Abstract
Background:
Deoxyuracil triphosphate nucleotide (dUTP) pyrophosphatase (dUTPase, DU) is an enzyme of caprine arthritis-encephalitis virus (CAEV) that minimizes incorporation of dUTP into the DNA. Caprine arthritis-encephalitis virus relies partly on its ability to escape from innate immunity to cause persistent infections. Interferon β (IFN-β) is an important marker for evaluating the innate immune system, and it has a broad spectrum of antiviral activity.
Aims:
This study was conducted to investigate the details of the IFN-β response to CAEV infection.
Methods:
The expression of IFN-β and the proliferation of Sendai virus (SeV) and vesicular stomatitis virus (VSV) were determined by real-time quantitative polymerase chain reaction (qPCR). The effect of DU on the IFN signaling pathway was evaluated using luciferase reporter assays.
Results:
In our study, the expression of IFN-β was significantly inhibited and the proliferation of SeV and VSV was promoted in cells overexpressing CAEV-DU. DU affected interferon stimulated response element (ISRE) and IFN-β promoter activities induced by RIG-I/MDA5/MAVS/TBK1 pathway, while did not affect them induced by interferon regulatory factor 3 (IRF3-5D).
Conclusion:
DU protein downregulated the production of IFN-β by inhibiting the activity of the signal transduction molecules upstream of IRF3, thereby, helping CAEV escape innate immunity. Findings of this work provide an evidence to understand the persistent infection and multiple system inflammation of CAEV.
Key Words: Caprine arthritis-encephalitis virus, dUTPase, Innate immunity, Interferon type I
Introduction
Caprine arthritis-encephalitis virus (CAEV) is classified as a small-ruminant lentivirus (SRLV) (Adedeji et al., 2013 ▶), and is an enveloped, single-stranded RNA virus, belonging to the genus Lentivirus in the family Retroviridae (Cheevers et al., 1981 ▶; Hess et al., 1986 ▶). Caprine arthritis-encephalitis virus causes caprine arthritis-encephalitis (CAE), which is a chronic progressive infectious disease in goats and other ruminants (Huang et al., 2012 ▶; Lamara et al., 2013 ▶; Nardelli et al., 2020 ▶). Caprine arthritis-encephalitis virus has a tropism for the monocyte-macrophage lineage, and the maturation of monocytes to macrophages can control the expression of the viral genome (Narayan et al., 1982 ▶; Narayan et al., 1983 ▶; Blacklaws, 2012 ▶; Crespo et al., 2012 ▶). Infection by CAEV usually results in chronic inflammatory diseases, such as interstitial pneumonia and leukoencephalomyelitis in lambs, interstitial mastitis, and chronic arthritis in adult goats (Li et al., 2013 ▶). Besides, the yield and quality of milk can be reduced due to CAEV infection (Peterhans et al., 2004 ▶; Kaba et al., 2012 ▶; Juste et al., 2020 ▶). Caprine arthritis-encephalitis virus infections have been identified worldwide (Greenwood et al., 1995 ▶; Bandeira et al., 2009 ▶; Oem et al., 2012 ▶; Tageldin et al., 2012 ▶). Since there is no effective treatment, CAEV has caused serious economic losses for goat farmers (Leitner et al., 2010 ▶; Tu et al., 2017 ▶).
Deoxyuracil triphosphate nucleotide (dUTP) pyrophosphatase (dUTPase, DU) is a ubiquitous enzyme that exists widely in prokaryotic cells, eukaryotic cells, and some groups of viruses (el-Hajj et al., 1988 ▶; Gadsden et al., 1993 ▶; Hizi and Herzig, 2015 ▶). The DU gene exists in most SRLV strains and other retroviruses, such as equine infectious anemia virus (EIAV), maedi visna virus (MVV), and CAEV (Payne and Elder, 2001 ▶; Crespo et al., 2012 ▶; Adedeji et al., 2013 ▶; Michiels et al., 2018 ▶). In lentivirus genomes, DU is encoded by the pol gene (Elder et al., 1992 ▶). DU hydrolyzes dUTP into Deoxyuracil monophosphate nucleotide (dUMP) and pyrophosphate (PPi). During reverse transcription, viral DU prevents the misincorporation of dUTP into the synthetic cDNA (McIntosh and Haynes, 1997 ▶). It has been determined that DU plays an important role in CAEV replication, pathogenesis, and genetic stability (Turelli et al., 1997 ▶). However, dUTPase is not essential for SRLV to infect or replicate (de Pablo-Maiso et al., 2018 ▶). Molecular clones of CAEV-Co lacking dUTPase are fully replicative (Saltarelli et al., 1990 ▶) and dUTPase is dispensable for MVV infection (Jonsson and Andresdottir, 2011 ▶). Even more, the whole SRLV genotype E does not encode a functional dUTPase and naturally infects goats with high efficiency (Juganaru et al., 2010 ▶).
Innate immune responses are prominent components of the host immune system, and inhibit viral infection through early detection of viral invasion and rapid initiation of defense mechanisms (Busca and Kumar, 2014 ▶; Sun and Lopez, 2017 ▶). In this process, Interferon β (IFN-β) is implicated in the activation of innate immune responses, and it has strong antiviral, antiproliferative, and immunomodulatory activities (Fensterl et al., 2015 ▶; Klotz et al., 2017 ▶). During RNA virus infection, cytoplasmic sensors, including retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), recognize viral RNAs and activate the mitochondrial antiviral signaling protein (MAVS) (Loo and Gale Jr, 2011 ▶). MAVS recruits tumor necrosis factor (TNF) receptor-associated factors (TRAFs), which subsequently activate IkB kinase (IKK) and TANK-binding kinase 1 (TBK1) complexes to phosphorylate IκBα and interferon regulatory factor 3 (IRF3), respectively. Then, nuclear factor (NF)-κB and phosphorylated IRF3 translocate into the nucleus to activate type I IFN signaling (Cui et al., 2014 ▶).
Recent findings have shown that sendai virus (SeV)-induced immune response can counteract SRLV infection (Pablo-Maiso et al., 2020 ▶). Therefore, SRLV may have strategies to escape innate immunity. Previous studies have demonstrated that the DU protein of CAEV can inhibit the production of IFN-β (Fu et al., 2020 ▶), but the molecular mechanism is still unclear. In this research, the relationship between DU and IFN-β was investigated.
