Abstract
Numerous examples of microbial phase-separated biomolecular condensates have now been identified following advances in fluorescence imaging and single molecule microscopy technologies. The structure, function, and potential applications of these microbial condensates are currently receiving a great deal of attention. By neatly compartmentalizing proteins and their interactors in membrane-less organizations while maintaining free communication between these macromolecules and the external environment, microbial cells are able to achieve enhanced metabolic efficiency. Typically, these condensates also possess the ability to rapidly adapt to internal and external changes. The biological functions of several phase-separated condensates in small bacterial cells show evolutionary convergence with the biological functions of their eukaryotic paralogs. Artificial microbial membrane-less organelles are being constructed with application prospects in biocatalysis, biosynthesis, and biomedicine. In this review, we provide an overview of currently known biomolecular condensates driven by liquid-liquid phase separation (LLPS) in microbial cells, and we elaborate on their biogenesis mechanisms and biological functions. Additionally, we highlight the major challenges and future research prospects in studying microbial LLPS.
Keywords: liquid-liquid phase separation, biomolecular condensates, membraneless organelles, multivalent interactions, crowded environments, cellular noise
Introduction
Recent developments in the field of liquid-liquid phase separation (LLPS) have led to a transformation in our understanding of the biogenesis of subcellular membrane-less compartments. As more and more phase-separated condensates are being discovered, there is considerable interest in exploring key factors (proteins) involved in the organizations and physiological functions of the compartments. However, we are still at an early stage of understanding the precise regulation and the biochemical processes inside the condensates, lacking a global view of the interactions within/among the compartments (and the environment).
For many years, the field of compartmentalization was limited to the study of membrane-bound organelles. The presence of these functionally and structurally distinct compartments is the essential feature of eukaryotic cells. In the 1980s, small granules that behaved as fluid droplets were identified in the cytosol, and these droplets were observed to fuse together into larger assemblies known as non-membranous organelles (Strome and Wood, 1982). High-resolution imaging studies (and other methods of determining molecular composition) have revealed that membrane-less compartments generally exhibit similar dynamics and similar assembly pathways, although their position, composition, and function may differ (Mitrea et al., 2018). P granules, a type of membrane-less compartment found in Caenorhabditis elegans, were the first biomolecular condensate observed to form via LLPS (Brangwynne et al., 2009). These early observations concerning P bodies greatly promoted the development of this field, furthering our understanding of the physical processes driving the formation of organelles. Subsequently, evidence was provided demonstrating the involvement of LLPS in the formation of additional membrane-less organelles, including nuclear Cajal bodies (in plant and animal cells, Frey et al., 1999; Sleeman et al., 2011; Riback et al., 2020) or the homologous nucleolar body (in budding yeast, Verheggen et al., 2001), nuclear speckles (Cotto et al., 1997; Chiodi et al., 2000; Brangwynne et al., 2011; Spector and Lamond, 2011; Tripathi et al., 2012), stress granules (Buchan and Parker, 2009; Kato et al., 2012; Youn et al., 2019; Yang P. et al., 2020), and the carboxysome (a well-studied subcellular compartment in cyanobacteria responsible for sequestering and concentrating Rubisco enzymes for CO2 fixation) (Wang et al., 2019). Moreover, evidence was presented that LLPS may be involved in forming bacterial inclusion bodies (IBs) (Baneyx and Mujacic, 2004; Singh and Panda, 2005; Sabate et al., 2010; Chebotareva et al., 2013; Azaldegui et al., 2021; Su et al., 2021). Although membrane-less organelles have no enclosing membrane, the condensates have been demonstrated to maintain (for hours to days) stable, coherent structures capable of compartmentalizing and concentrating specific sets of molecules and exchanging material with surrounding components (Shin and Brangwynne, 2017).
An understanding of the principles underlying the formation and function of biomolecular condensates is vital for any in-depth investigation of the physiology and pathophysiology of biological processes and systems. Using up-to-date imaging, structural, and computational methods, scientists have been able to study the features of LLPS in vitro and in vivo (Alberti et al., 2018; Bracha et al., 2018; Mitrea et al., 2018). However, an in-depth study of LLPS in the comparatively small prokaryotic cell remains technically challenging, limiting our understanding of the molecular basis and biological function of compartmentalization in microorganisms. Nevertheless, recent developments in this field have yielded an extraordinary leap in understanding. In this review, we highlight representative examples of phase-separated condensates observed in microbial cells. Using these examples, we summarize the underlying mechanisms accounting for the composition, function, and the assembly/disassembly of microbial membrane-less compartments. We have also highlighted a series of challenges and future perspectives in this exciting area.
Liquid Phase-Separated Organelles in Microorganisms
In Tiebackx (1911) firstly reported that coacervation was achieved through liquid-liquid phase separation (LLPS). Then, the concept of LLPS was applied in organic chemistry, especially in polymer chemistry (Jong and Kruyt, 1929). In this context, coacervation can be attained using either a mixture of oppositely charged polyelectrolytes (complex coacervation) or a polymer capable of self-association (self-coacervation) (Jong and Kruyt, 1929; Gabryelczyk et al., 2019). When a homogeneous polymer solution of macromolecules undergoes LLPS, two different phases are formed, a phase of concentrated molecules (dense phase) and a dilute molecule-depleted phase (dilute phase). The dense phase resembles liquid droplets (Alberti et al., 2019), and molecules in this phase can move quickly and are free to exchange interactions with multiple other molecules. Because molecules in the dense phase are highly likely to experience random molecule-molecule collisions, the potential for these molecules to complete biochemical reactions is high.
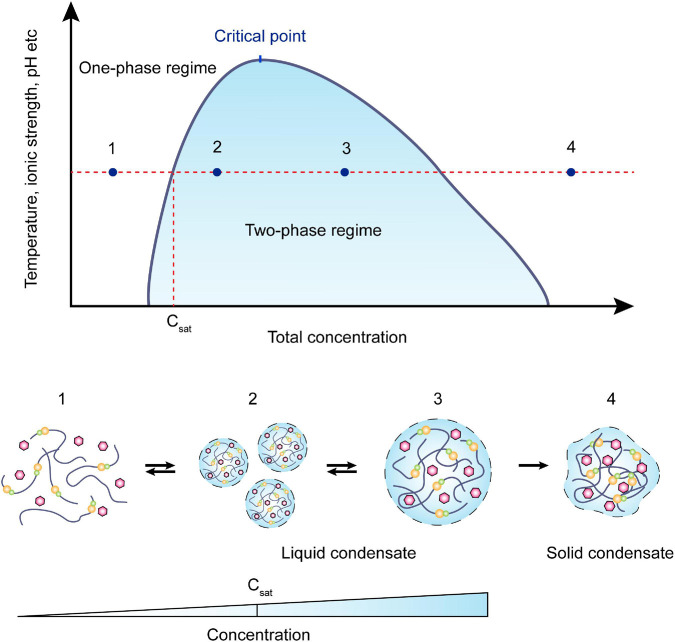
As early as 120 years ago, Wilson raised that protoplasm might be constructed by condensed liquid-droplet-like granules (Wilson, 1899). However, the commonly held view considered the cytoplasm as a fluid-like homogeneous mix of soluble proteins and compounds. It is only in the past decade, studies revealed that the cytosol does not act simply as a continuous medium but demonstrates complex rheological characteristics (Brangwynne et al., 2009; Guo et al., 2014; Shin and Brangwynne, 2017). The bacterial cytoplasm displays properties characteristic of glass-forming liquids and can solidify to resemble soft glass, depending on the metabolism, component sizes, and non-steric interactions (Parry Bradley et al., 2014; Xiang et al., 2021). Modern microscopy techniques reveal that the many proteins in bacteria tend to form large complexes targeted to specific regions within the cytosol (Abbondanzieri and Meyer, 2019). Some of the regions exhibit remarkable liquid droplet-like behaviors and undergo rapid assembly and disassembly in response to stress or cell signaling events (Sehgal et al., 2020). Evidence for this phenomenon includes the gathering of RNA degradosomes into bacterial ribonucleoprotein bodies (BR-bodies) displaying liquid-like behavior in Escherichia coli, Bacillus subtilis, and Caulobacter crescentus (Al-Husini et al., 2018, 2020; Hamouche et al., 2020). According to Hyman’s hypothesis (proposed by Hyman et al., 2014), the formation of phase-separated condensates in eukaryotes occurs through three main steps (Hyman et al., 2014; Strom et al., 2017; Peng and Weber, 2019): nucleation; rearrangement; and supersaturation (Figure 1). A saturation concentration, Csat, was defined such that: for C < Csat, the molecules are diffuse in solution; and for C > Csat, dense droplets form. If the concentration consistently increases, the liquid-like condensate may change into its gel-like or solid states (Figure 1; Nandana and Schrader, 2021). Notably, Csat values are not fixed, but vary with the concentration of condensate components (Dzuricky et al., 2020; Riback et al., 2020; Zhang et al., 2021). In yeast P bodies, seven proteins are present at high concentrations (5–15 mM), forming the “core” of the condensate, and 24 additional P-body proteins are present at lower concentrations (<2.6 mM) (Xing et al., 2020). It is important to note that Hyman’s hypothesis is not limited to eukaryotic cells (Azaldegui et al., 2021). It can also be applied to bacteria, which were once considered amorphous “bags of enzymes” lacking membrane-bound organelles (Al-Husini et al., 2018). For example, E. coli FtsZ, a well-studied tubulin homolog that is essential for cytokinesis, is capable of forming crowding-induced condensates (Monterroso et al., 2019). In vitro experiments indicate that FtsZ-rich droplets are formed only when FtsZ is in a complex with nucleoid-associated inhibitor SlmA (which antagonizes FtsZ polymerization while binding to specific sites on the E. coli chromosome), and that concentrations of SlmA greater than 40 μM (far above the physiological concentration) are required for FtsZ condensates to form (Han et al., 2012).
FIGURE 1.
Schematic view of a phase diagram. Phase separation is a function of molecular concentration under environmental conditions such as temperature, ionic strength, pH, etc. At a concentration below Csat, the system remains in the one-phase regime. As the concentration increases, two-phase regimes will coexist in the system, and the required concentration is effected by the environmental change as represented in the y-axis. Within the coexistence line (black), molecules often condense into smaller droplets and fuse into bigger droplets to lower the surface tension. These processes are usually reversible. When the concentration continuously increases, the droplets may irreversibly turn into gel-like or solid condensates.
Although the fundamental role played by phase separation in the spatiotemporal organization of essential microbial processes has recently been revealed (Table 1 and Supplementary Table 1), many of these processes have been scarcely explored in bacteria, largely owing to their small sizes and to resolution limits. Nevertheless, ten bacterial LLPS systems have already been identified (Azaldegui et al., 2021), and the number is increasing. These observations support the proposal that LLPS in microbes may be more the rule than the exception. In E. coli, aggregated proteins can be disaggregated during environmental stresses by chaperones, and their spatio-temporal localization is changed in the process (Winkler et al., 2010). In rod-shaped bacterial cells, cell poles are special regions for the localization of signaling and sensing proteins, and here proteins like MreB exhibit random movement (Lopian et al., 2010; Van Teeffelen et al., 2011; Govindarajan et al., 2013; Shi et al., 2018). Furthermore, a high-throughput tagging pipeline of C. crescentus proteins revealed 153 proteins with patchy or spotty subcellular localization patterns (Werner et al., 2009). Together, the above observations provide evidence that LLPS may be widely involved in subcellular organization across different microorganisms, although compartments remain to be discovered.
TABLE 1.
