Abstract
pp120 (Ceacam 1) undergoes ligand-stimulated phosphorylation by the insulin receptor, but not by the insulin-like growth factor 1 receptor (IGF-1R). This differential phosphorylation is regulated by the C terminus of the β-subunit of the insulin receptor, the least conserved domain of the two receptors. In the present studies, deletion and site-directed mutagenesis in stably transfected hepatocytes derived from insulin receptor knockout mice (IR−/−) revealed that Tyr1316, which is replaced by the nonphosphorylatable phenylalanine in IGF-1R, regulated the differential phosphorylation of pp120 by the insulin receptor. Similarly, the nonconserved Tyr1316 residue also regulated the differential effect of pp120 on IGF-1 and insulin mitogenesis, with pp120 downregulating the growth-promoting action of insulin, but not that of IGF-1. Thus, it appears that pp120 phosphorylation by the insulin receptor is required and sufficient to mediate its downregulatory effect on the mitogenic action of insulin. Furthermore, the current studies revealed that the C terminus of the β-subunit of the insulin receptor contains elements that suppress the mitogenic action of insulin. Because IR−/− hepatocytes are derived from liver, an insulin-targeted tissue, our observations have finally resolved the controversy about the role of the least-conserved domain of insulin and IGF-1Rs in mediating the difference in the mitogenic action of their ligands, with IGF-1 being more mitogenic than insulin.
The insulin receptor is essential to mediate insulin action on target cells (1, 27). It is a cell surface glycoprotein of a heterotetrameric structure that consists of two α- and two β-subunits. The extracellular α-subunits contain the insulin binding domains, and the transmembrane β-subunits contain the tyrosine kinase and the phosphorylation sites. Insulin binding to its receptor activates the tyrosine kinase to phosphorylate the receptor and other endogenous substrates, such as pp120 (Ceacam 1) (5a, 44), insulin receptor substrate proteins (IRS-1, -2, -3, and -4), Shc, and others (reviewed in references 65 and 66). Phosphorylation of different substrates is required to mediate the diverse effects of hormones on metabolism and growth (3, 60, 68).
Insulin and insulin-like growth factor 1 (IGF-1) receptors are structurally related, and all conserved tyrosine residues that are phosphorylated in the insulin receptor in response to insulin are also phosphorylated in the IGF-1 receptor in response to IGF-1 (10, 17, 23, 48, 71). Moreover, these receptors share many substrates, such as Shc and members of the IRS family, phosphorylation of which is regulated by the conserved Tyr960 in the juxtamembrane domain of the insulin receptor (18, 22, 67) and its corresponding residue in the IGF-1 receptor (8). Phosphorylated IRS-1 engages, in turn, in the formation of signaling complexes via phosphotyrosine-containing binding motifs with Src homology 2 (SH2) found in molecules like growth factor receptor binding protein (GRB2) (32, 56), Syp (SH PTP2) phosphotyrosine phosphatase (69), phosphatidylinositol (PI)-3′ kinase (4), and many others. By binding to GRB2 either directly or through Syp, IRS-1 couples GRB2 to insulin and IGF-1 receptors. Similarly, Shc couples these receptors to GRB2 even more predominantly than the IRS proteins (49, 53). GRB2 coupling to the receptors leads to its association with the Son of Sevenless (SOS) Ras GDP/GTP exchanger. This causes translocation of SOS to the plasma membrane in proximity to its p21ras substrate (16), activation of the Ras/mitogen-activated protein (MAP) kinase pathway, and regulation of cell growth, differentiation, and proliferation in response to insulin and IGF-1 (6, 9). Activation of the PI-3′ kinase–p70 ribosomal protein S6 kinase pathway also plays a significant role in mediating the mitogenic effects of insulin in many cell types, including hepatocytes (24, 52). PI-3′ kinase is coupled to the receptor via the IRS proteins, but can also directly bind, albeit less stably, to the receptor on the C terminus of the β-subunit of the receptor (57).
Because phosphorylation of substrates is required to mediate insulin and IGF-1 action, the common phosphorylation cascades that underlie the basic mechanism of insulin and IGF-1 action have failed to explain the different, albeit overlapping, physiologic functions mediated by the two receptors. The insulin receptor regulates metabolism (1), and the IGF-1 receptor mediates growth and differentiation (5, 31). Except for pp120 (41), most other insulin receptor substrates are similarly phosphorylated by the IGF-1 receptor. Moreover, pp120 phosphorylation is regulated by the least conserved C terminus of the β-subunit of the insulin receptor (41). Thus, the specificity of pp120 phosphorylation may serve as a biochemical marker for the physiologic differences between insulin and IGF-1 action. Therefore, delineation of the role of specific residues in the C terminus of the β-subunit of the insulin receptor in regulating pp120 phosphorylation may advance our understanding of the basic mechanism of the diverse physiologic functions of insulin and IGF-1.
The C terminus of the β-subunit of the insulin receptor contains two tyrosine residues that are phosphorylated in response to insulin: Tyr1316 and Tyr1322. Of these, Tyr1316 is not conserved in the IGF-1 receptor, where it is replaced by Phe1310. To address the role of these residues in pp120 phosphorylation, we examined the effect of abolishing their phosphorylation, by deletion or site-directed mutagenesis, on pp120 phosphorylation in stably transfected simian virus 40 (SV40)-transformed hepatocytes derived from the insulin receptor knockout (IR−/−) mouse (52). We observed that the nonconserved Tyr1316 in the β-subunit of the insulin receptor regulates the differential phosphorylation of pp120 by the insulin receptor.
The function of pp120 remains elusive. It may function as a tumor suppressor in colon, liver, and prostate (19, 20, 25, 26, 34, 46, 51, 62, 63) and as a downregulator of the mitogenic effects of insulin (15). pp120 may upregulate the transport of bile acids (55) and insulin (15) in the hepatocyte, as suggested by studies with transfected cells. Supportive evidence for a role in pp120 in cell adhesion has also emerged (7, 12). Because of the multiple functions ascribed to pp120, it has been referred to as pp120, C-CAM, and CBATP. Based on cDNA sequence analysis, pp120 has also been identified as Ca2+/Mg2+ ecto-ATPase (30, 36). Sequence analysis has also shown that pp120 is the rat homolog of the human biliary glycoprotein (BGP) (45).
pp120 is expressed as two alternative spliced isoforms, the shorter of which lacks most of the intracellular domain, including the phosphorylation sites (40). The short isoform has been known to function as a cell adhesion molecule, but not to play a significant role in the other functions attributed to pp120 (7, 12, 15, 55).
The basic mechanism of pp120 functions is not completely understood. However, pp120 phosphorylation is required for its function in insulin endocytosis (15), bile acid transport (55), and tumor suppression (20, 33). Dependence on an intact intracellular domain for the cell adhesion property of pp120 has also been reported (7). We have observed that inhibition of pp120 expression increased the mitogenic action of insulin in rat hepatoma H35 cells (15). Conversely, expression of pp120 decreased insulin mitogenesis in NIH 3T3 cells coexpressing insulin receptors compared to cells expressing insulin receptors alone (15). The mechanism of the downregulatory effect of pp120 on insulin-induced mitogenesis is not clear, but failure of the phosphorylation-defective isoforms (truncated and site-directed mutants) to decrease insulin-induced mitogenesis suggested that pp120 phosphorylation is required (15). Thus, in the present studies, we examined whether pp120 similarly regulates the mitogenic action of IGF-1 in IR−/− hepatocytes. In contrast to insulin, pp120 coexpression did not downregulate cell growth in response to IGF-1. Replacement of the C terminus of the β-subunit of the IGF-1 receptor with that of the insulin receptor restored the downregulatory effect of pp120 on cell growth in response to IGF-1. Furthermore, pp120 downregulation of insulin-induced mitogenesis required intact phosphorylation of the nonconserved Tyr1316 between insulin and IGF-1 receptors. Thus, pp120 phosphorylation by the insulin receptor appears to be required and sufficient to mediate its differential downregulation of insulin vis-à-vis IGF-1 mitogenesis.
MATERIALS AND METHODS
Materials.
All reagents for cell culture were from Mediatech, Inc. (Herndon, Va.). The plasmid carrying the hygromycin resistance gene Hygror, pREP4-Hygror, was purchased from Stratagene (La Jolla, Calif.). The bovine papillomavirus-based expression vector (pBPV) and all reagents for immunoblotting were from Amersham Pharmacia Biotech (Piscataway, N.J.). Lipofectamine reagent and protein A-agarose were purchased from Life Technologies, Inc. Hygromycin B was purchased from Calbiochem. Protease inhibitors were purchased from Boehringer Mannheim (Indianapolis, Ind.). Triton X-100 and other reagents used in cell lysis were purchased from Sigma (St. Louis, Mo.). All reagents for polyacrylamide gel electrophoresis (PAGE) were purchased from Bio-Rad Laboratories (Richmond, Calif.). Human insulin was purchased from Lilly, and insulin-free bovine serum albumin (BSA) was purchased from Intergen Co. (Des Plaines, Ill.). Recombinant human IGF-1, monoclonal antiphosphotyrosine (α-pTyr) antibodies, and polyclonal anti-IRS-1 (α-IRS-1) and anti-IRS-2 (α-IRS-2) antibodies were purchased from Upstate Biotechnology, Inc. (Lake Placid, N.Y.). The pp120 antibodies used in these studies were described previously (41). Briefly, the monoclonal antibody used to immunoprecipitate pp120 (α-HA4; an identical protein to pp120) was purified from ascites fluid from HA4 c19 cells purchased from the Developmental Studies Hybridoma Bank (Department of Biology, University of Iowa, Iowa City). The polyclonal antibody used to immunoblot pp120 (α-295) was raised in rabbit against a peptide (amino acids [aa] 51 to 64) in the extracellular domain of rat liver pp120.
Cells and cell culture.
The SV40-transformed hepatocytes were derived from the IR−/− mice (11, 52). As described previously (43), these cells were routinely maintained in complete medium A (alpha-modified Eagle's medium (α-MEM) containing 8% fetal calf serum, 1% glutamine, 200 nM dexamethasone, 100-U/ml penicillin, and 10-μg/ml streptomycin) at 33°C in 5% CO2.