Materials and Methods
Cells, virus, and antibody
Human embryonic kidney (HEK) 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) added with 10% fetal bovine serum (FBS) 100 U/ml penicillin and 10 μg/ml streptomycin sulfate in a 37°C, 5% CO2 incubator. Sendai virus and the vesicular stomatitis virus encoding green fluorescent protein (VSV-GFP) were cryopreserved in our laboratory (Shi et al., 2018 ▶). Internal reference antibody, anti-β-actin mouse monoclonal antibody, anti-Flag mouse monoclonal antibody, anti-GFP mouse monoclonal antibody, and goat anti-mouse IgG horseradish peroxidase (IgG HRP) conjugate were purchased from TransGen Co. (Beijing). SRE-luc/NF-κB-luc/IFN-β-luc plasmids and Flag-RIG-I/Flag-MDA5/Flag-MAVS/Flag-TBK1/IRF3-5D expression plasmids were kindly provided by Dr. Zexing Li from Tianjin Medical University. Poly (I:C) was purchased from Invitrogen.
Reverse transcription-polymerase chain reaction (RT-PCR) for complete coding sequence (CDS) amplification
Intracellular total RNAs were extracted from CAEV-infected goat joint synovial cells (GSM) cells using TRIzol reagent (Invitrogen, USA) as previously described (Huang et al., 2012 ▶). RNAs were reverse transcribed into cDNA using reverse transcriptase (TaKaRa, Dalian, China). A pair of oligonucleotide primers were designed based on the Shanxi strain of CAEV-SH (GU120138.1) to amplify the CDS region of DU protein and cloned into pcDNA3.1 vector (Clontech, China), as shown in Table 1.
Table 1.
Primers used for PCR amplification
| Primer name | GenBank No. | Sequence of primer (5´-3´) |
|---|---|---|
| pcDNA3.1-DU-F | GU120138.1 | CCACACTGGACTAGTGGATCCATGGCAGGATATGATTTAATATGT |
| pcDNA3.1-DU-R | CTTGGTACCGAGCTCGGATCCTAATATTAATTGTGCAAACTTT | |
| Flag-CMV2-DU-F | GU120138.1 | CGGAATTCAATGGCAGGATATGATTTAATATGT |
| Flag-CMV2-DU-R | GCTCTAGATCATAATATTAATTGTGCAAACTTT |
PCR: Polymerase chain reaction, and CMV2-DU: CMV promotor DU expression plasmid
Plasmid constructions
DU was amplified and cloned into the vector pEGFP-C1 and pFlag-CMV-2 to generate the fusion plasmid of pEGFP-DU and pFlag-CMV-DU. The primers were supplied in Table 1, EcoR I and Xba I sequences were marked by underline. The above sequences were verified by gene sequencing.
Western blot analysis
Cell lysates were heated for 10 min and isolated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Then, the separated proteins were transferred from SDS-PAGE to nitrocellulose (NC) fitter membranes (ExPro). After the transfer, the membrane was blocked with 5% skim milk in Tris buffer saline tween 20 (TBST) (0.05% Tween 20) for 1 h. The membrane was subsequently incubated with primary antibodies overnight at 4°C, and it was then washed 3 times with TBST and stained with HRP-conjugated secondary antibodies for 1 h. The membrane was detected using a chemiluminescence detection kit (Thermo Scientific, Waltham, MA, USA) and observed through Gel Imaging System (BIO-RAD, USA).
Real-time quantitative PCR (qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) and cDNA was synthesized using HiScript Q RT SuperMix (Vazyme, China). The comparative quantification of gene expression was analyzed by quantitative PCR (qPCR) carried out by ABI 7500 real-time PCR system (Applied Biosystems, Foster city, CA, USA). The thermal cycler program was made up of 95°C for 10 min, 40 cycles of 15 s at 95°C, then, 55°C for 30 s and 72°C for 30 s. This experiment used DBI Bioscience-2043 Bestar SybrGreen qPCR Mastermix. Relative levels of mRNA were normalized to the β-actin RNA levels in each sample. The relative transcript levels of the target gene were determined by the 2-ΔΔCt threshold method. The data were normalized to the expression level of the β-actin gene and analyzed using GraphPad Prism 6.0 software. All gene-specific primers are listed in Table 2.
Table 2.
Primers used for real-time qPCR amplification
| Primer name | GenBank No. | Sequence of primer (5´-3´) |
|---|---|---|
| SeV-F | NC_001552.1 | GAAAGAGATACCGAACCCAGAG |
| SeV-R | GCTTGAGGGAGTGTATTGTAGG | |
| IFN-β-F | NM_002176.3 | CAACAAGTGTCTGCTCGAAAT |
| IFN-β-R | TCTCCTCAGGGATGTCAAAG | |
| β-Actin-F | NM_001101.4 | GGAAATCGTGCGTGACATTAA |
| β-Actin-R | AGGAAGGAAGGCTGGAAGAG | |
| IRF3-F | NM_213770 | AGACGCTCACCACGCTACACC |
| IRF3-R | GCTTGAGGGAGTGTATTGTAGG | |
| NF-κB-F | NM_003998.3 | GAACCACACCCCTGCATATAG |
| NF-κB-R | GCATTTTCCCAAGAGTCATCC | |
| VSV-F | JX121109.2 | ATGTGAGCACTAAAGTAGCCCT |
| VSV-R | GTTCTTCCAAAGCTAACCCAGT | |
| Q-DU-F | GU 120138.1 | TAAAGAAATCACAATGGGCTA |
| Q-DU-R | ATTATTACCTGTATTTGTCCCT |
qPCR: Quantitative PCR, SeV: Sendai virus, IFN: Interferon, IRF: Interferon regulatory factor, NF: Nuclear factor, VSV: Vesicular stomatitis virus, DU: dUTPase, NC, NM, JX, and GU are marks of Genbank registered number
Luciferase reporter assay
293T cells were seeded in 24-well plates and co-transfected with 100 ng of IFNβ-luc, pFlag-DU (100 ng), and 100 ng of internal control -LacZ plasmid (Promega). Polyethylenimine (PEI) (Sigma, USA) was used for plasmid transfection, according to a previously described protocol (Yang et al., 2017). After 12 h of transfection, cells were inoculated with SeV or VSV at 0.5 multiplicity of infection (MOI) for another 16 h. The whole cell lysates were collected and luciferase activity was measured and normalized to the LacZ activity as previously described (Liu et al., 2019 ▶). HEK293T cells were co-transfected with the eukaryotic expression plasmid Flag-RIG-I, Flag-MDA5, Flag-MAVS, Flag-TBK1, or IRF3-5D (an IRF3 mutant that can activate downstream gene expression when expressed alone), and the related luciferase reporter plasmid to evaluate the activation of IFN signaling pathway as described previously (Fu et al., 2020 ▶).
