Abstract
Background
During the COVID-19 pandemic, thousands of pregnant women have been infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The implications of maternal SARS-CoV-2 infection on fetal and childhood well-being need to be characterized. We aimed to characterize the fetal immune response to maternal SARS-CoV-2 infection.
Methods
We performed single-cell RNA-sequencing and T cell receptor sequencing on cord blood mononuclear cells (CBMCs) from newborns of mothers infected with SARS-CoV-2 in the third trimester (cases) or without SARS-CoV-2 infection (controls).
Results
We identified widespread gene expression changes in CBMCs from cases, including upregulation of interferon-stimulated genes and major histocompatibility complex genes in CD14+ monocytes, transcriptional changes suggestive of activation of plasmacytoid dendritic cells, and activation and exhaustion of natural killer cells. Lastly, we observed fetal T cell clonal expansion in cases compared to controls.
Conclusions
As none of the infants were infected with SARS-CoV-2, our results suggest that maternal SARS-CoV-2 infection might modulate the fetal immune system in the absence of vertical transmission.
Impact
The implications of maternal SARS-CoV-2 infection in the absence of vertical transmission on fetal and childhood well-being are poorly understood.
Maternal SARS-CoV-2 infection might modulate the fetal immune system in the absence of vertical transmission.
This study raises important questions about the untoward effects of maternal SARS-CoV-2 on the fetus, even in the absence of vertical transmission.
Introduction
Millions of people worldwide have or will become infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing Coronavirus Disease 2019 (COVID-19), and the infection of pregnant women with SARS-CoV-2 infection has been widespread.1–6 Despite the prevalence of antepartum infection, we have a limited understanding of the implications of SARS-CoV-2 infection on fetal and offspring health. To date, there are limited case reports of vertical, mother-to-child transmission of SARS-CoV-2,7–12 and vertical transmission remains rare in most pregnancies complicated by maternal SARS-CoV-2 infection.1,6,13–17 Nonetheless, in the absence of direct fetal infection and toxicity, maternal SARS-CoV-2 infection may still affect fetal development. Maternal immune activation during pregnancy after viral infection without vertical transmission can have long-term consequences for the newborn, including abnormal neurologic18,19 or immune system development.20,21
Pregnancy is a complex immunologic state, and there is no data on the effect of SARS-CoV-2-dysregulated immune state during pregnancy on the fetus. Given the number of pregnant women infected with SARS-CoV-2 worldwide, it is important to determine the potential transgenerational implications of infection with SARS-CoV-2 during pregnancy beyond vertical transmission. Maternal SARS-CoV-2 test positivity has been significantly associated with admission for neonatal care and with neonatal morbidities such as respiratory distress syndrome and hyperbilirubinemia.22 To date, research on the implications of SARS-CoV-2 infection during pregnancy on the offspring immune system has been limited to antibody transplacental transfer6,23,24 and postnatal evaluation of infants born to mothers infected with SARS-CoV-2 during pregnancy without a nonexposed control group.25,26 The latter may be also confounded by ex utero determinants of immune development during the first week of life.27,28 In the present study, we characterize the composition and cell-type-specific transcriptional landscape of umbilical cord blood mononuclear cells (CBMCs) from term gestation infants (>37 weeks) born to mothers infected with SARS-CoV-2 in the third trimester without vertical transmission. This immunogenomic investigation provides evidence of both innate and adaptive fetal immune transcriptional changes in pregnancies complicated by SARS-CoV-2 infection. Our results suggest that even in the absence of vertical transmission, SARS-CoV-2 maternal infection in the third trimester might modulate the fetal immune system.
Results
To characterize the fetal immunologic landscape in pregnancies complicated by maternal SARS-CoV-2 infection, we performed droplet-based single-cell RNA-sequencing (scRNAseq) of CBMCs from infants born to mothers with SARS-CoV-2 infection during pregnancy (cases) and infants born to mothers without SARS-CoV-2 infection (controls). CBMCs from three cases and three controls were obtained from our biorepository.29 None of the three infants in this study born to mothers with SARS-CoV-2 were positive for SARS-CoV-2 postnatally, had detectable SARS-CoV-2 messenger RNA (mRNA) in the placenta, or developed any neonatal morbidity. All mothers with COVID-19 in the third trimester were classified as having mild disease without respiratory support.30 Infants born to mothers negative for SARS-CoV-2 and asymptomatic (universal screening at admission for labor) during the same epoch served as controls. Maternal comorbidities including well-controlled thyroid dysfunction, obesity, or gestational diabetes were matched between cases and controls as feasible. The time of maternal infection and birth in cases varied between 7 and 66 days. Table 1 displays demographic and clinical data from the cases and controls.
Table 1.
Clinical characteristics of cases and controls.
| Subjects | Onset of symptoms (GA) | SARS-CoV-2 PCR (GA) | Delivery (GA) | Days between the onset of symptoms and birth | Maternal symptoms at test | Delivery mode | Any labor? | Maternal comorbidities | Sex assigned at birth | Placental viral load by RT-PCR | 24 h nasopharyngeal viral load by RT-PCR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control 1 | NA | 39.9 | 40 | NA | VD | Yes | Male | 1 | NA | ||
| Control 2 | NA | 38.9 | 39w 4d | NA | CS | Yes | BMI > 30, thyroid disease | Female | 1 | NA | |
| Control 3 | NA | 38.4 | 38w 6d | NA | CS | Yes | BMI > 30 | Female | ND | NA | |
| Case 1 | 30w 3d | 30w 7d | 39w 7d | 66 | Fever/chills, nasal congestion, loss of taste/smell, sore throat, night sweats | VD | Yes | Thyroid disease | Male | 1 | Negative |
| Case 2 | 34w 4d | 35w 4d | 40w 1d | 40 | Cough, fever/chills, myalgias, headache, chest discomfort | CS | Yes | Diabetes/GDM, BMI > 30, thyroid disease | Male | 1 | Negative |
| Case 3 | 39w 0d | 39w 4d | 40w | 7 | Cough, fever/chills | VD | Yes | BMI > 30 | Female | 1 | Negative |
GA gestational age, VD vaginal delivery, CS cesarean section, ND not done, NA not applicable.