Construction of expression vectors.
Synthesis and subcloning into pBPV of the cDNA encoding the full-length isoform of rat pp120 (rFL) and the human insulin and IGF-1 receptors (hIR and hIGF-1R, respectively) were described previously (14, 44). Similarly, synthesis and subcloning into pBPV of recombinant cDNAs encoding the chimeric IGF-1 receptor (CHI), in which the entire C terminus (aa 1230 to 1337) of the β-subunit was replaced by the corresponding tail of the insulin receptor (aa 1245 to 1343), and the F1310Y IGF-1 receptor, in which Phe1310 was replaced by tyrosine, were described previously (13, 14). Synthesis and subcloning into pBPV of recombinant cDNA encoding the Δ43 hIR deletion mutant (Δ43 hIR) that lacks the terminal 43-aa tail of the β-subunit were also described previously (28) (Fig. 1).
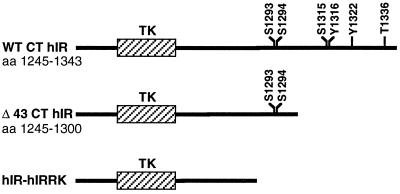
FIG. 1.
Insulin receptor deletion mutants. This diagram illustrates the tyrosine kinase (TK) and the C terminus (CT) of the β-subunit of the hIR. Wild-type hIR (WT hIR) contains several serine (S), threonine (T), and tyrosine (Y) phosphorylation sites. In the Δ43 CT hIR mutant, the terminal 43 aa containing both tyrosine residues were deleted from the C terminus of the β-subunit of the insulin receptor. hIR-hIRRK represents a heterologous insulin receptor in which the cytoplasmic domain of the insulin receptor was replaced with that of the orphan IRR.
The hIR-IRRK heterologous receptor, in which the intracellular domain of the hIR was replaced by the corresponding fragment of the insulin receptor-related receptor (IRR) was originally subcloned into the EcoRI and XbaI sites of the pECE expression vector (72). The intracellular portion of the IRR, an orphan receptor that belongs to the insulin receptor tyrosine kinase subfamily and for which a ligand has not yet been identified, lacks in its C terminus tyrosine and serine phosphorylation sites known to be present in the corresponding domain of the insulin receptor (54) (Fig. 1). The DNA fragment spanning nucleotides (nt) 1011 to 4043 was excised from the pECE construct by EcoRI-XbaI digestion and subcloned into the human elongation factor 1α promoter-based expression vector, pEF-1α Neo (43), at the same sites. The DNA fragment spanning nt 1 to 1011 from hIR was excised from the pECE construct by EcoRI-EcoRI digestion and subcloned into the pEF-1α construct 5′ of the hIR-hIRRK partial fragment.
To synthesize the cDNA encoding the Y1316F insulin receptor mutant (Y1316F hIR), two cDNA fragments were amplified by PCR with wild-type hIR (in the pGEM 4Z-WT hIR construct) as a template in the presence of Taq polymerase, as we have described previously (40). The first PCRa fragment (nt 4072 to 4360) was amplified by using sense (S1; 4072-AGCTTCGAGGAACACATCCCTTACACACAtATGAAC-4107) and antisense (α1; nt 4360 to 4331) primers. The S1 primer contained an A-to-T point mutation at nt 4076 (underlined) that encodes phenylalanine instead of tyrosine at aa 1316. The Ct mismatch (lowercase boldface letter) at nt 4101 was included in order to introduce a new NdeI site. The α1 primer spans the SpeI and PstI sites at nt 4334 and 4345, respectively. The second PCRb fragment (nt 2086 to 4110) was amplified by using wild-type sense S2 (nt 2086 to 2121 spanning the Tth111I site at nt 2100) and antisense α2 (complementary to S1) primers. PCR products were individually subcloned into the pCR II TA cloning plasmid per the manufacturer's instructions (Invitrogen). The DNA segment spanning nt 2100 to 4101 was isolated from the pCR IIb construct by XbaI-NdeI digestion and ligated into the pCR IIa construct at the same sites upstream of and in the same orientation as the PCRa DNA fragment (pCR IIa+b). The DNA fragment spanning nt 2100 to 4334 was then isolated from the pCR IIa+b by Tth111I-SpeI digestion and ligated into the pGEM4Z-WT hIR construct at the same sites in lieu of the wild-type DNA fragment. Following confirmation by enzyme digestion and sequence analysis, the full Y1316F hIR cDNA was excised from the pGEM4Z-hIR construct by XbaI-SpeI and ligated at the XbaI site of pEF-1α.
The recombinant cDNA encoding the double Y1316F/1322F hIR mutant was originally subcloned into the pCVSV expression vector (58). The DNA fragment carrying the double mutations and spanning nt 2100 to 4334 was removed from the pCVSV construct by Tth111I-SpeI digestion and ligated into the pGEM4Z-WT hIR construct at the same sites in lieu of the wild-type sequence. The cDNA encoding the entire cDNA encoding the Y1316F/Y1322F hIR mutant was then excised from the pGEM4Z-construct by SpeI-XbaI digestion for subcloning into the XbaI site of the pEF-1α expression vector.
Transfection.
Stable transfection of the SV40-transformed IR−/− hepatocytes in the presence of the pREP4-Hygror gene was achieved by the Lipofectamine method, as described previously (43). Individual clones were picked and expanded, and confluent cells were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM HEPES [pH 7.6], 1 mM phenylmethylsulfonyl fluoride, 10-μg/ml [each] protease inhibitors antipain dihydrochloride, pepstatin A, leupeptin, aprotinin, and bacitracin) for analysis on 7.5% sodium dodecyl sulfate (SDS)–PAGE gels and screening for pp120 expression by immunoblotting with a pp120 polypeptide antibody (α-295), as described previously (44). Screening for expression of insulin and IGF-1 receptors was achieved by measuring insulin binding in intact cells, as described previously (41, 44). The level of endogenous wild-type IGF-1 receptors in IR−/− hepatocytes was ∼1 × 105 to 2 × 105 IGF-1 receptors/cell (11). The level of mutant insulin and IGF-1 receptors in transfected IR−/− hepatocytes was ∼0.5 × 106 to 1.3 × 106 receptors per cell.
Phosphorylation of pp120 in intact cells.
IR−/− hepatocytes were expanded to confluence in 100-mm-diameter plates. Following overnight incubation in serum-free medium containing 0.1% insulin-free BSA and 25 mM HEPES (pH 7.4) for 8 h, cells were treated with either buffer alone or ligand (insulin or IGF-1) at 100 nM for 5 min prior to lysis in 1% Triton X-100 in the presence of phosphatase (EDTA, 4 mM; NaF, 100 mM; sodium pyrophosphate, 10 mM; sodium phosphate, 10 mM; ATP, 2 mM; sodium orthovanadate, 20 mM; N-ethylmaleimide, 5 mM; HEPES, 40 mM [pH 7.6]) and protease inhibitors (described above). Unless otherwise indicated, cell lysates were directly subjected to immunoprecipitation with either a monoclonal antibody against pp120/HA4 or α-pTyr prior to analysis by 7.5% SDS-PAGE and immunoblotting with horseradish peroxidase (HRP)-coupled α-pTyr antibody to detect phosphorylated proteins by the Amersham Enhanced Chemiluminescence (ECL) detection system (41). In some experiments, cell lysates were partially purified by wheat germ agglutinin affinity chromatography (44) prior to being subjected to immunoprecipitation with α-pTyr monoclonal antibody to immunoprecipitate phosphorylated pp120 and insulin receptors (42).
To examine the activation state of receptors in stable transfectants, cells were treated with ligand as mentioned above, and their lysates were subjected to immunoprecipitation with polyclonal antibodies against either IRS-1 (α-IRS-1) or IRS-2 (α-IRS-2). Following analysis by SDS-PAGE, proteins were transferred on nitrocellulose membranes and immunoblotted with HRP-coupled α-pTyr antibody to detect phosphorylated proteins by the ECL system (41). Experiments were carried out with at least two independent clones for each construct derived from the same transfection.
Quantitation of proteins.
Autoradiograms were scanned on an imaging densitometer (Bio-Rad model GS-670), and the proteins were quantitated by the Image NIH version 1.61 Macintosh software program.
Cell growth and proliferation.
The cell growth and proliferation assay was performed according to the method of Li et al. (29), with some modifications. Transfected IR−/− hepatocytes were seeded in triplicate into 12-well plates at a density of 3 × 103 cells per well. Cells were allowed to attach for 24 h in complete medium A prior to incubation in serum-free medium supplemented with 0.1% BSA and 25 mM HEPES for 24 h to reach quiescence. Insulin or IGF-1 at concentrations of 0, 0.1, 10, and 100 nM was added to triplicate wells, whereas complete medium A was added to some others. Following incubation for 24 and 48 h, cells were trypsinized and counted in a Coulter counter (Z1 model). Basal growth was measured as the number of cells grown in the absence of serum and hormones. Maximal growth was measured as the number of cells grown in the presence of serum. Hormone-induced cell growth was calculated as the percent maximal minus basal growth divided by the number of cells grown in complete medium (52). These experiments were repeated at least three times for each clone.
Statistical analysis.
Curves were compared by a multivariate analysis of variance, and individual points were compared by paired t tests. P values of less than 0.05 were considered statistically significant.
RESULTS
Phosphorylation of recombinant pp120 by deleted insulin receptors.
