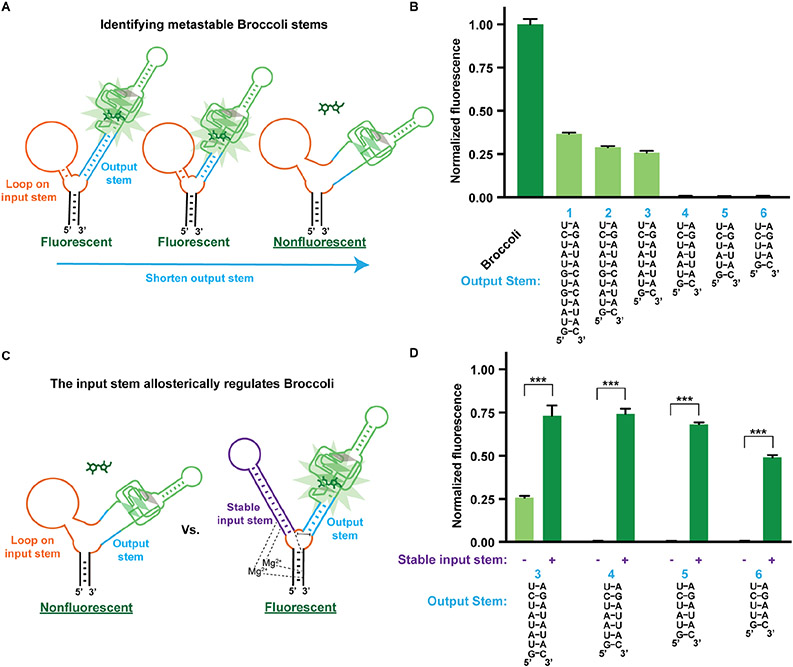
Figure 2. The input stem of the F30 three-way junction regulates Broccoli fluorescence on the output stem.
(A) A twelve-nucleotide loop (orange) was inserted on the input stem of the F30 three-way junction to prevent any stabilization effect of the input stem on the output stem. The length of the output stem was varied until Broccoli was destabilized. (B) Six different output stem variants were tested, and Broccoli alone was used as a positive control. RNA (0.5 μM) was incubated with 10 μM DFHBI-1T. Shown is the fluorescence (Ex 460 nm, Em 505 nm) normalized to Broccoli fluorescence. The mean and SEM values are shown (n=3). (C) Metastable output stems from (B) were assessed for fluorescence recovery when the loop was replaced with a 12 bp stable input stem (purple). The Broccoli aptamer was attached to the output stem of the F30 using variants 3-6 from (B). (D) Allosteric activation of Broccoli by the input stem. Broccoli was tested using four different output stem variants. RNA (0.5 μM) was incubated with 10 μM DFHBI-1T and the fluorescence was normalized to Broccoli fluorescence. The fluorescence of each variant is shown in the context of a stable input stem (+) or a loop in place of the input stem (−). The mean and SEM values are shown (n=3). *** P = 0.0002 (3), *** P = 2X10−6 (4), *** P = 4X10−8 (5), *** P = 4X10−7 (6).
We replaced the output stem with a series of helices of variable stability. We found that Broccoli fluorescence was lost when the output stem contained less than eight base pairs (Figure 2B). Thus, these output stems were sufficiently short that they had become metastable, i.e., they were not able to promote folding of Broccoli, but might be able to fold Broccoli with slight additional stabilization from the input stem.
We therefore asked if we could restore the stability of these metastable output stems by restoring the stabilizing effects of the input stem. The input stem positions the U-rich core domain in the three-way junction and thus stabilizes the output stem (Zhang et al., 2013). We therefore replaced the loop with a stable stem in the input stem position (Figure 2C). The stable input stem should restore the Mg2+ interactions and stabilize the positions of nucleotides in the core domain including the mismatch U•U base pair of the output stem. By inserting a stable duplex in the input stem, we recovered up to 50% of Broccoli fluorescence for different output stem sequences (Figure 2D). Thus, the input stem can allosterically regulate the output stem, and in turn activate Broccoli fluorescence.
Optimization of F30-Based SAM Sensors
We next asked if a ligand-binding aptamer in the input stem can control Broccoli fluorescence. For the ligand-binding aptamer, we chose the SAM-binding aptamer found in the SAM-III riboswitch (Lu et al., 2008). This SAM-binding aptamer exhibits a conformational change upon SAM binding (Smith et al., 2010). We previously found that this SAM-binding aptamer can regulate Spinach fluorescence when fused via a single transducer stem (Paige et al., 2012).
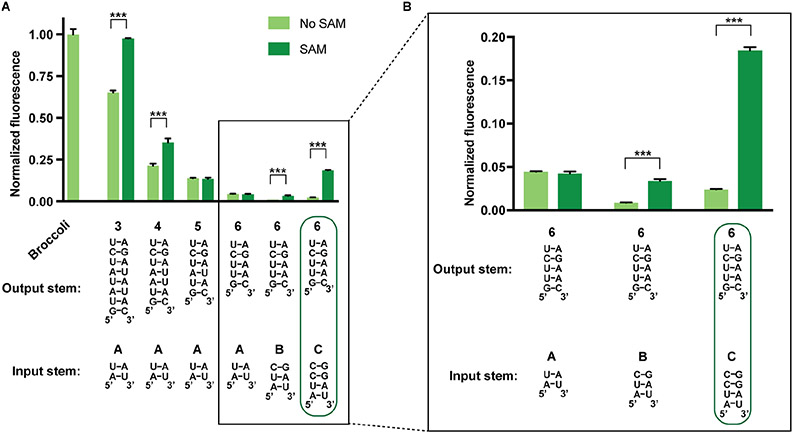
To generate an F30-based SAM sensor, we fused the SAM-binding aptamer to the input stem of the F30 three-way junction (Figure 1E). To optimize the F30-based sensor, we tested multiple variant stems for the input SAM aptamer and output Broccoli aptamer. Using this approach, we found an F30-based SAM sensor (6c) that showed a six-fold activation of fluorescence in the presence of SAM (Figure 3). Overall, with these different stems, we were able to develop an F30-based SAM sensor that induces Broccoli fluorescence in the presence of SAM.
Figure 3. Optimizing an F30-based SAM sensor.
(A) Optimization of input and output stems in F30-based SAM sensors. Seven different combinations of input and output stems were tested. RNA (0.5 μM) was incubated with 10 μM DFHBI-1T with or without 0.2 mM SAM. Boxed region is shown in greater detail in (B). Fluorescence of each sample was measured (Ex 460 nm, Em 505 nm) after 1 h incubation at room temperature. Values were normalized to the fluorescence of Broccoli. The mean and SEM values are shown (n=3). *** P = 2X10−6 (3A), *** P = 0.0009 (4A), *** P = 4X10−5 (6B), *** P = 3X10−7 (6C). (B) Boxed region from (A) shown in greater detail.
Although we chose Stem 1 as the output stem and Stem 2 as the input stem, it is possible that different stems could be used. We therefore tested other combinations, such as Stem 2 as the input stem (as above) and Stem 3 as the output stem, and Stem 2 as the output stem and Stem 3 as the input stem (Figure S1). We then performed the optimizations as above and identified output stems that could be regulated by the input stem. However, surprisingly neither alternative configuration of F30 showed induction of Broccoli fluorescence after addition of SAM (Figure S1). These results demonstrate that only some input/output configurations of the F30 three-way junction are readily converted into a SAM-based sensor.