Proposed phase-separated biomolecular condensates in microbial cells.
| Systems | Representative species | Biological processes | Functions | Molecular mechanisms | References |
| LLPS systems in eukaryotic microbes | |||||
| P body | Saccharomyces cerevisiae | Regulate gene transcription | Act against stresses | Defined modular domains (Modules), Intrinsically disordered regions (IDRs) | Decker and Parker, 2012; Jain and Parker, 2013; Hubstenberger et al., 2017; Loll-Krippleber and Brown, 2017; Luo et al., 2018 |
| Stress granules | Saccharomyces cerevisiae | Regulate translation | Act against stresses | IDRs | Buchan et al., 2011; Kato et al., 2012; Jain et al., 2016; Khong et al., 2017 |
| Large 1 (Lge1) protein | Saccharomyces cerevisiae | Accelerate the ubiquitination of histone | Regulate metabolic flux | IDRs | Turco et al., 2015; Kim et al., 2018; Gallego et al., 2020 |
| G body | Saccharomyces cerevisiae | Enhance glycolysis | Act against stresses | IDRs | Jin et al., 2017; Fuller et al., 2020 |
| Pyrenoid | Chlamydomonas reinhardtii | CO2 concentration | Regulate metabolic flux | IDRs | Mackinder et al., 2016; Freeman Rosenzweig et al., 2017; Wunder et al., 2018; He et al., 2020 |
| Yeast ataxin-2 protein (Pbp1) | Saccharomyces cerevisiae | Regulate cellular signaling and autophagy | Act against stresses | Modules | Kato et al., 2019; Yang et al., 2019 |
| DNA repair droplet | Saccharomyces cerevisiae | DNA repair | Act against stresses | IDRs | Lao et al., 2008; Oshidari et al., 2020 |
| Membrane invagination | Saccharomyces cerevisiae | Endocytosis | Act against stresses | IDRs | Bergeron-Sandoval et al., 2021; Lyon et al., 2021 |
| Prion protein | Saccharomyces cerevisiae | Regulate translation | Act against stresses | Modules, IDRs | Franzmann et al., 2018 |
| Heterochromatin protein 1 (HP1) | Schizosaccharomyces pombe | Chromatin compaction | Regulate metabolic flux | Modules | Canzio et al., 2013; Larson et al., 2017; Sanulli et al., 2019 |
| TBP associated factor 14 (Taf14) | Saccharomyces cerevisiae | Regulate gene transcription | Regulate metabolic flux | Modules | Schulze et al., 2010; Chen et al., 2020; Peil et al., 2020 |
| Cajal body homologs | Saccharomyces cerevisiae | Telomerase recruitment | Regulate metabolic flux | Modules | Verheggen et al., 2001; Mao et al., 2011 |
| LLPS systems in prokaryotic microbes | |||||
| Carboxysome | Synechococcus elongatus | CO2 concentration | Regulate metabolic flux | IDRs | Cameron et al., 2013; Chen et al., 2013; Sun et al., 2019; MacCready et al., 2020; Oltrogge et al., 2020 |
| BR-bodies | Caulobacter crescentus | Regulate RNA metabolism | Act against stresses | IDRs | Hardwick et al., 2011; Al-Husini et al., 2018, 2020; Bayas et al., 2018 |
| ParABS DNA segregation system | Escherichia coli | Regulate DNA segregation | Regulate metabolic flux | Modules | Sengupta et al., 2010; Graham et al., 2014; Sanchez et al., 2015; Debaugny et al., 2018; Guilhas et al., 2020 |
| RNA polymerase clusters | Escherichia coli | Control transcription | Regulate metabolic flux | Modules, IDRs | Cabrera and Jin, 2003; Weng et al., 2019; Ladouceur et al., 2020 |
| Pole-organizing protein (PopZ) | Caulobacter crescentus | Control spatial patterning | Regulate metabolic flux | IDRs | Dahlberg et al., 2020; Lasker et al., 2020 |
| Single-stranded DNA-binding protein (SSB) | Escherichia coli | DNA replication, repair, and recombination | Act against stresses | Modules, IDRs, Crowded environments | Zhao et al., 2019; Harami et al., 2020 |
| ATP-binding cassette transporter (Rv1747) | Mycobacterium tuberculosis | Cell growth | Regulate metabolic flux | IDRs | Spivey et al., 2011; Heinkel et al., 2019; Owen and Shewmaker, 2019 |
| Filamentous temperature-sensitive protein Z (FtsZ) assembly | Escherichia coli | Cell division | Regulate metabolic flux | Crowded Environments | Monterroso et al., 2016, 2019 |
| PolyP granules | Pseudomonas aeruginosa | Starvation response and regulation of DNA replication | Act against stresses | Modules, IDRs, Crowded environments | Kreuzer, 2013; Racki et al., 2017 |
| DNA-binding protein from starved cells (Dps) | Escherichia coli | Protect nucleoid from damage | Act against stresses | IDRs | Kim et al., 2004; Janissen et al., 2018 |
Control of Phase Separation in the Formation of Biomolecular Condensates
Although many key questions regarding the organizing principle and physicochemical driving forces of phase separation remain unanswered, in many cases, weak and reversible multivalent interactions between proteins and/or nucleic acids have been demonstrated to be important drivers of biomolecular condensates (Banani et al., 2017). Several different theories explaining phase separation in condensates have been proposed, positing a role for electrostatic interactions, cation-π interactions, aromatic interactions, volume exclusion/crowding, surface tension, or the permeability rate of molecules. With these theoretical frameworks, it may now be possible to explain how the assembly, composition, dynamics, physical properties, and biochemical functions of these biomolecular condensates are regulated. Here, we focus on known mechanisms involved in driving LLPS in microorganisms.
Multivalency-Driven Phase Separation
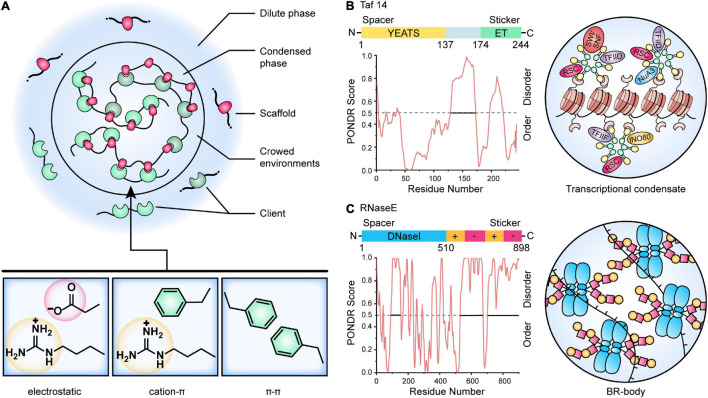
In Li et al. (2012) proposed that multivalent interactions are key factors involved in the phase separation of biomolecules. This view holds that biomolecular condensates are composed of large numbers of multivalent molecules, and thus they contain a variety of elements that control intramolecular or intermolecular interactions. For example, complex condensations can be built through the processes that receptors use to specifically combine with ligands. Therefore, increasing the number, valence, and interaction force of receptors and ligands may promote the formation of stable and large cell condensations. If these interactions occur among multivalent molecules, the molecules will form oligomers and condensations with large stoichiometric ratios (Jin et al., 2019). The essential proteins that drive reversible condensate formation are classified as “scaffolds,” and proteins that preferentially partition into the condensates have been classified as “clients” (Figure 2A). Notably, the two roles are not static or absolute, and it can be hard to unambiguously distinguish these roles in some biomolecular condensates under set environmental conditions (Banani et al., 2016, 2017). In cells, the diffusion speed of clients is much faster than that of scaffolds, and thus client/scaffold interactions are more transient than scaffold/scaffold interactions. The interactions are therefore often selective. For instance, bacterial polar protein PopZ has been shown to act as a selective scaffold that imposes a diffusion barrier to cytosolic proteins (such as the signaling protein CtrA) and constrains the mobility of these proteins at cell poles (Lasker et al., 2020). Furthermore, these interactions are frequently promoted by proteins composed of multiple-folded modular domains and/or intrinsically disordered regions (IDRs) (Gomes and Shorter, 2019).
FIGURE 2.
A model for the control of biomolecular condensates. (A) Multivalent interactions that drive LLPS. Scaffold molecules (red) that undergo LLPS are in stoichiometric excess (often in a crowding environment) and enriched for defined modular domains or intrinsically disordered regions. Client molecules (green) are recruited by binding to the free cognate sites in the scaffold. The critical scaffold/client or scaffold/scaffold interactions include electrostatic, cation-π, and π-π contacts. (B) Model of yeast Taf14-mediated transcriptional condensate. The Taf14 protein contains two main domains, an N-terminal YEATS (Yaf9, ENL, AF9, Taf14, Sas5) domain (yellow) that recognizes lysine acylation modification, as well as a C-terminal ET domain (green) that is reported as a protein-protein interaction domain and recognizes peptide substrates. The disordered regions of Taf14 were predicted by PONDR (Xue et al., 2010). Taf14 works as a scaffold protein that promotes phase separation of condensates and concentrates different transcriptional machinery to form Taf14-containing complexes, thereby enhancing transcription efficiency (Chen et al., 2020). (C) Model of Caulobacter RNase E BR-body assembly. The domain architecture for the RNase E protein is shown, and the disordered regions were predicted by PONDR (Xue et al., 2010). The N-terminal catalytic DNaseI domain (blue) and C-terminal disordered regions (yellow and red) are highlighted. The disordered regions contain positive-charged patches (Arg-rich RNA binding sites, yellow) and negative-charged patches (facilitating multivalent interactions with RNA, red), causing self-assembly of BR-bodies into condensates through electrostatic interactions (Al-Husini et al., 2018, 2020).
Proteins With Defined Modular Domains
Proteins with defined modular folding domains can assemble into higher-order oligomers via intermolecular interactions involving other proteins harboring compatible modular domains. These intermolecular interacting modular domains may be comprised of multiple folded domains or short linear motifs. A typical example from microorganisms is E. coli NusA, a transcription anti-termination factor that interacts directly with RNA polymerase (RNAP) (Ladouceur et al., 2020). NusA, working as a scaffold, contains six folded domains, including two C-terminal acidic repeat Arg-rich domains that recruit clients such as RNAP (and other anti-termination factors). After the scaffold has been built (scaffold proteins have been assembled), more molecules can be recruited to the system to complete the assembly of the condensates. The folded modular domains are often connected by IDRs or low complexity regions (LCRs), and these determine the material properties of the condensates (Harmon et al., 2017). TATA-binding protein-associated associated factor 14 (Taf14) from yeast was once thought as an exception that do not contain IDR or LCR (Chen et al., 2020), but sequence analysis using PONDR (Xue et al., 2010) and SMART (Letunic and Bork, 2018) show that it has two IDRs with Arg/Lys-rich and Glu-rich (Figure 2B). Taf14 is a well-studied, phase-separated transcriptional regulator that associates with a variety of other transcriptional regulators. It contains a YEATS (Yaf9, ENL, AF9, Taf14, and Sas5) domain as an effective reader of histone lysine crotonylation via a unique π–π stacking mechanism and an extra-terminal (ET) domain that recognizes a common motif in diverse transcriptional coactivator proteins such as RSC, SWI/SNF, NuA3, INO80, TFIID, and TFIIF (Andrews et al., 2016; Chen et al., 2020; Figure 2B). Meanwhile, some Taf14-binding partners (e.g., Tfg1) have a number of ET-binding sites that balance the stoichiometric ratio of different complexes in the compartmentalized transcriptional unit (Andrews et al., 2016; Chen et al., 2020).
