Abstract
The kinase PAK binds tightly to the SH3 domain of its partner PIX via a central proline-rich sequence. A different N-terminal sequence allows αPAK to bind an SH3 domain of the adaptor Nck. The Nck SH3[2] domain interacts equally with an 18-mer PAK-derived peptide and full-length αPAK. Detailed analysis of this binding by saturation substitution allows related Nck targets to be accurately identified from sequence characteristics alone. All Nck SH3[2] binding proteins, including PAK, NIK, synaptojanin, PRK2, and WIP, possess the motif PXXPXRXXS; in the case of PAK, serine phosphorylation at this site negatively regulates binding. We show that kinase autophosphorylation blocks binding by both Nck and PIX to αPAK, thus providing a mechanism to regulate PAK interactions with its SH3-containing partners. One cellular consequence of the regulatable binding of PAK is facilitation of its cycling between cytosolic and focal complex sites.
Signal transduction pathways often utilize protein-protein interaction modules whose domain structures are conserved at the primary or secondary structural level. Two domains frequently found on signaling molecules are Src homology 2 and 3 domains (41). In Src, these domains not only regulate association with other proteins but also intramolecular functions, including protein tyrosine kinase activity (35). The adaptor signaling molecules contain no catalytic domain (41). The best studied of these is Grb2, whose SH3 domains complex to the Ras guanine nucleotide exchange factor (GEF) SOS. Upon stimulation, tyrosine kinase receptors that engage Grb2 through binding to its SH2 domains recruit the Grb2-SOS complex, thus causing Ras activation (7).
We have recently described a new class of Rac1 GEF whose SH3 domain binds selectively to a nonconventional proline-rich binding sequence present in all mammalian PAKs (32). Because these PAK-interacting exchange (PIX) proteins are complexed to PAK, the kinase has roles both upstream and downstream of Rac and/or Cdc42 (39). Thus, recruitment of the complex via PAK leads to Rac activation, while PIX itself is known to play a role in localizing PAK to focal complexes (FCs) and activating the kinase (32). Although Cdc42 or Rac directly activate PAKs, the ubiquitous adaptor protein Nck, which binds to an N-terminally located proline-rich sequence (2, 5, 14), can also activate PAK by recruitment to the plasma membrane (27). PAK activation by Nck is mimicked when membrane-localizing signals are directly attached to PAK (30). Nck contains three tandem SH3 domains and a C-terminally located SH2 domain (8, 9, 25). The Drosophila Nck homologue, Dock, plays a role in axonal guidance: both DPak and Dock are highly expressed in the nervous system (15, 18). Membrane-tethered DPak acts as a dominant gain-of-function protein in dock mutants, restoring the normal pattern of R-cell connectivity; thus, DPak is a key downstream partner of Dock (19).
The structures of many SH3 domains have been determined by crystallographic and nuclear magnetic resonance protocols. These analyses reveal that the conserved aromatic residues form a hydrophobic patch on the surface of the SH3 domain (6, 20). Part of the binding affinity is contributed by hydrophobic interactions with conserved prolines, but it also involves ionic interactions, particularly with a basic residue positioned before or after the PXXP motif. The position of this basic residue determines the orientation (plus or minus) of the pseudosymmetric PXXP-containing ligand on the SH3 domain (13, 26). Since peptide binding requires the central portion of a polypeptide to adopt a type II polyproline helix conformation, the contexts of these target sequences play an important role in determining the relative affinity. Thus, it has been reported that the tight binding between Grb2 and the C-terminal region of SOS, with affinity in the submicromolar range, requires integrity of the SOS domain (47), whereas binding of SH3 domains to peptides derived from their target sequences occurs with affinities in the micromolar range (24).
In this study, we initially assessed SH3 binding of a number of domains related to PIX SH3 using a novel SH3 overlay protocol; stable complexes were detected with some, but not all, SH3 domains tested. Using the second SH3 domain (SH3[2]) of Nck, direct SH3 targets were then purified and identified by protein microsequencing. Of these, we chose PAK and NIK for further study, identifying 18-residue peptides within each that fulfill the binding function. Most of the identified targets contain related proline (and serine-threonine)-rich motifs. The interaction between Nck SH3[2] and PAK was also found to be negatively regulated by phosphorylation in vitro and in vivo. We propose that phosphorylation-mediated regulation of SH3 binding can play an important role in signaling through such adapter proteins. In the case of PAK, it appears that this effect, which occurs with both Nck and PIX, allows the kinase to cycle between FCs and the cytoplasm.
