Abstract
Genotypic variability among 96 Trichophyton rubrum strains which displayed different colony morphologies and were collected from four continents was investigated. Twelve markers representing 57 loci were analyzed by PCR fingerprinting, amplified fragment length polymorphism, and random amplified monomorphic DNA markers. Interestingly, none of the methods used revealed any DNA polymorphism, indicating a strictly clonal mode of reproduction and a strong adaptation to human skin.
Trichophyton rubrum (Castellani) Semon is the most common agent of dermatomycoses, primarily causing tinea pedis, onychomycosis, and tinea corporis. Nevertheless, it was discovered and described, first as Epidermophyton rubrum, in 1910 by Castellani (6), only after all other main dermatophytes had already been known for several decades. The species was suggested to have evolved in the late 19th century as a cause of chronic tinea corporis from areas of endemicity in Southeast Asia. The species has since spread throughout the world (23).
T. rubrum appears to be an obligatory anthropophilic species, being transmitted nearly exclusively from human to human. Animal infections have been rarely reported; all were caused by contact with humans. In the environment, T. rubrum can survive (without propagation) up to 18 months in its arthroconidial form (1). This suggests a strong adaptation to the human host.
The species' occurrence on humans is usually symptomatic. In about 2.5% of cases, the species has been isolated from apparently healthy patients. However, it remains unclear whether such patients are really free of dermatomycoses, as, e.g., Zaias (26) has reported the presence of microlesions in seemingly asymptomatic patients. This suggests that T. rubrum is unable to live as a keratinolytic saprophyte on human skin. Thus, it is likely that the same pool of strains is present on healthy and symptomatic patients.
T. rubrum strains can be phenotypically variable, e.g., in expression of the typical red colony reverse or in susceptibility to antimycotic drugs (10). However, even morphologically different dermatophyte species can be genetically closely related, as has been shown, e.g., by internal transcribed spacer sequencing (13, 14).
The aim of the present study was to investigate genotypic variability within a single species. Therefore, strains of T. rubrum were collected from epidemiologically unrelated patients from distant geographical areas in Europe, North America, Africa, and Asia, the latter being the suggested site of origin of the species. Several kinds of anonymous molecular markers were applied to detect strain-specific DNA polymorphisms.
The strains listed in Table 1 showed different colony morphologies and/or reduced sporulation. They were obtained from epidemiologically unrelated patients suffering from either tinea pedis (n = 38), onychomycosis (n = 29), tinea corporis (n = 15), tinea manuum (n = 3), or tinea cruris (n = 2). Two of the Japanese strains were the causative agents of tinea capitis and otitis externa. Reference strains were obtained from the Centraalbureau voor Schimmelcultures (CBS), Baarn, The Netherlands, or from the Faculty of Agriculture, University of Tokyo, Tokyo, Japan. Cultivation was performed on Sabouraud glucose agar for 2 weeks at room temperature prior to DNA isolation.
TABLE 1.