Alternative Configurations of Three-Way Junction-Based SAM Sensors
We next asked if other RNA three-way junctions could be converted into RNA-based devices. We chose to focus on RNA three-way junctions where the crystal structures have been solved in order to pick those with different and predictable allosteric stabilization mechanisms. In some cases, the three-way junctions contain stems in which the distal portion of the stems (i.e., the part away from the three-way junction) interact with each other. These three-way junctions are not useful for us since insertion of an aptamer would disrupt these potentially crucial tertiary interactions.
Therefore, we focused on three-way junctions where aptamer insertion would not be expected to disrupt folding. We selected four additional three-way junctions as potential transducers: a three-way junction of a phage Twort group 1 ribozyme (termed “Twort” here) (Figure S2A) (Golden et al., 2005); a three-way junction of helices 33, 34, and 35 from the Haloarcula marismortui 23S ribosomal RNA (termed “H33” here) (Figure S2B) (Klein et al., 2004); a three-way junction from Bacillus subtillis ribonuclease P B-type (termed “RNase P” here) (Figure S2C) (Krasilnikov et al., 2003); and a three-way junction from helices 99, 100, and 101 of the Haloarcula marismortui 23S ribosomal subunit (termed “H99” here) (Figure S2D) (Klein et al., 2004; Lescoute and Westhof, 2006). These three-way junctions contain interactions between one stem and at least one other stem that appear to allosterically stabilize the three-way junction structure.
For each of these three-way junctions, we examined the crystal structures of each to decide which stems would be the best for inserting the SAM aptamer and Broccoli. As with F30, there were too many potential configurations of aptamers in these three-way junctions to test each one. Thus, we selected the two stems that exhibit the most intramolecular interactions between each other to serve as the input and output stems for each sensor. We chose one of these two stems as the input stem and the other as the output stem. The remaining stem would serve as the “base” stem that contains the 5’ and 3’ ends of the RNA (Figure S2A-D).
We proceeded to test whether each three-way junction could be developed into a sensor using the approach used for F30 above. Thus, we first identified metastable stems that connect Broccoli to each three-way junction (Figure S3). As before, we converted the input stem into a loop to prevent allosteric stabilization from this stem. This approach led to the identification of metastable stems for Broccoli for each three-way junction except for the H99 three-way junction (Figure S3D). In the case of H99, we could not truncate the output stem more than 6 bp without deleting nucleotides that form intramolecular contacts with the input stem in the H99 three-way junction core. The shortest output stem in H99 tested did not greatly reduce Broccoli fluorescence. Based on this, H99 was not used further in this analysis.
Next, we asked if the folding of the input stem would allosterically stabilize the output stem in each three-way junction. To test this, we compared the fluorescence of Broccoli when the input stem was the loop sequence compared to a stable stem–loop hairpin. All three three-way junctions, i.e., H33, Twort, and RNase P, showed Broccoli fluorescence recovery when the loop on the input stem was replaced with a stable stem (Figure S3E-G). Taken together, these data demonstrate that the Twort, H33 and RNase P three-way junctions can act as transducers, where the input stem regulates Broccoli fluorescence.
To determine if the SAM aptamer could activate Broccoli, we fused the SAM-binding aptamer to the input stem of each three-way junction. We next tested different input and output stems to identify the maximal fold induction of Broccoli fluorescence upon addition of SAM (Figure S4). None of the RNase P three-way junction variants showed enhanced Broccoli fluorescence upon addition of SAM (Figure S4C). However, the best Twort and H33 three-way junction transducers led to 14-fold and 8-fold activation of Broccoli fluorescence, respectively (Figure S4A-B).
We selected two of the Twort-based sensors and one H33-based sensor along with the F30-based sensor for additional characterization since they exhibited the greatest SAM-dependent increase in fluorescence (Figure 4A). We selected the two Twort-based sensors (6D and 6B) because they exhibited different, potentially valuable properties. 6D is brighter than 6B, but 6B exhibits lower background fluorescence (Figure S4C). We renamed 6D and 6B to Twort A and Twort B, respectively.
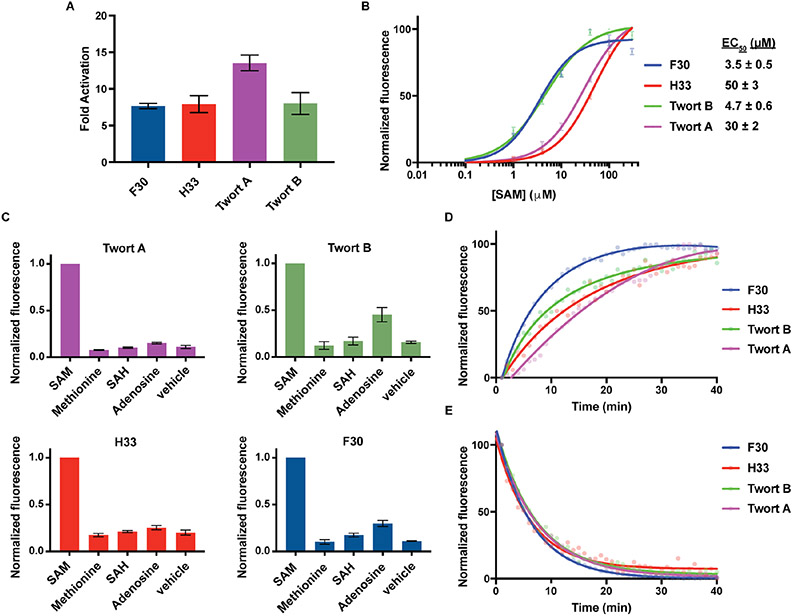
Figure 4. Characterizing different three-way junction-based SAM sensors.
(A) SAM-induced activation of three-way junction-based SAM sensors in vitro. Three-way junction-based SAM sensors (0.5 μM) were incubated with or without 0.2 mM SAM. Fluorescence was measured (Ex 460 nm, Em 505 nm) and the mean and SEM values are shown (n=3). (B) Dose-response curve for fluorescence activation of three-way junction-based SAM sensors by SAM. The mean EC50 and SEM values are shown (n=3). (C) Specificity of three-way junction-based SAM sensors for detecting SAM or the indicated metabolites (0.1 mM each). RNA (0.5 μM) was incubated for 1 h at 37°C with each metabolite. Mean and SEM values are shown (n=3). (D) Rates of three-way junction-based SAM sensor activation. The fluorescence signal of 0.5 μM sensor RNA was recorded every 1 min for 40 min after the addition of 0.1 mM SAM (Ex 460 nm, Em 505 nm). (E) The rates of three-way junction-based SAM sensor deactivation. Samples were prepared with 0.5 μM sensor RNA and 0.1 mM SAM and incubated for 1 h prior to SAM removal to initiate sensor deactivation. Fluorescence was recorded every 1 min (Ex 460 nm, Em 505 nm).
Binding Properties of Three-Way Junction-Based SAM Sensors
The three-way junction-based SAM sensors exhibit EC50 values from 3.5 μM to 50 μM (Figure 4B). Since this concentration range is similar to intracellular SAM levels (Hermes et al., 2005), this sensitivity range should be appropriate for imaging SAM in mammalian cells. The three-way junction-based SAM sensors also showed negligible fluorescence upon incubation with the metabolites related to SAM (Figure 4C). Although the Twort B and F30-based SAM sensors showed low levels of fluorescence enhancement when incubated with 0.1 mM adenosine, this concentration of adenosine is >50X the physiologic levels (Traut, 1994). These experiments demonstrate that three-way junction-based SAM sensors detect SAM with high specificity relative to similar metabolites.
The EC50 of the F30-based SAM sensor for SAH is greater than 300 μM (Figure S5A). Importantly, SAH-elicited fluorescence is much lower than SAM-induced fluorescence (Figure S5B). Therefore, we concluded that fluctuations in SAH levels would not interfere with SAM imaging using the F30-based SAM sensor.