MATERIALS AND METHODS
Bacterial expression vectors.
The pGEX-Ras vector was derived from pGEX-4T-1 as follows. The Ras(1-185) coding sequence was PCR amplified (Vent; New England Biolabs) to include a BglII site adjacent to codon 1 and, at the 3′ end (adjacent to codon 185), a BamHI-EcoRI linker (GGATCCCCGAATTC). This fragment was cloned into the pGEX-4T BamHI/EcoRI site, thereby regenerating a BamHI site downstream of the Ras sequence (the original BamHI site was lost; the new polylinker is identical to that in pGEX-4T-3). Various SH3 domains used in this study were derived by PCR with primers containing 5′ BamHI and 3′ XhoI cloning sites. The amplified human cDNA SH3 corresponds to the following codons (GenBank numbers are in parentheses): cortactin (A48063), 491 to 550; phospholipase Cγ (G4505869), 790 to 852; myosin 1C (U14391), 1052 to 1109; Nck SH3[1], 1 to 98; Nck SH3[2], 110 to 176; and Nck SH3[3], 167 to 266. The construct containing Nck SH3[1,2,3] contained residues 1 to 266. αPAK(1-250) is derived from a BamHI/BglII fragment as described previously (30). The αPIX SH3 domain was described previously (32).
Constructs encoding peptide sequences were derived from synthetic oligonucleotides containing the appropriate overhangs which were cloned into the BamHI and XhoI sites of pGEX 4T-1. (See Fig. 3 for the sequences of these peptides [which also contained 3′ termination codons].) The PTP-PEST sequence (328 to 344) encoded SKQDSPPPKPPRTRSCLV. Plasmids encoding the glutathione-S-transferase (GST) or GST-Ras fusion proteins were transformed into the Escherichia coli BL21 strain for protein expression as described previously (30).
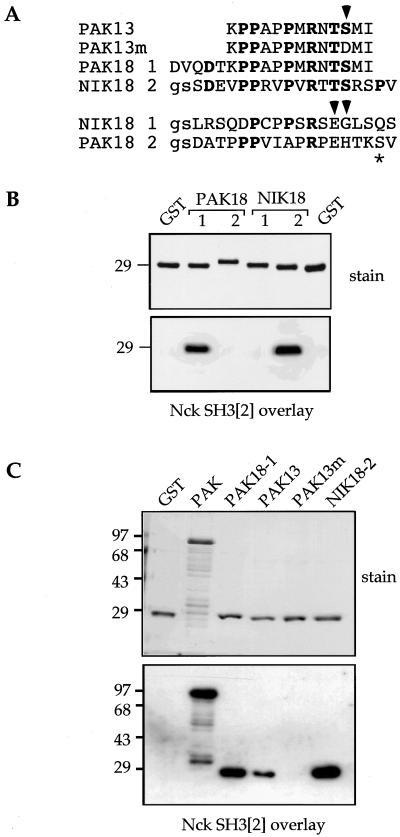
FIG. 3.
Identification of the Nck binding region in NIK and PAK. (A) Peptide sequences used to test Nck SH3[2] binding as indicated were expressed as GST fusion proteins. Residues conserved between PAK and NIK are in boldface; the arrowheads indicate the position of phosphorylated serine in PAK or nontolerated residues in NIK18 1. There is a phosphorylated serine in the Plx binding site (asterisk). (B) GST proteins displaying peptides (1 or 2) derived from PAK and NIK in panel A were analyzed by Nck SH3[2] overlay following separation of 2 μg of each protein on a 10% polyacrylamide gel. (C) PAK18 binds with efficiency similar to that of full-length PAK. A comparison of binding to 2 μg of αPAK(L404S) or 1 μg of the various peptides indicated in panel A is shown. The smaller peptide, PAK13, binds more weakly than PAK18; in this context, PAK13S21E fails to bind Nck. Numbers on the left indicate the molecular masses (in kilodaltons) of the markers.