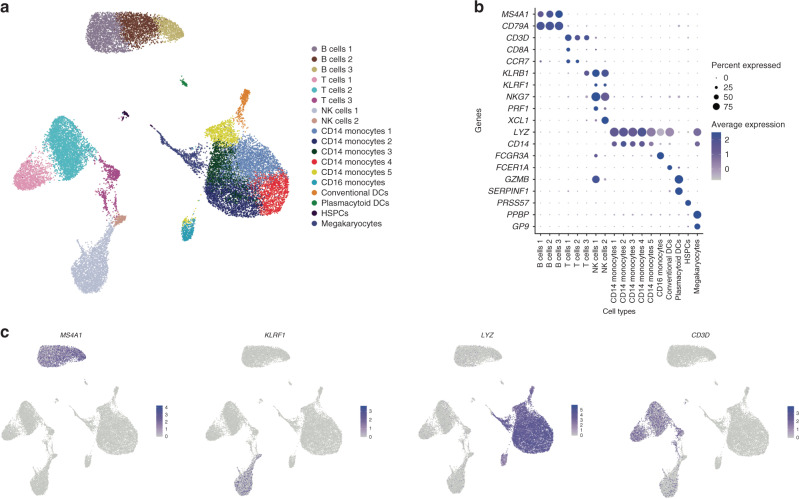
CBMCs were processed on the 10X Genomics Single-Cell Immune platform (see “Methods”). After quality control and doublet removal, we included 14,748 cells with high-quality single-cell transcriptomes from cases and 11,222 cells from controls in our dataset. (See quality control metrics in Supplemental Fig. 1A, B.) The cell population composition was visualized using uniform manifold approximation and projection (Fig. 1a), and cell types were inferred by cluster-specific canonical marker genes (Fig. 1b, c). We did not observe any differences in cell cluster composition between cases and controls (Supplemental Fig. 1c, d).
Fig. 1. Cell composition of cord blood mononuclear cells by scRNAseq.
a UMAP of all cases and control CBMCs, with cell populations labeled by color. N = 3 samples per group. b Dot plot of marker gene expression across cell clusters. Y-axis displays marker genes and X-axis displays cell clusters. Relative size of dots represents the percentage of gene expression within a cell cluster, and the relative color of dots represents average expression. c Marker gene plots for Ms4a1 (B cells), Klrf1 (NK cells), Lyz (monocytes), and CD3D (T cells). Colored dots indicate gene expression.
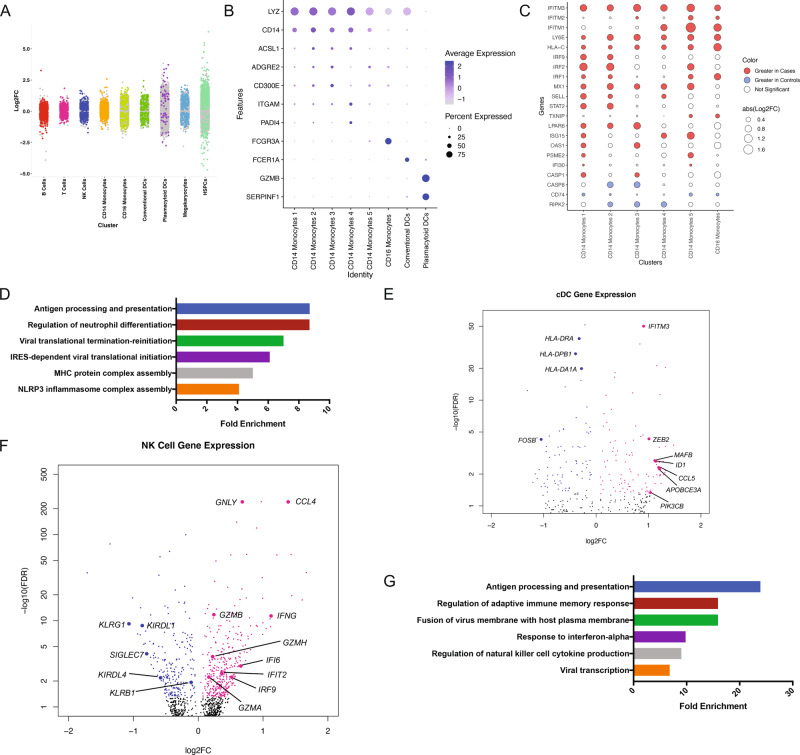
To explore transcriptional signatures in fetal immune cells associated with maternal SARS-CoV-2 infection, we performed differential gene expression (DGE) analysis within cell types comparing cases and controls. Genes with a false discovery rate (FDR) < 5% were considered statistically significant. We identified hundreds of genes across nearly all cell types with altered expression (Fig. 2a). We used gene ontology (GO) analysis to broadly classify genes significantly disrupted by maternal SARS-CoV-2 infection based on DGE (Supplemental Table 1).
Fig. 2. Genes expression differences between cord blood mononuclear cells from infants born to mothers with SARS-CoV2 infection (cases) or mothers without SARS-CoV-2 infection (controls).
a Strip plot displaying differential gene expression between cases and controls. Colored dots represent significant genes (FDR < 0.05). X-axis displays select CBMC cell types. N = 3 samples per group. b Dot plot of marker gene expression across monocyte cell clusters (CD14+ and CD16+). Y-axis displays marker genes and X-axis displays cell clusters. Relative size of dots represents the percentage of gene expression within a cell cluster, and the relative color of dots represents average expression. c Dot plot of highly variable genes in CD14+ and CD16+ monocytes between cases and controls. All significant genes FDR < 0.05. Red indicates an increase in expression in cases and blue indicates an increase in expression in controls. N = 3 samples per group. d Select Gene Ontology (GO; Biological Pathway) results for differentially expressed genes between cases and controls in CD14+ monocytes. GO categories have FDR < 0.05. X-axis indicates fold enrichment relative to the reference gene set of all expressed genes in the dataset. e Volcano plot of DGE between cases and controls in conventional dendritic cells (cDCs). Colored dots indicate statistical significance (FDR < 0.05). Positive log 2 FC (pink dots) indicates higher expression in cases, and negative log 2 FC (blue dots) indicates higher expression in controls. N = 3 samples per group. f Volcano plot of DGE between cases and controls in natural killer (NK) cells. Colored dots indicate statistical significance (FDR < 0.05). Positive log 2 FC (pink dots) indicates higher expression in cases, and negative log 2 FC (blue dots) indicates higher expression in controls. N = 3 samples per group. g Select Gene Ontology (GO; Biological Pathway) results for differentially expressed genes between cases and controls in NK cells. GO categories have FDR < 0.05. X-axis indicates fold enrichment relative to the reference gene set of all expressed genes in the dataset.