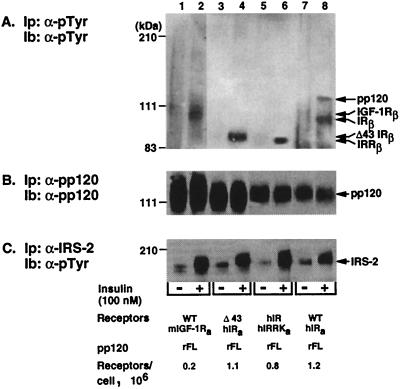
Deletion of the terminal 43 amino acids or replacement of the intracellular domain of the insulin receptor with that of the IRR impaired neither the affinity of the receptor to its ligand nor its tyrosine kinase activity (35, 72). Thus, we transfected IR−/− hepatocytes with mutant receptors to investigate the role of the C terminus of the β-subunit of the insulin receptor in pp120 phosphorylation. Cells transfected with full-length rat pp120 (rFL) alone or with comparable amounts of wild-type (WT) and mutant (Δ43 hIR and hIR-hIRRK) insulin receptors were incubated in the presence (Fig. 2, even lanes) or absence (Fig. 2, odd lanes) of insulin (100 nM) for 5 min at 33°C. Following partial purification of cell lysates, 50 μg of proteins was immunoprecipitated with α-pTyr monoclonal antibody prior to immunoblotting with α-pTyr (Fig. 2A) to detect tyrosine-phosphorylated proteins. To account for the amount of pp120 in the samples, amounts of proteins equal to those in panel A were immunoprecipitated with α-pp120/HA4 monoclonal antibody, analyzed by SDS-PAGE in parallel to the gel in panel A, and immunoblotted with α-pp120 polyclonal antibody (Fig. 2B). As expected, insulin at a high 100 nM concentration activated both IGF-1 and insulin receptors (Fig. 2A, even lanes). Because the level of endogenous IGF-1 receptors in IR−/− cells is lower than the level of recombinant insulin receptors, we exposed the immunoblot of lanes 1 to 2 longer than the rest of the immunoblot (lanes 3 to 8). As expected from our previous experiments (41), endogenous mouse (m) IGF-1 receptors failed to phosphorylate pp120 in cells transfected with rat pp120 alone (WT mIGF-1Ra/rFL pp120; “a” represents the clone used) (Fig. 2A, lane 2 versus 1). In contrast, insulin led to an ∼10-fold increase in the amount of phosphorylated tyrosine in pp120 derived from cells transfected with wild-type insulin receptors and pp120 (WT hIRa/rFL; “a” represents the clone used) (Fig. 2A, lane 8 versus 7). It is noteworthy that the low level of phosphotyrosines in the pp120 band is due to Tyr488 being the only residue in pp120 undergoing phosphorylation in response to insulin (44). Because endogenous IGF-1 receptors failed to phosphorylate pp120, pp120 phosphorylation in the WT hIRa/rFL pp120 clone was probably due to insulin activation of wild-type insulin receptors. However, when the distal 43 aa were removed from the C terminus of the β-subunit of the insulin receptor (Δ43 hIR) or when the C terminus was replaced with that of the IRR which lacks phosphorylation sites (hIR-hIRRK), insulin-stimulated pp120 phosphorylation was abolished (Fig. 2A, lanes 4 and 6, respectively). Thus, it appears that the distal 43 aa of the insulin receptor are required for pp120 phosphorylation by the insulin receptor.
FIG. 2.
Phosphorylation of recombinant pp120 by truncated insulin receptors in intact cells. IR−/− hepatocytes were stably transfected with cDNAs encoding the full-length isoform of rat pp120 (rFL) with either wild-type insulin receptors (WT hIRa/rFL) or with the Δ43 deletion mutant (Δ43 hIRa/rFL) or the heterologous hIR/hIRRK receptors (hIR/hIRRKa/rFL). Cells expressing full-length pp120 alone were used as controls (WT mIGF-1Ra/rFL, with m denoting endogenous mouse IGF-1 receptors). Subscripts denote the clone number. Cells were serum starved for 8 h prior to incubation in the presence (+ lanes) or absence (− lanes) of insulin (100 nM) for 5 min. Following partial purification on affinity chromatography, ∼50 μg of proteins was immunoprecipitated (Ip) with a monoclonal antibody against either phosphotyrosines (α-pTyr) (A) or pp120 (α-pp120) (B), analyzed in parallel by SDS-PAGE, and immunoblotted (Ib) with either HRP-coupled α-pTyr (A) or with a polyclonal antibody against pp120 (B). To examine IRS-2 phosphorylation in these cells, proteins derived from nonpurified cell lysates were immunoprecipitated with a polyclonal antibody against IRS-2 (α-IRS-2) and immunoblotted with α-pTyr antibody following electrophoresis (C). For optimal photographic visualization, the autoradiogram corresponding to lanes 1 to 2 was exposed threefold longer than the one corresponding to lanes 3 to 8. The number of receptors per cell expressed in each transfectant is shown at the bottom of the figure. Molecular mass markers are indicated on the left-hand side of the gel. Bands of ∼100, 95, and 90 kDa in panel A correspond to the β-subunits of the IGF-1R, insulin receptor, and Δ43 and IRR, respectively.
To investigate whether expressed receptors were capable of phosphorylating endogenous proteins in response to ligand, we examined phosphorylation of IRS-2 in transfected cells. Thus, serum-starved cells were treated with insulin (100 nM) prior to lysis and immunoprecipitation with α-IRS-2 antibody. Following transfer onto nitrocellulose membranes, proteins were probed with α-pTyr antibody to detect tyrosine-phosphorylated IRS-2. As shown in Fig. 2C, insulin-activated insulin and IGF-1 receptors phosphorylated IRS-2 in all transfectants, suggesting that IRS-2 is a common substrate of the tyrosine kinase of IGF-1, insulin, and IRRs. Moreover, these data support the notion that, in contrast to pp120, IRS-2 phosphorylation does not require the distal 43 aa of the C terminus of the β-subunit of the insulin receptor.
Phosphorylation of recombinant pp120 by site-directed insulin receptor mutants.
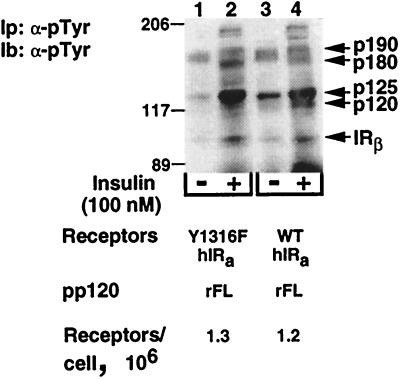
As Fig. 1 indicates, the distal 43 aa of the C terminus of the β-subunit of the insulin receptor include the tyrosine phosphorylation sites of this domain (Tyr1316 and Tyr1322). To determine which of these residues regulates pp120 phosphorylation by the insulin receptor, we mutated the nonconserved tyrosine to nonphosphorylatable phenylalanine either alone (Y1316F) or with Tyr1322 (Y1316F/Y1322F). Mutation of both residues to phenylalanine did not impair either the affinity of the receptor to its ligand or its tyrosine kinase activity (58). Similarly, replacing Tyr1316 with phenylalanine did not impair the affinity of the receptor to insulin (data not shown). To examine the tyrosine kinase activity of the Y1316F insulin receptor mutant, we subjected cell lysates derived from cells cotransfected with pp120 and comparable amounts of either wild-type or Y1316F insulin receptors to immunoprecipitation and immunoblotting with α-pTyr antibodies. As Fig. 3 reveals, insulin-induced tyrosine phosphorylation levels of the β-subunit (IRβ) of the receptors and of many unidentified proteins (p190, p180, and p125) were comparable in cells expressing Y1316F and those expressing wild-type receptors (lane 2 versus 4). This suggests that mutating Tyr1316 to phenylalanine did not alter the tyrosine kinase activity of the receptor. In contrast to wild-type insulin receptors, which induced tyrosine phosphorylation of a protein of Mr ∼120 kDa (p120) in response to insulin (Fig. 3, lane 4 versus 3), Y1316F receptors failed to phosphorylate this protein (Fig. 3, lane 2 versus 1). As expected, mutation of Tyr1322 in addition to Tyr1316 to phenylalanine led to the same observation (data not shown).
FIG. 3.
Phosphorylation of endogenous substrates by site-directed mutant insulin receptors in intact cells. IR−/− hepatocytes coexpressing full-length rat pp120 (rFL) with comparable numbers (1.2 × 106 to 1.3 × 106) of either wild-type (WT hIRa/rFL) or Y1316F (Y1316F hIRa/rFL) insulin receptors were treated with insulin as described in the legend to Fig. 2. Cell lysates were subjected to immunoprecipitation (Ip) with α-pTyr. Following analysis by SDS-PAGE, proteins were transferred on nitrocellulose membrane for immunoblotting (Ib) with HRP-coupled α-pTyr antibody and detection by the ECL system. Molecular mass markers are indicated on the left-hand side of the gel. Phosphorylated proteins of ∼190, 180, 125, and 120 kDa were referred to as p190, p180, p125, and p120, respectively. The ∼95-kDa band corresponds to the β-subunit of insulin receptors (IRβ).
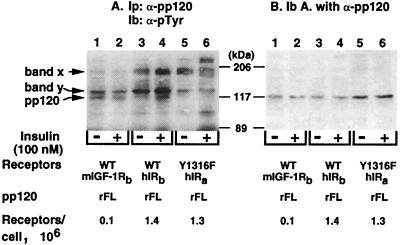
To determine whether the p120 protein is identical to pp120, the insulin receptor substrate, we treated transfected IR−/− hepatocytes with either buffer alone (Fig. 4, odd lanes) or 100 nM insulin (Fig. 4, even lanes) prior to lysis, immunoprecipitation with α-pp120/HA4 antibody, and immunoblotting with α-pTyr antibody (Fig 4A). The immunoblot was then reprobed with α-pp120 polyclonal antibody (Fig. 4B). Comparison of the immunoblot with α-pTyr antibody (Fig. 4A) to that with α-pp120 antibody (Fig. 4B) revealed the identity of the ∼120-kDa band as pp120. Moreover, insulin treatment of cells transfected with rat pp120 alone (WT mIGF-1Rb/rFL; “b” denotes a different clone from that of Fig. 2) did not increase tyrosine phosphorylation of pp120 by endogenous mouse IGF-1 receptors (Fig. 4A, lane 2 versus 1), as expected from our previous experiments (41) and from experiments shown in Fig. 2. In fact, pp120 phosphorylation in cells expressing IGF-1 receptors alone is decreased in response to insulin. Tyrosine phosphorylation of pp120 in the absence of ligand in cells expressing IGF-1 receptors alone is not at the present fully understood, but must certainly be related to the complexity of pp120 phosphorylation (44). For instance, pp120 phosphorylation is regulated not only by the activities of serine and tyrosine kinases, but also by a phosphatase activity associated with it (39). Additionally, since the IGF-1 receptor is not significantly phosphorylated in the absence of ligand (see below), it cannot be fully responsible for basal pp120 phosphorylation. Nonetheless, it is interesting that in contrast to cells expressing IGF-1 receptors alone, insulin treatment led to an ∼10-fold increase in tyrosine phosphorylation of pp120 in cells coexpressing wild-type insulin receptors (WT hIRb/rFL, where b denotes a different clone from that of Fig. 2) (Fig. 4A, lane 4 versus lane 3). Insulin treatment of cells coexpressing pp120 and Y1316F insulin receptors (Y1316F hIRa/rFL) did not increase pp120 phosphorylation (Fig. 4A, lane 6 versus lane 5). Thus, replacing the nonconserved Tyr1316 in the insulin receptor with the corresponding residue in the IGF-1 receptor abolished pp120 phosphorylation by the insulin receptor. Unidentified proteins of higher molecular weights were detected on this blot (Fig. 4A, bands x and y). However, their detection was not reproducible.