Strains analyzed in this study
| Strain | Diagnosis or comment(s) | Patient data
|
Origin | |||
|---|---|---|---|---|---|---|
| Sex | Age (yr) | |||||
| Clinical strainsb | ||||||
| R1 | Onychomycosis | M | 62 | Berlin | ||
| R2 | Tinea pedis | M | 54 | Berlin | ||
| R3 | Tinea cruris | M | 56 | Berlin | ||
| R4 | Onychomycosis | F | 76 | Berlin | ||
| R5 | Tinea manuum | M | 53 | Berlin | ||
| R6 | Tinea pedis | F | 39 | Berlin | ||
| R7 | Onychomycosis | M | 54 | Berlin | ||
| R8 | Onychomycosis | F | 15 | Berlin | ||
| R9 | Tinea pedis | F | 74 | Berlin | ||
| R10 | Onychomycosis | M | 56 | Berlin | ||
| R11 | Onychomycosis | F | 55 | Berlin | ||
| R12 | Onychomycosis | M | 61 | Berlin | ||
| R13 | Tinea pedis | M | 27 | Berlin | ||
| R14 | Onychomycosis | M | 59 | Berlin | ||
| R15 | Onychomycosis | M | 62 | Berlin | ||
| R18 | Onychomycosis | M | 27 | Berlin | ||
| R19 | Tinea pedis | M | 46 | Berlin | ||
| R20 | Onychomycosis | M | 27 | Berlin | ||
| R21 | Onychomycosis | F | 58 | Berlin | ||
| R22 | Tinea pedis | M | 35 | Berlin | ||
| R23 | Tinea corporis | F | 37 | Berlin | ||
| R24 | Onychomycosis | M | 36 | Berlin | ||
| R25 | Tinea corporis | M | 46 | Berlin | ||
| R26 | Tinea pedis | F | 68 | Berlin | ||
| R27 | Onychomycosis | M | 53 | Berlin | ||
| R28 | Tinea manuum | M | 61 | Berlin | ||
| R29 | Onychomycosis | M | 53 | Berlin | ||
| R30 | Tinea corporis | F | 57 | Berlin | ||
| R31 | Tinea pedis | F | 52 | Berlin | ||
| R32 | Tinea pedis | F | 67 | Berlin | ||
| R33 | Tinea pedis | M | 36 | Berlin | ||
| R34 | Onychomycosis | M | 16 | Berlin | ||
| R35 | Tinea pedis | M | 42 | Berlin | ||
| R36 | Tinea corporis | M | 71 | Berlin | ||
| R37 | Tinea pedis | F | 41 | Berlin | ||
| R38 | Onychomycosis | F | 77 | Berlin | ||
| R39 | Tinea pedis | M | 30 | Berlin | ||
| R40 | Tinea pedis | M | 64 | Berlin | ||
| R41 | Onychomycosis | F | 68 | Berlin | ||
| R42 | Tinea cruris | M | 53 | Berlin | ||
| R43 | Tinea corporis | M | 48 | Berlin | ||
| R44 | Tinea pedis | F | 53 | Berlin | ||
| R45 | Tinea pedis | M | 61 | Berlin | ||
| R46 | Tinea manuum | M | 48 | Berlin | ||
| R47 | Onychomycosis | F | 59 | Berlin | ||
| R48 | Tinea pedis | F | 32 | Berlin | ||
| R49 | Tinea corporis | F | 26 | Berlin | ||
| J11 | Tinea corporis | M | 60 | Hyougo | ||
| J15 | Tinea pedis | F | 39 | Tokyo | ||
| J16 | Unknown | —a | — | Nagasaki | ||
| J17 | Unknown | — | — | Nagasaki | ||
| J21 | Tinea pedis | F | 70 | Gifu | ||
| J22 | Unknown | — | — | Gifu | ||
| J23 | Unknown | — | — | Gifu | ||
| J24 | Tinea corporis | F | 52 | Shizuooka | ||
| J25 | Otitis externa | M | 14 | Kanagawa | ||
| J26 | Tinea pedis | M | 51 | Ishikawa | ||
| J27 | Tinea pedis | F | 44 | Ishikawa | ||
| J28 | Tinea pedis | F | 28 | Ishikawa | ||
| J29 | Tinea capitis | F | 83 | Ishikawa | ||
| J30 | Tinea pedis | F | 59 | Ishikawa | ||
| J31 | Tinea pedis | F | 64 | Ishikawa | ||
| J32 | Tinea pedis | M | 57 | Ishikawa | ||
| J33 | Tinea pedis | M | 73 | Ishikawa | ||
| J34 | Tinea pedis | F | 62 | Ishikawa | ||
| J35 | Tinea corporis | F | 84 | Ishikawa | ||
| J36 | Tinea pedis | M | 53 | Ishikawa | ||
| J37 | Tinea corporis | M | 68 | Ishikawa | ||
| J38 | Tinea pedis | M | 70 | Ishikawa | ||
| J39 | Unknown | F | 59 | Ishikawa | ||
| J40 | Tinea pedis | M | 65 | Ishikawa | ||
| J41 | Tinea pedis | F | 22 | Ishikawa | ||
| J42 | Tinea pedis | M | 46 | Ishikawa | ||