CD14+ monocytes were grouped into five clusters and CD16+ monocytes were grouped into one cluster (Fig. 2b). CD14+ subpopulations demonstrated variable expression of inflammatory genes, including ACSL1, ADGRE2, CD300E, and PADI4, which aligns with prior single-cell analysis showing monocyte diversity.31 Consistent with data from adult COVID-19 patients,32,33 we found that CD14+ monocytes from cases demonstrated increased expression of interferon (IFN)-stimulated genes (ISGs) (Fig. 2c) and concomitant IFNAR2 downregulation (Supplemental Fig. 2a), which could reflect exposure to IFN prenatally.34 Of note, we found that there was variable upregulation of ISGs among various CD14+ monocyte clusters in cases compared to controls (Fig. 2c), indicating that there is no homogeneous ISG regulation among fetal monocyte clusters secondary to maternal SARS-CoV2 infection. This finding is consistent with previous reports that upon stimulation with IFN-γ, fetal bone marrow monocytes have variable upregulation of ISGs when compared to adult bone marrow monocytes.35 GO analysis of DGE in CD14+ monocytes demonstrated enrichment of genes associated with antigen presentation and viral translational termination and reinitiation (Fig. 2d). CB CD14+ monocytes from cases also showed upregulation of major histocompatibility class (MHC) I and II genes (Supplemental Fig. 2A) suggesting activation in response to IFN signaling.36 However, some HLA genes were downregulated in CD14+ monocytes (HLA-E and HLA-B) (Supplemental Fig. 2A) consistent with decreased antigen presentation capacities in fetal bone marrow CD14+ monocytes compared to adult bone marrow counterparts upon in vitro stimulation with IFN-γ.35 Furthermore, CD14+ monocytes from cases showed upregulation of TLR receptor transcripts (TLR2, TLR4, and TLR5) paired with upregulation of FOS and downregulation of transcriptional inhibitors of nuclear factor kappa-light-chain-enhancer of activated B cells (NFKB), including NFKBIA and NFKBIE, all of which are associated with increased NFKB activation and cytokine production37 (Supplemental Fig. 2A). Of note, CD14+ monocytes from cases had decreased expression of autophagy (ATG14, ATG2A, and ATG3) and endoplasmic reticulum stress (XBP1 and HSPA5) genes, which may contribute to a defect in macrophage differentiation38 (Supplemental Fig. 2A). Expression of S100A Alarmins in CB monocytes has been associated with chorioamnionitis and fetal inflammatory response syndrome (FIRS).39 S100A8, S100A9, and S100A12 were found to be decreased in CD14+ monocytes in our cases compared to controls, which might suggest that the response elicited from SARS-CoV-2 maternal infection in monocytes differs from the changes elicited in FIRS secondary to chorioamnionitis (Supplemental Fig. 2A).
Similar to CD14+ monocytes, we identified induction of ISGs in nonclassical CB monocytes (CD16+) (Fig. 2c). In contrast to CD14+ monocytes, we found that there was decreased expression of cell adhesion genes (including PLAUR and THBS1), attenuation of immune activation signaling pathways genes (FOS, FOSB, MAP3K8, STAT6, and FCER1G), and decreased expression of inflammatory molecules like resistin (RETN) (Supplemental Fig. 2B). Together, these results suggest induction of ISGs in monocytes from cases compared to controls and differences in transcriptional changes in classical and nonclassical monocytes that might indicate preferential activation of classical monocytes in cases compared to controls.
We captured the transcriptomes of both plasmacytoid and conventional dendritic cells (pDC and cDC, respectively) in CB. In adults infected with SARS-CoV-2, both types of DCs are functionally impaired, and there is an increased ratio of cDCs to pDCs in severe patients.40 In our study, CB cDC from cases showed increased expression of ISGs like IFITM3 and APOBEC3A (Fig. 2e). Fetal cDC from cases showed a transcriptional profile suggestive of innate immune activation including increased expression of PIK3CB, which is downstream of TLR5 and TLR7,41 as well as increased transcription of CCL5, which can be upregulated after TLR3 stimulation (Fig. 2e).42 Evidence of impaired cDC maturation was suggested by upregulation of ID1, which antagonizes dendritic cell differentiation and antitumor immunity in mice,43 as well as increased MAFB transcription, which suppresses cDC maturation.44 cDCs from cases also demonstrated decreased expression of FOSB and many MHC II genes.45 pDCs in cases also showed markers of immune activation, including upregulation of RELB, which promotes DC activation through RelB-p50 dimer,46 upregulation of MHC class I and class II genes, and unfolded protein response activation, as shown by increased transcription of XBP147 (Supplemental Fig. 2C). Together, these transcriptional findings could be consistent with activation of pDC over cDC in the CB of cases, potentially through activation of TLRs.
In adults, SARS-CoV-2 infection is associated with fewer blood natural killer (NK) cells, but a higher activation state in circulating NK cells.48 We identified two clusters of CB NK cells. One population of NK cells (cluster 1) expressed higher levels of GZMB, while the second population of NK cells (cluster 2) expressed IL7R and XCL1, suggesting that cluster 1 corresponded to CD56dim and cluster 2 corresponded to CD56bright NK cells, as NCAM1 (CD56) is technically not well captured in scRNAseq.49 Similar to adult NK cells, CB NK cells from SARS-CoV-2-positive pregnancies showed signs of exposure to IFN, including induction of ISGs like IFI6, IFIT2, and IRF9 (Fig. 2f).48,50,51 We identified increased transcription of CCL4, expression of cytotoxic genes including GNLY, GZMA, GZMB, and GZMH, and increased transcription of IFNG, paired with decreased expression of NK inhibitory molecules (Fig. 2f).48,50,51 There were transcriptional changes associated with exhaustion, such as decreased expression of KLRG1 and SIGLEC7.52 DGE in NK cells between cases and controls were enriched for genes related to the IFN-α response, regulation of NK cell cytokine production, and viral transcription (Fig. 2g).