The three-way junction-based sensors exhibited a range of activation kinetics, with each sensor reaching >80% of the maximal fluorescence within 20 min (Figure 4D). Sensor deactivation kinetics was determined by removing SAM by rapid gel filtration. Each of the three-way junction-based sensors showed similar deactivation rates, with >80% fluorescence deactivation by 10 min (Figure 4E). Taken together, these data show the three-way junction-based SAM sensors exhibit reversible fluorescence activation and deactivation.
Three-Way Junction Sensors Designed Using Different Input and Output Aptamers
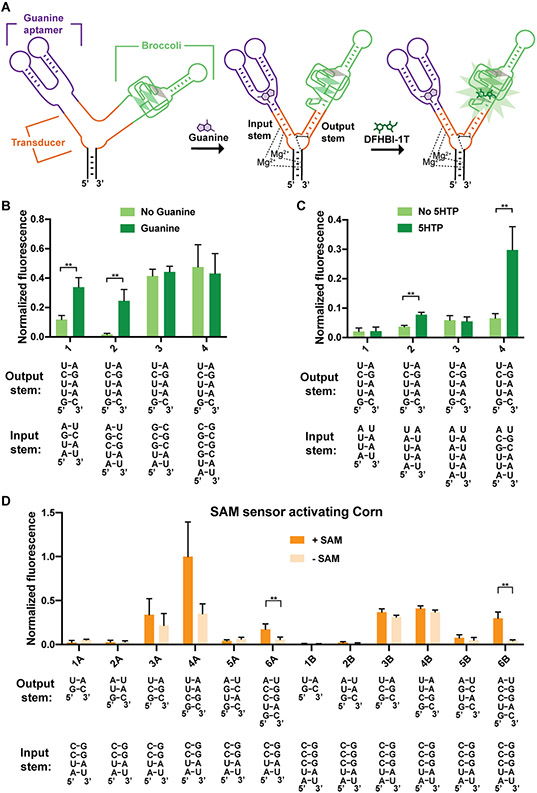
To test the versatility of this design, we tested a guanine-binding aptamer from the naturally occurring guanine riboswitch from the xpt-pbuX operon of B. subtilis. This aptamer contains a stem that is stabilized upon binding guanine (Batey et al., 2004). We also tested a 5-hydroxytryptophan (5HTP)-binding aptamer that was generated using SELEX (Porter et al., 2017). The 5HTP aptamer also contains a stem that is stabilized upon 5HTP binding (Porter et al., 2017).
We kept Broccoli in the output stem and fused the guanine or 5HTP aptamer to the input stem of F30 (Figure 5A). For both metabolite sensors we tested four variants of the input stem with the output stem variant that was previously optimized for F30-based SAM sensors above. We found F30-based guanine sensors and F30-based 5HTP sensors that showed significant fluorescence induction upon addition of their cognate metabolites (Figure 5B-C). Thus, F30-based sensors can be used with different ligand-binding input aptamers.
Figure 5. Optimizing F30-based sensors composed of different input and output aptamers.
(A) Schematic representation of an F30 three-way junction-based guanine sensor. Guanine binding to the guanine aptamer induces folding of the input stem, which facilitates the proper positioning of the U-rich core domain (shown are the H-bonds and the base stacking interactions of the core) and the output stem. The positioning of the U-rich core domain is expected to stabilize the output stem and promote the folding of Broccoli. (B) Optimization of F30-based guanine sensors. RNA (4 μM) was incubated with 40 μM DFHBI-1T with or without 0.2 mM guanine. Fluorescence of each sample was measured (Ex 460 nm, Em 505 nm) and normalized to Broccoli. The mean and SEM values are shown (n=3). **P = 0.005 (1), **P = 0.007 (2). (C) Optimization of input and output stems in F30-based 5HTP sensors. The mean and SEM values are shown (n=3). **P = 0.001 (2), ** P = 0.008 (4). (D) Optimization of F30-based SAM sensors with Corn as the output aptamer. RNA (0.5 μM) was incubated with 10 μM DFHO with or without 0.2 mM SAM. After 1 h, fluorescence was measured and normalized to the average fluorescence of combination 4A (the brightest combination tested). The mean and SEM values are shown (n=3). *P = 0.04 (6A), **P = 0.004 (6B).
We next asked if a different fluorogenic aptamer connected to the output stem could be allosterically activated upon SAM binding in F30-based SAM sensors. To test this, we focused on the yellow fluorogenic Corn aptamer (Song et al., 2017). Corn makes a fluorescent yellow complex by forming a homodimer that binds and activates the fluorophore DFHO (Warner et al., 2017).
To test whether Corn can be used as the output aptamer, we fused the 5’ and 3’ ends of Corn into the output stem in the F30-based SAM sensor. We developed twelve sensor constructs containing six stem variants that connect Corn with the F30 junction, and two variants of the input stem. One combination of input and output stems showed a ~6.8 fold increase in fluorescence induction upon addition of SAM (Figure 5D). Overall, these data show that the F30-based sensor is versatile with respect to both input and output aptamers.
Imaging of SAM Dynamics in Living Cells Using Three-Way Junction-Based SAM Sensors
In order for these sensors to be used in cells, they need to be expressed at sufficiently high levels to generate enough signal for detection using fluorescence microscopy. We therefore used the Tornado expression system (Litke and Jaffrey, 2019). In this method, the three-way junction sensor is first expressed as a linear transcript flanked by Twister ribozymes. The ribozymes facilitate autocatalytic cleavage and the resulting 5’ and 3’ ends are ligated by the endogenous RNA ligase RtcB. Circular RNAs expressed using the Tornado expression system reach low micromolar levels as opposed to low nanomolar levels typical for linear RNAs. Thus, Tornado-based expression of sensors can allow them to achieve the expression levels needed to generate sufficient fluorescence signal for fluorescent microscopy (Litke and Jaffrey, 2019).
We focused on imaging SAM. 5HTP and guanine are present at low levels in HEK293T cells (Traut, 1994), and therefore would not be sufficient for demonstrating sensor function.
First, we assessed whether the different three-way junction-based SAM sensors could be used to image SAM levels over time in live HEK293T cells. To test this, we expressed four different sensors (F30, H33, Twort A, and Twort B) in HEK293T cells. Expression of the circular RNAs was not toxic, since cells expressing the sensors did not show increased apoptosis (Figure S5C). Next we assessed the fluorescence of each sensor. Only the F30-based SAM sensor showed bright green fluorescence consistent with detection of SAM in untreated cells (Figure 6). The H33 and Twort-based SAM sensors exhibited low fluorescence in cells, despite showing higher brightness than the F30-based SAM sensors in vitro.
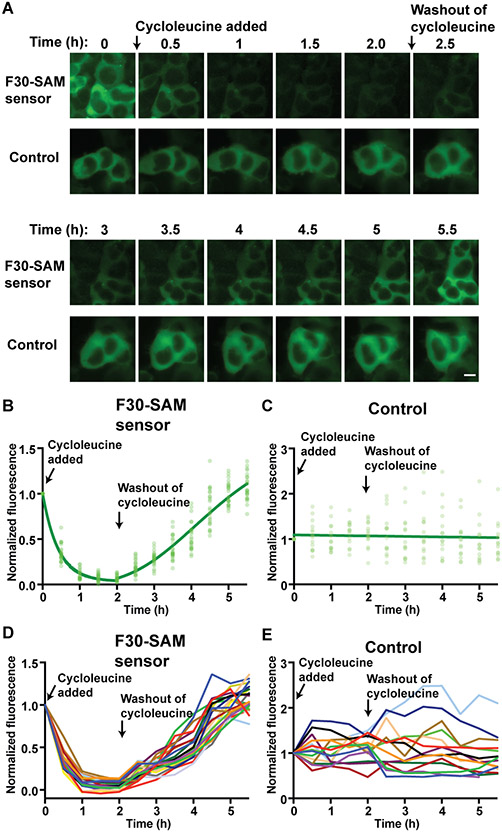
Figure 6. The F30-based SAM sensor enables live-cell imaging of SAM dynamics.