Proteins With Intrinsically Disordered Regions (IDRs)
In comparison to proteins with defined modular domains, proteins containing IDRs are characterized by more multi-valency and more flexible interaction modes, and therefore they represent the most abundant class of macromolecules that can drive phase separation under physiological conditions. By definition, IDRs lack a defined three-dimensional structure, and they encode multiple short-length amino acid motifs which can provide the basis for multivalent weakly adhesive intermolecular interactions. These motifs typically have a strong bias toward a limited number of amino acids, and are referred to as low complexity sequences (LCSs). They are classified as “stickers” because they demonstrate adhesive properties through π-π stacking, cation-π interactions, or charge-charge interactions (Figure 2A; Wang et al., 2018; Martin et al., 2019). Sequences between the motifs are referred to as “spacers.” Site-directed mutagenesis (or other modifications) in spacer residues can affect the thermophysical properties of the proteins, and thus change the material properties of condensates (Wang et al., 2018). A surprising degree of motional organization of IDPs (intrinsically disordered proteins) has been detected on the ps – ns scale, with IDPs demonstrating fast local vibrations and conformational sampling of backbone dihedral angles, and this may drive LLPS (Salvi et al., 2017).
Based on the examples already known, the biased amino acid compositions of IDRs in bacteria share common hallmarks with IDRs from higher eukaryotes. Thus, IDRs can be: (1) Rich in polar and uncharged amino acid residues such as Gln and Asn. Examples include the “prion-like” sequences in NIDR, the linker IDR of SARS-CoV-2 (Perdikari et al., 2020), and the Gln-rich region in McdB proteins that drives the positioning of the carboxysome (Cameron et al., 2013; MacCready et al., 2020); (2) Rich in charged residues such as Arg/Lys and Glu/Asp. Examples include the Arg-rich C-terminal domain of RNase E that is required for assembly of the core of the bacterial ribonucleoprotein body (BR-body) (Figure 2C; Al-Husini et al., 2018, 2020). Using these concepts, Wei et al. (2020) has developed an artificial membrane-less organelle in E. coli through heterologous overexpression of silk-like proteins using IDRs containing GGX (X = Lys, Tyr, Gln, or Ala) motifs, and this condensate is capable of catalyzing biochemical reactions (Yang et al., 2016; Wei et al., 2020).
Intrinsically disordered regions are notably scarce in bacterial proteomes (comprising less than 2–5% of the proteome) when compared with eukaryotic proteomes (where they comprise 30–40% of the human proteome) (Ward et al., 2004). However, this scarcity does not mean that phase-separation proteins are less relevant in bacteria. Indeed, evidence is accumulating to suggest that IDRs are key players within bacteria, and that these proteins drive LLPS to achieve cell divisions, metabolisms, and nucleoid organizations (Abbondanzieri and Meyer, 2019). Besides, several dedicated computational tools and resources have been published to serve as platforms for collecting, predicting, and annotating LLPS-associated proteins, providing convenient guides to study LLPS proteins in microbes. These databases include PhaSepDB (1You et al., 2020), LLPSDB (2Li et al., 2020), DrLLPS (3Ning et al., 2020), PhaSePro (4Mészáros et al., 2020), and so on. For a detailed review, see Pancsa et al. (2021).
Crowded Environments
The cytosol is a highly crowded environment in which macromolecules such as proteins, nucleic acids, and polysaccharides must push against and compete with each other to carry out their biological functions (McGuffee and Elcock, 2010; André and Spruijt, 2020). The macromolecule concentration in the cytosol of E. coli has been estimated to be ∼300 – 400 mg/mL. In vitro studies have demonstrated that the addition of “inert” crowding agents can induce or enhance LLPS in almost all cases. These agents help mimic a system with high viscosity and a low diffusion coefficient that is favorable for biochemical reactions. Using this system, the effects of pH, temperature, and ionic strength factors can be generally explored (André and Spruijt, 2020). For example, the addition of BSA facilitated the condensation of single-stranded DNA-binding protein (SSB) (Harami et al., 2020), whereas PEG/DNA enhanced the formation of phase-separated condensates composed of FtsZ-SlmA-SBS (Monterroso et al., 2019). In most of these cases, macromolecular crowding can be conceptualized as an “excluded volume effect” (i.e., different species cannot occupy the same space). Thus, inert crowding agents exclude other species from a definite volume (the excluded volume) (Minton, 1990; Ellis, 2001). In general, the total excluded volume depends on the size of the target biomolecules, their number, and their shape (Ellis, 2001). Using the example of the formation of FtsZ-SlmA-SBS droplets, the PEG/dextran system induced an asymmetrical distribution of the condensates (Monterroso et al., 2019). In general, the exclusion volume of macromolecules (such as proteins) is much larger than that of small molecules. As a consequence of this exclusion volume, there can be an accompanying increase in the effective concentration of biomolecules of several orders of magnitude, and this may alter the equilibrium, thermodynamic, and kinetic properties of biochemical reactions (Laurent, 1963). Moreover, this can lead to the formation of biomolecular condensates (Figure 2A; André and Spruijt, 2020). Notably, investigations of crowded environment effects were mainly performed in vitro by mimicking cytosol conditions. However, the excluded volume is affected by the crowders’ abundance, size, and polydispersity (Kim et al., 2015; Yang D. et al., 2020). For example, a 25% decrease in the crowding level from the physiological level was proposed to lead to an utterly diffuse chromosome. In contrast, a 30% increase in the crowder level could lead to a three-fold decrease in the volume of E. coli nucleoids (Yang D. et al., 2020). Besides, even the most widely used uncharged crowders (such as PEG) are usually not chemically inert. They may mediate non-steric interactions that contribute to folding proteins and chromosomes in vivo (Sheth and Leckband, 1997; Xiang et al., 2021). Thus, it is important to investigate the crowding effect in living cells.
Remarkably, there are two issues with the organizing principles that require further consideration, “nucleation” and “nuclear size control” (Hyman et al., 2014). During nucleation, molecules that can randomly assemble with the correct configuration are able to form new droplets. However, because of the limited time, homogeneous nucleation is extremely difficult (Malinovska et al., 2013). Nucleation can occur more favorably at pre-existing locations, such as ribosomes, RNA, etc., and thus cells may control the number and configuration of nucleation. Regarding nuclear size control, cells can control the nucleus size by stopping the merging process (Feric and Brangwynne, 2013). Using the surface effect, cells can utilize additional components that can only be dissolved in droplets to prevent droplets from Ostwald ripening (Webster and Cates, 1998; Zwicker et al., 2015; Bressloff, 2020). Currently, frameworks have been proposed for studying the non-equilibrium dynamics of the dense cellular aggregates, facilitating the link between phase separation and the gene regulatory processes inside the nucleus (Yamamoto et al., 2020; Kuan et al., 2021; Laghmach and Potoyan, 2021).
The Biological Function of Phase-Separated Condensates in Microorganisms
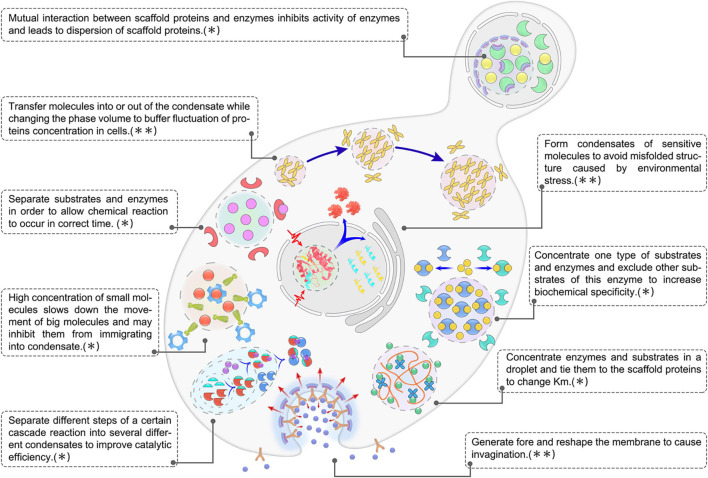
In cells, phase separation is controlled by the assembly and material state of a variety of chaperone proteins, posttranslational modifications (PTMs), and cellular factors, and these in turn determine the size, assembly rate, and material properties of protein condensates, ensuring that distinct cellular functions can be spatiotemporally coordinated (Wang and Zhang, 2019; Quiroz et al., 2020). The various biological activities coupled with LLPS include the classification of misfolded and unwanted proteins for degradation, chromatin organization, gene expression, the assembly of signaling clusters, actin- and microtubule-based cytoskeletal networks, the asymmetric segregation of cell fate determinants, and the formation of pre- and post-synaptic density signaling assemblies (Marrone et al., 2019; Zhang et al., 2020). The functional mechanisms of biomolecular condensates in microorganism are listed in Table 1. Here, we highlight the main functions and summarize them into two main categories.
Regulating Metabolic Flux
Enhancing Activities by Concentrating Enzymes and Substrates
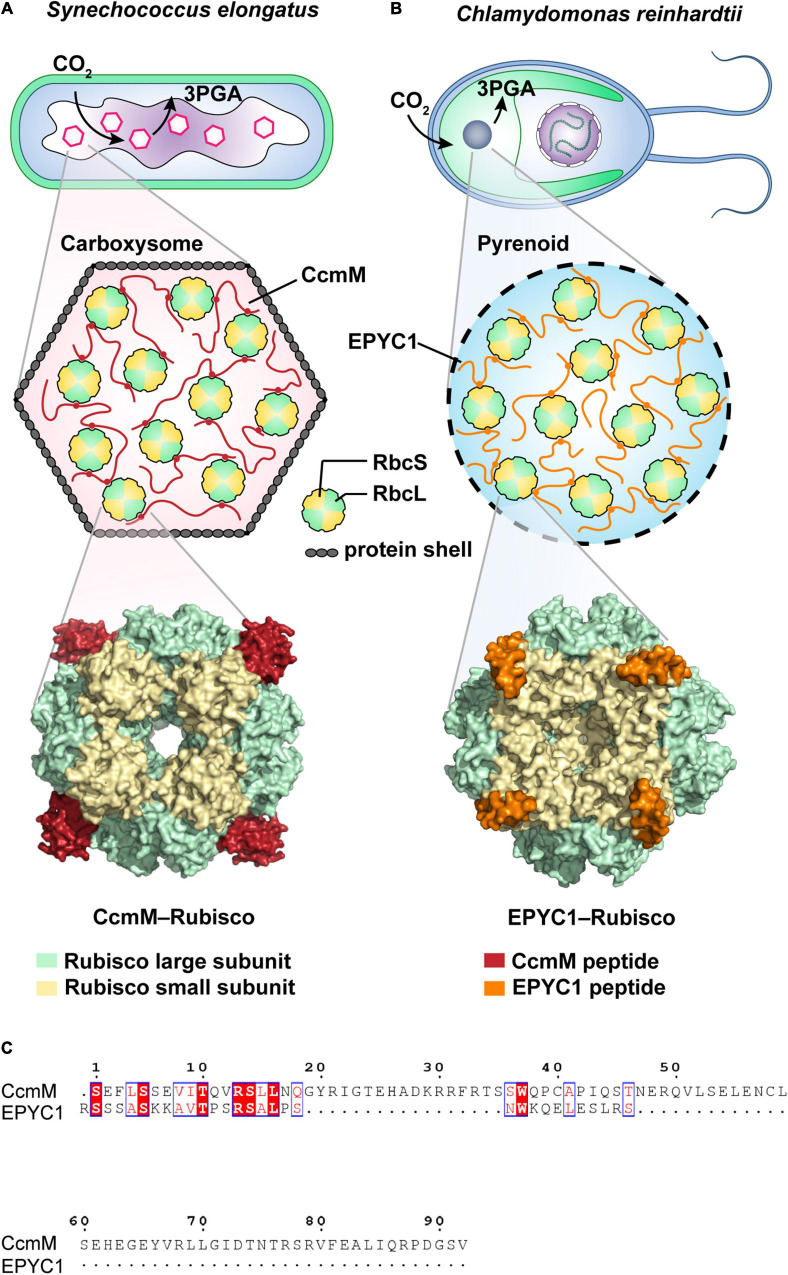
While membrane-bound organelles in eukaryotic cells are widely known to sequester biochemical pathways, membraneless organelles are also capable of organizing internal biochemical reactions. A classic example is the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) condensates found in both eukaryotic and prokaryotic photosynthetic microorganisms. In cyanobacteria and other chemoautotrophic bacteria, Rubisco (the most abundant protein on the planet and the first major enzyme in the Calvin cycle) is encapsulated in a specialized protein-encased micro-compartment termed the carboxysome (Figure 3A). This compartment facilitates HCO3– accumulation and conversion into CO2, known as the CO2 concentrating mechanism (CCM). Due to its proteinaceous shell, the carboxysome was previously believed to be para-crystalline in nature. However, recent discoveries have revealed that biogenesis of the β-carboxysome is achieved through LLPS by forming Rubisco-CcmM condensates (Figure 3A; Wang et al., 2019). In contrast, initiation of the a-carboxysome involves the coalescence of Rubiosco and CsoS2, a protein containing IDRs (Oltrogge et al., 2020). Furthermore, even distribution of the carboxysome is regulated by McdB, which is able to form pH-dependent droplets in vitro (MacCready et al., 2020).