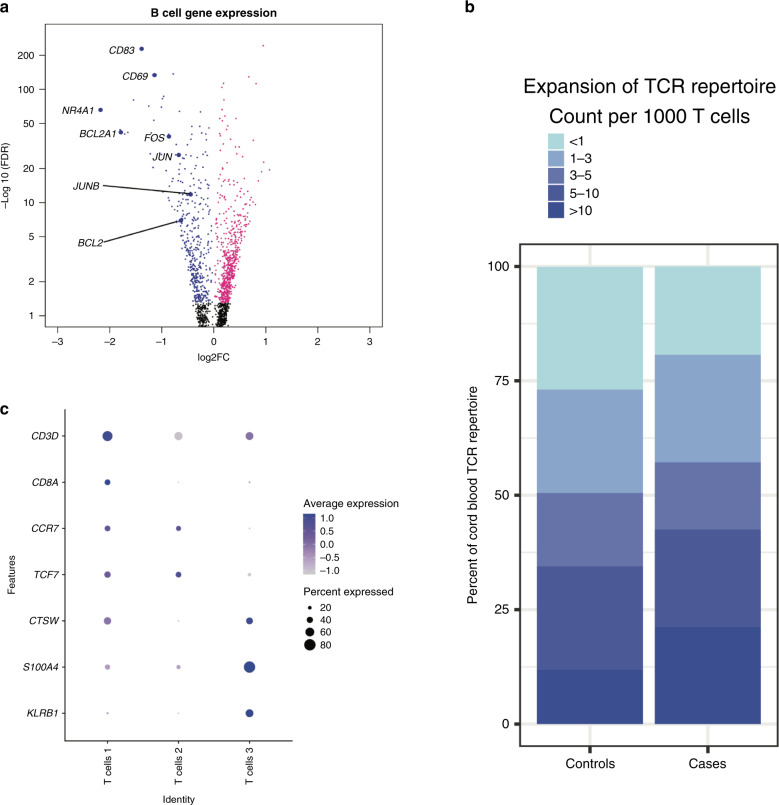
In adults with acute COVID-19, there is a heterogeneous adaptive immune response in peripheral blood, including B cell receptor and T cell receptor (TCR) arrangements specific to SARS-CoV-2.50 Given these findings, we evaluated whether maternal infection with SARS-CoV-2 had any effect on CB lymphocyte gene expression. In B cells from infants exposed to SARS-CoV-2 in utero, we identified three clusters of CB B cells corresponding to non-plasma/plasmablast (Clusters 1 and 2) and plasma/plasmablast cells (Cluster 3) based on MZB1 expression50 (Supplemental Fig. 2D). With the possible exception of germinal center (GC) B cells, all human B cell populations found in lymphoid tissues can also be demonstrated in peripheral blood53 with enrichment of mature, naive B cells and regulatory B cells in CB.54–56 Therefore, transcripts of genes related to B cell receptor activation are detected in circulating B cells57–59 and their products might participate in tonic B cell signaling under homeostatic conditions in peripheral blood.60 Human CB B cell transcriptional program differs from adult peripheral B cell and confers accelerated but transient responsiveness to stimulation.56 Surprisingly, in B cells from infants born to mothers infected with SARS-CoV-2, we identified decreased transcription of genes downstream of B cell receptors in all clusters compared to controls. Specifically, we found a decreased transcription of NR4A1, CD69, and CD83 in all B cells (Fig. 3a). NR4A1 encodes Nur77, an orphan nuclear receptor that is induced upon B cell activation in peripheral blood in humans.61 CD83 is expressed in peripheral B cells and correlates with the engagement of BCR, TLRs, or CD40.62,63 Concordant with transcriptional evidence of defective B cell activation in CB lymphocytes from cases, we also found decreased expression of CD69,64 activator protein-1, and nuclear factor of activated T cells genes,65 as well as anti-apoptotic genes, including BCL2 and BCL2A166 (Fig. 3a). These transcriptional changes suggestive of potential B cell dysfunction, combined with our prior findings of decreased transplacental transmission of IgG against SARS-CoV-2 compared to IgG against other antigens,6,23 might translate into potential impairments in antibody-mediated immunity to SARS-CoV-2 in neonates born to mothers with COVID-19. The humoral defect might be secondary to impaired passive immunity against SARS-CoV2 combined with potential impaired B cell activation in cases compared to controls, but this phenomenon requires further functional characterization.
Fig. 3. Effects of maternal SARS-CoV-2 in B and T cord blood lymphocytes.
a Volcano plot of DGE between cases and controls in B cells. Colored dots indicate statistical significance (FDR < 0.05). Positive log 2 FC (pink dots) indicates higher expression in cases, and negative log 2 FC (blue dots) indicates higher expression in controls. N = 3 samples per group. b Dot plot of marker gene expression across T cell clusters. Y-axis displays marker genes, and X-axis displays cell clusters. The relative size of dots represents the percentage of gene expression within a cell cluster, and the relative color of dots represents average expression. c Stacked bar plot of T cell receptor clones in cases and controls, with colors indicating the number of clones in a particular category. The difference in the distribution of T cell clones between cases and controls is statistically significant (Kolmogorov–Smirnov test p value 2.2e − 16).
We also identified three clusters of T cells (Fig. 3b). Cluster 1 corresponded to cytotoxic (CD8+) T cells, while Clusters 2 and 3 corresponded to helper T cells.57 Naive helper T cells predominate in CB67–69 and retain a partial expression of a fetal-associated regulatory T cell signature.67,70–72 Increased expression of CCR7 in T cell Cluster 2 suggests that this cluster includes either naive T cells or central memory T cells,73 and increased CTSW and KLRB1 in Cluster 3 suggest that this cluster includes effector and memory T cells.74,75 In adults with COVID-19, CD8+ T cells show decreased cytotoxic potential and exhaustion driven by interleukin-6 (IL-6).76 Similar to adults, CB CD8+ T cells from cases demonstrated transcriptional signatures suggestive of impaired function including decreased expression of GZMA, FOS, and JUN77–79 (Supplemental Fig. 3A). Furthermore, there was increased expression of KLRB1 and CCR7 (Supplemental Fig. 3A). CCR7 is expressed in naive and central memory T cells and lost in effector, effector memory, and terminally differentiated T cells.73,80 KLRB1 encodes CD161.75 CD161 is associated with IL-17-secreting T cells81–85 and is found in CB-naive T cells that become IL-17-secreting T cells.82,85 CD161 is also expressed in innate-like T cells including mucosal-associated invariant T cells,86 gamma-delta-positive T cells,82 invariant NK T cells,87 and promyelocytic leukemia zinc-finger protein-positive CD4+ T cells.88 In innate-like T cells, CD161 may track with infections or inflammatory complications of pregnancy.88 GO analysis of DGEs in CD8+ T cells demonstrated enrichment for genes associated with T cell tolerance, proliferation, and the response to IFN-γ (Supplemental Fig. 3B). In T cell Cluster 2, we found increased expression of IL-6–IL-17 axis genes including RORA, ARID5A, RBPJ, and IL6ST in cases compared to controls (Supplemental Fig. 3C). The IL-6–IL-17 axis has been implicated in mediating the neurodevelopmental effects of maternal immune activation in mice89–92 and coordinating tissue inflammatory responses.93
T cell antigen receptor (TCR) repertoire in T cells reflects selection by self and foreign antigens. To investigate the repertoire of TCRs in CB from SARS-CoV-2-exposed pregnancies and controls, we performed single-cell TCR sequencing. A total of 1943 T cells were analyzed, and T cells with TCR information were well equally distributed between subject and T cell populations (Supplemental Fig. 3D, E). In CB from pregnancies complicated by maternal SARS-CoV-2 infection, there was significantly greater number of T cells with >5 clones compared to controls (40.4% in cases vs 30.9% in controls, Kolmogorov–Smirnov test p value 2.2e − 16) (Fig. 3d). The T cell clonal expansion in CB from cases is consistent with results of T cell repertoire analysis from adults infected with SARS-CoV-2.50 However, an analysis of T cell clonal diversity across four metrics (Shannon, inverse Simpson, Chao1, and abundance-based coverage estimator) revealed variability between samples in clonal diversity and no clear differences between cases and controls (Supplemental Fig. 3F).