FIG. 4.
Phosphorylation of recombinant pp120 by site-directed mutant insulin receptors in intact cells. IR−/− hepatocytes stably transfected with cDNAs encoding the full-length isoform of rat pp120 (rFL) either alone (WT mIGF-1Rb/rFL, where b denotes the clone number and represents a different clone from that of Fig. 2) or with comparable numbers (1.3 × 106 to 1.4 × 106) of wild-type (WT hIRb/rFL) and Y1316F (Y1316F hIRa/rFL) insulin receptors were treated with insulin as described in the legend to Fig. 2. Cell lysates were subjected to immunoprecipitation (Ip) with a monoclonal antibody against pp120 (α-pp120) (A), analyzed by SDS-PAGE, and immunoblotted (Ib) with HRP-coupled α-pTyr (A). To assess the amount of pp120 in the immunopellets, the immunoblot in panel A was reprobed with α-pp120 polyclonal antibody (B). Molecular mass markers are indicated on the right-hand side of the gel. Bands x and y were not identified, but their detection was not reproducible.
These data suggest that, in contrast to other substrates of the insulin receptor, pp120 phosphorylation requires an intact nonconserved Tyr1316 in the C terminus of the β-subunit of the receptor.
Phosphorylation of recombinant pp120 by IGF-1 receptor mutants.
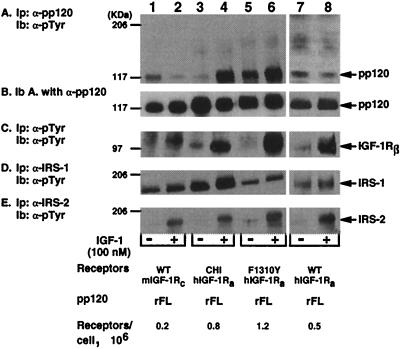
We then examined whether mutation of the nonconserved Phe1310 in the C terminus of the β-subunit of the IGF-1 receptor to tyrosine (the corresponding residue in the insulin receptor) restored pp120 phosphorylation by the receptor. This mutation impaired neither the affinity of the receptor to its ligand nor its tyrosine kinase activity in transfected NIH 3T3 cells (13). Thus, IR−/− hepatocytes were transfected with rat pp120 alone (WT mIGF-1Rc/rFL) or with either wild-type (WT hIGF-1Ra/rFL), F1310Y (F1310Y hIGF-1Ra/rFL), or chimeric IGF-1 receptors in which the C terminus of the β-subunit was replaced with the corresponding fragment of the insulin receptor (CHI hIGF-1Ra/rFL). Transfected cells were treated with IGF-1 (100 nM) prior to lysis and immunoprecipitation with antibodies against either pp120 (Fig. 5A; α-pp120), phosphotyrosines (Fig. 5C; α-pTyr), IRS-1 (Fig. 5D; α-IRS-1), or IRS-2 (Fig. 5E; α-IRS-2). To account for the amount of pp120 in the immunopellets, the immunoblot in Fig. 5A was reprobed with α-pp120 antibody (Fig. 5B). IGF-1 treatment caused comparable phosphorylation of the β-subunit (IGF-1Rβ) of the wild type receptor (Fig. 5C, lanes 2 versus 1 and 8 versus 7) and IGF-1 receptor mutants (Fig. 5C, chimeric, lane 4 versus 3, and F1310Y, lane 6 versus 5), supporting previous observations that these mutations did not impair the tyrosine kinase activity of the receptor and its autophosphorylation in transfected NIH 3T3 cells (13, 14). As expected from our previous experiments (41) and from experiments in Fig. 2 and 4, activated wild-type IGF-1 receptors failed to stimulate pp120 phosphorylation in cells transfected with pp120 alone (WT mIGF-1Rc/rFL) (Fig. 5A, lane 2 versus 1) or with pp120 and wild-type IGF-1 receptors (WT hIGF-1Ra/rFL) (Fig. 5A, lane 8 versus 7). As in our previous reports (41), replacing the C terminus of the β-subunit of the IGF-1 receptor with that of the insulin receptor restored pp120 phosphorylation by the chimeric IGF-1 receptor, as indicated by the ∼15-fold increase in the amount of phosphorylated tyrosine in pp120 in cells coexpressing chimeric receptors (CHI hIGF-1Ra/rFL) (Fig. 5A, lane 4 versus 3). Similarly, mutating Phe1310 to tyrosine in the IGF-1 receptor resulted in an approximately five-fold increase in the amount of phosphorylated tyrosines in pp120 (Fig. 5A, lane 6 versus 5), suggesting that replacing the nonconserved Phe1310 residue with the corresponding residue in the insulin receptor restored pp120 phosphorylation by the IGF-1 receptor. Interestingly, basal pp120 phosphorylation was higher in cells overexpressing the F1310Y mutant (Fig. 5A, lane 5) than that in cells overexpressing either wild-type (Fig. 5A, lane 7) or chimeric (Fig. 5A, lane 3) IGF-1 receptors. Because the F1310Y IGF-1 receptor was not basally phosphorylated (Fig. 5C, lane 5), it is not probably the kinase responsible for pp120 in the absence of ligand (Fig. 5A, lane 5). This lends more credence to the notion that other kinases may cause pp120 phosphorylation in the absence of ligand.
FIG. 5.
Phosphorylation of recombinant pp120 by site-directed mutant IGF-1 receptors in intact cells. IR−/− hepatocytes were stably transfected with cDNAs encoding the full-length isoform of rat pp120 (rFL) either alone (WT mIGF-1Rc/rFL) or with wild-type (WT hIGF-1Ra/rFL), chimeric (CHI hIRa/rFL), and F1310Y IGF-1 (F1310Y hIGF-1Ra/rFL) receptors. Transfectants expressed 0.2 × 106, 0.5 × 106, 0.8 × 106, and 1.2 × 106 receptors per cell, respectively. Cells were serum starved for 8 h prior to incubation in the presence (+ lanes) or absence (− lanes) of IGF-1 (100 nM) for 5 min. Cell lysates were then subjected to immunoprecipitation (Ip) with antibodies against pp120 (A), phosphotyrosines (C), IRS-1 (D), or IRS-2 (E) prior to analysis by SDS-PAGE and immunoblotting (Ib) with HRP-coupled α-pTyr. To assess the amount of pp120 in the immunopellets, the immunoblot in panel A was reprobed with a polyclonal antibody against pp120 (B). Molecular mass markers are indicated on the left-hand side of the gel.
As indicated in Fig. 5D and E, transfecting pp120 in the hepatocytes did not impair the ability of IGF-1 to activate its receptor to phosphorylate other substrates, such as IRS-1 (Fig. 5D, lanes 2 versus 1 and 8 versus 7) and IRS-2 (Fig. 5E, lanes 2 versus 1 and 8 versus 7). As opposed to previous experiments with IR−/− hepatocytes (52), the level of basal IRS-1 phosphorylation was high (Fig. 5D, lane 1). The insignificant basal phosphorylation of IRS-2 in these cells (Fig. 5E, odd lanes) suggests that the high basal phosphorylation of IRS-1 is not largely due to intrinsic activation of these cells, perhaps upon their transformation with SV40. Because basal IRS-1 phosphorylation in cells transfected with the Hygror plasmid alone was similarly high (data not shown), we conclude that pp120 transfection did not alter IRS-1 phosphorylation in these cells. Thus, the high basal IRS-1 phosphorylation in our experiments, compared to that in previous reports (52), is probably due to differences in the amount of proteins immunoprecipitated and in the α-IRS-1 antibodies used, among other technical variabilities. Nonetheless, replacing the C-terminus domain of the IGF-1 receptor with that of the insulin receptor or mutating its Phe1310 to the corresponding residue in the insulin receptor did not impair either IRS-1 phosphorylation or IRS-2 phosphorylation by IGF-1 receptors in response to ligand (Fig. 5D and E, lanes 4 versus 3 and 6 versus 5, respectively). This is in agreement with previous observations that phosphorylation of IRS-1, Shc, and Crk-II by these mutant IGF-1 receptors was intact in transfected NIH 3T3 cells (13). Moreover, our data indicate that these IGF-1 mutations did not impair the tyrosine activity of the receptor in transfected IR−/− hepatocytes.
Proliferation and growth of cells expressing insulin receptor mutants.