FIGURE 3.
Schematic illustrations of CO2-fixing phase-separated liquid organelles in prokaryotic or eukaryotic cells. (A) Carboxysome-based Rubisco condensate found in the prokaryotic cyanobacterium Synechococcus elongatus PCC7942. As a scaffold protein, CcmM peptide (red) binds the Rubisco large subunit (RbcL, green) and Rubisco large subunit (RbcS, yellow) via salt bridges and van der Waals contacts to form the CcmM-Rubisco complex. It includes the condensates in the carboxysome covering with protein shells. As shown in the cryo-EM structure (6hbc, Wang et al., 2019), CcmM fills a pocket between the RbcL dimers and the loop of RbcS. (B) Pyrenoid-based Rubisco condensate found in the eukaryotic microalgae Chlamydomonas reinhardtii. The pyrenoid matrix is predominantly composed of Rubisco-EPYC1 complexes, forming by the multivalent interactions of EPYC1 peptide (orange) and Rubisco (green and yellow) (Freeman Rosenzweig et al., 2017; Wunder et al., 2018; Meyer et al., 2020; Barrett et al., 2021). Cryo-EM supported a structural model (7jsx, He et al., 2020), showing that EPYC1 binds close to the equator of the Rubisco cylinder and forms a codependent network of the specific low-affinity bonds (Mackinder et al., 2016; He et al., 2020). (C) Alignment of the Rubisco-binding regions from both CcmM and EYPC1 peptides by using Clustal Omega (Sievers and Higgins, 2021) and ESPript 3.0 (Robert and Gouet, 2014).
In eukaryotic microalgae, liquid-like Rubisco-EPYC1 (Essential Pyrenoid Component 1) condensates display functional similarity to Rubisco-CcmM condensates, they are found compartmentalized in an analogous chloroplast-like CCM compartment called the pyrenoid (Figure 3B; Freeman Rosenzweig et al., 2017; Wunder et al., 2018; Barrett et al., 2021). Co-expression of EPYC1 and a plant-algal hybrid Rubisco in higher plant Arabidopsis chloroplasts can lead to phase-separated condensation of Rubisco in chloroplasts (Atkinson et al., 2020). Unlike carboxysome, the pyrenoid lacks proteinaceous shells. Interestingly, cryo-electron tomography (cryo-ET) revealed that the packing of the pyrenoid-based Rubisco condensates in microalgae resembles the hexagonal lattice found in cyanobacterial Rubisco condensates (Engel et al., 2015). Cryo-ET also showed that both the algal EPYC1 and cyanobacterial CcmM bind close to the equator of the Rubisco cylinder (Wang et al., 2019; He et al., 2020), although the binding sites are different. Specifically, EPYC1 binds uniquely to the Rubisco small subunit (RbcS) via electrostatic and hydrophobic interactions (Figure 3B; He et al., 2020; Meyer et al., 2020), while CcmM contacts both Rubisco large subunit (RbcL, Wang et al., 2019) and RbcS via salt bridges and van der Waals contacts (Figure 3A). Both EPYC1 and CcmM have repeat regions and intrinsically disordered proteins, and as has been side, the Rubisco condensation events appear to be regulated in a similar multivalent mechanism. However, their amino acid compositions (15.2% identity in the Rubisco-binding regions, Figure 3C) are considerably divergent (Mackinder et al., 2016; Wang et al., 2019). Altogether, these observations strongly suggest an evolutionary convergence of Rubisco condensates for the vital biological process of CO2 fixation. Likewise, convergent evolution has also been revealed in the formation of ribonucleoprotein (RNP) condensates aiding RNA metabolism in eukaryotic and bacterial cells, both depending on the modulation of DEAD Box ATPases. For a detailed review, see Nandana and Schrader (2021).
A phase-separated condensate may also contain multiple dense phases, so that different enzymes in the cascade can be concentrated in different compartments. In each separation phase, weak interactions between proteins and/or substrates are strongly amplified (Zeng et al., 2018), and the substrates undergo a vectorial transfer from one dense phase to another one while being enzymatically modified in each phase. The best biochemical example of vectorial organization within a biomolecule condensation is the production of ribosomes in nucleoli, where ribosomal RNA is transcribed in the innermost layer, and then processed and assembled as the ribosomal proteins pass through the outer phase (Feric et al., 2016). By concentrating one specific protein with its potential interacting molecules (and excluding other molecules), the condensates can control the specificity of the reaction. In a process akin to the classic Ostwald Ripening, larger condensates can grow bigger while smaller condensates lose molecules (Alexandrov, 2014). In Mycobacterium tuberculosis, ABC transporter Rv1747 undergoes controllable phase separation by acting in conjunction with several cluster-promoting factors that function as serine/threonine protein kinases (STPKs). The majority of these STPKs facilitate specific multivalent interactions by phosphorylation and Rv1747 clustering, whereas the remaining STPKs are involved in extensive signaling cross-talk and serve to dissolve the Rv1747 droplets via dephosphorylation (Heinkel et al., 2019). Thus, STPKs comprise a “multi-valency dial” which allows rapid and reversible differentiation of Rv1747 condensates in response to intracellular signaling (Glass et al., 2017; Heinkel et al., 2019).
Inhibition of Activities Through Sequestration
However, it should be noted that condensation does not always result in the acceleration of reaction velocity. For example, guide RNA (gRNA), the basic modification element for small nuclear RNA (snRNA), is usually concentrated in Cajal bodies. However, suppression of Cajal bodies does not appear to impact the modification effectiveness of snRNA, even though the gRNA is scattered as a consequence (Davis et al., 2015). The reasons behind the activity inhibition are manifold. Firstly, enzymes and the high concentrated scaffold proteins may interfere with each other. The scaffold may inhibit (via covalent modification) the activities of enzymes that disperse condensates (Kuznetsova et al., 2015; Banani et al., 2017). The reduction in available volume associated with high molecular condensation (molecular crowding) is also likely to influence allosteric modulation of enzymes and their binding affinity for substrates, consequently affecting enzyme activity (Kuznetsova et al., 2015). In addition, the condensates are porous structures, and the high concentration of small molecules in solution will slow down the movement of other molecules. For instance, free volume between the concentrated scaffold components may be used as a pore through which small proteins will move (as if the polymer does not exist). In contrast, the movement of macromolecules that cannot access these pores is restricted. This effect may be especially significant for condensates containing ribonucleic acid. Finally, variances in the viscoelasticity of condensates, caused by the degree of IDR maturity, the interaction dynamics of multi-domain scaffolds, RNA composition, or energy consumption processes, may affect molecular dynamics within and on the boundaries.
Acting Against Noise and Stress
Buffering Cellular Noise
Liquid-liquid separation may reduce intracellular protein condensation fluctuations (protein noise) caused by the stochastic nature of gene expression in prokaryotic and eukaryotic cells. In a phase-separating system, protein concentrations inside and outside the droplets are constrained by the solubility threshold. In response to a change in the total concentration of protein, the number and size of the droplets are adjusted to reduce these fluctuations in protein concentration, thus increasing the robustness of cellular processes (Klosin et al., 2020). For example, the amount of bacterial single-stranded DNA binding protein (SSB), an essential protein in genome metabolism, is considerably higher than the amount required during replication (Bobst et al., 1985). The excess SSB and its interacting proteins are dynamically phase-separated within droplets and stored at the cell membrane. In the event of DNA damage, the droplets are rapidly (half-time, ∼70 ms) dissolved and SSB is released to protect the exposed ssDNA and repair the damage (Harami et al., 2020).
Sensing Stimuli and Switching
Macromolecules inside the condensates can communicate freely with external environmental factors, making it possible for the macromolecules to respond rapidly when cells sense external stimuli (Riback et al., 2017; Ruff et al., 2018). Hence, cellular functions may be turned on/off by controlling the formation or dissolution of condensates. For example, the budding yeast translation termination factor Sup35 can form reversible liquid-like condensates in response to sudden stress, ensuring that the function of the translation termination factor is retained, while the condensates can subsequently solidify to form protective protein gels. During this process, negatively charged amino acids in the prion-domain of Sup35 (which are at a high density) function as a pH sensor involved in regulating condensate formation. Upon release from stress, the gel-like condensates are dissolved (Franzmann et al., 2018). Similar processes explain the fitness advantages of yeast P-bodies, stress granules, and bacterial BR-bodies during cell stress (Shah et al., 2013; Wheeler et al., 2016; Al-Husini et al., 2018).
Another active response of condensates to stimuli involves the modulation of polymer folding states. For example, in response to heat stress, heat-labile proteins migrate into the nucleus where they bind with nucleolar protein and form condensates that protect the protein from irreversible aggregation. When the heat stress is removed, these proteins can fold into the correct conformation (Frottin et al., 2019). Likewise, mRNA poly(A) binding protein Pab1 in budding yeast undergoes rapid condensation following heat shock (Riback et al., 2017). In bacteria, similar processes were observed with the DNA-binding protein from starved cells (Dps). In response to stress, Dps binds DNA to change its topology, compacting the DNA into a dense condensate. However, RNA polymerase can freely access the “buried” genes (while other proteins are blocked) (Janissen et al., 2018). This “one-size fits all” approach protects the genome from damage and helps bacteria survive over a diverse range of stress conditions, including heat shock and oxidative stress (Karas et al., 2015; Janissen et al., 2018).
In the face of stresses, biomolecular condensates can even generate and transduce force and thus reshape the cellular architecture. A typical example in yeast is the formation of condensates at the sites of clathrin-mediated endocytosis (CME). The endocytic coat protein Sla1 at the hub of the condensates can bind with both membrane and cytosol proteins (Bergeron-Sandoval et al., 2021). By balancing condensate-membrane and condensate-cytosol interaction energies, the force is exerted sufficiently to drive membrane invagination (Bergeron-Sandoval et al., 2021; Lyon et al., 2021).