Discussion
While the application of scRNAseq analysis to CBMC in the context of maternal illness is novel, our study is exploratory and has several limitations. Importantly, the small number of samples limits the generalizability of our conclusions. However, few studies have evaluated CB immune populations by single-cell transcriptomics,94,95 and our results illustrate an important and potentially underrecognized effect of the COVID-19 pandemic that should be further studied. Interpretation of our findings in light of prior known literature of CB leukocytes is challenging given the lack of detailed characterization of CBMC populations using scRNAseq as has been done in peripheral blood mononuclear cells from adults.94,95 Future studies using simultaneous epitope and transcriptome measurement in single cells will make these correlations possible.96 All cases included in this study were classified as mild maternal SARS-CoV-2 infection; more severe maternal infection could result in more dramatic or different fetal immune genomic signatures, as it has been demonstrated that infection during pregnancy increases the possibility of intensive care unit admission.5,97–99 Furthermore, the time from infection to delivery and CB collection likely affects the immune phenotype observed in CB. As the time of maternal infection and birth in our cohort varied between 7 and 66 days, more pronounced findings may be found in samples with more homogeneous timing between infection and collection. Lastly, all mothers affected with SARS-CoV-2 in our cohort had comorbidities including well-controlled thyroid dysfunction, obesity, or gestational diabetes. Although we included mothers with similar comorbidities in the control population (except for gestational diabetes) and all these comorbidities were medically managed, it is possible that our results are influenced by the comorbidities of the mothers. However, thyroid disease, obesity, or gestational diabetes in the mother have not been reported to trigger the transcriptional response patterns we observed in cases compared to controls.100–103 Lastly, we did not functionally test the CB leukocytes to evaluate the implications of the transcriptional observations we report.
Despite these limitations, the present study identifies transcriptional changes suggestive of a fetal immune response after maternal infection with SARS-CoV-2 in the absence of vertical transmission and suggests potential transplacental immune implications of maternal SARS-CoV-2 infection in the absence of mother-to-child transmission. The source of signals promoting transcriptional changes in neonatal monocytes and other immune cells in the absence of vertical transmission is unknown. Ex vivo studies have shown that transplacental transfer of IL-1β, IL-6, and tumor necrosis factor-α is limited.104 Type I IFNs are increased in the peripheral circulation of patients with mild COVID-19,32 but the ability of IFN to cross the human placenta is unclear.105 Our results raise the possibility that pro-inflammatory signaling in the mother in response to SARS-CoV-2 might promote IFN signaling at the feto-maternal interface, and placental barrier dysfunction could result in loss of selective placental permeability to circulating maternal factors. Consistent with this possibility, a recent report has identified placental inflammatory responses in SARS-CoV-2-infected mothers compared to controls in the absence of viral mRNA in the placenta.106 It is widely accepted that different viral infections, such as severe acute respiratory syndrome by influenza H1N1 virus or human immunodeficiency virus (HIV) in pregnant women, can compromise the offspring’s fetal immune system even when the infection is limited to the mother or placental bed.19,21,107–109 Furthermore, even without fetal infection, maternal HIV and H1N1 infections can be associated with neurological and behavioral diseases in offspring.19,21,107–109 To our knowledge, this is the first time that scRNAseq has been used to explore the effect of maternal infection on the fetal immune system. Therefore, it is possible that the effects observed in our cohort are not SARS-COV-2-specific. However, S100A genes in monocytes were downregulated in our cohort in response to SARS-CoV2, while they are upregulated in monocytes of infants exposed to chorioamnionitis,39 which might suggest, at minimum, differences in the response of fetal monocytes to maternal infection with SARS-CoV2 compared to chorioamnionitis.
Further experimental and functional data need to be collected to clarify how maternal infection with SARS-CoV-2 influences the fetal immune system, whether similar changes occur during other maternal infections without vertical transmission and the potential effect in the newborn as reported for HIV and influenza.19,21,107,108 Given the extensive literature linking maternal immune dysregulation and abnormal fetal development in viral infections, this study raises important questions about untoward effects of maternal SARS-CoV-2 on the fetus, even in the absence of vertical transmission, and highlights the need for further studies to better characterize the fetal immune response in pregnancies affected by SARS-CoV-2 infection.
Methods
Sample collection, cryopreservation, and placental viral load
The subjects were six infants born at term to mothers with or without SARS-CoV-2 infection in the third trimester. Parents of the infants provided informed consent before sample collection and study participation. The study was approved by the Institutional Review Board of the Mass General Brigham (IRB 2020P001478 and IRB2020P000804). Cord mononuclear cells were collected using Ficoll and cryopreserved as described.29 We used dimethyl sulfoxide as our cryopreservant agent as it adequately conserves gene expression profiles in cryopreserved cells compared to fresh cells in droplet-based single-cell RNA-sequencing.110 We excluded preterm infants, as a strong pro-inflammatory signature in CB has been reported in infants born preterm.111 None of the infants was exposed to prenatal steroids, was diagnosed with intrauterine growth restriction, or had any neonatal morbidities. Placental viral load was measured as previously reported.112
Single-cell RNA-sequencing
CBMC aliquots were thawed in a 37 °C water bath and resuspended in RPMI-1640 with 10% fetal bovine serum (FBS) (Thermo Fisher). Samples were centrifuged at 350 × g for 7 min at 4 °C. Cells were resuspended in 100 μl of 1× phosphate-buffered saline with 2.5% FBS and 2 mM EDTA.
Dead cells and red blood cells were depleted using the EasySep Dead Cell Depletion Kit and EasySep RBC Depletion Reagent (STEMCELL), according to the manufacturer’s instructions. Cells were resuspended in RPMI/10% FBS and counted. Cells were loaded onto the 10X Chromium controller at a targeted recovery density of 10,000 cells per sample. Samples were processed and sequencing libraries were created using the 10X the Chromium Next GEM single-cell V(D)J Reagent Kit v1.1 with human TCR V(D)J enrichment following the manufacturer’s instructions.
Single-cell RNA-sequencing data analysis
Sequencing data were aligned to the genome and processed using the 10X Genomics Cell Ranger software, version 4.0.0. All cells were combined into a single dataset. Doublets were removed using Scrublet version 0.2.1, and the remaining cells were reclustered. Mitochondrial genes were filtered from the dataset. Cells with fewer than 250 or more than 2500 unique genes were excluded. Cells were then clustered using the Seurat R package (version 3.2.3). Specifically, the SCT functionality of Seurat was used to identify cell types that did not depend upon unique aspects of individual samples. Clustering resolution was set to 0.8, and the first 15 principal components were used. The data were log normalized and scaled to 10,000 transcripts per cell. The expression of known marker genes was used to assign each cluster to one of the main cell types. The Seurat FindMarkers function was used to identify genetic markers of cellular subtypes.
Identification of differentially expressed genes between cases and controls
To identify differentially expressed genes by cell type, we performed a differential gene expression analysis using Monocle2. The analysis was conducted on each cell type and also certain unions of cell types with common traits. The data were modeled and normalized using a negative binomial distribution and counts data were normalized for gene length and read depth. Genes whose FDR was <5% were considered statistically significant. GO analysis was performed using gprofiler2 version 0.2.0, and terms were selected from the Biological Process category of GO terms.
TCR sequencing
TCR sequencing data were analyzed using the R package scRepertoire (version 3.12).
Supplementary information
Acknowledgements
We thank Dr. Pablo J. Patino (Department of Immunology, School of Medicine, Universidad de Antioquia) for helpful discussions.