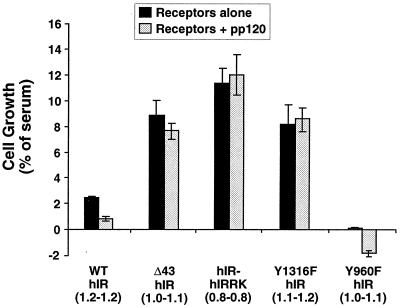
Because hepatocytes constitute a major site of the insulin receptor's expression, we aimed at transfecting IR−/− hepatocytes with insulin receptor mutants to investigate the role of the C terminus of the β-subunit of the insulin receptor in insulin mitogenesis. Because expression of the IGF-1 receptors in these hepatocytes was slightly elevated to ∼0.1 × 105 to 0.2 × 105 receptors/cell (11), we treated cells with insulin at the low concentration of 0.1 nM in order to avoid potential activation of endogenous IGF-1 receptors and measured cell growth and proliferation as a marker of mitogenesis. Stable transfectants expressing comparable numbers of receptors per cell in each clonal pair (with or without pp120) and among the different receptor types were used in these studies. As Fig. 6 reveals, deletion of the 43 aa from the C terminus or replacement of the intracellular tail of the insulin receptor with that of the IRR increased insulin-induced cell growth in comparison with that of cells expressing wild-type insulin receptors (Δ43 hIR, 8.88 ± 2.53, and hIR-hIRRK, 11.3 ± 1.26, versus WT hIR, 2.46 ± 0.10; p < 0.05). Similarly, mutation of Tyr1316 to phenylalanine resulted in an approximately fourfold increase in insulin-induced cell growth in comparison with that of cells expressing wild-type insulin receptors (Y1316F hIR, 8.16 ± 1.55, versus WT hIR, 2.46 ± 0.10; P < 0.05). Although not shown, double Y1316F and Y1322F mutations produced the same effect as the single Y1316F mutation. Increased insulin-induced cell growth upon removing the C terminus of the β-subunit of the insulin receptor or abolishing its tyrosine phosphorylation sites in a cell derived from hepatocytes suggests that this domain contains elements that suppress the mitogenic action of insulin. Because a similar observation was made in cells that do not express endogenous pp120 (2, 47, 58), the increase in the growth-promoting action of insulin in IR−/− hepatocytes upon mutation of the C terminus of the β-subunit of the insulin receptor is not regulated by the endogenous expression of pp120 in hepatocytes.
FIG. 6.
Cell proliferation in response to insulin. IR−/− hepatocytes were transfected with the wild-type insulin receptor (WT hIR) or insulin receptor mutants (Δ43, hIR-hIRRK, Y1316F and Y960F), either alone (solid bars) or in addition to pp120 (shaded bars). Following incubation for 24 h in serum-containing complete medium (to determine maximum growth) or in serum-free medium supplemented with 0.1% BSA either alone (to determine basal growth) or with 0.1 nM insulin, cells were trypsinized and counted. Numbers in parentheses denote the number of receptors per cell in millions in transfectants with receptors alone (first number) or with receptors and pp120 (second number). These experiments were performed in triplicate and repeated at least three times. Data represent the mean ± standard deviation of these repeated experiments.
In agreement with our previous thymidine uptake assays (15), pp120 expression decreased the effect of insulin on the growth of cells expressing wild-type insulin receptors by approximately threefold (Fig. 6; WT hIR/pp120, 0.82 ± 0.12, versus WT hIR, 2.46 ± 0.10; P < 0.05). In contrast, pp120 expression did not alter the mitogenic action of insulin in cells coexpressing insulin receptors with a mutated C terminus (Δ43 hIR/pp120, 7.67 ± 0.50, versus Δ43 hIR, 8.88 ± 2.53; hIR-hIRRK/pp120, 12.0 ± 1.57, versus hIR-hIRRK, 11.3 ± 1.26; and Y1316F hIR/pp120, 8.46 ± 1.09, versus Y1316F hIR, 8.16 ± 1.55; P > 0.05). These data suggest that the downregulatory effect of pp120 on insulin mitogenesis is mediated by the C terminus of the β-subunit of the insulin receptor and, in particular, by its Tyr1316 residue, a nonconserved residue between the insulin and IGF-1 receptors.
In contrast to mutation on the C-terminus domain of the insulin receptor, mutation of Tyr960 in the juxtamembrane domain markedly decreased cell growth in response to insulin (Y960F hIR, 0.12 ± 0.01, versus WT hIR, 2.46 ± 0.10; P < 0.05). This suggests that phosphorylation on Tyr960 in the juxtamembrane domain of the insulin receptor promotes the mitogenic action of insulin. Because IRS-2 phosphorylation requires intact Tyr960 in the insulin receptor, the marked decrease in cell growth upon abolishing phosphorylation of this residue is not surprising in light of the observation that the IRS-2–PI-3′ kinase pathway mediated insulin mitogenesis in IR−/− hepatocytes (52). Moreover, pp120 expression was correlated with decreased growth of cells transfected with Y960F receptors (Fig. 6; −1.89 ± 0.24). Whether this is due to a proapoptotic effect of pp120 is not clear at the present time. Nonetheless, as in cells expressing wild-type insulin receptors, pp120 expression markedly decreased insulin-mediated growth of cells expressing Y960F insulin receptors compared to that of cells expressing Y960F receptors alone (Y960F hIR/pp120, −1.89 ± 0.24, versus Y960F hIR, 0.12 ± 0.01; P < 0.05). This suggests that the downregulatory effect of pp120 on the growth-promoting action of insulin does not require intact Tyr960 in the juxtamembrane domain of the receptor.
Proliferation and growth of cells expressing IGF-1 receptor mutants.
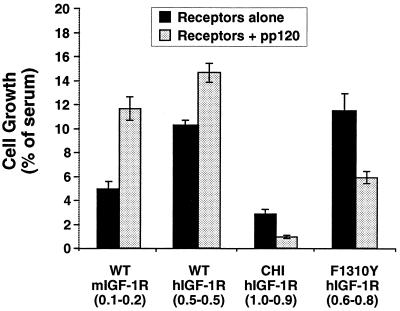
Next, we investigated the role of pp120 on the growth-promoting action of IGF-1 in transfected IR−/− hepatocytes. To this end, we used transfectants expressing comparable numbers of receptors per cell in each clonal pair (with or without pp120). In marked contrast to insulin, pp120 expression did not decrease the mitogenic action of IGF-1 in IR−/− hepatocytes that were cotransfected with either wild-type IGF-1 receptors (Fig. 7; WT hIGF-1R/pp120) or with Hygror (Fig. 7; WT mIGF-1R/pp120). The effect of pp120 on IGF-1 mitogenesis ranged from a modest increase (Fig. 7; WT hIGF-1R/pp120, 14.6 ± 0.85 versus 10.3 ± 0.45; P < 0.05) to a twofold increase (Fig. 7; WT mIGF-1R/pp120, 11.8 ± 0.89, versus WT mIGF-1R, 5.07 ± 0.65; P < 0.05). The level of increase is perhaps inversely related to the level of phosphorylation of pp120 in response to IGF-1 in cells expressing IGF-1 receptors (Fig. 5). Replacing the C terminus of the IGF-1 receptor with the corresponding fragment of the insulin receptor decreased the effect of IGF-1 on cell growth compared to that in cells expressing wild-type receptors (Fig. 7; CHI hIGF-1R, 2.91 ± 0.37, versus WT mIGF-1R, 5.07 ± 0.65, or WT hIGF-1R, 10.3 ± 0.45; P < 0.05). The level of decrease is marked in light of the fact that the transfectants express about twofold more chimeric than wild-type receptors (∼1.0 × 106 versus 0.5 × 106 receptors/cell). These data suggest that the C terminus of the β-subunit of the insulin receptor contains elements that suppress the mitogenic action of hormones. Expressing pp120 further decreased the effect of IGF-1 on the growth of cells coexpressing chimeric IGF-1 receptors (Fig. 7; CHI hIGF-1R/pp120, 0.97 ± 0.13, versus CHI hIGF-1R, 2.91 ± 0.37; P < 0.05). Similarly, pp120 expression decreased the effect of IGF-1 on the growth of IR−/− hepatocytes coexpressing F1310Y IGF-1 receptors (Fig. 7; F1310Y hIGF-1R/pp120, 6.04 ± 0.39, versus F1310Y hIGF-1R, 11.4 ± 1.53; P < 0.05). Because replacing the C terminus of the β-subunit of the IGF-1 receptor with the corresponding fragment of the insulin receptor and replacing its Phe1310 with tyrosine (the corresponding residue in the insulin receptor) restored pp120 phosphorylation in response to IGF-1 (Fig. 5), it appears that pp120 phosphorylation is required for its downregulatory effect on mitogenesis.
FIG. 7.
Cell proliferation in response to IGF-1. WT mIGF-1R represents IR−/− hepatocytes transfected with Hygror plasmid alone, but expressing endogenous IGF-1 receptors. These cells were transfected with either pp120 alone (shaded bars) or with wild-type (WT), chimeric (CHI), or F1310Y hIGF-1 receptors either alone (solid bars) or in addition to pp120 (shaded bars). Following incubation with 0.1 nM IGF-1, as described in the legend to Fig. 5, cells were trypsinized and counted. Numbers in parentheses denote the number of receptors per cell in millions in transfectants with receptors alone (first number) or with receptors and pp120 (second number). These experiments were performed in triplicate and repeated at least three times. Data represent the mean ± standard deviation of these repeated experiments.
Growth of cells expressing F1310Y IGF-1 receptors in response to IGF-1 was comparable to that of cells expressing wild-type receptors (Fig. 7; F1310Y hIGF-1R, 11.4 ± 1.53, versus WT hIGF-1R, 10.3 ± 0.45; P > 0.05). This suggests that elements other than intact Phe1310 in the C terminus of the IGF-1 receptor play a significant role in the growth-promoting action of IGF-1, in agreement with previous observations with NIH 3T3 fibroblasts (13). However, in light of the endogenous expression of IGF-1 receptors in IR−/− hepatocytes, it is hard to conclude from the current studies the precise effect of the Phe1310-to-tyrosine mutation on IGF-1 mitogenesis. Despite the reasonably elevated expression of IGF-1 receptors in IR−/− hepatocytes derived from the insulin receptor knockout mice, the expression of IGF-1 receptors in hepatocytes is usually much less significant (37). Thus, physiologic cells derived from the IGF-1 receptor knockout mice, for example, would constitute a better system to address the exact role of the nonconserved Phe1310 in the IGF-1 receptor in the differential mitogenic action of insulin and IGF-1. Nonetheless, mutating Phe1310 in the IGF-1 receptor to tyrosine (the corresponding residue in the insulin receptor) restored the downregulatory effect of pp120 on IGF-1 mitogenesis. Conversely, replacing Tyr1316 of the insulin receptor with phenylalanine (the corresponding residue in the IGF-1 receptor) abolished the downregulatory effect of pp120 on insulin mitogenesis. Taken together, these data suggest that the differential effect of pp120 on insulin vis-à-vis IGF-1 mitogenesis is regulated by the nonconserved Tyr1316 residue of the insulin receptor.