(A) Single-cell imaging of circular F30-based SAM sensor fluorescence in response to cycloleucine treatment. HEK293T cells expressing the circular F30-based SAM sensor were compared to cells expressing circular F30-Broccoli, which serves as a control due to its constitutive fluorescence. Cells were imaged for 2 h after addition of 50 mM cycloleucine, and then 3.5 h following replacement with cycloleucine-free culture media. Cells showed a drop in F30-based SAM sensor fluorescence after cycloleucine treatment, and recovery of fluorescence after washing out cycloleucine. Control cell fluorescence stayed consistently high over the course of the experiment. Image acquisition time 500 ms. Scale bar 10 μm. (B) Quantification of fluorescence from individual cells expressing F30-based SAM sensor was calculated from images taken every 30 min. Fluorescence values for each cell were normalized to the fluorescence at time t=0. Light green circles show normalized fluorescence values for individual cells (n=23). The dark green curve shows the results of nonlinear regression of the data fit with a one phase exponential decay (0-2 h) and a specific binding function (2-5.5). (C) Total cellular fluorescence from individual cells expressing F30-Broccoli was calculated as in (B). (D) Individual fluorescence trajectories from cells expressing the F30-based SAM sensor. Normalized cellular fluorescence from individual cells expressing the F30-based SAM sensor was calculated as described above. Normalized fluorescence of each individual cell is shown over the course of the experiment (different color for each cell) (n=23). (E) Individual fluorescence trajectories from cells expressing the control RNA, F30-Broccoli. Fluorescence was measured as in (D), n=14.
Conceivably, variable expression levels could explain the variation in signal between different three-way junction-based SAM sensors in mammalian cells. To test this, we harvested the cells and resolved total RNA by denaturing polyacrylamide gel electrophoresis. Staining these gels with SYBR Gold showed similarly intense bands representing the sensor RNA (Figure S5D). Thus, different expression levels do not account for the different brightness of each sensor in cells.
We next asked whether variable Mg2+ dependence could explain the variation in signal between different three-way junction-based SAM sensors in mammalian cells. The levels of free Mg2+ are low (0.25 - 1 mM) in mammalian cells (Grubbs, 2002), which can reduce the function of Mg2+-dependent RNA devices. However, each sensor showed a similar EC50 for Mg2+ (Figure S5E). Therefore, differences in Mg2+ dependence did not explain the poor performance of H33 and Twort-based SAM sensors in cells. Thus, the enhanced cellular fluorescence of the F30-based SAM sensor compared to Twort and H33-based SAM sensors might be due to other factors, such as improved folding in the cellular milieu.
We next asked whether the F30-based SAM sensor can image SAM levels inside mammalian cells. We inhibited SAM production with cycloleucine, an inhibitor of SAM biosynthesis (Lombardini and Talalay, 1971). Cells expressing the F30-based SAM sensor showed greater than 80% fluorescence loss within 1 h of cycloleucine treatment (Figure 6A-B, D). As a control, we used an RNA with constitutive Broccoli fluorescence — this RNA contains a stable stem in F30 in place of the SAM aptamer so SAM cannot regulate its fluorescence. The fluorescence of this control F30-Broccoli RNA was unaffected by cycloleucine treatment (Figure 6A, C, E). As an additional control, we used an F30-based SAM sensor containing a point mutation in the SAM-binding domain, that was previously shown to disrupt SAM binding (Lu et al., 2011). This mutant showed less dependence on SAM levels and lower overall brightness in vitro (Figure S5F-G). Its fluorescence was too low to be visualized in living cells (Figure S5F-G).
After 2 h of cycloleucine treatment, we removed the cycloleucine from cells by replacing the media with fresh cycloleucine-free media. Cells expressing the F30-based SAM sensor recovered cellular fluorescence within 3 h after cycloleucine removal (Figure 6A-B, D). Cells expressing the constitutively fluorescent F30-Broccoli control construct exhibited high fluorescence throughout these experiments (Figure 6A, C, E). Overall, these data show that the F30-based SAM sensor enables real-time monitoring of SAM dynamics in individual mammalian cells.
We next wanted to determine the relationship between fluorescence levels of the F30-based SAM sensor in living cells and the intracellular concentration of SAM. To test this, we measured intracellular SAM levels at several time points after cycloleucine using a biochemical SAM assay (Figure S6A). Overall, the average cellular fluorescence levels linearly correlated with the measured SAM levels (p<0.05) (Figure S6B-D).
Lastly, we asked how the F30-based sensor SAM sensor compares to the previously described Broccoli-SAM sensor (Litke and Jaffrey, 2019). Overall both sensors showed similar brightness and dynamic range (Figure S6E-F). These data suggest that the three-way junction approach developed here can provide similar performance as a conventional allosteric approach.
DISCUSSION
Strategies for constructing RNA-based devices are limited, with ligand-based regulation of simple quasi-stable transducer stems being the most widely used approach since their original use in allosteric ribozymes (Soukup and Breaker, 1999; Tang and Breaker, 1997). Here we describe an approach for converting highly stable three-way junctions into ligand-regulated RNA-based biosensors. This approach takes advantage of the diverse interactions that occur between helical stems in three-way junctions. We show that the SAM aptamer can be inserted into various three-way junctions such that SAM binding allosterically regulates another stem that contains the Broccoli aptamer. This overall approach is highly versatile. For example, we show that different three-way junctions can be similarly converted into sensors. Additionally, sensors can be constructed with different metabolite-binding aptamers and fluorogenic aptamers. Notably, a SAM biosensor developed from the F30 three-way junction was also functional in mammalian cells, enabling live imaging of SAM levels in HEK293T cells. Overall, our studies reveal a general approach for converting highly stable three-way junctions into ligand-regulated RNA-based biosensors.
The concept behind the three-way junction-based sensors is that the structural features that confer the stability of three-way junctions can be regulated by a ligand-binding aptamer. The structural features include Mg2+ interactions between phosphates of different stems and H-bonding interactions between bulge sequences within the junction core domain. In the latter case, the folding of one stem would be required for properly positioning these bulge sequences so that they can interact with adjacent stems. By making these interactions dependent on a ligand-binding aptamer, these aptamers likely regulate the overall folding of the three-way junction in a ligand-dependent manner.
The three-way junction-based sensors described here require input and output aptamers that both contain a base-paired stem. These aptamers can be derived from riboswitches (e.g. SAM aptamer and guanine aptamer) or SELEX experiments (Broccoli, Corn, and 5HTP aptamer). For the aptamers used here, the stems are formed by hybridization of the 5’ and 3’ sequences of these aptamers. Additionally, these stems need to be connected to important structural domains in the aptamer. In the case of Broccoli, the stem is directly connected to a U•A•U base triple that forms the base of the fluorophore-binding pocket (Huang et al., 2014; Warner et al., 2014). Thus, the folding of this stem is required for fluorophore binding and subsequent fluorescence activation. In the case of the SAM aptamer, the stem is directly linked to the SAM-binding domain (Lu et al., 2008). Many metabolite-binding aptamers contain stems since they are often derived from SELEX experiments which utilize stem-containing libraries (Davis and Szostak, 2002). Riboswitch-derived aptamers also typically contain a stem that is essential for riboswitch function (Serganov and Nudler, 2013). Thus, many aptamers will be suitable for incorporation in the RNA devices described here.