Discussion
Evidence is accumulating that phase transitions may be a general mechanism through which microorganisms regulate cellular functions and rapidly adapt to a changing environment. The functions are presented as a model in Figure 4. However, several issues remain unresolved: What mechanisms regulate the specific recruitment of macromolecules in membrane-less organelles? In particular, why are some molecules allowed entry into these organelles while other molecules are selectively excluded? How (and under what circumstances) are these condensates assembled and disassembled? How can the biochemical reactions inside the condensates be scrutinized? By what mechanisms do some condensates divide further into additional compartments (or structured regions) that perform specialized functions? These questions may be addressed by studying the behavior of microbial cells at length over a relative long time scale, and by studying the structural, dynamic, and thermodynamic aspects of these condensates.
FIGURE 4.
Overview of biological functions of biomolecular condensates in microbial cells. The image shown is representative of nine main functions of LLPS in microbial condensates, which could be further summarized into two categories: ∗,condensates play a role in regulating metabolic flux. ∗∗,condensates play a role in acting against noise and stress.
Numerous technical challenges need to be overcome to solve these problems. In microbial condensates (especially condensates from prokaryotic cells), the major hurdle is condensate size (Alberti et al., 2019). While traditional microscopic approaches can be used to detect LLPS in eukaryotic cells, LLPS in prokaryotic cells are typically an order of magnitude smaller (McSwiggen et al., 2019). Thus, prokaryotic condensates in vivo are typically <100–300 nm in diameter (Cho et al., 2018) which is beyond the spatial resolution of light microscopy (∼300 nm, Wenger et al., 2007), resulting in all the condensates appearing spherical. In vitro studies in simulation systems, can be a viable alternative to the in vivo assays. To date, almost all understanding of the protein structures and dynamics involved in bacterial condensates have been garnered from in vitro studies using recombinant proteins. However, these systems are comprised of only one or (at most) two components, and are considerably less complex than in vivo systems, which have properties that are determined by the coexistence of hundreds of thousands of macromolecules and small molecules in a highly confined volume. To mimic the crowded subcellular environment, crowding agents can be added to the in vitro systems. As mentioned above, however, it is not a simple matter to mimic typical condensate viscosity or viscoelasticity. Recently, single-molecule tracking/single-particle tracking (SMT/SPT) super-resolution microscopy and fluorescence correlation spectroscopy (FCS) have proved to be promising tools for investigating the properties of condensates in microorganisms (Gahlmann and Moerner, 2014; Tuson and Biteen, 2015; Sahoo et al., 2016; Jiang et al., 2017; Wei et al., 2017; Maharana et al., 2018; Sieben et al., 2018; Coelho et al., 2020; Gwosch et al., 2020; Peng et al., 2020), while proximity-dependent labeling approaches have been applied to map the protein interactome within the condensates (Govers et al., 2017; Markmiller et al., 2018; Ramanathan et al., 2018). Fluorescence recovery after photobleaching (FRAP), the “gold standard” assay in eukaryotic cells for measuring condensate fluidity and the dynamics of protein exchanges, may also be applied to study bacterial condensates, although model choice and data analysis need to be carefully considered (McSwiggen et al., 2019; Taylor et al., 2019). Together, the application of new fluorescence microscopy techniques to the study of microbial LLPS may prove ground-breaking, creating exciting new perspectives (Cambre and Aertsen, 2020).
To mimic subcellular compartmentalization and control micro-reactions in space and time, artificial membraneless organelles with liquid-like properties have been successfully constructed.
For example, artificial intracellular condensates were de novo designed in E. coli basing on a simple repeat sequence of (Gly-Arg-Gly-Asp-Ser-Pro-Tyr-Ser)XX (where XX is the number of repeats, between 20 and 80). They exhibited controllable dynamics by modulating the molecular weights (number of the repeats, Dzuricky et al., 2020). Protein/RNA coacervates, spider silk protein, and elastic-like protein were also engineered in E. coli with reversible formations, tunable dynamics, and selective enrichments in components, depending on the protein levels and the ratio of charged residues (Mushnikov et al., 2019; Wei et al., 2020; Yeong et al., 2020). In a recently engineered condensate comprised of small ubiquitin-like modifier (SUMOylation), enzyme activity increased approximately 36-fold in the droplets (compared with the surrounding bulk solution) (Peeples and Rosen, 2021). These studies have paved the way for the construction of synthetic membraneless organelles with designer functions in prokaryotes. These synthetic membraneless organelles have broad application prospects in biocatalysis, synthetic biology, and metabolic engineering (Deshpande et al., 2001). Multi-stimuli-responsive carriers (thermal or pH-responsive reversible coacervate droplets) can also be imbued with the ability to package and deliver drugs (Gabryelczyk et al., 2019). Furthermore, microfluidic techniques can be employed to create monodisperse coacervate droplets, making it possible to mimic diverse intracellular activities within uniform unilamellar lipid vesicles (Deshpande et al., 2001; Van Swaay et al., 2015; Deng and Huck, 2017; Love et al., 2020; Zhao et al., 2021). However, the intrinsic properties and functions of these coacervate droplets may differ dramatically as a function of size, and it remains unclear how large a condensate must grow before specific functions can arise (Lyon et al., 2021). One of the biggest challenges is spatiotemporal control over the time-programmed condensation and disassembly of the coacervate. The time-programmed phase behavior is currently available by changes in pH, temperature, ionic strength, light (UV), and more recently by enzyme-mediated catalytic activity (Shin et al., 2017; Schuster et al., 2018; Martin et al., 2019; Wheeler et al., 2019; Garabedian et al., 2021). Furthermore, by converting chemical fuels, the coacervate droplet could behave like a protocell capable of self-division, making it an ideal model for approaching the dynamic complexity of living cells (Zwicker et al., 2017; Donau et al., 2020; Karoui et al., 2021).
In summary, the number of liquid-like condensates identified in microorganisms has grown rapidly during the last few years. While the fundamental role of LLPS in membrane-less compartmentalization has drawn intense interest, new questions and hypotheses concerning the molecular mechanisms and biological processes associated with these microbial condensates have been raised. These gaps in knowledge may be filled through the development of multiscale and interdisciplinary approaches. As the field moves forward, new applications for microbial condensates will be explored.
Author Contributions
GZ and GY contributed to conception, revision, and wrote sections of the manuscript. ZG and WZ wrote the first draft of the manuscript. GZ and ZG designed and made the figures and the table. RC and SZ wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This study was funded by the Natural Science Foundation of Shandong Province (Grant No. ZR2020MC002) and the National Natural Science Foundation of China (Grant Nos. 31970367 and 31640002).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.751880/full#supplementary-material
References
- Abbondanzieri E. A., Meyer A. S. (2019). More than just a phase: the search for membraneless organelles in the bacterial cytoplasm. Curr. Genet. 65 691–694. 10.1007/s00294-018-00927-x [DOI] [PubMed] [Google Scholar]
- Alberti S., Gladfelter A., Mittag T. (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176 419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S., Saha S., Woodruff J. B., Franzmann T. M., Wang J., Hyman A. A. (2018). A user’s guide for phase separation assays with purified proteins. J. Mol. Biol. 430 4806–4820. 10.1016/j.jmb.2018.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov D. V. (2014). On the theory of ostwald ripening: formation of the universal distribution. J. Phys. A 48 035103. 10.1088/1751-8113/48/3/035103 [DOI] [Google Scholar]
- Al-Husini N., Tomares D. T., Bitar O., Childers W. S., Schrader J. M. (2018). Alpha-proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71 1027–1039. 10.1016/j.molcel.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Husini N., Tomares D. T., Pfaffenberger Z. J., Muthunayake N. S., Samad M. A., Zuo T., et al. (2020). BR-bodies provide selectively permeable condensates that stimulate mRNA decay and prevent release of decay intermediates. Mol. Cell 78 670–682. 10.1016/j.molcel.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André A. A. M., Spruijt E. (2020). Liquid-liquid phase separation in crowded environments. Int. J. Mol. Sci. 21:5908. 10.3390/ijms21165908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews F. H., Shinsky S. A., Shanle E. K., Bridgers J. B., Gest A., Tsun I. K., et al. (2016). The Taf14 yeats domain is a reader of histone crotonylation. Nat. Chem. Biol. 12 396–398. 10.1038/nchembio.2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N., Mao Y., Chan K. X., McCormick A. J. (2020). Condensation of Rubisco into a proto-pyrenoid in higher plant chloroplasts. Nat. Commun. 11:6303. 10.1101/2020.10.26.354332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaldegui C. A., Vecchiarelli A. G., Biteen J. S. (2021). The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 120 1123–1138. 10.1016/j.bpj.2020.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S. F., Lee H. O., Hyman A. A., Rosen M. K. (2017). Biomolecular condensates, organizers of cellular biochemistry. Nat. Rev. Mol. Cell. Biol. 18 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., et al. (2016). Compositional control of phase-separated cellular bodies. Cell 166 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F., Mujacic M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22 1399–1408. 10.1038/nbt1029 [DOI] [PubMed] [Google Scholar]
- Barrett J., Girr P., Mackinder L. C. M. (2021). Pyrenoids, CO2-fixing phase separated liquid organelles. Biochim. Biophys. Acta- Mol. Cell Res. 1868:118949. 10.1016/j.bbamcr.2021.118949 [DOI] [PubMed] [Google Scholar]
- Bayas C. A., Wang J., Lee M. K., Schrader J. M., Shapiro L., Moerner W. E. (2018). Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 115 E3712–E3721. 10.1073/pnas.1721648115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron-Sandoval L. P., Kumar S., Heris H. K., Chang C., Cornell C. E., Keller S. L., et al. (2021). Proteins with prion-like domains can form viscoelastic condensates that enable membrane remodeling and endocytosis. bioRxiv [Preprint] 10.1101/145664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobst E. V., Bobst A. M., Perrino F. W., Meyer R. R., Rein D. C. (1985). Variability in the nucleic acid binding site size and the amount of single-stranded DNA-binding protein in Escherichia coli. FEBS Lett. 181 133–137. 10.1016/0014-5793(85)81128-5 [DOI] [PubMed] [Google Scholar]
- Bracha D., Walls M. T., Wei M. T., Zhu L., Kurian M., Avalos J. L., et al. (2018). Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175 1467–1480. 10.1016/j.cell.2018.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., et al. (2009). Granules are liquid droplets that localize by controlled dissolution/condensation. Science 324 1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brangwynne C. P., Mitchison T. J., Hyman A. A. (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108 4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressloff P. C. (2020). Active suppression of Ostwald ripening: beyond mean-field theory. Phys. Rev. E 101:042804. 10.1103/PhysRevE.101.042804 [DOI] [PubMed] [Google Scholar]
- Buchan J. R., Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36 932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Yoon J. H., Parker R. (2011). Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell. Sci. 124 228–239. 10.1242/jcs.078444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera J. E., Jin D. J. (2003). The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 50 1493–1505. 10.1046/j.1365-2958.2003.03805.x [DOI] [PubMed] [Google Scholar]
- Cambre A., Aertsen A. (2020). Bacterial vivisection: how fluorescence-based imaging techniques shed a light on the inner workings of bacteria. Microbiol. Mol. Biol. Rev. 84 e00008–e20. 10.1128/MMBR.00008-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J. C., Wilson S. C., Bernstein S. L., Kerfeld C. A. (2013). Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155 1131–1140. 10.1016/j.cell.2013.10.044 [DOI] [PubMed] [Google Scholar]