Mammalian expression vectors.
The pXJ-HA and pXJ-Flag mammalian vectors used for transfection and microinjection experiments have been described previously (30). The full-length Nck sequence was amplified with oligonucleotides containing 5′ BamHI and 3′ XhoI sites. Full-length rat GIT1 (42) was derived from a cDNA library and cloned, also using a BamHI site flanking the initiator methionine.
Purification of Nck binding proteins.
GST-Nck SH3[2] proteins were dialyzed overnight against phosphate-buffered saline (PBS) and coupled to cyanogen bromide (CNBr)-activated Sepharose (Sigma) to yield 2 mg of affinity matrix/ml. Rat tissues were homogenized in buffer A (25 mM HEPES [pH 7.3], 0.5% Triton X-100, 1 mM EDTA, 25 mM NaF, 1 mM sodium orthovanadate), freshly added 5 mM dithiothreitol, and 10 μg each of leupeptin and aprotinin/ml. The supernatants from centrifugation (100,000 × g; 40 min) were diluted to 5 mg/ml with buffer B (PBS plus 50 mM Tris-HCl, pH 7.8, 0.1% Triton X-100, 0.5 mM MgCl2) and passed through affinity columns (a ratio of 20 ml of extract per ml of matrix). After washing with 10 column volumes of buffer B, bound proteins were released by heating them (100°C; 10 min) in a 1/5 dilution of sodium dodecyl sulfate (SDS) sample buffer (0.4% SDS) and concentrated. Sepharose Q (Pharmacia) ion-exchange chromatography was carried out manually using a ratio of 20 mg of brain extract/ml of Sepharose. After the sample was loaded, elution was carried out in buffer A containing 100 mM incremental NaCl steps (up to 500 mM). The 150 to 350 mM fraction was collected by washing it with 2 column volumes of 150 mM NaCl buffer and eluting with 2 column volumes of 350 mM buffer. This fraction was immediately loaded onto a 1-ml Nck SH3[2]-Sepharose column, washed, and processed as described above. The relevant bands were excised from the stained gels and processed for protein microsequencing as described previously (30).
Overlays with [γ-32P]GTP labeled proteins.
The Pepspot filter and the PAK9-23 and PAK9-23(PS21) phosphopeptides were synthesized and purified by Jerini Biotools. The purified proteins were separated on 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) (NEN) membranes. The filters were blocked for 2 to 16 h (4°C) in PBS containing 10% skim milk prior to overlay analysis. The GST-Ras fusion proteins (10 μg) were incubated for 4 min with 10 μCi of [γ-32P]GTP in 50 μl of exchange buffer (25 mM HEPES [pH 7.3], 50 mM KCl, 2.5 mM EDTA). This mixture was immediately added to 3 ml of binding and wash buffer (PBS containing 25 mM HEPES, pH 7.3, 5 mM MgCl2, and 0.05% Triton X-100) containing 0.1 mM GTP and added to a roller bottle containing the PVDF membrane. Following a 1-h incubation at 4°C, the filters were washed (three times for 10 min each time) with binding and wash buffer and exposed to PhosphorImager plates (Molecular Dynamics) for quantification or to X-ray film.
Biacore analysis.
The Nck SH3[2] domain was cloned into the pMal vector (New England Biolabs) in order to express maltose-binding protein (MBP) fusions. Proteins were eluted from the amylose resin in PBS containing 50 mM Tris-HCl, 5% glycerol, 0.1% Triton X-100, 0.5 mM MgCl2, and 20 mM maltose and stored at −70°C prior to use. For Biacore studies, the GST-PAK or GST-peptide proteins were dialyzed overnight against PBS and coupled to CM5 sensor chips (Pharmacia-LKB) under standard immobilization conditions in 10 mM sodium acetate (pH 5.0). The sensorgrams were collected at 10-μl/min flow rates at 24°C using Pharmacia HEPES-buffered saline (HBS) buffer–0.05% NP-40. Regeneration cycles were carried out by treating the surface with HBS plus 0.1% SDS for 4 min.