DISCUSSION
The physiologic functions of insulin and IGF-1 are initiated upon binding to their receptors followed by activation of multiple phosphorylation cascades. Because insulin and IGF-1 receptors are related and they share many signaling mechanisms, it has been difficult to depict the molecular basis of the different functions elicited by their ligands (1, 5, 31).
Differential phosphorylation of pp120 by the insulin receptor is regulated by Tyr1316, a nonconserved phosphorylation site in the C terminus of the insulin receptor.
Using stably transfected NIH 3T3 fibroblasts (41) and hepatocytes (current studies), we have shown that pp120 is unique among other substrates of the insulin receptor insofar as it does not undergo ligand-stimulated phosphorylation by the IGF-1 receptor kinase. Additionally, its insulin-stimulated phosphorylation is regulated by the C terminus of the β-subunit of the insulin receptor, as opposed to other major substrates, such as IRS-1 and Shc (2, 38). Instead, phosphorylation of these substrates is regulated by Tyr960 in the juxtamembrane domain of the insulin receptor (22, 67) and its corresponding residue in the IGF-1 receptor (8). The present studies revealed that deleting the distal 43 aa from the C terminus of the β-subunit of the insulin receptor and, in particular, mutating the Tyr1316 residue therein contained to phenylalanine, as is the case in the IGF-1 receptor, abolished insulin-induced pp120 phosphorylation by the insulin receptor without significantly altering the phosphorylation state of the receptor. Conversely, mutating the corresponding Phe1310 in the IGF-1 receptor to tyrosine, its corresponding residue in the insulin receptor, restored pp120 phosphorylation by the IGF-1 receptor in response to IGF-1. This suggests that differential pp120 phosphorylation by the insulin receptor requires intact Tyr1316, a nonconserved residue in the two receptors. These data represent the first evidence of a single amino acid regulating differential phosphorylation of a substrate by two closely related receptors with ∼84% homology in their tyrosine kinase domains.
Differential pp120 phosphorylation by the insulin receptor regulates its specific downregulatory effect on insulin-induced mitogenesis.
Because the C termini of the β-subunits of insulin and IGF-1 receptors are the least conserved, it has long been postulated that they regulate functional diversity between these two related receptors. However, there has been no experimental evidence to support this hypothesis. Despite the recent evidence supporting a role for the C terminus of the β-subunit of the insulin receptor in regulating the metabolic action of insulin (50), most reports agree that this domain does not regulate the metabolic action of insulin or its receptor-mediated endocytosis (2, 38, 59). More controversial is the role of the C terminus of the β-subunit of the insulin receptor in regulating the mitogenic action of insulin (2, 38, 59). For instance, insulin-induced thymidine uptake and MAP kinase activity were either normal (38, 70) or enhanced (2, 47, 58) in cells transfected with insulin receptor mutants depleted of phosphorylation sites in the C terminus. The controversy has been attributed to transfection of nonphysiologic cells in these experiments. Our current studies are the first to invoke transfection of hepatocytes, which physiologically express high levels of insulin receptors, to address the role of the C terminus of the β-subunit of the insulin receptor in the growth-promoting action of insulin. Despite the fact that transforming the IR−/− hepatocytes with SV40 may decrease their physiologic state, their derivation from the liver of the insulin receptor knockout mouse rendered them ideal to study the regulation of insulin signaling by the insulin receptor. Insulin treatment of IR−/− hepatocytes overexpressing wild-type insulin receptors induced cell growth at a lower level than that elicited by IGF-1 treatment of hepatocytes transfected with wild-type IGF-1 receptors. This supports the notion that IGF-1 is more mitogenic than insulin (1, 5, 31). Replacement of the C terminus of the β-subunit of the IGF-1 receptor with the corresponding fragment in the insulin receptor decreased cell growth in response to IGF-1. Thus, the C terminus of the β-subunit of the insulin receptor contains negative regulators of the growth-promoting action of insulin in hepatocytes. Our data are in disagreement with those from previous reports in which expression of identical chimeric IGF-1 receptors in NIH 3T3 cells resulted in either unchanged or slightly increased thymidine uptake (14) and MAP kinase activity (61) in response to IGF-1. The discrepancy between those results and ours may be attributed to the different cell lines used. Nonetheless, abolishing tyrosine phosphorylation in the C terminus of the β-subunit of the insulin receptor, either by deletion or by site-directed mutagenesis, enhanced the growth-promoting action of insulin in IR−/− hepatocytes. This suggests that tyrosine phosphorylation in the C terminus of the β-subunit of the insulin receptor regulates the low mitogenic action of insulin. Because we transfected cells derived from insulin-targeted tissues, we believe that our data have finally resolved the controversy over the downregulation of the mitogenic action of insulin by the C terminus of the β-subunit of its receptor.
In contrast to insulin (Fig. 6) (15), pp120 expression was not correlated with a decrease in the mitogenic action of IGF-1 in IR−/− hepatocytes (Fig. 7). In light of the low abundance of IGF-1 receptors compared to that of insulin receptors in hepatocytes (37), the differential downregulatory effect of pp120 on the growth-promoting action of insulin is probably physiologic. Because pp120 phosphorylation was increased by ligand-activated insulin receptors, but not by IGF-1 receptors (Fig. 4 and 5) (41), pp120 phosphorylation appears to be required for its downregulation of the growth-promoting action of insulin. This conclusion is supported by our observations that (i) restoring pp120 phosphorylation by chimeric and F1310Y IGF-1 receptors (Fig. 5) was correlated with decreased IGF-1 mitogenesis by pp120 (Fig. 7), and (ii) abolishing pp120 phosphorylation by mutating tyrosine phosphorylation sites on either the C terminus of the β-subunit of the insulin receptor (Fig. 2, 3, and 4) or on pp120 (15) eliminated the effect of pp120 on insulin mitogenesis. Furthermore, mutating Tyr960 in the juxtamembrane domain of the β-subunit of the insulin receptor did not alter either the effect of pp120 on insulin mitogenesis (Fig. 6) or its phosphorylation by the insulin receptor (42). Taken together, these data suggest that pp120 phosphorylation by the insulin receptor is required and sufficient to regulate its differential downregulatory effect on the mitogenic effects of insulin vis-à-vis IGF-1.
How pp120 phosphorylation regulates its effect on insulin mitogenesis is not clear. However, Syp has recently been found to bind to Tyr488 and Tyr515 of BGP1, the human homolog of rat pp120 (21). Moreover, we have recently observed that pp120 binds to Shc and that this association is increased upon insulin-stimulated pp120 phosphorylation by the insulin receptor tyrosine kinase (M. N. Poy, M. Fernström, and S. M. Najjar, Diabetes 48[Suppl. 1], abstr. A33, 1999). Given the positive role of Shc and Syp in insulin mitogenesis, it is possible that pp120 binding to these molecules sequesters them and limits their availability for GRB2 coupling to the receptor. This would downregulate the Ras/MAP kinase pathway, leading to decreased cell growth, proliferation, and mitogenesis. Insulin-induced growth of hepatocytes expressing Y960F insulin receptors supports this model. Given the regulation of phosphorylating Shc and the IRS proteins by Tyr960 and their role in activating the Ras/MAP kinase and the PI-3′ kinase pathways, it is expected that mutating Tyr960 in the insulin receptor to nonphosphorylatable phenylalanine would markedly decrease cell growth in response to insulin (Fig. 6). When coexpressed with Y960F insulin receptors, pp120 binds to Syp. This reduces the Syp pool available to associate with Tyr1322 in the receptor (57, 64) and decreases mitogenesis and cell growth in response to insulin compared to that of cells expressing Y960F receptors alone.
Downregulation of the mitogenic effects of insulin appears to be unique to pp120 in comparison to that of other substrates of the insulin receptor, such as IRS-1, genetic ablation of which resulted in growth retardation in mice (3, 60). Because IRS-1 phosphorylation requires intact Tyr960 in the juxtamembrane, whereas that of pp120 requires intact Tyr1316 in the C terminus of the insulin receptor, phosphorylation of pp120 by the insulin receptor appears to provide an alternative signaling pathway via which the insulin receptor kinase modulates the biologic action of insulin. In view of the relationship between the regulation of pp120 phosphorylation by the C terminus of the β-subunit of the insulin receptor and the implication of this domain in the mitogenic response to insulin, an extension of our hypothesis is that abnormal pp120 expression is associated with abnormal growth and development. Further studies are required to shed light on this possibility.
ACKNOWLEDGMENTS
We thank Richard A. Roth (Stanford University), Jerrold M. Olefsky (University of California, San Diego), Derek LeRoith (NIDDK, NIH), and Domenico Accili (NICHD, NIH) for providing recombinant hIR-hIRRK, Y1316F/Y1322F hIR, F1310Y hIGF-1R, and Δ43 hIR mutant receptors, respectively. We also thank D. Accili for providing us with the IR−/− hepatocytes, Myrna Saouda for her technical assistance with cloning experiments, and Curtis V. Choice and Yan Yang for their technical assistance in transfection experiments.
This work was supported by the National Science Foundation (grant MCB-9601427 to S.M.N.)