- Canzio D., Liao M., Naber N., Pate E., Larson A., Wu S., et al. (2013). A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 496 377–381. 10.1038/nature12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebotareva N. A., Eronina T. B., Roman S. G., Poliansky N. B., Muranov K. O., Kurganov B. I. (2013). Effect of crowding and chaperones on self-association, aggregation and reconstitution of apophosphorylase B. Int. J. Biol. Macromol. 60 69–76. 10.1016/j.ijbiomac.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Chen A. H., Robinson-Mosher A., Savage D. F., Silver P. A., Polka J. K. (2013). The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLoS One 8:e76127. 10.1371/journal.pone.0076127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wang D., Wu B., Yan F., Xue H., Wang Q., et al. (2020). Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation. Nat. Commun. 11:4206. 10.1038/s41467-020-18021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodi I., Biggiogera M., Denegri M., Corioni M., Weighardt F., Cobianchi F., et al. (2000). Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J. Cell Sci. 113 4043–4053. 10.1242/jcs.113.22.4043 [DOI] [PubMed] [Google Scholar]
- Cho W. K., Spille J. H., Hecht M., Lee C., Li C., Grube V., et al. (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho S., Baek J., Graus M. S., Halstead J. M., Nicovich P. R., Feher K., et al. (2020). Ultraprecise single-molecule localization microscopy enables in situ distance measurements in intact cells. Sci. Adv. 6:eaay8271. 10.1126/sciadv.aay8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto J., Fox S., Morimoto R. (1997). HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci. 110 2925–2934. 10.1242/jcs.110.23.2925 [DOI] [PubMed] [Google Scholar]
- Dahlberg P. D., Saurabh S., Sartor A. M., Wang J., Mitchell P. G., Chiu W., et al. (2020). Cryogenic single-molecule fluorescence annotations for electron tomography reveal in situ organization of key proteins in Caulobacter. Proc. Natl. Acad. Sci. U.S.A. 117 13937–13944. 10.1073/pnas.2001849117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. W., Aumiller W. M., Jr., Hashemian N., An S., Armaou A., Keating C. D. (2015). Colocalization and sequential enzyme activity in aqueous biphasic systems: experiments and modeling. Biophys. J. 109 2182–2194. 10.1016/j.bpj.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaugny R. E., Sanchez A., Rech J., Labourdette D., Dorignac J., Geniet F., et al. (2018). A conserved mechanism drives partition complex assembly on bacterial chromosomes and plasmids. Mol. Syst. Biol. 14:e8516. 10.15252/msb.20188516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R. (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4:a012286. 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N. N., Huck W. T. S. (2017). Microfluidic formation of monodisperse coacervate organelles in liposomes. Angew Chem. Int. Ed. Engl. 56 9736–9740. 10.1002/anie.201703145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S., Brandenburg F., Lau A., Last M. G. F., Spoelstra W. K., Reese L., et al. (2001). Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11 114–119. 10.1016/S0959-440X(00)00172-X [DOI] [PubMed] [Google Scholar]
- Donau C., Späth F., Sosson M., Kriebisch B. A. K., Schnitter F., Tena-Solsona M., et al. (2020). Active coacervate droplets as a model for membraneless organelles and protocells. Nat. Commun. 11:5167. 10.1038/s41467-020-18815-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzuricky M., Rogers B. A., Shahid A., Cremer P. S., Chilkoti A. (2020). De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 12 814–825. 10.1038/s41557-020-0511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J. (2001). Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26, 597–604. 10.1016/s0968-0004(01)01938-7 [DOI] [PubMed] [Google Scholar]
- Engel B. D., Schaffer M., Cuellar L. K., Villa E., Plitzko J. M., Baumeister W. (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. Elife 4:e04889. 10.7554/eLife.11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M., Brangwynne C. P. (2013). A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell. Biol. 15 1253–1259. 10.1038/ncb2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., et al. (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165 1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann T. M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A. S., Nüske E., et al. (2018). Phase separation of a yeast prion protein promotes cellular fitness. Science 359:eaao5654. 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- Freeman Rosenzweig E. S., Xu B., Kuhn Cuellar L., Martinez-Sanchez A., Schaffer M., Strauss M., et al. (2017). The Eukaryotic CO(2)-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171 148–162. 10.1016/j.cell.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M. R., Bailey A. D., Weiner A. M., Matera A. G. (1999). Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 9 126–135. 10.1016/S0960-9822(99)80066-9 [DOI] [PubMed] [Google Scholar]
- Frottin F., Schueder F., Tiwary S., Gupta R., Körner R., Schlichthaerle T., et al. (2019). The nucleolus functions as a phase-separated protein quality control compartment. Science 365 342–347. 10.1126/science.aaw9157 [DOI] [PubMed] [Google Scholar]
- Fuller G. G., Han T., Freeberg M. A., Moresco J. J., Ghanbari Niaki A., Roach N. P., et al. (2020). RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. Elife 9:e48480. 10.7554/eLife.48480.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabryelczyk B., Cai H., Shi X., Sun Y., Swinkels P. J. M., Salentinig S., et al. (2019). Hydrogen bond guidance and aromatic stacking drive liquid-liquid phase separation of intrinsically disordered histidine-rich peptides. Nat. Commun. 10:5465. 10.1038/s41467-019-13469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann A., Moerner W. E. (2014). Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat. Rev. Microbiol. 12 9–22. 10.1038/nrmicro3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego L. D., Schneider M., Mittal C., Romanauska A., Gudino Carrillo R. M., Schubert T., et al. (2020). Phase separation directs ubiquitination of gene-body nucleosomes. Nature 579 592–597. 10.1038/s41586-020-2097-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M. V., Wang W., Dabdoub J. B., Tong M., Caldwell R. M., Benman W., et al. (2021). Designer membraneless organelles sequester native factors for control of cell behavior. Nat. Chem. Biol. 17 998–1007. 10.1038/s41589-021-00840-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass L. N., Swapna G., Chavadi S. S., Tufariello J. M., Mi K., Drumm J. E., et al. (2017). Mycobacterium tuberculosis universal stress protein Rv2623 interacts with the putative ATP binding cassette. (ABC) transporter Rv1747 to regulate mycobacterial growth. PLoS Pathog. 13:e1006515. 10.1371/journal.ppat.1006515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E., Shorter J. (2019). The molecular language of membraneless organelles. J. Biol. Chem. 294 7115–7127. 10.1074/jbc.TM118.001192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers S. K., Gayan E., Aertsen A. (2017). Intracellular movement of protein aggregates reveals heterogeneous inactivation and resuscitation dynamics in stressed populations of Escherichia coli. Environ. Microbiol. 19 511–523. 10.1111/1462-2920.13460 [DOI] [PubMed] [Google Scholar]
- Govindarajan S., Elisha Y., Nevo-Dinur K., Amster-Choder O. (2013). The general phosphotransferase system proteins localize to sites of strong negative curvature in bacterial cells. mBio 15 e443–e413. 10.1128/mBio.00443-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T. G., Wang X., Song D., Etson C. M., van Oijen A. M., Rudner D. Z., et al. (2014). ParB spreading requires DNA bridging. Genes Dev. 28 1228–1238. 10.1101/gad.242206.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhas B., Walter J. C., Rech J., David G., Walliser N. O., Palmeri J., et al. (2020). ATP-driven separation of liquid phase condensates in bacteria. Mol. Cell 79 293–303. 10.1016/j.molcel.2020.06.034 [DOI] [PubMed] [Google Scholar]
- Guo M., Ehrlicher A. J., Jensen M. H., Renz M., Moore J. R., Goldman R. D., et al. (2014). Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell 158 822–832. 10.1016/j.cell.2014.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwosch K. C., Pape J. K., Balzarotti F., Hoess P., Ellenberg J., Ries J., et al. (2020). MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 17 217–224. 10.1038/s41592-019-0688-0 [DOI] [PubMed] [Google Scholar]
- Hamouche L., Billaudeau C., Rocca A., Chastanet A., Ngo S., Laalami S., et al. (2020). Dynamic membrane localization of RNase Y in Bacillus subtilis. mBio 11 e3337–e3319. 10.1128/mBio.03337-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149 768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Harami G. M., Kovács Z. J., Pancsa R., Pálinkás J., Baráth V., Tárnok K., et al. (2020). Phase separation by ssDNA binding protein controlled via protein-protein and protein-DNA interactions. Proc. Natl. Acad. Sci. U.S.A. 117 26206–26217. 10.1073/pnas.2000761117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick S. W., Chan V. S., Broadhurst R. W., Luisi B. F. (2011). An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res. 39 1449–1459. 10.1093/nar/gkq928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon T. S., Holehouse A. S., Rosen M. K., Pappu R. V. (2017). Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6:e30294. 10.7554/eLife.30294.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Chou H. T., Matthies D., Wunder T., Meyer M. T., Atkinson N., et al. (2020). The structural basis of Rubisco phase separation in the pyrenoid. Nat. Plants 6 1480–1490. 10.1038/s41477-020-00811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinkel F., Abraham L., Ko M., Chao J., Bach H., Hui L. T., et al. (2019). Phase separation and clustering of an ABC transporter in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 116 16326–16331. 10.1073/pnas.1820683116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., et al. (2017). P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68 144–157. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Hyman A. A., Weber C. A., Jülicher F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30 39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Jain S., Parker R. (2013). The discovery and analysis of P Bodies. Adv. Exp. Med. Biol. 768 23–43. 10.1007/978-1-4614-5107-5_3 [DOI] [PubMed] [Google Scholar]
- Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A., Parker R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janissen R., Arens M. M. A., Vtyurina N. N., Rivai Z., Sunday N. D., Eslami-Mossallam B., et al. (2018). Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell 174 1188–1199. 10.1016/j.cell.2018.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Pryse K. M., Melnykov A., Genin G. M., Elson E. L. (2017). Investigation of nanoscopic phase separations in lipid membranes using inverse FCS. Biophys. J. 112 2367–2376. 10.1016/j.bpj.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F. L., Zhao M., Park M., Park S. J. (2019). Recent trends of foaming in polymer processing: a review. Polymers (Basel) 11:953. 10.3390/polym11060953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Fuller G. G., Han T., Yao Y., Alessi A. F., Freeberg M. A., et al. (2017). Glycolytic enzymes coalesce in g bodies under hypoxic stress. Cell Rep. 20 895–908. 10.1016/j.celrep.2017.06.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong H. G., Kruyt H. R. (1929). Coacervation (partial miscibility in colloid systems). Proc. K Ned. Akad Wet 32 849–856. [Google Scholar]
- Karas V. O., Westerlaken I., Meyer A. S. (2015). The DNA-binding protein from starved cells (Dps). utilizes dual functions to defend cells against multiple stresses. J. Bacteriol. 197 3206–3215. 10.1128/JB.00475-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoui H., Seck M. J., Martin N. (2021). Self-programmed enzyme phase separation and multiphase coacervate droplet organization. Chem. Sci. 12 2794–2802. 10.1039/D0SC06418A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149 753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Yang Y. S., Sutter B. M., Wang Y., McKnight S. L., Tu B. P. (2019). Redox state controls phase separation of the yeast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell 177 711–721. 10.1016/j.cell.2019.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A., Matheny T., Jain S., Mitchell S. F., Wheeler J. R., Parker R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68 808–820. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., An Y. K., Park S., Lee J. S. (2018). Bre1 mediates the ubiquitination of histone H2B by regulating Lge1 stability. FEBS Lett. 592 1565–1574. 10.1002/1873-3468.13049 [DOI] [PubMed] [Google Scholar]