Author contributions
J.D.M. and B.T.K. conceived and designed the study. J.D.M., B.T.K., D.P., X.A., J.Z.L., and A.G.E. performed experiments and acquired data. A.-C.V. provided essential protocols. J.D.M., B.T.K., B.F., N.P.S., A.G.E. and A.-C.V. analyzed data. J.D.M. and B.T.K. drafted the manuscript, and all authors edited the manuscript. J.D.M., X.A., A.G.E. and P.H.L. contributed to clinical sample collection. P.H.L. and B.T.K. cosupervised the study.
Funding
This work was supported by Research Fellowship Award # 707702 from the Crohn’s and Colitis Foundation (J.D.M.), NINDS K08 NS112338-02 (B.T.K.), UM-1 AI069412-15S1 (J.Z.L.), and HD100022 and 1R01HD100022 and 3R01HD100022-02S2 (A.G.E.).
Data availability
Sequencing data have been deposited in the Gene Expression Omnibus under accession no. GSE165193.
Informed consent
Parents of the infants provided informed consent before sample collection and study participation. The study was approved by the Institutional Review Board of the Mass General Brigham (IRB 2020P001478 and IRB2020P000804).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Paul H. Lerou, Brian T. Kalish.
Contributor Information
Juan D. Matute, Email: jmatute@mgh.harvard.edu
Brian T. Kalish, Email: brian.kalish@sickkids.ca
Supplementary information
The online version contains supplementary material available at 10.1038/s41390-021-01793-z.
References
- 1.Chen L, et al. Clinical characteristics of pregnant women with Covid-19 in Wuhan, China. N. Engl. J. Med. 2020;382:e100. doi: 10.1056/NEJMc2009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prabhu M, et al. Pregnancy and postpartum outcomes in a universally tested population for SARS‐CoV‐2 in New York City: a prospective cohort study. Bjog Int. J. Obstet. Gynaecol. 2020;127:1548–1556. doi: 10.1111/1471-0528.16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton D, Fuchs K, D’Alton M, Goffman D. Universal screening for SARS-CoV-2 in women admitted for delivery. N. Engl. J. Med. 2020;382:2163–2164. doi: 10.1056/NEJMc2009316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huntley BJF, et al. Rates of maternal and perinatal mortality and vertical transmission in pregnancies complicated by severe acute respiratory syndrome coronavirus 2 (SARS-Co-V-2) infection: a systematic review. Obstet. Gynecol. 2020;136:303–312. doi: 10.1097/AOG.0000000000004010. [DOI] [PubMed] [Google Scholar]
- 5.Ellington S, et al. Characteristics of women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status—United States, January 22–June 7, 2020. Morb. Mortal. Wkly. Rep. 2020;69:769–775. doi: 10.15585/mmwr.mm6925a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edlow AG, et al. Assessment of maternal and neonatal SARS-CoV-2 viral load, transplacental antibody transfer, and placental pathology in pregnancies during the COVID-19 pandemic. JAMA Netw. Open. 2020;3:e2030455. doi: 10.1001/jamanetworkopen.2020.30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raschetti R, et al. Synthesis and systematic review of reported neonatal SARS-CoV-2 infections. Nat. Commun. 2020;11:5164. doi: 10.1038/s41467-020-18982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotlyar, A. et al. Vertical transmission of COVID-19: a systematic review and meta-analysis. Am. J. Obstet. Gynecol. 10.1016/j.ajog.2020.07.049 (2020).
- 9.Vivanti AJ, et al. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenizia C, et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat. Commun. 2020;11:5128. doi: 10.1038/s41467-020-18933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong L, et al. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA. 2020;323:1846–1848. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascio, D. D. et al. Risk factors associated with adverse fetal outcomes in pregnancies affected by coronavirus disease 2019 (COVID-19): a secondary analysis of the WAPM study on COVID-19. J. Perinat. Med. 49, 111–115 (2020). [DOI] [PubMed]
- 14.Dumitriu, D. et al. Outcomes of neonates born to mothers with severe acute respiratory syndrome coronavirus 2 infection at a large medical center in New York City. JAMA Pediatr. 175, 157–167 (2021). [DOI] [PMC free article] [PubMed]
- 15.Walker K, et al. Maternal transmission of SARS‐COV‐2 to the neonate, and possible routes for such transmission: a systematic review and critical analysis. Bjog. Int. J. Obstet. Gynaecol. 2020;127:1324–1336. doi: 10.1111/1471-0528.16362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury R, et al. Characteristics and outcomes of 241 births to women with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection at five New York City Medical Centers. Obstet. Gynecol. 2020;136:273–282. doi: 10.1097/AOG.0000000000004025. [DOI] [PubMed] [Google Scholar]
- 17.Knight M, et al. Characteristics and outcomes of pregnant women admitted to hospital with confirmed SARS-CoV-2 infection in UK: national population based cohort study. BMJ. 2020;369:m2107. doi: 10.1136/bmj.m2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Haddad BJS, et al. Long-term risk of neuropsychiatric disease after exposure to infection in utero. JAMA Psychiat. 2019;76:594. doi: 10.1001/jamapsychiatry.2019.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Haddad BJS, et al. The fetal origins of mental illness. Am. J. Obstet. Gynecol. 2019;221:549–562. doi: 10.1016/j.ajog.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abu-Raya B, Kollmann TR, Marchant A, MacGillivray DM. The immune system of HIV-exposed uninfected infants. Front. Immunol. 2016;7:383. doi: 10.3389/fimmu.2016.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apostol AC, Jensen KDC, Beaudin AE. Training the fetal immune system through maternal inflammation—a layered hygiene hypothesis. Front. Immunol. 2020;11:123. doi: 10.3389/fimmu.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norman M, et al. Association of maternal SARS-CoV-2 infection in pregnancy with neonatal outcomes. JAMA. 2021;325:2076–2086. doi: 10.1001/jama.2021.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atyeo, C. et al. Compromised SARS-CoV-2-specific placental antibody transfer. Cell10.1016/j.cell.2020.12.027 (2020). [DOI] [PMC free article] [PubMed]
- 24.Beharier, O. et al. Efficient maternal to neonatal transfer of antibodies against SARS-CoV-2 and BNT162b2 mRNA COVID-19 vaccine. J. Clin. Invest. 10.1172/jci150319 (2021). [DOI] [PMC free article] [PubMed]
- 25.Liu P, et al. The immunologic status of newborns born to SARS-CoV2-infected mothers in Wuhan, China. J. Allergy Clin. Immun. 2020;146:101–109.e1. doi: 10.1016/j.jaci.2020.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng H, et al. Antibodies in infants born to mothers with COVID-19 pneumonia. JAMA. 2020;323:1848–1849. doi: 10.1001/jama.2020.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henrick BM, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 28.Olin A, et al. Stereotypic immune system development in newborn children. Cell. 2018;174:1277–1292.e14. doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shook LL, et al. Rapid establishment of a COVID-19 perinatal biorepository: early lessons from the first 100 women enrolled. BMC Med. Res. Methodol. 2020;20:215. doi: 10.1186/s12874-020-01102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.NIH. COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines (NIH, 2020).