Naturally occurring three-way junctions have evolved to undergo efficient folding in living cells. We previously found that many aptamers, such as the original Spinach fluorogenic aptamer, fold poorly in cells and require folding scaffolds for optimal function (Filonov et al., 2015; Strack et al., 2013). Since three-way junctions have evolved to fold in a cellular context they may be more likely to fold and function in living cells, even if they are modified to contain aptamers. Additionally, since each three-way junction is derived from bacterial or phage RNA, their endogenous protein-binding partners, if they exist, would not be present in mammalian cells.
Notably, only the F30 three-way junction-based sensors functioned in HEK293T cells. Some RNA-based devices derived from bacteria require high concentrations of Mg2+ (e.g. > 1 mM) for their folding or function (Alatossava et al., 1985; Draper, 2004; Kulshina et al., 2010), which is substantially higher than the free Mg2+ concentration in mammalian cells (0.25-1 mM) (Grubbs, 2002; Romani, 2013). However, the F30-based sensor was not substantially different from the other sensors in terms of its Mg2+ requirement. Thermal instability is unlikely to be a reason for the lack of function of the other three-way junctions since all the three-way junction sensors were tested in vitro at 37°C, which is the same temperature that the sensors were examined in live cells. Instead, other factors such as binding to mammalian proteins, endogenous helicases, or misfolding that occurs in the mammalian cytosol may be responsible. Thus, sensors have to be tested in mammalian cells even if they exhibit highly efficient function in vitro.
The F30-based SAM sensor described here, as well as the Broccoli SAM sensor (Litke and Jaffrey, 2019) and Corn SAM sensor (Kim and Jaffrey, 2019) can all be used to image SAM in living cells. Each of these SAM sensors has its own advantages and disadvantages. Both the F30-based SAM sensor and the Broccoli SAM sensor use the Broccoli aptamer, which makes them significantly brighter than the Corn SAM sensor. Although the Corn SAM sensor shows high photostability, the development of the BI fluorophore for imaging Broccoli can potentially allows Broccoli-based sensors to achieve similar photostability (Li et al., 2020).
SIGNIFICANCE
Here we describe an approach to make genetically encoded RNA-based biosensors by repurposing naturally occurring, highly stable RNAs that form three-way junction structures. Three-way junctions are unusually stable due to intramolecular interactions involving multiple helical stems that mutually stabilize the overall three-way junction structure. We took advantage of these interactions, but made these interactions conditional, rather than constitutive. We show that a small molecule-binding aptamer and a fluorogenic aptamer can be inserted into different arms of a three-way junction in such a way that the small molecule can bind and allosterically induce the folding and fluorescence of the fluorogenic aptamer. We show that different metabolite-binding aptamers and fluorogenic aptamers can be connected via three-way junctions to make a variety of allosteric metabolite sensors. These studies present a versatile approach for designing conditionally regulated RNA devices.
STAR Methods
RESOURCE AVAILIBILITY
Lead Contact
Lead Contact, Samie R. Jaffrey (srj2003@med.cornell.edu).
Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact.
Data and Code Availability
The study did not generate/analyze datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Transfection
HEK293T/17 (human embryonic kidney, female, ATCC CRL-11268) or HeLa (human epithelial, female, ATCC CCL-2) cell lines were cultured in full DMEM (Thermo Fisher Scientific 11995-065) with 10% FBS, 100 U/ml penicillin and 100 μg/ml of streptomycin under standard tissue culture conditions (at 37°C and 5% CO2). Cells were detached for splitting and plating using TrypLE Express (Life Technologies) according to the manufacturer’s instructions. Cells were screened for mycoplasma contamination before passaging using Hoechst 33258, according to ATCC recommendations. The cell lines were recently purchased prior to use, but not additionally authenticated. To transfect cells with plasmids, FuGENE HD (Promega 2311) was used as transfection reagent. 1 d before transfection, cells were seeded onto 24-well plates (Corning CLS3527) for microscopy experiments or 6-well plates (Corning CLS3516) for in vitro experiments.
METHOD DETAILS
Structural Information.
The following resolved crystal structures were used in this paper to design RNA based sensors: PDB 4KZ2 (Zhang et al., 2013), PDB 1Y0Q (Golden et al., 2005), PDB 1S72 (Klein et al., 2004), PDB 1NBS (Krasilnikov et al., 2003). Structural images were made using PyMol (Schrödinger).
Preparation of RNA.
Double-stranded DNA templates were designed to contain a 5’ T7 promoter to be used for in vitro transcription. Double-stranded DNA templates were prepared from single-stranded DNA oligos (Integrated DNA Technologies). Templates were amplified by PCR with Taq DNA polymerase (NEB M02373) and gel purified using QIAquick Gel Extraction Kit (Qiagen 28704).
In vitro transcription reactions were performed at 37°C for >10 h with the AmpliScribe™ T7-Flash™ transcription kit (Lucigen ASF3507). RNase-Free DNase I (Lucigen ASF3507) was used at 37°C for 15 min to terminate the reactions. RNA was purified from reactions using Zymo Research RNA Clean & Concentrator™-25 columns (Zymo Research R1707). The quality of RNA samples was checked with polyacrylamide gel electrophoresis using a precast 10% TBE-Urea Gel (Invitrogen EC62752BOX) at 250 V for 30 min. RNA bands were imaged using a ChemiDoc MP (Bio-Rad) with a preset channel (302 nm excitation and 590/110 nm emission) after staining with SYBR Gold (ThermoFisher S11494) diluted 1:10,000 in TBE buffer.
Cloning of Three-Way Junction-Based SAM Sensors.
To prepare pAV-Tornado-F30-Based SAM sensor, the NotI and SacII restriction sites were added to a DNA template containing the three-way junction-based SAM sensors. The pAV-Tornado vector which contains a U6+27 promoter, a U6 terminator, SV40 origin and a Tornado expression cassette (Litke and Jaffrey, 2019; Paul et al., 2002) was used for cloning. F30-based SAM sensor was inserted into the pAV-Tornado expression cassette (Litke and Jaffrey, 2019) of a pAV vector using cloning with NotI and SacII restriction sites. One shot Stbl3 chemical competent E. coli (ThermoFisher C737303) were used for cloning of plasmids. Bacteria were grown on LB:Agar plates supplemented with carbenacillin in a 37°C incubator. Individual colonies were inoculated and in LB supplemented with carbacillin grown in a 37°C incubator with shaking overnight.
Measurements of RNA Fluorescence In Vitro.
Fluorescence measurements of three-way junction RNAs in vitro.
Purified RNAs were diluted to 1 μM in buffer containing 100 mM KCl, 1 mM MgCl2, 40 mM HEPES (pH 7.5). RNAs were then heated up to 75°C for 5 min and then cooled to room temperature. RNA solutions were then mixed with an equal volume of the same buffer containing 20 μM DFHBI-1T or DFHO (synthesized (Song et al., 2014, 2017) or Lucerna 410-1mg). Samples contained a final concentration 0.5 μM RNA and 10 μM DFHBI-1T or DFHO, unless otherwise noted. After 1 h incubation, fluorescence signal of each sample was measured using a Fluoromax-4C (Horiba Scientific) with 460 nm excitation and 505 nm emission, 5 nm slit widths, and 0.1 s integration time. Background signal was detected with the RNA-free sample containing the same concentration of DFHBI-1T in 100 mM KCl, 1 mM MgCl2, 40 mM K-HEPES (pH 7.5). The detected background signal was subtracted from the signal obtained from each RNA-containing sample. Some fluorescence data was collected using a SpectraMax M-series plate reader (Molecular Devices) (figures 5 B and C, S1 B, C, F, and G, S3, S4, and S5 A and B). Results are shown for n=3 biological replicates, unless otherwise stated. No blinding was used in these experiments.