- Kim J., Jeon C., Jeong H., Jung Y., Ha B. Y. (2015). A polymer in a crowded and confined space: effects of crowder size and poly-dispersity. Soft Matt. 11 1877–1888. 10.1039/C4SM02198C [DOI] [PubMed] [Google Scholar]
- Kim J., Yoshimura S. H., Hizume K., Ohniwa R. L., Ishihama A., Takeyasu K. (2004). Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 32 1982–1992. 10.1093/nar/gkh512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosin A., Oltsch F., Harmon T., Honigmann A., Jülicher F., Hyman A. A., et al. (2020). Phase separation provides a mechanism to reduce noise in cells. Science 367 464–468. 10.1126/science.aav6691 [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N. (2013). DNA damage responses in prokaryotes: regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb. Perspect. Biol. 5:a012674. 10.1101/cshperspect.a012674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan H. S., Pönisch W., Jülicher F., Zaburdaev V. (2021). Continuum theory of active phase separation in cellular aggregates. Phys. Rev. Lett. 126:018102. 10.1103/PhysRevLett.126.018102 [DOI] [PubMed] [Google Scholar]
- Kuznetsova I. M., Zaslavsky B. Y., Breydo L., Turoverov K. K., Uversky V. N. (2015). Beyond the excluded volume effects: mechanistic complexity of the crowded milieu. Molecules (Basel, Switzerland) 20 1377–1409. 10.3390/molecules20011377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladouceur A. M., Parmar B. S., Biedzinski S., Wall J., Tope S. G., Cohn D., et al. (2020). Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc. Natl. Acad. Sci. U.S.A. 117 18540–18549. 10.1073/pnas.2005019117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laghmach R., Potoyan D. A. (2021). Liquid-liquid phase separation driven compartmentalization of reactive nucleoplasm. Phys. Biol. 18:015001. 10.1088/1478-3975/abc5ad [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao J. P., Oh S. D., Shinohara M., Shinohara A., Hunter N. (2008). Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29 517–524. 10.1016/j.molcel.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., et al. (2017). Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K., von Diezmann L., Zhou X., Ahrens D. G., Mann T. H., Moerner W. E., et al. (2020). Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat. Microbiol. 5 418–429. 10.1038/s41564-019-0647-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent T. C. (1963). Interaction between polysaccharides and other macromolecules: 5. The solubility of proteins in the presence of dextran. Biochem. J. 89 253–257. 10.1042/bj0890253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46 D493–D496. 10.1093/nar/gkx922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Peng X., Li Y., Tang W., Zhu J., Huang J., et al. (2020). LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 48 D320–D327. 10.1093/nar/gkz778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll-Krippleber R., Brown G. W. (2017). P-body proteins regulate transcriptional rewiring to promote DNA replication stress resistance. Nat. Commun. 8:558. 10.1038/s41467-017-00632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopian L., Elisha Y., Nussbaum-Shochat A., Amster-Choder O. (2010). Spatial and temporal organization of the E. coli PTS components. EMBO J. 29 3630–3645. 10.1038/emboj.2010.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C., Steinkühler J., Gonzales D. T., Yandrapalli N., Robinson T., Dimova R., et al. (2020). Reversible pH-responsive coacervate formation in lipid vesicles activates dormant enzymatic reactions. Angew Chem. Int. Ed Engl. 59 5950–5957. 10.1002/anie.201914893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Na Z., Slavoff S. A. (2018). P-bodies: composition, properties, and functions. Biochemistry 57 2424–2431. 10.1021/acs.biochem.7b01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon A. S., Peeples W. B., Rosen M. K. (2021). A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22 215–235. 10.1038/s41580-020-00303-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCready J. S., Basalla J. L., Vecchiarelli A. G. (2020). Origin and evolution of carboxysome positioning systems in Cyanobacteria. Mol. Biol. Evol. 37 1434–1451. 10.1093/molbev/msz308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder L. C. M., Meyer M. T., Mettler-Altmann T., Chen V. K., Mitchell M. C., Caspari O., et al. (2016). A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl. Acad. Sci. U.S.A. 113 5958–5963. 10.1073/pnas.1522866113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D. K., Richter D., Pozniakovsky A., Poser I., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360 918–921. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Alberti S. (2013). Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim. Biophys. Acta 1834 918–931. 10.1016/j.bbapap.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Mao Y. S., Zhang B., Spector D. L. (2011). Biogenesis and function of nuclear bodies. Trends Genet. 27 295–306. 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S., Soltanieh S., Server K. L., Mak R., Jin W., Fang M. Y., et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172 590–604. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone L., Drexler H. C. A., Wang J., Tripathi P., Distler T., Heisterkamp P., et al. (2019). FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 138 67–84. 10.1007/s00401-019-01998-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N., Tian L., Spencer D., Coutable-Pennarun A., Anderson J. L. R., Mann S. (2019). Photoswitchable phase separation and oligonucleotide trafficking in DNA coacervate microdroplets. Angew Chem. Int. Ed Engl. 58 14594–14598. 10.1002/anie.201909228 [DOI] [PubMed] [Google Scholar]
- McGuffee S. R., Elcock A. H. (2010). Diffusion crowding and protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 6:e1000694. 10.1371/journal.pcbi.1000694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen D. T., Mir M., Darzacq X., Tjian R. (2019). Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 33 1619–1634. 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros B., Erdős G., Szabó B., Schád E., Tantos A., Abukhairan R., et al. (2020). PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 48 D360–D367. 10.1093/nar/gkz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. T., Itakura A. K., Patena W., Wang L., He S., Emrich-Mills T., et al. (2020). Assembly of the algal CO2-fixing organelle, the pyrenoid, is guided by a Rubisco-binding motif. Sci. Adv. 6:eabd2408. 10.1126/sciadv.abd2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A. P. (1990). Holobiochemistry: the effect of local environment upon the equilibria and rates of biochemical reactions. Int. J. Biochem. 22 1063–1067. 10.1016/0020-711X(90)90102-9 [DOI] [PubMed] [Google Scholar]
- Mitrea D. M., Chandra B., Ferrolino M. C., Gibbs E. B., Tolbert M., White M. R., et al. (2018). Methods for physical characterization of phase-separated bodies and membrane-less organelles. J. Mol. Biol. 430 4773–4805. 10.1016/j.jmb.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterroso B., Zorrilla S., Sobrinos-Sanguino M., Keating C. D., Rivas G. (2016). Microenvironments created by liquid-liquid phase transition control the dynamic distribution of bacterial division FtsZ protein. Sci. Rep. 6:35140. 10.1038/srep35140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterroso B., Zorrilla S., Sobrinos-Sanguino M., Robles-Ramos M. A., López-Álvarez M., Margolin W., et al. (2019). Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 20:e45946. 10.15252/embr.201845946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushnikov N. V., Fomicheva A., Gomelsky M., Bowman G. R. (2019). Inducible asymmetric cell division and cell differentiation in a bacterium. Nat. Chem. Biol. 15, 925–931. 10.1038/s41589-019-0340-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandana V., Schrader J. M. (2021). Roles of liquid-liquid phase separation in bacterial RNA metabolism. Curr. Opin. Microbiol. 61 91–98. 10.1016/j.mib.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning W., Guo Y., Lin S., Mei B., Wu Y., Jiang P., et al. (2020). DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 48 D288–D295. 10.1093/nar/gkz1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltrogge L. M., Chaijarasphong T., Chen A. W., Bolin E. R., Marqusee S., Savage D. F. (2020). Multivalent interactions between CsoS2 and Rubisco mediate alpha-carboxysome formation. Nat. Struct. Mol. Biol. 27 281–287. 10.1038/s41594-020-0387-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshidari R., Huang R., Medghalchi M., Tse E. Y. W., Ashgriz N., Lee H. O., et al. (2020). DNA repair by Rad52 liquid droplets. Nat. Commun. 11:695. 10.1038/s41467-020-14546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen I., Shewmaker F. (2019). The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int. J. Mol. Sci. 20:5501. 10.3390/ijms20215501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancsa R., Vranken W., Mészáros B. (2021). Computational resources for identifying and describing proteins driving liquid–liquid phase separation. Briefings Bioinform. 10.1093/bib/bbaa408 [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry Bradley R., Surovtsev Ivan V., Cabeen Matthew T., O’Hern Corey S., Dufresne Eric R., Jacobs-Wagner C. (2014). The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156 183–194. 10.1016/j.cell.2013.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples W., Rosen M. K. (2021). Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17 693–702. 10.1038/s41589-021-00801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil K., Jürgens H., Luige J., Kristjuhan K., Kristjuhan A. (2020). Taf14 is required for the stabilization of transcription pre-initiation complex in Saccharomyces cerevisiae. Epigenet. Chromatin 13:24. 10.1186/s13072-020-00347-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A., Weber S. C. (2019). Evidence for and against liquid-liquid phase separation in the nucleus. Non-coding RNA 5:50. 10.3390/ncrna5040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Li W., Yao Y., Xing W., Li P., Chen C. (2020). Phase separation at the nanoscale quantified by dcFCCS. Proc. Natl. Acad. Sci. U.S.A. 117 27124–27131. 10.1073/pnas.2008447117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdikari T. M., Murthy A. C., Ryan V. H., Watters S., Naik M. T., Fawzi N. L. (2020). SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 39:e106478. 10.15252/embj.2020106478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz F. G., Fiore V. F., Levorse J., Polak L., Wong E., Pasolli H. A., et al. (2020). Liquid-liquid phase separation drives skin barrier formation. Science 367:eaax9554. 10.1126/science.aax9554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki L. R., Tocheva E. I., Dieterle M. G., Sullivan M. C., Jensen G. J., Newman D. K. (2017). Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114 E2440–E2449. 10.1073/pnas.1615575114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., Majzoub K., Rao D. S., Neela P. H., Zarnegar B. J., Mondal S., et al. (2018). RNA–protein interaction detection in living cells. Nat. Methods 15 207–212. 10.1038/nmeth.4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback J. A., Katanski C. D., Kear-Scott J. L., Pilipenko E. V., Rojek A. E., Sosnick T. R., et al. (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168 1028–1040. 10.1016/j.cell.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback J. A., Zhu L., Ferrolino M. C., Tolbert M., Mitrea D. M., Sanders D. W., et al. (2020). Composition-dependent thermodynamics of intracellular phase separation. Nature 581 209–214. 10.1038/s41586-020-2256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 42, W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff K. M., Roberts S., Chilkoti A., Pappu R. V. (2018). Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J. Mol. Biol. 430 4619–4635. 10.1016/j.jmb.2018.06.031 [DOI] [PubMed] [Google Scholar]
- Sabate R., de Groot N. S., Ventura S. (2010). Protein folding and aggregation in bacteria. Cell Mol. Life Sci. 67 2695–2715. 10.1007/s00018-010-0344-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo B., Drombosky K. W., Wetzel R. (2016). Fluorescence correlation spectroscopy: a tool to study protein oligomerization and aggregation in vitro and in vivo. Methods Mol. Biol. 1345 67–87. 10.1007/978-1-4939-2978-8_5 [DOI] [PubMed] [Google Scholar]
- Salvi N., Abyzov A., Blackledge M. (2017). Analytical description of NMR relaxation highlights correlated dynamics in intrinsically disordered proteins. Angew Chem. Int. Ed Engl. 56 14020–14024. 10.1002/anie.201706740 [DOI] [PubMed] [Google Scholar]