| U0 | Tinea pedis | M | 29 | United States | ||
| U1 | Onychomycosis | F | — | Bronxville, N.Y. | ||
| U2 | Tinea corporis | F | 31 | Utica, N.Y. | ||
| U3 | Tinea pedis | F | 54 | Auburn, Maine | ||
| U4 | Onychomycosis | M | 70 | Bronxville, N.Y. | ||
| U6 | Tinea corporis | M | 35 | Plattsburg, N.Y. | ||
| U7 | Onychomycosis | M | 61 | Auburn, Maine | ||
| U8 | Onychomycosis | M | 65 | Auburn, Maine | ||
| U9 | Onychomycosis | M | 60 | Utica, N.Y. | ||
| U10 | Tinea pedis | F | 28 | Auburn, Maine | ||
| U11 | Tinea pedis | M | 38 | Auburn, Maine | ||
| U12 | Onychomycosis | M | 38 | Lewiston, Maine | ||
| U13 | Tinea corporis | M | 54 | Plattsburg, N.Y. | ||
| U14 | Onychomycosis | F | 48 | Elmira, N.Y. | ||
| U15 | Tinea corporis | M | 73 | Rochester, N.Y. | ||
| U16 | Onychomycosis | M | 62 | Plattsburg, N.Y. | ||
| U17 | Tinea corporis | M | 78 | N. Tarrytown, N.Y. | ||
| U18 | Tinea pedis | M | 49 | Lewiston, Maine | ||
| U19 | Onychomycosis | M | 38 | Auburn, Maine | ||
| U20 | Onychomycosis | F | 50 | Plattsburg, N.Y. | ||
| U21 | Tinea pedis | — | — | Bronxville, N.Y. | ||
| A17 | Tinea pedis | M | 40 | Africa | ||
| Reference strains | ||||||
| CS1 (CBS 303.38) | AUT of T. per-vesei | Unknown | ||||
| CS2 (CBS 392.58) | Hemispherical colony; red | The Netherlands | ||||
| CS3 (CBS 304.60) | var. granular; flat colony | The Netherlands | ||||
| CS4 (A.T.U. TR9) | var. nigricans; hemispherical | Unknown | ||||
| CS5 (A.T.U. TR10) | var. nigricans; melanoid | Unknown | ||||
—, unknown.
Designations indicate geographical regions, as follows: R, Germany; J, Japan; U, United States, A, Africa.
DNA extraction, PCR fingerprinting using the simple repeat sequence (AC)10 (21), and amplified fragment length polymorphism (AFLP) analysis were performed as described previously (14). The following combinations of primer pairs, with three selective nucleotides each (indicated by boldface type), were used for AFLP analysis: (i) EcoRI-TAA (5′-GAC TGC GTA CCA ATT CTA A) and MseI-TAA (5′-GAT GAG TCC TGA GTA ATA A), (ii) EcoRI-TGC (5′-GAC TGC GTA CCA ATT CTG C) and MseI-TGC (5′-GAT GAG TCC TGA GTA ATG C), (iii) EcoRI-TGC (5′-GAC TGC GTA CCA ATT CTG C) and MseI-CTG (5′-GAT GAG TCC TGA GTA ACT G), and (iv) EcoRI-TAA (5′-GAC TGC GTA CCA ATT CTA A) and MseI-CTC (5′-GAT GAG TCC TGA GTA ACT C). Random amplified monomorphic DNA (RAMD) markers for population analysis were developed as described previously (12) and screened for single-strand conformation polymorphisms (SSCP). The following primer pairs were designed for amplification of eight locus-specific DNA fragments: (i) B6/20-1 (5′-GCA AAA CAA ACG CCA AGT AA) and B6/20-2 (5′-ACC CAC ACA TTG AGG AAA AC) (800 bp), (ii) B8/11-1 (5′-TGC CAA ACT ACA CGA ACA TA) and B8/11-2 (5′-CCC AGG TAG TCA GGA GGT AA) (900 bp), (iii) B7/10-1 (5′-AAA CTT GGA GGG CAG GAG AG) and B7/10-2 (5′-TCA CAC TGG GAA CTG AAC AG) (850 bp), (iv) B7/18-1 (5′-CAA GTT TGT GCT CAG TTA TG) and B7/18-2 (5′-AGG ACA GGA CCC AGA GAA TG) (500 bp), (v) B4/14-1 (5′-TCA TCC TTC TTC CCA ACC TC) and B4/14-2 (5′-ATC ACG GAC TAC GGT TTA GC) (800 bp), (vi) B3/20a-1 (5′-CAA AGC AAA CCA ACG ATG TC) and B3/20a-2 (5′-ATT ATG GCA AGG GAT TCA T) (650 bp), (vii) B3/20b-1 (5′-TGC CAG GGC TGA TGG TTT TT) and B3/20b-2 (5′-ACC CAA GGC ACC AGG AAC CC) (850 bp), and (viii) B3/11-1 (5′-ACC CTC GCT TCG TGC CAG TT) and B3/11-2 (5′-GCA AAA ACG AGC AGA GCA CT).