Cell microinjection, transfection, and staining.
Subconfluent HeLa cells were microinjected into the nucleus with 50 ng of each expression plasmid DNA/ml using an Eppendorf microinjector. After 2 h, the cells were fixed in 3% paraformaldehyde for 20 min and stained as described previously (30). COS-7 cells were transfected using 3 μg of plasmid plus 30 μl of DOSPER (Roche) per 100-mm-diameter dish and harvested 16 h after the addition of DNA. Antibody sources were as follows: anti-Flag and anti-Flag M2 Sepharose were from Sigma, anti-hemagglutinin was from Roche, anti-paxillin was from Transduction Laboratories, and anti-αPAK was as described previously (30).
RESULTS
A subset of SH3 domains identifies target proteins in overlays.
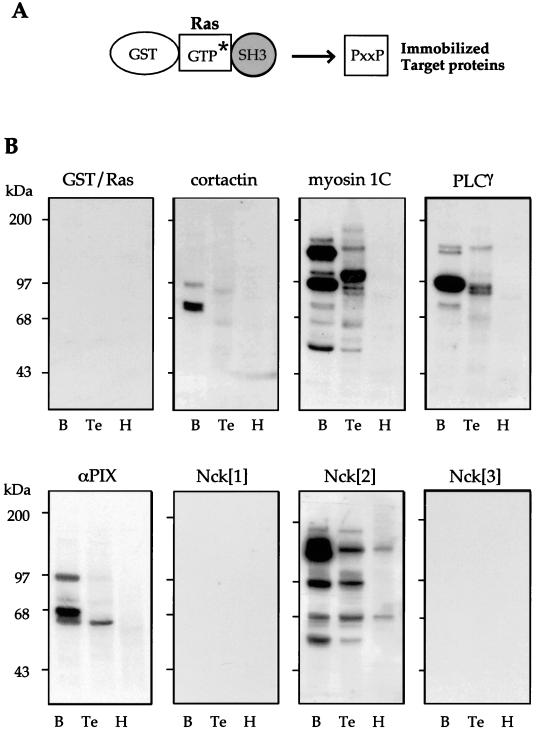
PIX binds with high affinity to PAK-derived peptides (32) and overlays by using the PIX SH3 domain to selectively detect PAKs (i.e., the bona fide targets). This prompted us to determine whether related SH3 domains exhibited similar properties. The ligand overlay method has been used in the analysis of SH3 binding (45, 54) and is most sensitive when γ-32P-labeled SH3 probes are employed. To facilitate rapid and efficient labeling of fusion proteins, we have exploited the ability of the small G protein Ras to rapidly sequester [γ-32P]GTP in an Mg2+-dependent manner with a Kd of ∼10−12. The GST-Ras-SH3 fusion system shown in Fig. 1A allows efficient (>90%) incorporation; this contrasts with labeling methods such as the use of kinase phosphorylation with [γ-32P]ATP, which in our hands is ∼10% efficient. GST-Ras itself does not produce signals after proteins are subjected to SDS-polyacrylamide gel electrophoresis (Fig. 1B). We then probed rat tissue extracts for binding proteins using the SH3 domains of αPIX, phospholipase Cγ (PLCγ), myosin 1C, cortactin, and Nck (designated [1], [2], and [3]). Interestingly those SH3 domains more closely related to PIX (32) formed stable complexes with specific proteins enriched in brain and testis (Fig. 1). The patterns of binding proteins detected with myosin 1C, PLCγ, and Nck[2] SH3 domains were distinct. The ∼100-kDa brain protein detected by most probes corresponds to dynamin, which binds many SH3 domains in vitro (16).
FIG. 1.
Protein overlays define multiple target proteins for certain SH3 domains. (A) Schematic diagram of the GST-Ras construct showing the arrangements of structural domains. [γ-32P]GTP (asterisk) is introduced into the fusion protein by incubating the protein for 4 min in buffer containing 2.5 mM EDTA. Binding and washes are performed in buffers containing 5 mM MgCl2. (B) Labeled SH3 domains as shown were overlaid onto blots (on PVDF) containing 80 μg of soluble proteins derived from rat tissues: brain (B), testis (Te), and heart (H). The filters were exposed for 4 h at −70°C. As a control, GST-Ras alone produces no signals. In addition to dynamin (∼100-kDa band in brain), the αPIX SH3 domain detects the three isoforms of PAK (α, β, and γ), with the level of αPAK being highest.