Statistical analysis
The above experiments were done with at least three independent replicates. Statistical significance among different results was determined by GraphPad Prism software using Student’s t-test. P-values less than 0.05 were considered statistically significant (* P<0.05, ** P<0.01, *** P<0.001, and NS: No significant).
Results
DU is a negative regulator of IFN-β
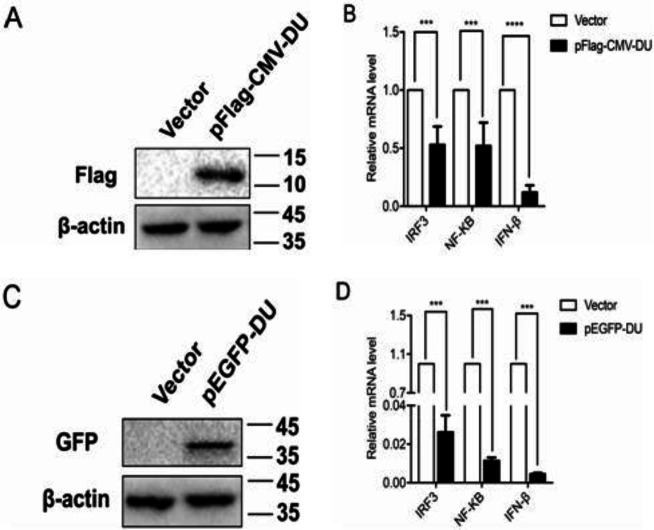
To investigate whether the expression of DU affects the IFN-β production, pFlag-CMV-DU expression vector was constructed and the expression was identified by Western blot (Fig. 1A). Next, 293T cells were transfected with pFlag-CMV-DU or empty vector for 24 h before SeV stimulation, and the mRNA levels of IFN-β and its transcription factors IRF3 and NF-κB were detected by RT-qPCR. Consistent with the previous study (Fu et al., 2020 ▶), our results suggested that overexpression of DU significantly reduced the mRNA levels of IFN-β, IRF3, and NF-κB, as compared with controls (Fig. 1B).
Fig. 1.
Label proteins do not affect the inhibitory effect of DU on IFN-β. (A and C) 293T cells were transfected with pFlag-CMV-DU (0.5 μg) (A), and pEGFP-DU (0.5 μg) (C) and empty vector. The cells were collected at 24 h post-transfection and tested by Western blot. Molecular weight markers in kDa are shown on the right. (B and D) 293T cells were transfected with pFlag-CMV-DU (B), pEGFP-DU (D), and empty vector. Twelve hours after transfection, cells were infected with 0.5 MOI SeV. The infected cells were collected at 24 h post-infection, mRNA levels of IRF3, NF-κB, and IFN-β were tested by real time quantitative reverse transcription-polymerase chain reaction (RT-qPCR). Differences in data considered statistically significant at p-value less than 0.05 (* P<0.05, ** P<0.01, *** P<0.001, and NS: No significant). CMV: Cytomegalovirus, DU: dUTPase, IRF3: Interferon regulatory factor 3, NF: Nuclear factor, IFN-β: Interferon beta, pEGFP: Enhanced green fluorescence protein plasmid, MOI: Multiplicity of infection, and SeV: Sendai virus
Considering that the molecular weight of DU protein is about 10 kDa, which is relatively small, to exclude the effect of tag protein on DU function, we constructed a DU expression plasmid with GFP tag (pEGFP-DU). Western blot analysis showed that the pEGFP-DU expression vector was successfully constructed (Fig. 1C). Consistently, transfection of the pEGFP-DU expression vector also dramatically decreased the mRNA levels of SeV-activated IFN-β, IRF3, and NF-κB compared to the pFlag-CMV-DU results (Fig. 1D). All over, these data demonstrate that DU is a negative regulator of IFN-β.
DU facilitates SeV and VSV proliferation
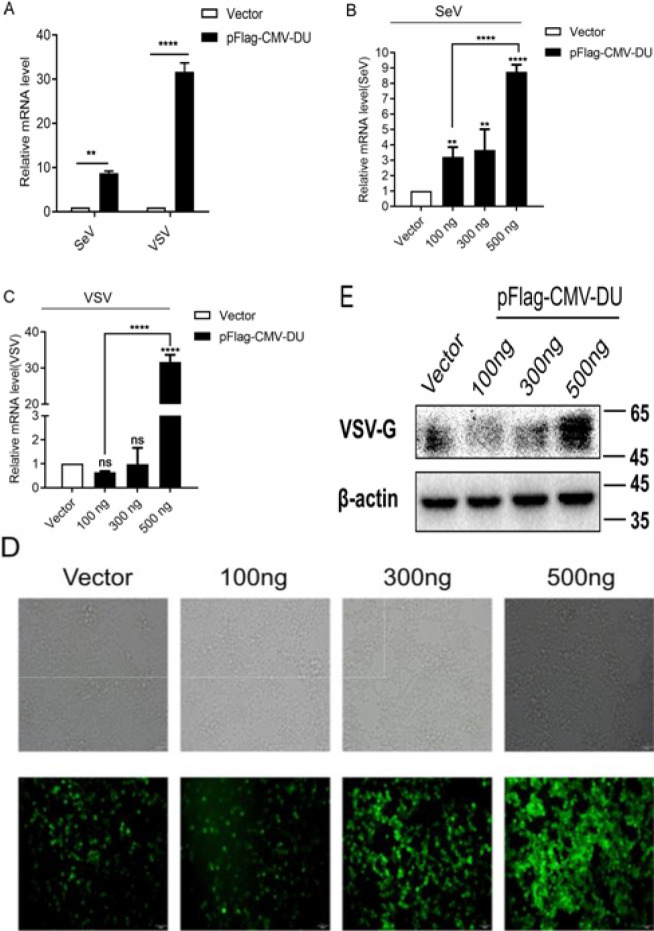
Since we have determined that DU inhibits IFN-β mRNA level, it prompted us to investigate the effect of DU on viral proliferation. 293T cells were transfected with pFlag-CMV-DU expression plasmids or empty vector and infected with SeV or VSV-GFP at 24 h post transfection. Then, total RNA was extracted from the cells at 16 h post infection and analyzed for the mRNA levels of SeV and VSV by RT-qPCR. As shown in Figs. 2A-C, the overexpression of DU upregulated the mRNA levels of SeV and VSV in a dose-dependent manner. Correspondingly, the results of immunofluorescence and Western blot analyses further confirmed that DU dose dependently promoted VSV replication (Figs. 2D and E). Together, these observations suggest that DU facilitates the proliferation of SeV and VSV in a dose-dependent manner.