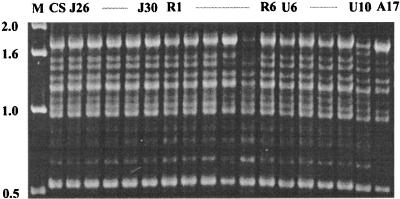
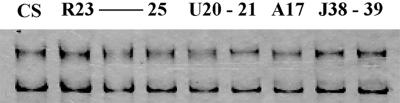
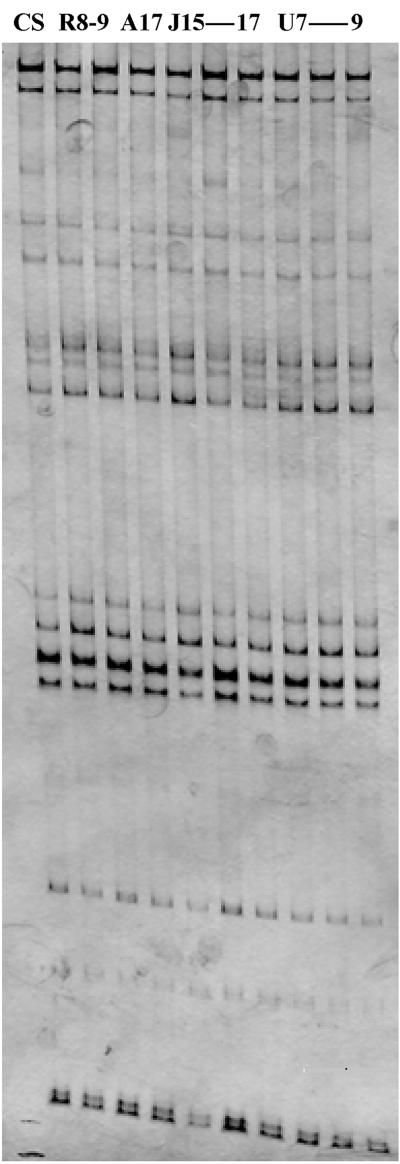
The different techniques applied are known to reveal variabilities among closely related strains. In total, 57 DNA loci representing 12 markers were analyzed (Table 2). With PCR fingerprinting using the simple repeat primer (AC)10 and with the SSCP technique using eight RAMD markers, not a single polymorphism was detected. Similar results were yielded when 53 strains were tested with the PCR fingerprinting primer M13 (data not shown). DNA profiles of 95 strains from four continents were strictly identical (Table 2; Fig. 1 to 3). The same multilocus genotype was also generated for five reference strains; AFLP primer pair TAA-TAA was not used. The reference strains were isolated over a period of 60 years and included two morphological variants (Table 1 [var. granular and var. nigricans]). Some polymorphism was revealed with one of the three AFLP primer pairs in that one strain from Germany (R29) showed two additional bands in the DNA profile with TGC-CTG. The American strains were analyzed with AFLP primer pair TAA-CTG instead of TAA-TAA; all patterns were identical.
TABLE 2.