Nck SH3[2] was selected for further study, since among its binding partners are the serine and threonine PAK kinases that are targets for Cdc42 and Rac1 (31). The Nck SH3[1,2,3] binding pattern indicated that no further targets were detected (not shown), suggesting these flanking SH3[1] and SH3[3] domains do not add significantly to binding stability under these assay conditions. To establish the identities of the other proteins that also bind robustly to Nck SH3[2], we then undertook purification of these proteins from rat brain.
Purification and microsequencing of Nck SH3[2] binding proteins.
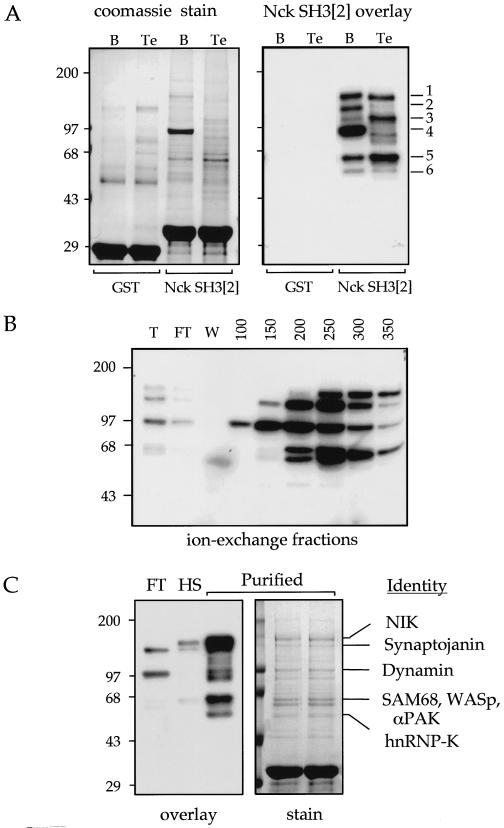
GST-SH3 affinity chromatography has been used to purify and identify many target proteins. Using total cytosolic extracts, numerous bands were eluted (compared with GST alone), although only a subset of these represent direct targets for Nck SH3[2] (Fig. 2A). This confirmed that proteins detected in overlays do interact in solution; only the ∼100-kDa brain protein (dynamin) was sufficiently pure for identification. Ion-exchange fractionation prior to affinity chromatography reduced nonspecific components (compare Fig. 2A and C), with the fractions monitored by SH3 overlay (Fig. 2B). The p140 and p160 bands were clearly separated under these conditions. The relative intensity of the p60 band increased during purification, possibly due to proteolysis of the p68 band (see Discussion). The Q-Sepharose brain fraction (150 to 350 mM NaCl fraction), depleted of dynamin, was applied to a 1-ml Nck SH3[2]-CNBr Sepharose column, and SDS-released material was separated on a 7.5% polyacrylamide gel. Sufficient protein was obtained for conventional tryptic peptide microsequencing. Corresponding peptide sequences matched the proteins shown in Fig. 2C. Of these, p145 synaptojanin had not previously been identified as a Nck partner. From comparison of signals obtained by overlays with their Coomassie blue-stained counterparts, we conclude that in brain extracts NIK, synaptojanin, and the p68s (SAM68, N-WASP, and PAK) bind most tightly.
FIG. 2.
Purification of Nck binding proteins. (A) Detection of binding proteins from brain (B) and testis (Te) extracts. Total cytosolic extracts were passed through columns containing immobilized GST or GST-Nck SH3[2] as indicated. Proteins eluted with SDS treatment (left) were analyzed by GST–Ras-Nck SH3[2] binding. Differences between brain and testis samples are the p140 brain and the testis p110 binders. Numbers on the left are the molecular masses (in kilodaltons) of the markers, and those on the right indicate the positions of the six Nck binding proteins. (B) Sepharose Q ion-exchange fractionation of brain proteins was carried out as described in Materials and Methods. T, total extract; FT, flowthrough fraction; W, wash fraction; HS, high-salt fraction. The Nck SH3[2] binding proteins were assayed using 20 μl of material from each fraction. (C) Identification of Nck binding proteins by protein microsequencing. Bands were excised from the Coomassie blue-stained gels as shown and processed for tryptic digestion, high-performance liquid chromatography separation, and Edman microsequencing. At least three peptides with unambiguous sequences were used to identify the proteins as shown.