Fig. 2.
Overexpression of DU promotes SeV/VSV proliferation. (A) 293T cells were transfected with pFlag-CMV-DU expression plasmid or empty vector, 12 h after transfection, cells were infected with 0.5 MOI SeV or VSV. The mRNA level of SeV or VSV was examined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis at 16 h post-infection. (B and C) 293T cells were transfected with different doses of pFlag-CMV-DU expression plasmids (100 ng/300 ng/500 ng), 12 h after transfection, cells were infected with SeV or VSV and the mRNA level of SeV or VSV was tested by RT-qPCR analysis at 16 h post-infection. (D and E) 293T cells were transfected with pFlag-CMV-DU expression plasmid or control plasmid, after 12 h, cells were infected with VSV-GFP, immunofluorescence microscope imaging (D), and Western blot (E) used for observing viral proliferation at 16 h post-infection. Molecular weight markers in kDa are shown on the right. Results were normalized to those of the control gene β-actin and are presented relative to those of control cells. Differences in data considered statistically significant at p-value less than 0.05 (* P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, and NS: No significant). CMV: Cytomegalovirus, DU: dUTPase, MOI: Multiplicity of infection, SeV: Sendai virus, and VSV-GFP: Vesicular stomatitis virus harbored green fluorescence protein gene
DU inhibits IFN-β production in a dose-dependent manner
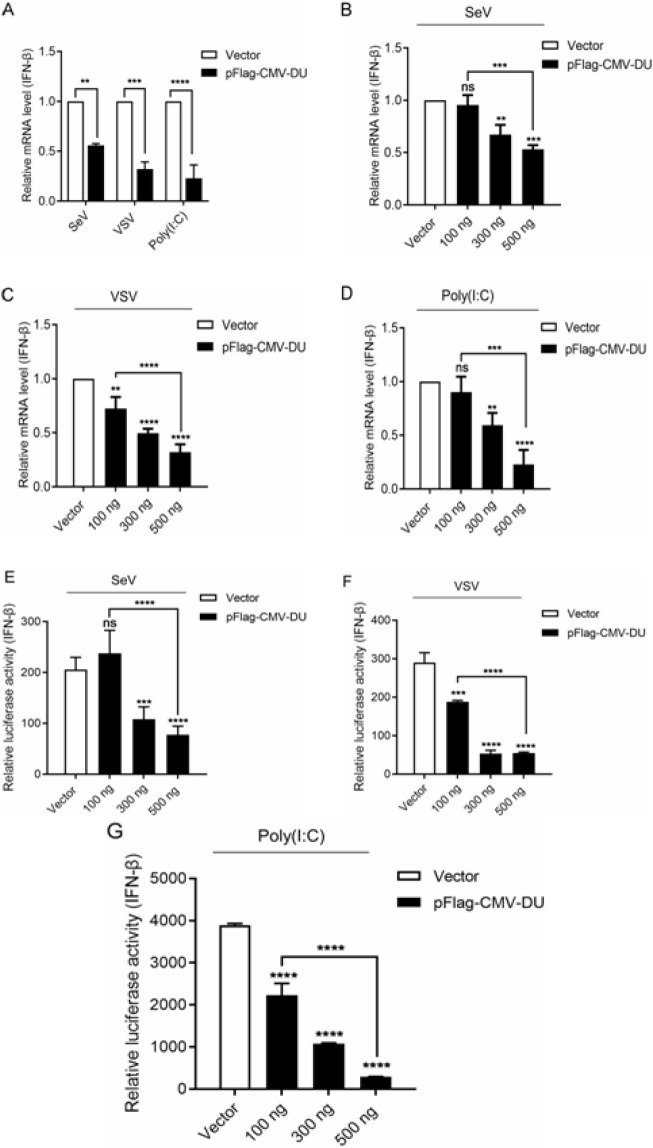
To further confirm the inhibitory effect of DU on the expression of IFN-β, the transcription level of the IFN-β in cells treated with SeV, VSV-GFP or poly (I:C) (1 μg/ml) were compared between transfection plasmid of DU and blank vector cells. As shown in Fig. 3A, overexpression of DU significantly reduced the mRNA level of IFN-β induced by SeV, VSV-GFP, and poly (I:C). This inhibitory effect was dose dependent (Figs. 3B-D).
Fig. 3.
DU inhibits IFN-β expression in a dose-dependent manner. (A-D) 293T cells were transfected with pFlag-CMV-DU or empty vector. At 12 h post-transfection, cells were infected with SeV/VSV-GFP/poly (I:C). The infected cells were collected at 16 h post-infection and mRNA levels of IFN-β were tested by quantitative reverse transcription-polymerase chain reaction (RT-qPCR). (E-G) 293T cells were co-transfected with IFN-β-luc and pFlag-CMV-DU and then, inoculated with SeV/VSV-GFP/poly (I:C), finally, the luciferase activity of IFN-β promotor was tested. Differences in data considered statistically significant at p-value less than 0.05 (* P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, and NS: No significant). CMV: Cytomegalovirus, DU: dUTPase, SeV: Sendai virus, VSV-GFP: Vesicular stomatitis virus harbored green fluorescence protein gene, and IFN: Interferon
Next, 293T cells were co-transfected with the luciferase reporter plasmid of IFN-β (IFN-β-luc) and different doses of pFlag-CMV-DU expression vector to verify the above RT-qPCR assay results. The results demonstrated that DU inhibited SeV, VSV, and poly (I:C)-activated IFN-β promoter activity in a dose dependent manner (Figs. 3E-G). Collectively, the DU dose dependently inhibits the production of IFN-β to promote viral proliferation.