Results of DNA fragment analyses with T. rubrum strains
| Assay method and primer set | DNA fragment profile for strain origina
|
No. of loci | No. of genotypes | Genetic diversity | ||||
|---|---|---|---|---|---|---|---|---|
| R (n = 48) | J (n = 26) | U (n = 21) | CS (n = 5) | A (n = 1) | ||||
| RAMD | ||||||||
| B7/10 | A | A | A | A | A | 1 | 1 | 0 |
| B7/18 | B | B | B | B | B | 1 | 1 | 0 |
| B8/11 | C | C | C | C | C | 1 | 1 | 0 |
| B4/14 | D | D (2) | D (4) | D | D | 1 | 1 | 0 |
| B6/20 | E | E (2) | E | E | E | 1 | 1 | 0 |
| B3/20a | F | F (1) | F (2) | F | F | 1 | 1 | 0 |
| B3/20b | G | G | G | G | G | 1 | 1 | 0 |
| B3/11 | H | H (1) | H (4) | H | H | 1 | 1 | 0 |
| AFLP | ||||||||
| TAA-TAA | K (7) | K | −(U) | − | K | 8 | 1 | 0 |
| TGC-TGC | L (4) | L | L (1) | L | L | 15 | 1 | 0 |
| TGC-CTG | M (3) | M (1) | M | M | M | 12 | 1 | 0 |
| PCR | ||||||||
| AC10 | S | S | S | S | S | 14 | 1 | 0 |
| Overall | 57 | 1 | 0 | |||||
Strain origins: R, Germany; J, Japan; U, United States; CS, reference strains; A, Africa. Values in parentheses are numbers of strains not assayed. −(U), TAA-CTG was used instead of TAA-TAA.
FIG. 1.
PCR fingerprinting patterns of representative strains of T. rubrum obtained with primer (AC)10. Lane M, 1-kb ladder; lane CS, T. rubrum CBS 392.58.
FIG. 3.
SSCP patterns of representative strains of T. rubrum obtained with RAMD marker B7/18. CS, T. rubrum CBS 392.58.
Only limited genetic investigations were performed, showing dermatophytes to be haploid. To address the population genetics in diploid organisms, e.g., Candida albicans, codominant locus-specific markers have to be used. Methods such as PCR fingerprinting or AFLP analysis are usually dominant, and null alleles are not detectable in heterozygous individuals.
The codominant RAMD markers used confirmed the haploid genotype of T. rubrum. Not a single SSCP was generated among the populations investigated, and only one allele type was detected when representatives of each marker were sequenced (data not shown). Thus, in the continuation of our study we used DNA fragment analysis methods which are easier to perform.
In a study on the population structure of C. albicans, a facultative human pathogenic fungus with a primarily clonal modus of propagation and for which no teleomorph is known, we detected at least in six RAMD markers 12 polymorphic loci (12). In this study, none of the eight markers displayed any variability. Similarly, none of the PCR fingerprinting loci showed any polymorphism. In contrast, in a population of 50 C. albicans isolates collected in a restricted area (Durham, N.C.) and analyzed by the same technique, the same strains showed a large diversity of banding patterns (12). The slight variation in T. rubrum R29 generated by one of the three AFLP markers could be correlated neither with source or locality nor with clinical pictures or phenotypic characteristics of the strain. A genetic diversity of 0, one overrepresented genotype (Table 2), and the observed linkage disequilibrium among the loci investigated are in agreement with Tibayrenc's criteria for clonality in microorganisms (25).
The most parsimonious explanation of our results is a strictly clonal mode of reproduction among populations of this species. Even the population obtained from Japan did not exhibit a distinct genotype. A single multilocus genotype has probably spread worldwide since its emergence in the areas of Southeast Asia in which it is endemic. This suggests a highly specialized lifestyle for the organisms, as they are adapted not only to the human host but also to a specialized body site, i.e., the skin.