REFERENCES
- 1.Accili D, Drago J, Lee E J, Johnson M D, Cool M H, Salvatore P, Asico L D, Jose P A, Taylor S I, Westphal H. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 2.Ando A, Momomura K, Tobe K, Yamamoto-Honda R, Sakura H, Tamori Y, Kaburagi Y, Koshio O, Akanuma Y, Yazaki Y, et al. Enhanced insulin-induced mitogenesis and mitogen-activated protein kinase activities in mutant insulin receptors with substitution of two COOH-terminal tyrosine autophosphorylation sites by phenylalanine. J Biol Chem. 1992;267:12788–12796. [PubMed] [Google Scholar]
- 3.Araki E, Lipes M A, Patti M E, Bruning J C, Haag B R, Johnson R S, Kahn C R. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 4.Backer J M, Schroeder G G, Kahn C R, Myers M J, Wilden P A, Cahill D A, White M F. Insulin stimulation of phosphatidylinositol 3-kinase activity maps to insulin receptor regions required for endogenous substrate phosphorylation. J Biol Chem. 1992;267:1367–1374. [PubMed] [Google Scholar]
- 5.Baker J, Liu J P, Robertson E J, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 5a.Beauchemin N, et al. Nomenclature announcement. Exp Cell Res. 1999;252:243–249. doi: 10.1006/excr.1999.4610. [DOI] [PubMed] [Google Scholar]
- 6.Blenis J. Signal transduction via the MAP kinases: proceed at your own RSK. Proc Natl Acad Sci USA. 1993;90:5889–5892. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung P H, Thompson N L, Earley K, Culic O, Hixson D, Lin S H. Cell-CAM105 isoforms with different adhesion functions are coexpressed in adult rat tissues and during liver development. J Biol Chem. 1993;268:6139–6146. [PubMed] [Google Scholar]
- 8.Craparo A, O'Neill T J, Gustafson T A. Non-SH2 domains within insulin receptor substrate-1 and SHC mediate their phosphotyrosine-dependent interaction with the NPEY motif of the insulin-like growth factor I receptor. J Biol Chem. 1995;270:15639–15643. doi: 10.1074/jbc.270.26.15639. [DOI] [PubMed] [Google Scholar]
- 9.Crews C M, Erikson R L. Extracellular signals and reversible protein phosphorylation: what to Mek of it all. Cell. 1993;74:215–217. doi: 10.1016/0092-8674(93)90411-i. [DOI] [PubMed] [Google Scholar]
- 10.Danielsen A G, Liu F, Hosomi Y, Shii K, Roth R A. Activation of protein kinase C alpha inhibits signaling by members of the insulin receptor family. J Biol Chem. 1995;270:21600–21605. doi: 10.1074/jbc.270.37.21600. [DOI] [PubMed] [Google Scholar]
- 11.Di Cola G, Cool M H, Accili D. Hypoglycemic effect of insulin-like growth factor-1 in mice lacking insulin receptors. J Clin Investig. 1997;99:2538–2544. doi: 10.1172/JCI119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edlund M, Öbrink B. Evidence for calmodulin binding to the cytoplasmic domains of two C-CAM isoforms. FEBS Lett. 1993;327:90–94. doi: 10.1016/0014-5793(93)81046-3. [DOI] [PubMed] [Google Scholar]
- 13.Esposito D L, Blakesley V A, Koval A P, Scrimgeour A G, LeRoith D. Tyrosine residues in the C-terminal domain of the insulin-like growth factor-I receptor mediate mitogenic and tumorigenic signals. Endocrinology. 1997;138:2979–2988. doi: 10.1210/endo.138.7.5281. [DOI] [PubMed] [Google Scholar]
- 14.Faria T N, Blakesley V A, Kato H, Stannard B, LeRoith D, Roberts C T., Jr Role of the carboxyl-terminal domains of the insulin and insulin-like growth factor I receptors in receptor function. J Biol Chem. 1994;269:13922–13928. [PubMed] [Google Scholar]
- 15.Formisano P, Najjar S M, Gross C N, Philippe N, Oriente F, Kern-Buell C I, Accili D, Gorden P. Receptor-mediated internalization of insulin. Potential role of pp120/HA4, a substrate of the insulin receptor kinase. J Biol Chem. 1995;270:24073–24077. doi: 10.1074/jbc.270.41.24073. [DOI] [PubMed] [Google Scholar]
- 16.Gale N W, Kaplan S, Lowenstein E J, Schlessinger J, Bar-Sagi D. Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras. Nature. 1993;363:88–92. doi: 10.1038/363088a0. [DOI] [PubMed] [Google Scholar]
- 17.Grønborg M, Wulff B S, Rasmussen J S, Kjeldsen T, Gammeltoft S. Structure-function relationship of the insulin-like growth factor-I receptor tyrosine kinase. J Biol Chem. 1993;268:23435–23440. [PubMed] [Google Scholar]
- 18.Gustafson T A, He W, Craparo A, Schaub C D, O'Neill T J. Phosphotyrosine-dependent interaction of SHC and insulin receptor substrate 1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol Cell Biol. 1995;15:2500–2508. doi: 10.1128/mcb.15.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hixson D C, McEntire K D, Obrink B. Alterations in the expression of a hepatocyte cell adhesion molecule by transplantable rat hepatocellular carcinomas. Cancer Res. 1985;45:3742–3749. [PubMed] [Google Scholar]
- 20.Hsieh J-T, Luo W, Song W, Wang Y, Kleinerman D I, Van N T, Lin S-H. Tumor suppressive role of an androgen-regulated epithelial cell adhesion molecule (C-CAM) in prostate carcinoma cell revealed by sense and antisense approaches. Cancer Res. 1995;55:190–197. [PubMed] [Google Scholar]
- 21.Huber M, Izzi L, Grondin P, Houde C, Kunath T, Veillette A, Beauchemin N. The carboxy-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J Biol Chem. 1999;274:335–344. doi: 10.1074/jbc.274.1.335. [DOI] [PubMed] [Google Scholar]
- 22.Kaburagi Y, Momomura K, Yamamoto H R, Tobe K, Tamori Y, Sakura H, Akanuma Y, Yazaki Y, Kadowaki T. Site-directed mutagenesis of the juxtamembrane domain of the human insulin receptor. J Biol Chem. 1993;268:16610–16622. [PubMed] [Google Scholar]
- 23.Kato H, Faria T N, Stannard B, Roberts C T, Jr, LeRoith D. Essential role of tyrosine residues 1131, 1135, and 1136 of the insulin-like growth factor-I (IGF-I) receptor in IGF-I action. Mol Endocrinol. 1994;8:40–50. doi: 10.1210/mend.8.1.7512194. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M, Ogihara M. Proliferation of adult rat hepatocytes in primary culture induced by insulin is potentiated by cAMP-elevating agents. Eur J Pharmacol. 1997;327:87–95. doi: 10.1016/s0014-2999(97)89682-3. [DOI] [PubMed] [Google Scholar]
- 25.Kleinerman D I, Dinney C P, Zhang W W, Lin S H, Van N T, Hsieh J T. Suppression of human bladder cancer growth by increased expression of C-CAM1 gene in an orthotopic model. Cancer Res. 1996;56:3431–3425. [PubMed] [Google Scholar]
- 26.Kleinerman D I, Troncoso P, Lin S H, Pisters L L, Sherwood E R, Brooks T, von Eschenbach A C, Hsieh J T. Consistent expression of an epithelial cell adhesion molecule (C-CAM) during human prostate development and loss of expression in prostate cancer: implication as a tumor suppressor. Cancer Res. 1995;55:1215–1220. [PubMed] [Google Scholar]
- 27.Lee J, Pilch P F. The insulin receptor: structure, function and signalling. Am J Physiol. 1994;266:C319–C334. doi: 10.1152/ajpcell.1994.266.2.C319. [DOI] [PubMed] [Google Scholar]
- 28.Levy-Toledano R, Caro L H, Accili D, Taylor S I. Investigation of the mechanism of the dominant negative effect of mutations in the tyrosine kinase domain of the insulin receptor. EMBO J. 1994;13:835–842. doi: 10.1002/j.1460-2075.1994.tb06326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Resnicoff M, Baserga R. Effect of mutations at serines 1280–1283 on the mitogenic and transforming activities of the insulin-like growth factor I receptor. J Biol Chem. 1996;271:12254–12260. doi: 10.1074/jbc.271.21.12254. [DOI] [PubMed] [Google Scholar]
- 30.Lin S H, Guidotti G. Cloning and expression of a cDNA coding for a rat liver plasma membrane ecto-ATPase. The primary structure of the ecto-ATPase is similar to that of the human biliary glycoprotein I. J Biol Chem. 1989;264:14408–14414. [PubMed] [Google Scholar]
- 31.Liu J P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 32.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 33.Luo W, Earley K, Tantingco V, Hixson D C, Liang T C, Lin S H. Association of an 80 kDa protein with C-CAM1 cytoplasmic domain correlates with C-CAM1-mediated growth inhibition. Oncogene. 1998;16:1141–1147. doi: 10.1038/sj.onc.1201619. [DOI] [PubMed] [Google Scholar]
- 34.Luo W, Wood C G, Earley K, Hung M C, Lin S H. Suppression of tumorigenicity of breast cancer cells by an epithelial cell adhesion molecule (C-CAM1): the adhesion and growth suppression are mediated by different domains. Oncogene. 1997;14:1697–1704. doi: 10.1038/sj.onc.1200999. [DOI] [PubMed] [Google Scholar]
- 35.Maegawa H, McClain D A, Freidenberg G, Olefsky J M, Napier M, Lipari T, Dull T J, Lee J, Ullrich A. Properties of a human insulin receptor with a COOH-terminal truncation. II. Truncated receptors have normal kinase activity but are defective in signaling metabolic effects. J Biol Chem. 1988;263:8912–8917. [PubMed] [Google Scholar]
- 36.Margolis R N, Schell M J, Taylor S I, Hubbard A L. Hepatocyte plasma membrane ecto-ATPase (pp120/HA4) is a substrate for tyrosine kinase activity of the insulin receptor. Biochem Biophys Res Commun. 1990;166:562–566. doi: 10.1016/0006-291x(90)90845-e. [DOI] [PubMed] [Google Scholar]