DU has an inhibitory effect on the upstream of IRF3 in the signaling pathway
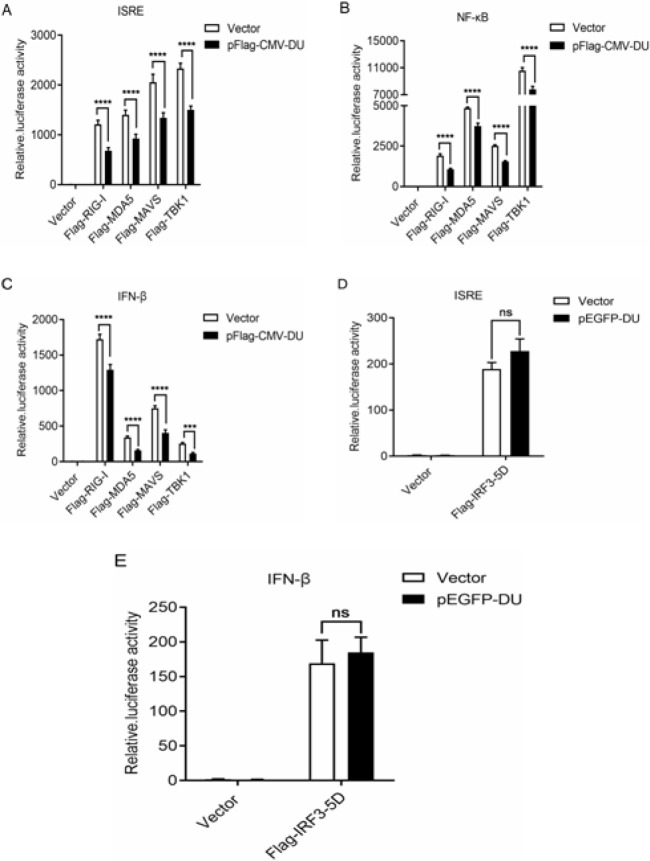
Previous studies have shown that SeV, VSV, and poly (I:C) activate downstream IFN signaling pathways through activating MAVS, RIG-I, and MDA5, respectively. To further clarify the specific mechanism by which DU inhibits the production of IFN-β, we explored the role of DU in the IFN-β signaling pathway. The luciferase reporter plasmids of interferon stimulated response element (ISRE), NF-κB and IFN-β (ISRE-luc, NF-κB-luc, and IFN-β-luc), and eukaryotic expression plasmids of RIG-I, MDA5, MAVS, and TBK1 (Flag-RIG-I, Flag-MDA5, Flag-MAVS, and Flag-TBK1), and pEGFP-DU or empty vector were co-transfected into 293T cells. The effect of DU on the signaling pathway was determined by detecting the luciferase activity of the reporters. As shown in Figs. 4A-C, overexpression of RIG-I, MDA5, MAVS, and TBK1 induced the promoter activation of ISRE, NF-κB, and IFN-β, however, co-transfection with DU significantly attenuated this activation, suggesting that DU was a negative regulator of the IFN-β signaling pathway.
Fig. 4.
DU is an inhibitory factor in IFN-β signaling pathway. (A-C) 293T cells were co-transfected with ISRE-luc/NF-κB-luc/IFN-β-luc plasmids (100 ng), Flag-RIG-I/Flag-MDA5/Flag-MAVS/Flag-TBK1, and pEGFP-DU expression plasmids or empty vector, then, the luciferase activity of ISRE, NF-κB, and IFN-β promoters was examined. (D and E) 293T cells were co-transfected with ISRE-luc/IFN-β-luc plasmids and pEGFP-DU expression plasmids or empty vector, then, the luciferase activity of ISRE and IFN-β promotors was tested. Differences in data considered statistically significant at p-value less than 0.05 (* P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, and NS: No significant). Data are representative of three independent experiments. ISRE: Interferon stimulated response element, CMV: Cytomegalovirus, RIG: Retinoic acid-inducible gene I, MDA5: Melanoma differentiation-associated gene 5, MAVS: Mitochondrial antiviral signaling protein, TBK1: TANK-Binding kinase 1, NF: Nuclear factor, IFN: Interferon, IRF3: Interferon regulatory factor 3, and pEGFP-DU: dUTPase
IRF3 is an important downstream transcription factor of MAVS and TBK1, and it can be stimulated by upstream signals to undergo phosphorylation and dimerization, then, it enters the nucleus and activates transcription (Liu et al., 2015 ▶; Bakshi et al., 2017 ▶; He et al., 2017 ▶). To verify the effect of DU on the transcription activity of IRF3 IRF3-5D plasmid, a constitutively active form of IRF3 (Sen et al., 2010 ▶), was co-transfected into 293T cells with DU plasmid. It was found that the ISRE and IFN-β promoter activities activated by IRF3-5D were not affected by DU (Figs. 4D and E). The results indicated that the inhibitory effect of DU on the IFN signaling pathway was located at upstream of IRF3 transcription factor.
Discussion
DU is a ubiquitous enzyme regulated by the cell cycle and is abundant both in differentiated and undifferentiated cells (Turelli et al., 1997 ▶). When the ratio of dUTP/dTTP (Deoxyuracil triphosphate nucleotide/Thymidine triphosphate deoxynucleotide) in the virus-infected cells increases, the reverse transcription of the virus can proceed normally (Hizi and Herzig, 2015 ▶), but the process of viral DNA integration into host cell DNA and the expression of most viral proteins are blocked (Weil et al., 2013 ▶). As a dUTPase, DU can catalyze the conversion of dUTP to dUMP and PPi, help to maintain a low ratio of dUTP/dTTP (McGeoch, 1990 ▶; Payne and Elder, 2001 ▶; Vertessy and Toth, 2009 ▶), thereby, reducing the possibility of dUTP incorporation into cDNA, providing an advantageous environment for viral replication (Chen et al., 2002 ▶; Kato et al., 2014 ▶).