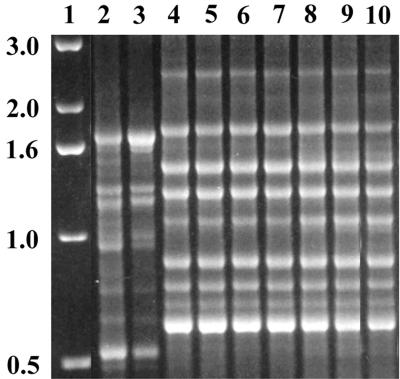
Our results agree with our earlier studies on Trichophyton tonsurans (9) and Trichophyton verrucosum (17) as well as with those of other authors (18, 19). These authors detected no intraspecies DNA polymorphisms among 8 and 29 clinical isolates of T. rubrum or Trichophyton mentagrophytes var. interdigitale, respectively. In contrast to these results, some authors seemed to be able to discriminate T. rubrum strains by molecular typing methods. Nishio et al. (22) reported on two different mitochondrial DNA (mtDNA) genotypes in 92 T. rubrum strains investigated. We tested a few representatives of their type I and II strains by using PCR fingerprinting and found that a misidentification had been made. The DNA fragment patterns of the type I strains obtained with primers M13 and (AC)10 (Fig. 4) correspond to the reference strains of T. rubrum, whereas the DNA profiles of the type II strains were identical to those of T. mentagrophytes var. goetzii (CBS 845.73) and to Arthroderma vanbreuseghemii (RV 27960 = CBS 646.73). Excluding their misidentified strains, Nishio et al. (22) revealed no heterogeneity in the mtDNA pattern of T. rubrum. The six T. rubrum strains investigated by de Bièvre et al. (8) were not available for study, but Nishio et al. (22) mentioned that two isolates belonged to the above mtDNA type II strains and are likely also to be misidentifications. Colony morphologies atypical of T. rubrum, e.g., nonpigmented colony reverse or isolates with reduced sporulation, may lead to misidentifications when no physiological tests for identification are performed. For reliable diagnosis, growth on bromcresol purple-milk solids-glucose agar and urease tests are particularly recommended (16).
FIG. 4.
PCR fingerprinting patterns obtained with primer (AC)10. Lane 1, 1-kb ladder; lane 2, T. rubrum CBS 392.58; lane 3, T. rubrum type I strain (KMU 3332); lanes 4 to 8, T. rubrum type II strains (KMU Tp88, −92, −131, −132, and −133, respectively); lane 9, T. mentagrophytes var. goetzii CBS 845.73; lane 10, A. vanbreuseghemii CBS 646.73. (KMU, Kanazawa Medical University.)
In conclusion, human pathogenic dermatophyte species exhibit a uniformity in their genetic makeup which is as yet unknown for clinical strains of other fungi (2–5, 7, 11, 20, 24). This might be an expression of a general survival strategy for those fungi which propagate in a very specialized ecological niche.
FIG. 2.
AFLP patterns of representative strains of T. rubrum obtained with primer pair CTG-CTG. CS, T. rubrum CBS 392.58.
Acknowledgments
For providing and identifying clinical strains, we thank H.-J. Tietz, Department of Dermatology, Charité Hospital (Berlin, Germany); M. Kawasaki, Department of Dermatology, Kanazawa Medical University (Ishikawa, Japan); K. Kitamura and H. Ishizaki, National Kanazawa Hospital (Ishikawa, Japan); and V. Chaturvedi, New York State Department of Health. We thank G. S. de Hoog for critical reading of the manuscript.
Funding was provided by the Deutsche Forschungsgemeinschaft, GR 1147/1-1 and GR 1147/1-2, to H.-J. Tietz and Y. Gräser.