- Sanchez A., Cattoni D. I., Walter J. C., Rech J., Parmeggiani A., Nollmann M., et al. (2015). Stochastic self-assembly of ParB proteins builds the bacterial DNA segregation apparatus. Cell Syst. 1 163–173. 10.1016/j.cels.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Sanulli S., Trnka M. J., Dharmarajan V., Tibble R. W., Pascal B. D., Burlingame A. L., et al. (2019). HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575 390–394. 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. M., Kane C. M., Ruiz-Manzano A. (2010). The YEATS domain of Taf14 in Saccharomyces cerevisiae has a negative impact on cell growth. Mol. Genet. Genomics 283 365–380. 10.1007/s00438-010-0523-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B. S., Reed E. H., Parthasarathy R., Jahnke C. N., Caldwell R. M., Bermudez J. G., et al. (2018). Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9:2985. 10.1038/s41467-018-05403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal P. B., Westley J., Lerea K. M., DiSenso-Browne S., Etlinger J. D. (2020). Biomolecular condensates in cell biology and virology: Phase-separated membraneless organelles (MLOs). Anal. Biochem. 597:11369. 10.1016/j.ab.2020.113691 [DOI] [PubMed] [Google Scholar]
- Sengupta M., Nielsen H. J., Youngren B., Austin S. (2010). P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation. J. Bacteriol. 192 1175–1183. 10.1128/JB.01245-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K. H., Zhang B., Ramachandran V., Herman P. K. (2013). Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193 109–123. 10.1534/genetics.112.146993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth S. R., Leckband D. (1997). Measurements of attractive forces between proteins and end-grafted poly(ethylene glycol) chains. Proc. Natl. Acad. Sci. U.S.A. 94 8399–8404. 10.1073/pnas.94.16.8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Bratton B. P., Gitai Z., Huang K. C. (2018). How to build a bacterial cell: MreB as the foreman of E. coli construction. Cell 172 1294–1305. 10.1016/j.cell.2018.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Berry J., Pannucci N., Haataja M. P., Toettcher J. E., Brangwynne C. P. (2017). Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168 159.e114–171.e114. 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C. P. (2017). Liquid phase condensation in cell physiology and disease. Science 357:eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Sieben C., Banterle N., Douglass K. M., Gönczy P., Manley S. (2018). Multicolor single-particle reconstruction of protein complexes. Nat. Methods 15 777–780. 10.1038/s41592-018-0140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D. G. (2021). The clustal omega multiple alignment package. Methods Mol. Biol. 2231, 3–16. 10.1007/978-1-0716-1036-7_1 [DOI] [PubMed] [Google Scholar]
- Singh S. M., Panda A. K. (2005). Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 99 303–310. 10.1263/jbb.99.303 [DOI] [PubMed] [Google Scholar]
- Sleeman J. E., Trinkle-Mulcahy L., Prescott A. R., Ogg S. C., Lamond A. I. (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. J. Cell Sci. 116 2039–2050. 10.1242/jcs.00400 [DOI] [PubMed] [Google Scholar]
- Spector D. L., Lamond A. I. (2011). Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3:a000646. 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivey V. L., Molle V., Whalan R. H., Rodgers A., Leiba J., Stach L., et al. (2011). Forkhead-associated. (FHA) domain containing ABC transporter Rv1747 is positively regulated by Ser/Thr phosphorylation in Mycobacterium tuberculosis. J. Biol. Chem. 286 26198–26209. 10.1074/jbc.M111.246132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., Karpen G. H. (2017). Phase separation drives heterochromatin domain formation. Nature 547 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W. B. (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 79 1558–1562. 10.1073/pnas.79.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J. M., Wilson M. Z., Samuel C. E., Ma D. (2021). Formation and function of liquid-like viral factories in negative-sense single-stranded RNA virus infections. Viruses 13:126. 10.3390/v13010126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wollman A. J. M., Huang F., Leake M. C., Liu L. N. (2019). Single-organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell 31 1648–1664. 10.1105/tpc.18.00787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. O., Wei M. T., Stone H. A., Brangwynne C. P. (2019). Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys. J. 117 1285–1300. 10.1016/j.bpj.2019.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiebackx F. W. (1911). Gleichzeitige Ausflockung zweier Kolloide. Z Chem. Ind. Kolloide 8 198–201. 10.1007/BF01503532 [DOI] [Google Scholar]
- Tripathi V., Song D. Y., Zong X., Shevtsov S. P., Hearn S., Fu X. D., et al. (2012). SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell. 23 3694–3706. 10.1091/mbc.e12-03-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco E., Gallego L. D., Schneider M., Köhler A. (2015). Monoubiquitination of histone H2B is intrinsic to the Bre1 RING domain-Rad6 interaction and augmented by a second Rad6-binding site on Bre1. J. Biol. Chem. 290 5298–5310. 10.1074/jbc.M114.626788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson H. H., Biteen J. S. (2015). Unveiling the inner workings of live bacteria using super-resolution microscopy. Anal. Chem. 87 42–63. 10.1021/ac5041346 [DOI] [PubMed] [Google Scholar]
- Van Swaay D., Tang T. Y., Mann S., de Mello A. (2015). Microfluidic formation of membrane-free aqueous coacervate droplets in water. Angew Chem. Int. Ed Engl. 54 8398–8401. 10.1002/anie.201502886 [DOI] [PubMed] [Google Scholar]
- Van Teeffelen S., Wang S., Furchtgott L., Huang K. C., Wingreen N. S., Shaevitz J. W., et al. (2011). The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. U.S.A. 108 15822–15827. 10.1073/pnas.1108999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Mouaikel J., Thiry M., Blanchard J. M., Tollervey D., Bordonné R., et al. (2001). Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. Embo J. 20 5480–5490. 10.1093/emboj/20.19.5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yan X., Aigner H., Bracher A., Nguyen N. D., Hee W. Y., et al. (2019). Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566 131–135. 10.1038/s41586-019-0880-5 [DOI] [PubMed] [Google Scholar]
- Wang J., Choi J. M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., et al. (2018). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174 688–699. 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang H. (2019). Phase separation, transition, and autophagic degradation of proteins in development and pathogenesis. Trends Cell Biol. 29 417–427. 10.1016/j.tcb.2019.01.008 [DOI] [PubMed] [Google Scholar]
- Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004). Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337 635–645. 10.1016/j.jmb.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Webster A. J., Cates M. E. (1998). Stabilisation of emulsions by trapped species. Langmuir 14 2068–2079. 10.1021/la9712597 [DOI] [Google Scholar]
- Wei M. T., Elbaum-Garfinkle S., Holehouse A. S., Chen C. C., Feric M., Arnold C. B., et al. (2017). Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9 1118–1125. 10.1038/nchem.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S. P., Qian Z. G., Hu C. F., Pan F., Chen M. T., Lee S. Y., et al. (2020). Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 16 1143–1148. 10.1038/s41589-020-0579-9 [DOI] [PubMed] [Google Scholar]
- Weng X., Bohrer C. H., Bettridge K., Lagda A. C., Cagliero C., Jin D. J., et al. (2019). Spatial organization of RNA polymerase and its relationship with transcription in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 116 20115–20123. 10.1073/pnas.1903968116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J., Conchonaud F., Dintinger J., Wawrezinieck L., Ebbesen T. W., Rigneault H., et al. (2007). Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys. J. 92 913–919. 10.1529/biophysj.106.096586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J. N., Chen E. Y., Guberman J. M., Zippilli A. R., Irgon J. J., Gitai Z. (2009). Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U.S.A. 106 7858–7863. 10.1073/pnas.0901781106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J. R., Matheny T., Jain S., Abrisch R., Parker R. (2016). Distinct stages in stress granule assembly and disassembly. Elife 5:e18413. 10.7554/eLife.18413.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R. J., Lee H. O., Poser I., Pal A., Doeleman T., Kishigami S., et al. (2019). Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv [Preprint] 10.1101/721001 [DOI] [Google Scholar]
- Wilson E. B. (1899). The structure of protoplasm. Science 10 33–45. 10.1126/science.10.237.33 [DOI] [PubMed] [Google Scholar]
- Winkler J., Seybert A., König L., Pruggnaller S., Haselmann U., Sourjik V., et al. (2010). Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29 910–923. 10.1038/emboj.2009.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T., Cheng S. L. H., Lai S. K., Li H. Y., Mueller-Cajar O. (2018). The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 9:5076. 10.1038/s41467-018-07624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Surovtsev I. V., Chang Y., Govers S. K., Parry B. R., Liu J., et al. (2021). Interconnecting solvent quality, transcription, and chromosome folding in Escherichia coli. Cell 184 3626–3642. 10.1016/j.cell.2021.05.037 [DOI] [PubMed] [Google Scholar]
- Xing W., Muhlrad D., Parker R., Rosen M. K. (2020). A quantitative inventory of yeast P body proteins reveals principles of composition and specificity. Elife 9:e56525. 10.7554/eLife.56525.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., Uversky V. N. (2010). PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804 996–1010. 10.1016/j.bbapap.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Yamazaki T., Hirose T. (2020). Phase separation driven by production of architectural RNA transcripts. Soft Matt. 16 4692–4698. 10.1039/C9SM02458A [DOI] [PubMed] [Google Scholar]
- Yang D., Männik J., Retterer S. T., Männik J. (2020). The effects of polydisperse crowders on the compaction of the Escherichia coli nucleoid. Mol. Microbiol. 113 1022–1037. 10.1111/mmi.14467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Mathieu C., Kolaitis R. M., Zhang P., Messing J., Yurtsever U., et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181 325–345. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. S., Kato M., Wu X., Litsios A., Sutter B. M., Wang Y., et al. (2019). Yeast Ataxin-2 forms an intracellular condensate required for the inhibition of TORC1 signaling during respiratory growth. Cell 177 697–710. 10.1016/j.cell.2019.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. X., Qian Z. G., Zhong J. J., Xia X. X. (2016). Hyper-production of large proteins of spider dragline silk MaSp2 by Escherichia coli via synthetic biology approach. Process Biochem. 51 484–490. 10.1016/j.procbio.2016.01.006 [DOI] [Google Scholar]
- Yeong V., Werth E. G., Brown L. M., Obermeyer A. C. (2020). Formation of biomolecular condensates in bacteria by tuning protein electrostatics. ACS Cent Sci. 6 2301–2310. 10.1021/acscentsci.0c01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You K., Huang Q., Yu C., Shen B., Sevilla C., Shi M., et al. (2020). PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 48 D354–D359. 10.1093/nar/gkz847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J. Y., Dyakov B. J. A., Zhang J., Knight J. D. R., Vernon R. M., Forman-Kay J. D., et al. (2019). Properties of stress granule and P-Body proteomes. Mol. Cell 76 286–294. 10.1016/j.molcel.2019.09.014 [DOI] [PubMed] [Google Scholar]
- Zeng M., Chen X., Guan D., Xu J., Wu H., Tong P., et al. (2018). Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell 174 1172–1187. 10.1016/j.cell.2018.06.047 [DOI] [PubMed] [Google Scholar]
- Zhang H., Ji X., Li P., Liu C., Lou J., Wang Z., et al. (2020). Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci. 63 953–985. 10.1007/s11427-020-1702-x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu B., Weiner B. G., Meir Y., Wingreen N. S. (2021). Decoding the physical principles of two-component biomolecular phase separation. Elife 10:e62403. 10.7554/eLife.62403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Xu W., Zhang X., Wang X., Ge Y., Yuan E., et al. (2021). Understanding the phase separation characteristics of nucleocapsid protein provides a new therapeutic opportunity against SARS-CoV-2. Protein Cell 12, 734–740. 10.1007/s13238-021-00832-z:1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Liu Y., Wang Z., He R., Zhang J. X., Xu F., et al. (2019). Super-resolution imaging reveals changes in Escherichia coli SSB localization in response to DNA damage. Genes Cells 24 814–826. 10.1111/gtc.12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker D., Hyman A. A., Jülicher F. (2015). Suppression of Ostwald ripening in active emulsions. Phys. Rev. E Stat. Nonlin Soft Matt. Phys. 92:012317. 10.1103/PhysRevE.92.012317 [DOI] [PubMed] [Google Scholar]
- Zwicker D., Seyboldt R., Weber C. A., Hyman A. A., Jülicher F. (2017). Growth and division of active droplets provides a model for protocells. Nat. Phys. 13 408–413. 10.1038/nphys3984 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.