IFN-β is an important antiviral cytokine in innate immunity that inhibits the proliferation of most viruses. On contrast, many viruses downregulation IFN expression by disturbing IFN signaling pathway using their coding proteins to escape from innate immunity. Small-ruminant lentiviruses weakly induce type-I IFN (Zink and Narayan, 1989 ▶). Besides, dUTPase is dispensable for MVV infection (Jonsson and Andresdottir, 2011 ▶). The inhibition effects of dUTPase in IFN-β production were also detected in our previous report (Fu et al., 2020 ▶) CAEV with a point mutation in the gene coding dUTPase has been shown to revert to the wild type (WT) in an infected goat (Turelli et al., 1997 ▶). All those results showed that dUTPase prompts clinical signs and pathogenesis of retrovirus infection. Accordingly, dUTPase is not essential for SRLV infection, SRLV Roccaverano strain (genotype E) lacking the entire dUTPase and Vpr-like genes replicates efficiently in non-dividing cells (Juganaru et al., 2010 ▶), which has been described as a low pathogenic strain in goats (Reina et al., 2009 ▶). Thus, we speculated that the inhibition of IFN-β by DU is one of the probable reasons for the low production of IFN-β. To further validate the inhibitory effect of DU on IFN-β production, we overexpressed different doses of plasmid encoding dUTPase (100, 300, 500 ng, respectively) in 293T cells. DU showed a dose-dependent inhibition of IFN-β expression. Correspondingly, DU could promote the replication of SeV and VSV.
SeV/VSV/Poly (I:C) can be recognized by the organism’s pattern recognition receptors (PRRs), which activate the downstream signal transduction pathway and activate IFN-β. dUTPase can be recognized by RIG-I, which induces the downstream activation of IFN-β; poly (I:C) induces the activation of MDA5 (Kato et al., 2008 ▶); SeV infection leads to the activation and aggregation of MAVS on mitochondria, and then MAVS activates IFN-β signaling pathway. Although SeV/VSV/Poly (I:C) are recognized by different receptors, these receptors ultimately activate IFN-β through the downstream “MAVS-TBK1-IRF3” signaling pathway. In our study, we found that CAEV-DU can inhibit the RIG-I/MDA5/MVAS/TBK1-activated IFN-β signaling pathway, but it has no influence on the activity of IRF3-induced IFN-β signaling pathway, indicating that the target molecule for DU to inhibit IFN-β production is located upstream of IRF3.
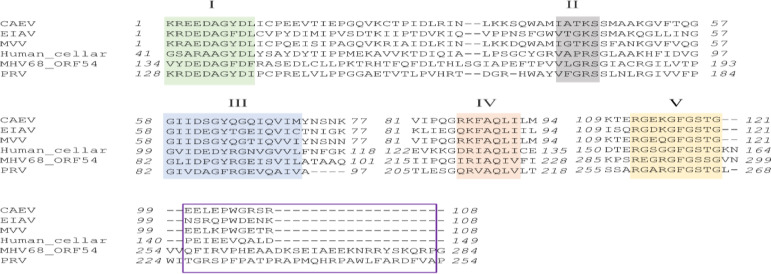
Pseudorabies virus dUTPase UL50 and mouse herpesvirus 68 (MHV68) ORF54 are two kinds of herpesviral dUTPases. Recent studies have shown that MHV68 ORF54 and PRV dUTPase can antagonize type I IFN signaling independent of their dUTPase activity (Leang et al., 2011 ▶; Zhang et al., 2017 ▶). Deoxyuracil triphosphate nucleotide (dUTP) pyrophosphatase usually have five conserved amino acid (aa) motifs. Multiple sequence alignment of dUTPases between different viruses (SRLV and herpesvirus) and humans is shown in Fig. 5. The study by Zhang et al. (2017) ▶ showed that the region between motifs IV and V is critical for PRV UL50 inhibition of type I IFN signaling. However, this region is almost absent in several SRLV dUTPases and cellular dUTPase but appears in MHV68 ORF54, which can also inhibit type I IFN signaling. Considering that CAEV is a kind of retrovirus and herpesvirus belonging to DNA viruses. It is probably an important reason for the sequence difference between CAEV dUTPase and herpesviral dUTPases. Hence, we speculated that CAEV dUTPase may have different mechanisms compared to herpesviral dUTPases to help the virus evade host type I IFN response, and this requires further investigation.
Fig. 5.
Multiple sequence alignment of dUTPases between different viruses and humans. The five highly-conserved domains typical of dUTPases are indicated above the sequences. The functional region of PRV UL50 in charge of anti-IFN activity reported by Zhang (2017) is marked by a purple rectangle. The sequences of the following dUTPases are as follows: MVV (GI:9626549), CAEV (GI:266706151), EIAV (GI:323836), MHV68 (GI:114782444), PRV (GI: 51557490), and humans (GI:4503423). CAEV: Caprine arthritis-encephalitis virus, EIAV: Equine infectious anemia virus, MVV: Maedi-visna virus, MHV: Mouse hepatitis virus, ORF: Open reading frame, PRV: Pseudorabies virus, and IFN: Interferon
In summary, our study preliminarily determines the mechanism by which DU negatively regulates the production of IFN-β by inhibiting the upstream of IRF3 in the IFN-β signaling pathway, thereby inhibiting the transcriptional expression of IFN-β. This work presents a theoretical basis for the CAEV to evade innate immunity. However, more details about the interactions between DU and the IFN-β signaling pathway also demand further exploration.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFD0500500), and the National Natural Science Foundation in China (No. 30771613).
Conflict of interest
The authors declare no financial and/or non-financial competing interests.