REFERENCES
- 1.Baer R L, Rosenthal S A, Furnari D. Survival of dermatophytes applied on the feet. J Investig Dermatol. 1955;24:619–662. doi: 10.1038/jid.1955.83. [DOI] [PubMed] [Google Scholar]
- 2.Bart-Delabesse E, Humbert J-F, Delabesse E, Bretagne S. Microsatellite markers for typing Aspergillus fumigatus isolates. J Clin Microbiol. 1998;36:2413–2418. doi: 10.1128/jcm.36.9.2413-2418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlotti A, Chaib F, Couble A, Bourgeois N, Blanchard V, Villard J. Rapid identification and fingerprinting of Candida krusei by PCR-based amplification of the species-specific repetitive polymorphic sequence CKRS-1. J Clin Microbiol. 1997;35:1337–1343. doi: 10.1128/jcm.35.6.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter D A, Burt A, Taylor J W, Koenig G L, White T J. Clinical isolates of Histoplasma capsulatum from Indianapolis, Indiana, have a recombining population structure. J Clin Microbiol. 1996;34:2577–2584. doi: 10.1128/jcm.34.10.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellani A. Observation on a new species found in Tinea cruris. Br J Dermatol. 1910;5:148–150. [Google Scholar]
- 7.Debeaupuis J P, Sarfati J, Chazalet V, Latge J P. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Bièvre C, Dauguet C, Nguyen V H, Ibrahim-Granet O. Polymorphism in mitochondrial DNA of several Trichophyton rubrum isolates from clinical specimens. Ann Inst Pasteur/Microbiol. 1987;138:719–727. doi: 10.1016/0769-2609(87)90149-9. [DOI] [PubMed] [Google Scholar]
- 9.El Fari, M., Y. Gräser, W. Presber, and H.-J. Tietz. An epidemic of tinea corporis caused by Trichophyton tonsurans among children (wrestlers) in Germany. Myloses, in press. [DOI] [PubMed]
- 10.Fachin A L, Maffei C M L, Martinez-Rossi N M. In vitro susceptibility of Trichophyton rubrum isolates to griseovulvin and tioconazole. Induction and isolation of a resistant mutant to both antimycotic drugs. Mycopathologia. 1996;135:141–143. doi: 10.1007/BF00632334. [DOI] [PubMed] [Google Scholar]
- 11.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gräser Y, El Fari M, Vilgalys R, Kuijpers A F A, de Hoog G S, Presber W, Tietz H J. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med Mycol. 1999;37:105–114. [PubMed] [Google Scholar]
- 14.Gräser Y, Kuijpers A, Presber W, de Hoog G S. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol. 1999;37:315–330. doi: 10.1046/j.1365-280x.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 15.Huey B, Hall J. Hypervariable DNA fingerprinting in Escherichia coli: minisatellite probe from bacteriophage M13. J Bacteriol. 1989;171:2528–2532. doi: 10.1128/jb.171.5.2528-2532.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane J, Summerbell R, Sigler L, Krajden S, Land G. Laboratory handbook of dermatophytes. Belmont, Calif: Star Publishing Co.; 1997. [Google Scholar]
- 17.Kielstein P, Wolf H, Gräser Y, Buzina W, Blanz P. On the variability of Trichophyton verrucosum isolates from vaccinated herds with ringworm of cattle. Mycoses. 1998;41:58–64. doi: 10.1111/j.1439-0507.1998.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, Coloe S, Pedersen J, Baird R. Use of arbitrarily primed polymerase chain reaction to differentiate Trichophyton dermatophytes. FEMS Microbiol Lett. 1996;136:147–150. doi: 10.1111/j.1574-6968.1996.tb08040.x. [DOI] [PubMed] [Google Scholar]
- 19.Mochizuki T, Watanabe S, Uehara M. Genetic homogeneity of Trichophyton mentagrophytes var. interdigitale isolated from geographically distant regions. J Med Vet Mycol. 1996;34:139–143. [PubMed] [Google Scholar]
- 20.Moriello K A, De Boer D J. Fungal flora of the coat of pet cats. Am J Vet Res. 1991;52:602–606. [PubMed] [Google Scholar]
- 21.Niesters H G M, Goessens W H F, Meis J M F G, Quint W G V. Rapid polymerase chain reaction-based identification assay for Candida species. J Clin Microbiol. 1993;31:904–910. doi: 10.1128/jcm.31.4.904-910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishio K, Kawasaki M, Ishizaki H. Phylogeny of the genera Trichophyton using mitochondrial DNA analysis. Mycopathologia. 1992;117:127–132. doi: 10.1007/BF00442772. [DOI] [PubMed] [Google Scholar]
- 23.Rippon J W, editor. Medical mycology: the pathogenic fungi and pathogenic actinomycetes. Philadelphia, Pa: W. B. Saunders Co.; 1988. p. 178. [Google Scholar]
- 24.Soares C M, Madlun E E, da Silva S P, Pereira M, Felipe M S. Characterization of Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:505–507. doi: 10.1128/jcm.33.2.505-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere S F, Darde M L, Ayala F J. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaias, N. Personal communication.