- 31.Villani A-C, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadjadj, J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science10.1126/science.abc6027 (2020). [DOI] [PMC free article] [PubMed]
- 33.Schulte-Schrepping J, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arunachalam PS, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krow-Lucal ER, Kim CC, Burt TD, McCune JM. Distinct functional programming of human fetal and adult monocytes. Blood. 2014;123:1897–1904. doi: 10.1182/blood-2013-11-536094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keskinen P, Ronni T, Matikainen S, Lehtonen A, Julkunen I. Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology. 1997;91:421–429. doi: 10.1046/j.1365-2567.1997.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorrington MG, Fraser IDC. NF-κB signaling in macrophages: dynamics, crosstalk, and signal integration. Front. Immunol. 2019;10:705. doi: 10.3389/fimmu.2019.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Germic N, Frangez Z, Yousefi S, Simon H-U. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell. Death Differ. 2019;26:715–727. doi: 10.1038/s41418-019-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golubinskaya V, et al. Expression of S100A alarmins in cord blood monocytes is highly associated with chorioamnionitis and fetal inflammation in preterm infants. Front. Immunol. 2020;11:1194. doi: 10.3389/fimmu.2020.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou R, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavalieri D, et al. DC-ATLAS: a systems biology resource to dissect receptor specific signal transduction in dendritic cells. Immunome Res. 2010;6:10. doi: 10.1186/1745-7580-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinzierl AO, et al. Effective chemokine secretion by dendritic cells and expansion of cross-presenting CD4−/CD8+ dendritic cells define a protective phenotype in the mouse model of Coxsackievirus myocarditis. J. Virol. 2008;82:8149–8160. doi: 10.1128/JVI.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papaspyridonos M, et al. Id1 suppresses anti-tumour immune responses and promotes tumour progression by impairing myeloid cell maturation. Nat. Commun. 2015;6:6840. doi: 10.1038/ncomms7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Li R, Xiang S, Xiao W. MafB, a target of microRNA-155, regulates dendritic cell maturation. Open Life Sci. 2016;11:46–54. doi: 10.1515/biol-2016-0006. [DOI] [Google Scholar]
- 45.Belz GT, Nutt SL. Transcriptional programming of the dendritic cell network. Nat. Rev. Immunol. 2012;12:101–113. doi: 10.1038/nri3149. [DOI] [PubMed] [Google Scholar]
- 46.Shih VF-S, et al. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways. Nat. Immunol. 2012;13:1162–1170. doi: 10.1038/ni.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beisel C, et al. TLR7-mediated activation of XBP1 correlates with the IFNα production in humans. Cytokine. 2017;94:55–58. doi: 10.1016/j.cyto.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Maucourant C, et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5:eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 2019;10:3931. doi: 10.1038/s41467-019-11947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J-Y, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 51.Wilk AJ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1–7. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varchetta, S. et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 10.1038/s41423-020-00557-9, 1–9 (2020). [DOI] [PMC free article] [PubMed]
- 53.Sanz I, et al. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Front. Immunol. 2019;10:2458. doi: 10.3389/fimmu.2019.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarvaria A, et al. IL-10+ regulatory B cells are enriched in cord blood and may protect against cGVHD after cord blood transplantation. Blood. 2016;128:1346–1361. doi: 10.1182/blood-2016-01-695122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esteve-Solé A, et al. Characterization of the highly prevalent regulatory CD24hiCD38hi B-cell population in human cord blood. Front. Immunol. 2017;8:201. doi: 10.3389/fimmu.2017.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Budeus B, et al. Human cord blood B cells differ from the adult counterpart by conserved ig repertoires and accelerated response dynamics. J. Immunol. 2021;206:2839–2851. doi: 10.4049/jimmunol.2100113. [DOI] [PubMed] [Google Scholar]
- 57.Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 10.1038/s41587-020-0465-8, 1–10 (2020). [DOI] [PMC free article] [PubMed]
- 58.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uhlen M, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366:eaax9198. doi: 10.1126/science.aax9198. [DOI] [PubMed] [Google Scholar]
- 60.Myers DR, Zikherman J, Roose JP. Tonic signals: why do lymphocytes bother? Trends Immunol. 2017;38:844–857. doi: 10.1016/j.it.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashouri JF, Weiss A. Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol. 2017;198:657–668. doi: 10.4049/jimmunol.1601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kretschmer B, Kühl S, Fleischer B, Breloer M. Activated T cells induce rapid CD83 expression on B cells by engagement of CD40. Immunol. Lett. 2011;136:221–227. doi: 10.1016/j.imlet.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 63.Grosche L, et al. The CD83 molecule—an important immune checkpoint. Front. Immunol. 2020;11:721. doi: 10.3389/fimmu.2020.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmermann M, et al. Antigen extraction and B cell activation enable identification of rare membrane antigen specific human B cells. Front. Immunol. 2019;10:829. doi: 10.3389/fimmu.2019.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorter DJJ, de, Vos JCM, Pals ST, Spaargaren M. The B cell antigen receptor controls AP-1 and NFAT activity through Ras-mediated activation of Ral. J. Immunol. 2007;178:1405–1414. doi: 10.4049/jimmunol.178.3.1405. [DOI] [PubMed] [Google Scholar]
- 66.Slomp A, Peperzak V. Role and regulation of pro-survival BCL-2 proteins in multiple myeloma. Front. Oncol. 2018;8:533. doi: 10.3389/fonc.2018.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bunis DG, et al. Single-cell mapping of progressive fetal-to-adult transition in human naive T cells. Cell Rep. 2021;34:108573. doi: 10.1016/j.celrep.2020.108573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D’Arena G, et al. Flow cytometric characterization of human umbilical cord blood lymphocytes: immunophenotypic features. Haematologica. 1998;83:197–203. [PubMed] [Google Scholar]
- 69.Zhang X, et al. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci. Transl. Med. 2014;6:238ra72–238ra72. doi: 10.1126/scitranslmed.3008748. [DOI] [PubMed] [Google Scholar]
- 70.Ng MSF, Roth TL, Mendoza VF, Marson A, Burt TD. Helios enhances the preferential differentiation of human fetal CD4+ naïve T cells into regulatory T cells. Sci. Immunol. 2019;4:eaav5947. doi: 10.1126/sciimmunol.aav5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mold JE, et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 2010;330:1695–1699. doi: 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michaëlsson J, Mold JE, McCune JM, Nixon DF. Regulation of T cell responses in the developing human fetus. J. Immunol. 2006;176:5741–5748. doi: 10.4049/jimmunol.176.10.5741. [DOI] [PubMed] [Google Scholar]