References
- Adedeji AO, Barr B, Gomez-Lucia E, Murphy B. A polytropic caprine arthritis encephalitis virus promoter isolated from multiple tissues from a sheep with multisystemic lentivirus-associated inflammatory disease. Viruses. 2013;5:2005–2018. doi: 10.3390/v5082005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Taylor J, Strickson S, McCartney T, Cohen P. Identification of TBK1 complexes required for the phosphorylation of IRF3 and the production of interferon beta. Biochem. J. 2017;474:1163–1174. doi: 10.1042/BCJ20160992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira DA, de Castro RS, Azevedo EO, de Souza Seixas Melo L, de Melo CB. Seroprevalence of caprine arthritis-encephalitis virus in goats in the Cariri region, Paraiba state, Brazil. Vet. J. 2009;180:399–401. doi: 10.1016/j.tvjl.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Blacklaws BA. Small ruminant lentiviruses: immunopathogenesis of visna-maedi and caprine arthritis and encephalitis virus. Comp. Immunol. Microbiol. Infect. Dis. 2012;35:259–269. doi: 10.1016/j.cimid.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Busca A, Kumar A. Innate immune responses in hepatitis B virus (HBV) infection. Virol. J. 2014;7:11–22. doi: 10.1186/1743-422X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheevers WP, Roberson S, Klevjer-Anderson P, Crawford TB. Characterization of caprine arthritis-encephalitis virus: a retrovirus of goats. Arch. Virol. 1981;67:111–117. doi: 10.1007/BF01314610. [DOI] [PubMed] [Google Scholar]
- Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- Crespo H, Jauregui P, Glaria I, Sanjosé L, Polledo L, García-Marín JF, Luján L, de Andrés D, Amorena B, Reina R. Mannose receptor may be involved in small ruminant lentivirus pathogenesis. Vet. Res. 2012;43:1–6. doi: 10.1186/1297-9716-43-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum. Vaccin Immunother. 2014;10:3270–3285. doi: 10.4161/21645515.2014.979640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pablo-Maiso L, Domenech A, Echeverria I, Gomez-Arrebola C, de Andres D, Rosati S, Gomez-Lucia E, Reina R. Prospects in innate immune responses as potential control strategies against non-primate lentiviruses. Viruses. 2018;10:435–468. doi: 10.3390/v10080435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JH, Lerner DL, Hasselkus-Light CS, Fontenot DJ, Hunter E, Luciw PA, Montelaro RC, Phillips TR. Distinct subsets of retroviruses encode dUTPase. J. Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Hajj HH, Zhang H, Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriolo. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, Chattopadhyay S, Sen GC. No love lost between viruses and interferons. Annu. Rev. Virol. 2015;2:549–572. doi: 10.1146/annurev-virology-100114-055249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Lu D, Su Y, Chi H, Wang J, Huang J. The Vif protein of caprine arthritis encephalitis virus inhibits interferon production. Arch. Virol. 2020;165:1557–1567. doi: 10.1007/s00705-020-04637-z. [DOI] [PubMed] [Google Scholar]
- Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. The EMBO J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood PL, North RN, Kirkland PD. Prevalence, spread and control of caprine arthritis-encephalitis virus in dairy goat herds in New South Wales. Austr. Vet. J. 1995;72:341–345. doi: 10.1111/j.1751-0813.1995.tb07538.x. [DOI] [PubMed] [Google Scholar]
- He X, Ma S, Tian Y, Wei C, Zhu Y, Li F, Zhang P, Wang P, Zhang Y, Zhong H. ERRalpha negatively regulates type I interferon induction by inhibiting TBK1-IRF3 interaction. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Pyper JM, Clements JE. Nucleotide sequence and transcriptional activity of the caprine arthritis-encephalitis virus long terminal repeat. J. Virol. 1986;60:385–393. doi: 10.1128/jvi.60.2.385-393.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A, Herzig E. dUTPase: the frequently overlooked enzyme encoded by many retroviruses. Retrovirology. 2015;12:70–77. doi: 10.1186/s12977-015-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sun Y, Liu Y, Xiao H, Zhuang S. Development of a loop-mediated isothermal amplification method for rapid detection of caprine arthritis-encephalitis virus proviral DNA. Arch. Virol. 2012;157:1463–1469. doi: 10.1007/s00705-012-1322-y. [DOI] [PubMed] [Google Scholar]
- Jonsson SR, Andresdottir V. Propagating and detecting an infectious molecular clone of maedi-visna virus that expresses green fluorescent protein. J. Vis. Exp. 2011;9:3483–3485. doi: 10.3791/3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juganaru M, Reina R, Grego E, Profiti M, Rosati S. LTR promoter activity of SRLV genotype E, strain Roccaverano. Vet. Res. Comm. 2010;34:47–51. doi: 10.1007/s11259-010-9390-5. [DOI] [PubMed] [Google Scholar]
- Juste RA, Villoria M, Leginagoikoa I, Ugarte E, Minguijon E. Milk production losses in Latxa dairy sheep associated with small ruminant lentivirus infection. Prev. Vet. Med. 2020;176:104886–104892. doi: 10.1016/j.prevetmed.2020.104886. [DOI] [PubMed] [Google Scholar]
- Kaba J, Strzałkowska N, Jóźwik A, Krzyżewski J, Bagnicka E. Twelve-year cohort study on the influence of caprine arthritis-encephalitis virus infection on milk yield and composition. J. Dairy Sci. 2012;95:1617–1622. doi: 10.3168/jds.2011-4680. [DOI] [PubMed] [Google Scholar]
- Kato A, Hirohata Y, Arii J, Kawaguchi Y. Phosphorylation of herpes simplex virus 1 dUTPase upregulated viral dUTPase activity to compensate for low cellular dUTPase activity for efficient viral replication. J. Virol. 2014;88:7776–7785. doi: 10.1128/JVI.00603-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz D, Baumgartner W, Gerhauser I. Type I interferons in the pathogenesis and treatment of canine diseases. Vet. Immunol. Immunopathol. 2017;191:80–93. doi: 10.1016/j.vetimm.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Lamara A, Fieni F, Chatagnon G, Larrat M, Dubreil L, Chebloune Y. Caprine arthritis encephalitis virus (CAEV) replicates productively in cultured epididymal cells from goats. Comp. Immunol. Microbiol. Infect. Dis. 2013;36:397–404. doi: 10.1016/j.cimid.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Leang RS, Wu TT, Hwang S, Liang LT, Tong L, Truong JT, Sun R. The anti-interferon activity of conserved viral dUTPase ORF54 is essential for an effective MHV-68 infection. PLoS Pathog. 2011;7:e1002292. doi: 10.1371/journal.ppat.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner G, Krifucks O, Weisblit L, Lavi Y, Bernstein S, Merin U. The effect of caprine arthritis encephalitis virus infection on production in goats. Vet. J. 2010;183:328–331. doi: 10.1016/j.tvjl.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou F, Li X, Wang J, Zhao X, Huang J. Development of TaqMan-based qPCR method for detection of caprine arthritis-encephalitis virus (CAEV) infection. Arch. Virol. 2013;158:2135–2141. doi: 10.1007/s00705-013-1728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630–1-2630-11. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu C, Huang S, Wang X, Wen M, Zheng J, Wang W, Fu Y, Tian S, Li L, Li Z andWang X. The otubain YOD1 suppresses aggregation and activation of the signaling adaptor MAVS through Lys63-linked deubiquitination. J. Immunol. 2019;202:2957–2970. doi: 10.4049/jimmunol.1800656. [DOI] [PubMed] [Google Scholar]