Fluorescence measurements of the three-way junction-based SAM sensors incubated with different SAM analogs.
Samples were prepared as described above. SAM (Sigma-Aldrich A7007), SAH (Sigma-Aldrich A9384), adenosine (Sigma-Aldrich 9251), or methionine (Sigma-Aldrich M9625) was added to RNA samples at 0.1 mM. Then, the samples were incubated at 37°C for 1 h. Fluorescence signal was measured at room temperature as described above. Results are from n=3 biological replicates. No blinding was used in these experiments.
Measuring rates of the three-way junction-based SAM sensor activation and deactivation.
To measure the activation rate, 0.5 μM RNA sensor was incubated with 10μM DFHBI-1T in a buffer containing 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5). After the addition of 0.1 mM SAM to the prepared RNA sample, fluorescence emission was recorded each min over a 40-min period. The fluorescence measurement was normalized to the maximum intensity (100) and the minimum intensity (0). Results are from single replicates. No blinding was used in these experiments.
To measure the deactivation rate, a solution of 0.5 μM RNA sensor and 10 μM DFHBI-1T was first incubated with 0.1 mM SAM (Sigma-Aldrich A7007) at 37°C for 1 h. When the sensor reached the maximal fluorescence, the solution was transferred to a Micro Bio-Spin Column with Bio-Gel P-30 (Bio-Rad 7326223). This column was buffer-exchanged with buffer containing 100 mM KCl, 0.5 mM MgCl2, 40 mM K-HEPES (pH 7.5). The sample was loaded on the buffer-exchanged column and centrifuged at 1,000 × g for 4 min. The flow-through after gel filtration contained the sensor RNA in a SAM-free buffer. 10 μM DFHBI-1T was newly added to the collected flow-through. Then, the fluorescence emission of the flow-through was recorded over a 30-min period at 1 min interval at 37°C. The fluorescence measurement was normalized to the maximum intensity (100) and the minimum intensity (0). Results are from single replicates. No blinding was used in these experiments.
Each fluorescence emission measurement was plotted. One site-Total model of nonlinear regression analysis in GraphPad Prism 8 was used to draw a curve fitting the activation rate measurements. Dissociation-One phase exponential decay model of nonlinear regression analysis in GraphPad Prism 8 was used to draw a curve fitting the deactivation rate measurements.
Fluorescence measurements for dose-response relationships.
50 nM sensor RNA and 1 μM DFHBI-1T was incubated with SAM (Sigma-Aldrich A4377) or SAH (Sigma-Aldrich A9384), at indicated concentrations in the buffer (100 mM KCl, 1 mM MgCl2, 40 mM HEPES, pH 7.5) at 37°C for 1h. The fluorescence emission of each sample was measured as described above. Agonist vs. response-variable slope (four parameters) model of nonlinear regression analysis in GraphPad Prism 8 was used to draw a curve and calculate a hill coefficient from the measurements. Results are shown for n=3 biological replicates, unless otherwise stated. No blinding was used in these experiments.
Fluorescence measurements of F30-based guanine and 5HTP sensors.
RNA samples were prepared as described above. Samples contained a final concentration of 2 μM RNA, 20 μM DFHBI-1T, and 0.2 mM guanine (Sigma 51030-10G) or 2 mM 5HTP (Santa Cruz SC-278210) in the same buffer described above (100 mM KCl, 1 mM MgCl2, 40 mM K-HEPES (pH 7.5). Fluorescence of samples without guanine or 5HTP were also measured for comparison. Background signal was detected with the RNA-free sample containing 20 μM DFHBI-1T and 0.2 mM guanine or 2 mM 5HTP in 100 mM KCl, 1 mM MgCl2, 40 mM K-HEPES (pH 7.5). The detected background signal was subtracted from the signal obtained from each RNA-containing sample. Results are shown for n=3 biological replicates, unless otherwise stated. No blinding was used in these experiments.
Cellular Experiments, Microscopy and Image Processing.
To image cells, we used glass-bottomed 24-well plates (MatTek Corporation P24G-1.5-13-F), coated with poly-D-lysine (Cultrex, 3429-100-01) overnight and rinsed once in water. These plates were additionally coated with Cultrex Mouse Laminin I (Thermo Fisher 340001002) for at least 1 h and rinsed twice with water. Cells were cultured onto pre-treated glass-bottomed plates 1 d prior to transfection. Cell culture media was changed to FluoroBrite DMEM (A1896701) supplemented with 10% FBS and 1× GlutaMax-I (Thermo Fisher Scientific) 6 h before imaging. HEK293T cells expressing the circular F30-based-SAM sensor or the circular F30-Broccoli were treated with 10 μMBI (Li et al., 2020) and 2 μg/mL of Hoechst 33342 (Thermo Fisher H3570) 1 h before imaging. Live-cell fluorescence images were acquired with a CoolSnap HQ2 CCD camera through a 40x oil objective (ND 0.75) mounted on a Nikon Eclipse TE2000-E microscope and analyzed with the NIS-Elements software. Conditions were maintained at 37°C and 5% CO2 during live-cell imaging. The filter set used for Broccoli detection was a filter cube with excitation filter 470 ± 20 nm, dichroic mirror 495 nm (long pass), and emission filter 525 ± 25 nm. Hoechst-stained nuclei were imaged with 350 ± 25 nm excitation filter, 400 nm (long pass) dichroic mirror, and 460 ± 25 nm emission filter (all filters are from Chroma Technology). Cell mean fluorescence intensity was computed using Fiji (Schindelin et al., 2012) by measuring the total fluorescence signal in a region of interest (ROI) divided by ROI area (μm2) and subtracting background based on the mean fluorescence intensity of an untransfected cell. Data in Figure S7E is shown after subtracting background using Image J automatic background subtraction with a rolling ball radius of 300 pixels. ROI was defined as cell cytoplasm area. Cell nucleus area was excluded from ROI.
The timeframe for collecting images for (Figure 6 and Figure S7E) was determined based on previous studies using genetically encoded SAM sensors. In vitro data was included if the RNA sample was homogeneous and of high quality. Data from living cells were included if cells appeared healthy throughout the course of the experiment and did not show significant changes in shape. Imaging data was taken from healthy cells in three separate wells each for F30-control cells and F30-SAM sensor cells. The objective of these experiments was to engineer RNA three-way junction-based sensors and apply the sensors to metabolite imaging in living cells. All hypotheses were predetermined prior to the experiments. Blinding was not used at any stage of the study. No strategies were used for randomization or stratification.
Intracellular SAM imaging.
HEK293T cells were plated onto coated glass-bottom plates. Cells were transfected 24 h later with Tornado plasmids encoding the F30-based-SAM sensor and F30-Broccoli and cultured 2 days prior to imaging. Using the live cell conditions described above, we imaged cells for 2 h after adding cycloleucine (Sigma-Aldrich A48105) to 50 mM (25 mM for Figure S7E) at 15 min intervals. Then, we withdrew cycloleucine by changing cell culture media into fresh media and continued to image cells every 15 min for 6 additional h. Fiji (Schindelin et al., 2012) was used for processing images and for measure the cell mean fluorescence intensity at each time point. Acquisition time: 500 ms.