- 37.Morgan D, Jarnagin K, Roth R. Purification and characterization of the receptor for insulin-like growth factor 1. Biochemistry. 1986;25:5560–5564. doi: 10.1021/bi00367a032. [DOI] [PubMed] [Google Scholar]
- 38.Murakami M S, Rosen O M. The role of insulin receptor autophosphorylation in signal transduction. J Biol Chem. 1991;266:22653–22660. [PubMed] [Google Scholar]
- 39.Najjar S M. pp120, a substrate of the insulin receptor tyrosine kinase, is associated with phosphatase activity. Biochem Biophys Res Commun. 1998;247:457–461. doi: 10.1006/bbrc.1998.8822. [DOI] [PubMed] [Google Scholar]
- 40.Najjar S M, Accili D, Philippe N, Jernberg J, Margolis R, Taylor S I. pp120/ecto-ATPase, an endogenous substrate of the insulin receptor tyrosine kinase, is expressed as two variably spliced isoforms. J Biol Chem. 1993;268:1201–1206. [PubMed] [Google Scholar]
- 41.Najjar S M, Blakesley V A, Li Calzi S, Kato H, LeRoith D, Choice C V. Differential phosphorylation of pp120 by insulin and insulin-like growth factor-1 receptors: role for the C-terminal domain of the beta-subunit. Biochemistry. 1997;36:6827–6834. doi: 10.1021/bi962634h. [DOI] [PubMed] [Google Scholar]
- 42.Najjar S M, Choice C V, Soni P, Whitman C M, Poy M N. Effect of pp120 on receptor-mediated insulin endocytosis is regulated by the juxtamembrane domain of the insulin receptor. J Biol Chem. 1998;273:12923–12928. doi: 10.1074/jbc.273.21.12923. [DOI] [PubMed] [Google Scholar]
- 43.Najjar S M, Lewis R E. Persistent expression of foreign genes in cultured hepatocytes: expression vectors. Gene. 1999;230:41–45. doi: 10.1016/s0378-1119(99)00052-9. [DOI] [PubMed] [Google Scholar]
- 44.Najjar S M, Philippe N, Suzuki Y, Ignacio G A, Formisano P, Accili D, Taylor S I. Insulin-stimulated phosphorylation of recombinant pp120/HA4, an endogenous substrate of the insulin receptor tyrosine kinase. Biochemistry. 1995;34:9341–9349. doi: 10.1021/bi00029a009. [DOI] [PubMed] [Google Scholar]
- 45.Nédellec P, Dveksler G S, Daniels E, Turbide C, Chow B, Basile A A, Holmes K V, Beauchemin N. Bgp2, a new member of the carcinoembryonic antigen-related gene family, encodes an alternative receptor for mouse hepatitis viruses. J Virol. 1994;68:4525–4537. doi: 10.1128/jvi.68.7.4525-4537.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neumaier M, Paululat S, Chan A, Matthaes P, Wagener C. Biliary glycoprotein, a potential human cell adhesion molecule, is down-regulated in colorectal carcinomas. Proc Natl Acad Sci USA. 1993;90:10744–10748. doi: 10.1073/pnas.90.22.10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pang L, Milarski K L, Ohmichi M, Takata Y, Olefsky J M, Saltiel A R. Mutation of two carboxyl-terminal tyrosines in the insulin receptor results in enhanced activation of mitogen-activated protein kinase. J Biol Chem. 1994;269:10604–10608. [PubMed] [Google Scholar]
- 48.Pillay T S, Whittaker J, Lammers R, Ullrich A, Siddle K. Multisite serine phosphorylation of the insulin and IGF-I receptors in transfected cells. FEBS Lett. 1991;288:206–211. doi: 10.1016/0014-5793(91)81035-7. [DOI] [PubMed] [Google Scholar]
- 49.Pronk G J, McGlade J, Pelicci G, Pawson T, Bos J I. Insulin-induced phosphorylation of the 46- and 52-kDa Shc proteins. J Biol Chem. 1993;268:5748–5753. [PubMed] [Google Scholar]
- 50.Reusch J E, Hsieh P, Bhuripanyo P, Carel K, Leitner J W, Olefsky J M, Draznin B. Insulin inhibits nuclear phosphatase activity: requirement for the C-terminal domain of the insulin receptor. Endocrinology. 1995;136:2464–2469. doi: 10.1210/endo.136.6.7750468. [DOI] [PubMed] [Google Scholar]
- 51.Rosenberg M, Nedellec P, Jothy S, Fleiszer D, Turbide C, Beauchemin N. The expression of mouse biliary glycoprotein, a carcinoembryonic antigen-related gene, is down-regulated in malignant mouse tissues. Cancer Res. 1993;53:4938–4945. [PubMed] [Google Scholar]
- 52.Rother K I, Imai Y, Caruso M, Beguinot F, Formisano P, Accili D. Evidence that IRS-2 phosphorylation is required for insulin action in hepatocytes. J Biol Chem. 1998;273:17491–17497. doi: 10.1074/jbc.273.28.17491. [DOI] [PubMed] [Google Scholar]
- 53.Sasaoka T, Rose D W, Saltiel A R, Draznin B, Olefsky J M. Evidence for a functional role of Shc proteins in mitogenic signaling induced by insulin, insulin-like growth factor-I, and epidermal growth factor. J Biol Chem. 1994;269:13689–13694. [PubMed] [Google Scholar]
- 54.Shier P, Watt V M. Tissue-specific expression of the rat insulin receptor-related receptor gene. Mol Endocrinol. 1992;6:723–729. doi: 10.1210/mend.6.5.1603082. [DOI] [PubMed] [Google Scholar]
- 55.Sippel C J, Fallon R J, Perlmutter D H. Bile acid efflux mediated by the rat liver canalicular bile acid transport/ecto-ATPase protein requires serine 503 phosphorylation and is regulated by tyrosine 488 phosphorylation. J Biol Chem. 1994;269:19539–19545. [PubMed] [Google Scholar]
- 56.Skolnik E Y, Batzer A, Li N, Lee C H, Lowenstein E, Mohammadi M, Margolis B, Schlessinger J. The function of GRB2 in linking the insulin receptor to Ras signaling pathways. Science. 1993;260:1953–1955. doi: 10.1126/science.8316835. [DOI] [PubMed] [Google Scholar]
- 57.Staubs P A, Reichart D R, Saltiel A R, Milarski K L, Maegawa H, Berhanu P, Olefsky J M, Seely B L. Localization of the insulin receptor binding sites for the SH2 domain proteins p85, Syp, and GAP. J Biol Chem. 1994;269:27186–27192. [PubMed] [Google Scholar]
- 58.Takata Y, Webster N J, Olefsky J M. Mutation of the two carboxyl-terminal tyrosines results in an insulin receptor with normal metabolic signaling but enhanced mitogenic signaling properties. J Biol Chem. 1991;266:9135–9139. [PubMed] [Google Scholar]
- 59.Takata Y, Webster N J G, Olefsky J M. Intracellular signaling by a mutant human insulin receptor lacking the carboxyl-terminal tyrosine autophosphorylation sites. J Biol Chem. 1992;267:9065–9070. [PubMed] [Google Scholar]
- 60.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 61.Tartare S, Mothe I, Kowalski-Chauvel A, Breittmayer J P, Ballotti R, Van Obberghen E. Signal transduction by a chimeric insulin-like growth factor-1 (IGF-1) receptor having the carboxyl-terminal domain of the insulin receptor. J Biol Chem. 1994;269:11449–11455. [PubMed] [Google Scholar]
- 62.Thompson N L, Lin S H, Panzica M A, Hixson D C. Cell CAM 105 isoform RNA expression is differentially regulated during rat liver regeneration and carcinogenesis. Pathobiology. 1994;62:209–220. doi: 10.1159/000163912. [DOI] [PubMed] [Google Scholar]
- 63.Turbide C, Kunath T, Daniels E, Beauchemin N. Optimal ratios of biliary glycoprotein isoforms required for inhibition of colonic tumor cell growth. Cancer Res. 1997;57:2781–2788. [PubMed] [Google Scholar]
- 64.Van Horn D J, Myers M G, Jr, Backer J M. Direct activation of the phosphatidylinositol 3-kinase by the insulin receptor. J Biol Chem. 1994;269:29–32. [PubMed] [Google Scholar]
- 65.White M F. The IRS-signalling system: a network of docking proteins that mediate insulin and interleukin signalling. Mol Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 66.White M F, Kahn C R. The insulin signaling system. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 67.White M F, Livingston J N, Backer J M, Lauris V, Dull T J, Ullrich A, Kahn C R. Mutation of the insulin receptor at tyrosine 960 inhibits signal transmission but does not affect its tyrosine kinase activity. Cell. 1988;54:641–649. doi: 10.1016/s0092-8674(88)80008-4. [DOI] [PubMed] [Google Scholar]
- 68.Withers D J, Gutierrez J S, Towery H, Burks D J, Ren J M, Previs S, Zhang Y, Bernal D, Pons S, Shulman G I, Bonner-Weir S, White M F. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 69.Xiao S, Rose D W, Sasaoka T, Maegawa H, Burke T R J, Roller P P, Shoelson S E, Olefsky J M. Syp (SH-PTP2) is a positive mediator of growth factor-stimulated mitogenic signal transduction. J Biol Chem. 1994;269:21244–21248. [PubMed] [Google Scholar]
- 70.Yamamoto-Honda R, Kadowaki T, Momomura K, Tobe K, Tamori Y, Shibasaki Y, Mori Y, Kaburagi Y, Koshio O, Akanuma Y, et al. Normal insulin receptor substrate-1 phosphorylation in autophosphorylation-defective truncated insulin receptor. Evidence that phosphorylation of substrates might be sufficient for certain biological effects evoked by insulin. J Biol Chem. 1993;268:16859–16865. [PubMed] [Google Scholar]
- 71.Yamasaki H, Prager D, Gebremedhin S, Melmed S. Human insulin-like growth factor I receptor 950 tyrosine is required for somatotroph growth factor signal transduction. J Biol Chem. 1992;267:20953–20958. [PubMed] [Google Scholar]
- 72.Zhang B, Roth R A. The insulin receptor-related receptor. Tissue expression, ligand binding specificity, and signaling capabilities. J Biol Chem. 1992;267:18320–18328. [PubMed] [Google Scholar]