- Loo YM, Gale Jr M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ. Protein sequence comparisons show that the ‘pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nuc. Acid Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh EM, Haynes RH. dUTP pyrophosphatase as a potential target for chemotherapeutic drug development. Acta Biochim. Polo. 1997;44:159–171. [PubMed] [Google Scholar]
- Michiels R, Van Mael E, Quinet C, Welby S, Cay AB, De Regge N. Seroprevalence and risk factors related to small ruminant lentivirus infections in Belgian sheep and goats. Prev. Vet. Med. 2018;151:13–20. doi: 10.1016/j.prevetmed.2017.12.014. [DOI] [PubMed] [Google Scholar]
- Narayan O, Kennedy-Stoskopf S, Sheffer D, Griffin DE, Clements JE. Activation of caprine arthritis-encephalitis virus expression during maturation of monocytes to macrophages. Infect. Imm. 1983;41:67–73. doi: 10.1128/iai.41.1.67-73.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan O, Wolinsky JS, Clements JE, Strandberg JD, Griffin DE, Cork LC. Slow virus replication: the role of macrophages in the persistence and expression of visna viruses of sheep and goats. J. Gen. Virol. 1982;59:345–356. doi: 10.1099/0022-1317-59-2-345. [DOI] [PubMed] [Google Scholar]
- Nardelli S, Bettini A, Capello K, Bertoni G, Tavella A. Eradication of caprine arthritis encephalitis virus in the goat population of South Tyrol, Italy: analysis of the tailing phenomenon during the 2016-2017 campaign. J. Vet. Diagn. Invest. 2020;32:589–593. doi: 10.1177/1040638720934055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oem JK, Chung JY, Byun JW, Kim HY, Kwak D, Jung BY. Large-scale serological survey of caprine arthritis-encephalitis virus (CAEV) in Korean black goats (Capra hircus aegagrus) J. Vet. Med. Sci. 2012;74:1657–1659. doi: 10.1292/jvms.12-0103. [DOI] [PubMed] [Google Scholar]
- Pablo-Maiso LD, Echeverría I, Rius-Rocabert S, Luján L, Garcin D, Andrés DD, Nistal-Villán E, Reina R. Sendai virus, a strong inducer of anti-lentiviral state in ovine cells. Vaccines. 2020;8:206–221. doi: 10.3390/vaccines8020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SL, Elder JH. The role of retroviral dUTPases in replication and virulence. Curr. Protein Pept. Sci. 2001;2:381–388. doi: 10.2174/1389203013381008. [DOI] [PubMed] [Google Scholar]
- Peterhans E, Greenland T, Badiola J, Harkiss G, Bertoni G, Amorena B, Eliaszewicz M, Juste RA, Kraßnig R, Lafont JP. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 2004;35:257–274. doi: 10.1051/vetres:2004014. [DOI] [PubMed] [Google Scholar]
- Reina R, Grego E, Bertolotti L, De Meneghi D, Rosati S. Genome analysis of small-ruminant lentivirus genotype E: a caprine lentivirus with natural deletions of the dUTPase subunit, vpr-like accessory gene, and 70-base-pair repeat of the U3 region. J. Virol. 2009;83:1152–1155. doi: 10.1128/JVI.01627-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltarelli M, Querat G, Konings DA, Vigne R, Clements JE. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology. 1990;179:347–364. doi: 10.1016/0042-6822(90)90303-9. [DOI] [PubMed] [Google Scholar]
- Sen N, Sommer M, Che X, White K, Ruyechan WT, Arvin AM. Varicella-zoster virus immediate-early protein 62 blocks interferon regulatory factor 3 (IRF3) phosphorylation at key serine residues: a novel mechanism of IRF3 inhibition among herpesviruses. J. Virol. 2010;84:9240–9253. doi: 10.1128/JVI.01147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Su Y, Li R, Zhang L, Chen C, Zhang L, Faaberg K, Huang J. Dual regulation of host TRAIP post-translation and nuclear/plasma distribution by porcine reproductive and respiratory syndrome virus non-structural protein 1α promotes viral proliferation. Front. Immunol. 2018;9:3023. doi: 10.3389/fimmu.2018.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lopez CB. The innate immune response to RSV: Advances in our understanding of critical viral and host factors. Vaccine. 2017;35:481–488. doi: 10.1016/j.vaccine.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tageldin MH, Johnson EH, Al-Busaidi RM, Al-Habsi KR, Al-Habsi SS. Serological evidence of caprine arthritis-encephalitis virus (CAEV) infection in indigenous goats in the Sultanate of Oman. Trop. Anim. Health Prod. 2012;44:1–3. doi: 10.1007/s11250-011-9883-4. [DOI] [PubMed] [Google Scholar]
- Tu PA, Shiu JS, Lee SH, Pang VF, Wang DC, Wang PH. Development of a recombinase polymerase amplification lateral flow dipstick (RPA-LFD) for the field diagnosis of caprine arthritis-encephalitis virus (CAEV) infection. J. Virol. Methods. 2017;243:98–104. doi: 10.1016/j.jviromet.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Turelli P, Guiguen F, Mornex JF, Vigne R, Querat G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J. Virol. 1997;71:4522–4530. doi: 10.1128/jvi.71.6.4522-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertessy BG, Toth J. Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 2009;42:97–106. doi: 10.1021/ar800114w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil AF, Ghosh D, Zhou Y, Seiple L, McMahon MA, Spivak AM, Siliciano RF, Stivers JT. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl. Acad. Sci. USA. 2013;110:E448–E457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhou X, Li R, Fu X, Sun P. Optimized PEI-based transfection method for transient transfection and lentiviral production. Curr. Protoc. Chem. Biolo. 2017;9:147–157. doi: 10.1002/cpch.25. [DOI] [PubMed] [Google Scholar]
- Zhang R, Xu A, Qin C, Zhang Q, Chen S, Lang Y, Wang M, Li C, Feng W, Zhang R, Jiang Z, Tang J. Pseudorabies virus dUTPase UL50 induces lysosomal degradation of type I interferon receptor 1 and antagonizes the alpha interferon response. J. Virol. 2017;91:e01148–17. doi: 10.1128/JVI.01148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink M, Narayan O. Lentivirus-induced interferon inhibits maturation and proliferation of monocytes and restricts the replication of caprine arthritis-encephalitis virus. J. Virol. 1989;63:2578–2584. doi: 10.1128/jvi.63.6.2578-2584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]