- 73.Campbell JJ, et al. CCR7 expression and memory T cell diversity in humans. J. Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 74.Cano-Gamez E, et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat. Commun. 2020;11:1801. doi: 10.1038/s41467-020-15543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fergusson JR, Fleming VM, Klenerman P. CD161-expressing human T cells. Front. Immunol. 2011;2:36. doi: 10.3389/fimmu.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mazzoni, A. et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest.10.1172/jci138554 (2020). [DOI] [PMC free article] [PubMed]
- 77.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Z, et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368:eaaz7548. doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 79.Papavassiliou AG, Musti AM. The multifaceted output of c-Jun biological activity: focus at the junction of CD8 T cell activation and exhaustion. Cells. 2020;9:2470. doi: 10.3390/cells9112470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 81.Ramesh R, et al. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 2014;211:89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maggi L, et al. CD161 is a marker of all human IL‐17‐producing T‐cell subsets and is induced by RORC. Eur. J. Immunol. 2010;40:2174–2181. doi: 10.1002/eji.200940257. [DOI] [PubMed] [Google Scholar]
- 83.Billerbeck E, et al. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc. Natl Acad. Sci. USA. 2010;107:3006–3011. doi: 10.1073/pnas.0914839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kondo T, Takata H, Matsuki F, Takiguchi M. Cutting edge: phenotypic characterization and differentiation of human CD8+ T cells producing IL-17. J. Immunol. 2009;182:1794–1798. doi: 10.4049/jimmunol.0801347. [DOI] [PubMed] [Google Scholar]
- 85.Cosmi L, et al. Human interleukin 17–producing cells originate from a CD161+CD4+ T cell precursor. J. Exp. Med. 2008;205:1903–1916. doi: 10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walker LJ, et al. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 2012;119:422–433. doi: 10.1182/blood-2011-05-353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lalla Cde, et al. Innate-like effector differentiation of human invariant NKT cells driven by IL-7. J. Immunol. 2008;180:4415–4424. doi: 10.4049/jimmunol.180.7.4415. [DOI] [PubMed] [Google Scholar]
- 88.Halkias, J. et al. CD161 contributes to prenatal immune suppression of IFNγ-producing PLZF+ T cells. J. Clin. Invest. 10.1172/jci125957 (2019). [DOI] [PMC free article] [PubMed]
- 89.Yockey LJ, Lucas C, Iwasaki A. Contributions of maternal and fetal antiviral immunity in congenital disease. Science. 2020;368:608–612. doi: 10.1126/science.aaz1960. [DOI] [PubMed] [Google Scholar]
- 90.Choi GB, et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yim YS, et al. Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature. 2017;549:482–487. doi: 10.1038/nature23909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalish, B. T. et al. Maternal immune activation in mice disrupts proteostasis in the fetal brain. Nat. Neurosci. 10.1038/s41593-020-00762-9, 1–10 (2020). [DOI] [PMC free article] [PubMed]
- 93.McGeachy MJ, Cua DJ, Gaffen SL. The IL-17 family of cytokines in health and disease. Immunity. 2019;50:892–906. doi: 10.1016/j.immuni.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao, Y. et al. Single-cell transcriptomic landscape of nucleated cells in umbilical cord blood. Gigascience8, giz047 (2019). [DOI] [PMC free article] [PubMed]
- 95.Jin X, et al. Characterization of dendritic cell subtypes in human cord blood by single-cell sequencing. Biophys. Rep. 2019;5:199–208. doi: 10.1007/s41048-019-00096-5. [DOI] [Google Scholar]
- 96.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zambrano, L. D. et al. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status—United States, January 22–October 3, 2020. Morb. Mortal. Wkly. Rep. 69, 1641–1647 (2020). [DOI] [PMC free article] [PubMed]
- 98.Lokken EM, et al. Higher severe acute respiratory syndrome coronavirus 2 infection rate in pregnant patients. Am. J. Obstet. Gynecol. 2021;225:75.e1–75.e16. doi: 10.1016/j.ajog.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kubiak JM, et al. Severe acute respiratory syndrome coronavirus 2 serology levels in pregnant women and their neonates. Am. J. Obstet. Gynecol. 2021;225:73.e1–73.e7. doi: 10.1016/j.ajog.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mason E, et al. Maternal influences on the transmission of leukocyte gene expression profiles in population samples from Brisbane, Australia. PLoS ONE. 2010;5:e14479. doi: 10.1371/journal.pone.0014479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Svensson J, et al. Maternal autoimmune thyroid disease and the fetal immune system. Exp. Clin. Endocrinol. Diabetes. 2011;119:445–450. doi: 10.1055/s-0031-1279741. [DOI] [PubMed] [Google Scholar]
- 102.Wilson RM, et al. Maternal obesity alters immune cell frequencies and responses in umbilical cord blood samples. Pediatr. Allergy Immunol. 2015;26:344–351. doi: 10.1111/pai.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yanai S, et al. Diabetic pregnancy activates the innate immune response through TLR5 or TLR1/2 on neonatal monocyte. J. Reprod. Immunol. 2016;117:17–23. doi: 10.1016/j.jri.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 104.Aaltonen R, Heikkinen T, Hakala K, Laine K, Alanen A. Transfer of proinflammatory cytokines across term. Placenta Obstet. Gynecol. 2005;106:802–807. doi: 10.1097/01.AOG.0000178750.84837.ed. [DOI] [PubMed] [Google Scholar]
- 105.Yockey LJ, Iwasaki A. Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity. 2018;49:397–412. doi: 10.1016/j.immuni.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu-Culligan A, et al. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med. 2021;2:591–610.e10. doi: 10.1016/j.medj.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cavalcante MB, Cavalcante CT, de MB, Sarno M, Barini R, Kwak-Kim J. Maternal immune responses and obstetrical outcomes of pregnant women with COVID-19 and possible health risks of offspring. J. Reprod. Immunol. 2020;143:103250. doi: 10.1016/j.jri.2020.103250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Borges-Almeida E, et al. The impact of maternal HIV infection on cord blood lymphocyte subsets and cytokine profile in exposed non-infected newborns. BMC Infect. Dis. 2011;11:38. doi: 10.1186/1471-2334-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones CI, et al. Maternal HIV status skews transcriptomic response in infant cord blood monocytes exposed to Bacillus Calmette–Guerín. AIDS. 2020;35:23–32. doi: 10.1097/QAD.0000000000002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wohnhaas CT, et al. DMSO cryopreservation is the method of choice to preserve cells for droplet-based single-cell RNA sequencing. Sci. Rep. 2019;9:10699. doi: 10.1038/s41598-019-46932-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matoba N, et al. Differential patterns of 27 cord blood immune biomarkers across gestational age. Pediatrics. 2009;123:1320–1328. doi: 10.1542/peds.2008-1222. [DOI] [PubMed] [Google Scholar]
- 112.Fajnzylber J, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Gene Expression Omnibus under accession no. GSE165193.