PARP Cleavage Assay
For detection of apoptosis, HeLa cells were either treated with 1 μg/ml doxorubicin (Sigma Aldrich D1515) 24 h before lysis or transfected using FuGENE HD 48 h before lysis. Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM sodium orthovanadate, 1 mM NaF, prepared at 25°C) containing Halt protease and phosphatase inhibitor cocktail (ThermoFisher 78440) and quantified using a Pierce BCA Protein Assay Kit (ThermoFisher 23225). Proteins were separated by PAGE, transferred to polyvinylidene difluoride membranes and blocked using 5% milk in TBS-T for 1 h at 25°C. Blots were incubated overnight in TBS-T containing 5% BSA at 4 °C with Cleaved PARP (Asp214) XP rabbit mAb (Cell Signaling Technology D64E10). Control blots were incubated with GAPDH mouse mAb (Cell Signaling Technology 97166). Blots were then probed using HRP-linked mouse IgG antibodies (GE Healthcare NA931V) or rabbit IgG (GE Healthcare NA9314V) as secondary antibodies, then developed using Pierce ECL Substrate (ThermoFisher 32106). Chemiluminescence signal was detected using a ChemiDoc MP (Bio-Rad).
Cellular SAM Concentration Measurements.
SAM was isolated from HEK293T cells using a previously described method with slight modification (Mentch et al., 2015). Briefly, HEK293T cells were plated on 6-well plate at a density of 0.5x106 per well on day 0. On day 1, cells were either transfected with the sensor plasmid or they were left un-transfected. SAM was isolated approximately 36 hours after transfection. Two hours before addition of cycloleucine, the media from each well was replaced with fresh media. Cycloleucine (50 mM final) was added at indicted time points. For cycloleucine withdrawal, the media was aspirated and the cells were washed with 1x PBS once. Then cycloleucine free fresh media was added to each well. At the end of each time points, cells were washed with cold 1x PBS. Then 1mL of cold 80% methanol (chilled at −80°C for 1 hour) was added to each well immediately and the plates were transferred to −80°C freezer and kept overnight. Next day, the plate was placed on top of dry ice and 4μL each of formic acid and β-mercaptoethanol was added to each well. Then the cells were carefully scraped from the bottom of the plates. The liquid from each well was placed in a 1.7 mL Eppendorf tube and centrifuged @ 16000xg for 15 mins at 4°C. The supernatant was carefully transferred to a new 1.7 mL tube and the liquid was evaporated using a speedvac system. The cell extract was finally dissolved in 25 μL of water and mixed well. Then they were centrifuged @ 16000xg for 15 mins at 4°C and the supernatant was carefully transferred to a new tube. The SAM concentration was then determined according to the manufacturer's protocol (Bridge-It® SAM Fluorescence Assay Kit (Mediomics, USA). Briefly, a standard curve was generated using known concentration of SAM and the assay reagent. Then 10 μL of the cell extract was used for each time points for measuring the SAM concentration at each time points. Concentration measurements were converted to cellular concentrations using the assumption that cells were spherical, diameter 20 μm. All measurements were performed in triplicate and the error bar represents standard deviation.
QUANTIFICATION AND STATISTICAL ANALYSIS
In vitro experiments (Figures 2B and D, 3A and B, 4 A-C, 5 B-D, S6A, B, F, G, and S7A) were performed in triplicate. The mean and SEM values are shown for plotted graphs (n=3). Significance was determined by unpaired t test provided in Prism 8 (Graphpad). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Curves were prepared using nonlinear regression (Figures 4, 6B, S6B and G) and linear regression (Figure 6C) in Prism 8 (Graphpad). Pearson Correlation was calculated (Figure S7B-D) with 2-sided p-values using Prism 8 (Graphpad). No particular methods were used to determine that data met assumptions of the statistical approaches.
Supplementary Material
Sequences of DNA templates and primers (related to STAR Methods)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cleaved PARP (Asp214) XP rabbit mAb | Cell Signaling Technology | Cat#D64E10 |
| GAPDH mouse mAb | Cell Signaling Technology | Cat#97166 |
| HRP-linked mouse IgG | GE Healthcare | Cat#NA931V |
| HRP-linked rabbit IgG | GE Healthcare | Cat#NA9314V |
| Bacterial and virus strains | ||
| One shot™ Stbl3™ chemical competent E.coli | Thermo Fisher | Cat#C737303 |
| Chemicals, peptides, and recombinant proteins | ||
| BI | (Li et al., 2020) or Lucerna | Cat#600 |
| DFHBI-1T | (Song et al., 2014) or Lucerna | Cat#410 |
| DFHO | (Song et al., 2017) or Lucerna | Cat#500 |
| S-adenosyl-methionine chloride dihydrochloride | Sigma-Aldrich | Cat#A7007 |
| S-adenosyl-methionine iodide | Sigma-Aldrich | Cat#A4377 |
| S-adenosyl-homocysteine | Sigma-Aldrich | Cat#A9384 |
| adenosine | Sigma-Alrdrich | Cat#9251 |
| methionine | Sigma-Aldrich | Cat#M9625 |
| guanine | Sigma-Aldrich | Cat#51030 |
| 5-hydroxytryptophan | Santa Cruz | Cat#278210 |
| cycloleucine | Sigma-Aldrich | Cat#A48105 |
| TRIzol™ LS Reagent | Invitrogen | Cat#10296010 |
| doxorubicin | Sigma-Aldrich | Cat#D1515 |
| Halt protease and phosphatase inhibitor cocktail | ThermoFisher | Cat#78440 |
| Pierce ECL Substrate | ThermoFisher | Cat#32106 |
| Critical commercial assays | ||
| AmpliScribe™ T7-Flash™ transcription kit | Lucigen | Cat#ASF3507 |
| Micro Bio-Spin Columns with Bio-Gel P-30 | Bio-Rad | Cat#7326223 |
| FuGENE HD | Promega | Cat#2311 |
| Bridge-It® SAM Fluorescence Assay Kit | Mediomics | Cat#1-1-1003 |
| Pierce BCA Protein Assay Kit | ThermoFisher | Cat#23225 |
| Deposited Data | ||
| Bacteriophage Phi-29 RNA structure | (Zhang et al., 2013) | PDB: 4KZ2 |
| Phage Twort group 1 ribozyme structure | (Golden et al., 2005) | PDB: 1Y0Q |
| Haloarcula marismortui 23S ribosomal RNA structure | (Klein et al., 2004) | PDB: 1S72 |
| Bacillus subtillis ribonuclease P B-type structure | (Krasilnikov et al., 2003) | PDB: 1NBS |
| Experimental models: cell lines | ||
| HEK293T/17 | ATCC | Cat#CRL-11268 |
| HeLa | ATCC | Cat#CCL-2 |
| Oligonucleotides | ||
| RiboRuler Low Range RNA Ladder | ThermoFisher | Cat#SM1831 |
| Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pAV-Tornado-Broccoli-SAM Sensor | (Litke and Jaffrey, 2019) | N/A |
| pAV-Tornado-F30-Broccoli-SAM sensor | This paper | N/A |
| pAV-Tornado-Broccoli-SAM Sensor Mutant | This paper | N/A |
| pAV-Tornado-Broccoli-Stable Control | This paper | N/A |
| Software and algorithms | ||
| Fiji | NIH | fiji.sc |
| NIS-Element | Nikon | www.nikon.com |
| Prism 8 | GraphPad | www.graphpad.com |
| PyMol | Schrödinger | pymol.org |
Highlights.
Naturally occurring RNA three-way junctions are engineered to undergo inducible folding
Three-way junction-based metabolite sensors are constructed using multiple aptamers
The three-way junction sensor platform is a versatile approach for making RNA sensors
Three-way junction sensors detect SAM levels in live mammalian cells
Acknowledgements
We thank members of the Jaffrey lab for helpful comments and suggestions. This work was supported by NIH grant R01NS064516, R35NS111631, and the American Diabetes Association Pathway to Stop Diabetes Grant 1-18-VSN-02 to S.R.J. J.D.M. was supported by a Medical Scientist Training Program grant from NIH/NIGMS, award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. J.L.L. was supported by NIH F31AI134100. H.K. was supported by Kwanjeong Educational Foundation.
Footnotes
Declaration of Interests
S.R.J. is the co-founder of Lucerna Technologies and has equity in this company. Lucerna has licensed technology related to Spinach, Broccoli, and other RNA-fluorophore complexes. S.R.J. and J.L.L. are founders of Chimerna Therapeutics and have equity in this company.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alatossava T, Jütte H, Kuhn A, and Kellenberger E (1985). Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol 162, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, and Montange RK (2004). Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432, 411–415. [DOI] [PubMed] [Google Scholar]
- Bose D, Su Y, Marcus A, Raulet DH, and Hammond MC (2016). An RNA-Based Fluorescent Biosensor for High-Throughput Analysis of the cGAS-cGAMP-STING Pathway. Cell Chem. Biol 23, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K, Veetil AT, Jaffrey SR, and Krishnan Y (2016). Nucleic Acid–Based Nanodevices in Biological Imaging. Annu. Rev. Biochem 85, 349–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, and Szostak JW (2002). Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci 99, 11616–11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, and Ke A (2011). Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc. Natl. Acad. Sci. U. S. A 108, 7357–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE (2004). A guide to ions and RNA structure. RNA 10, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Moon JD, Svensen N, and Jaffrey SR (2014). Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc 136, 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Kam CW, Song W, and Jaffrey SR (2015). In-Gel Imaging of RNA Processing Using Broccoli Reveals Optimal Aptamer Expression Strategies. Chem. Biol 22, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden BL, Kim H, and Chase E (2005). Crystal structure of a phage Twort group I ribozyme–product complex. Nat. Struct. Mol. Biol 12, 82–89. [DOI] [PubMed] [Google Scholar]
- Grubbs RD (2002). Intracellular magnesium and magnesium buffering. BioMetals 15, 251–259. [DOI] [PubMed] [Google Scholar]
- Hallberg ZF, Su Y, Kitto RZ, and Hammond MC (2017). Engineering and In Vivo Applications of Riboswitches. Annu. Rev. Biochem 86, 515–539. [DOI] [PubMed] [Google Scholar]
- Hermes M, von Hippel S, Osswald H, and Kloor D (2005). S-Adenosylhomocysteine Metabolism in Different Cell Lines: Effect of Hypoxia and Cell Density. Cell. Physiol. Biochem 15, 233–244. [DOI] [PubMed] [Google Scholar]
- Hill AC, and Schroeder SJ (2017). Thermodynamic stabilities of three-way junction nanomotifs in prohead RNA. RNA 23, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Suslov NB, Li N-S, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, and Piccirilli JA (2014). A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol 10, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Sales-Lee J, and Hammond MC (2013). RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic di-GMP and Cyclic AMP-GMP. J. Am. Chem. Soc 135, 4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Chen C, Whiteley AT, Portnoy DA, and Hammond MC (2015). RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP. J. Am. Chem. Soc 137, 6432–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, and Jaffrey SR (2019). A Fluorogenic RNA-Based Sensor Activated by Metabolite-Induced RNA Dimerization. Cell Chem. Biol 26, 1725–1731.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, and Steitz TA (2004). The Roles of Ribosomal Proteins in the Structure Assembly, and Evolution of the Large Ribosomal Subunit. J. Mol. Biol 340, 141–177. [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, and Mondragón A (2003). Crystal structure of the specificity domain of ribonuclease P. Nature 421, 760–764. [DOI] [PubMed] [Google Scholar]
- Kulshina N, Edwards TE, and Ferré-D’Amaré AR (2010). Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA 16, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, and Westhof E (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, and Westhof E (2006). Topology of three-way junctions in folded RNAs. RNA 12, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kim H, Litke JL, Wu J, and Jaffrey SR (2020). Fluorophore-Promoted RNA Folding and Photostability Enables Imaging of Single Broccoli-Tagged mRNAs in Live Mammalian Cells. Angew. Chemie - Int. Ed 59, 4511–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke JL, and Jaffrey SR (2019). Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol 37, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardini JB, and Talalay P (1971). Formation, functions and regulatory importance of S-adenosyl-l-methionine. Adv. Enzyme Regul 9, 349–384. [DOI] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, and Ke A (2008). Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. Mol. Biol 15, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, and Ke A (2011). Variable sequences outside the sam-binding core critically influence the conformational dynamics of the SAM-III/SMK box riboswitch. J. Mol. Biol 409, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gómez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. (2015). Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 22, 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, and Breaker RR (2002). Genetic control by a metabolite binding mRNA. Chem. Biol 9, 1043. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Luo Y, Zhou J, Dayie TK, and Sintim HO (2012). Nanomolar fluorescent detection of c-di-GMP using a modular aptamer strategy. Chem. Commun 48, 9059. [DOI] [PubMed] [Google Scholar]
- Paige JS, Wu KY, and Jaffrey SR (2011). RNA mimics of green fluorescent protein. Science 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Nguyen-Duc T, Song W, and Jaffrey SR (2012). Fluorescence imaging of cellular metabolites with RNA. Science 335, 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, and Engelke DR (2002). Effective expression of small interfering RNA in human cells. Nat. Biotechnol 20, 505–508. [DOI] [PubMed] [Google Scholar]
- Porter EB, Polaski JT, Morck MM, and Batey RT (2017). Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat. Chem. Biol 13, 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MP, and Ellington A (1999). In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol 17, 62–66. [DOI] [PubMed] [Google Scholar]
- Romani AMP (2013). Magnesium Homeostasis in Mammalian Cells. In Metallomics and the Cell. Metal Ions in Life Sciences, (Springer, Dordrecht: ), pp. 69–118. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Fuchs RT, Grundy FJ, and Henkin TM (2010). The SAM-responsive SMK box is a reversible riboswitch. Mol. Microbiol 78, 1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Strack RL, and Jaffrey SR (2013). Imaging bacterial protein expression using genetically encoded RNA sensors. Nat. Methods 10, 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Strack RL, Svensen N, and Jaffrey SR (2014). Plug-and-Play Fluorophores Extend the Spectral Properties of Spinach. J. Am. Chem. Soc 136, 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Filonov GS, Kim H, Hirsch M, Li X, Moon JD, and Jaffrey SR (2017). Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol 13, 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, and Breaker RR (1999). Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Structure 7, 783–791. [DOI] [PubMed] [Google Scholar]
- Strack RL, Disney MD, and Jaffrey SR (2013). A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat–containing RNA. Nat. Methods 10, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Nguyen T, McAuliffe K, You M, Sun Z, Nguyen T, McAuliffe K, and You M (2019). Intracellular Imaging with Genetically Encoded RNA-based Molecular Sensors. Nanomaterials 9, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, and Breaker RR (1997). Rational design of allosteric ribozymes. Chem. Biol 4, 453–459. [DOI] [PubMed] [Google Scholar]
- Traut TW (1994). Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem 140, 1–22. [DOI] [PubMed] [Google Scholar]
- Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR, and Ferré-D’Amaré AR (2014). Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol 21, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner KD, Sjekloća L, Song W, Filonov GS, Jaffrey SR, and Ferré-D’Amaré AR (2017). A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat. Chem. Biol 13, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, and Breaker RR (2002). Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956. [DOI] [PubMed] [Google Scholar]
- You M, Litke JL, and Jaffrey SR (2015). Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci 112, E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, and Chi Y-I (2013). Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 19, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of DNA templates and primers (related to STAR Methods)
Data Availability Statement
The study did not generate/analyze datasets or code.