Sequence requirement for efficient Nck SH3[2] binding.
The smallest region of NIK previously shown by yeast two-hybrid analysis to bind Nck encompassed residues 443 to 620, which contains two proline-rich regions apparently involved in binding Nck (53). We expressed these sequences individually as 18-amino-acid peptides fused to GST (Fig. 3A) and tested them by overlay for binding to Nck SH3[2] (Fig. 3B). NIK18-2 resembles the proline-rich sequence PAK18-1 (Fig. 3A) that binds Nck SH3[2] (49), and both contain the PXXPXRXXS consensus sequence previously derived for Nck SH3[2] (43). Interestingly, PAK18-1 binds as efficiently as full-length PAK (Fig. 3C), suggesting that its binding was not further stabilized by the peptide being present within a larger domain. A smaller, 13-residue peptide, PAK13, also bound Nck SH3[2], albeit with fourfold-reduced signal (Fig. 3C); thus, the five residues N terminal to PAK13, though not essential, do contribute to binding. Significantly, PAK13m, in which an acidic residue replaces S21, gave no signal.
Stable complexes between Nck SH3[2] and target peptides in vitro.
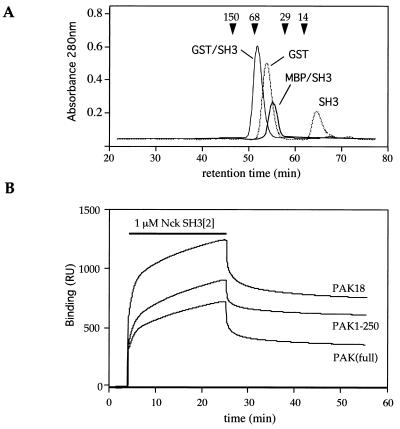
To directly assess the binding of Nck SH3[2] to target sequences, we next used surface plasmon resonance analysis. Since the dimerization behavior of GST fusion proteins in solution can contribute to spurious Biacore measurements (21), Nck SH3[2] was purified as a fusion with MBP. The relative sizes of these proteins were determined by gel filtration (Fig. 4A). The MBP-SH3 protein behaved as a monomer under these conditions. This fusion protein was then tested against GST-PAK, GST-PAK1-250, and GST-PAK18 immobilized to the biosensor chip (approximately equal mass coupling). The trace shows binding at 1 μM MBP-Nck SH3[2], which exhibits biphasic kinetics (Fig. 4B). Considering the coupling efficiency and molecular mass differences, the extents and rates of binding were essentially identical. The binding showed a component with an unusually slow dissociation phase. Similar results were obtained with NIK18 (data not shown).
FIG. 4.
Biacore analysis of the interaction of SH3 domains with target sequences. (A) Superdex200 gel filtration analysis of GST-SrcSH3. The thrombin-cleaved material (dotted line) was treated with 10 U of thrombin per ml for 1 h at room temperature to release free SH3. The GST proteins are present exclusively as dimers, while MBP-Nck SH3[2] exhibits retention corresponding to a monomeric ∼50-kDa protein. Analytical gel filtration was conducted on the Smart system (Pharmacia) in HBS at 30 μl/min at 20°C. Standard protein markers (Sigma) are indicated by arrowheads: lysozyme, 14 kDa; carbonic anhydrase, 29 kDa; serum albumin, 66 kDa; and alcohol dehydrogenase 150 kDa. (B) Coupling (rU values) to CM5 channels was as follows: control channel GST, 2,110; GST-PAK18, 1,650; GST-PAK1-250, 1,760; and GST-PAKL404S, 2,280. The sensorgram shows the addition of 1 μM MBP-Nck SH3[2] at a flow rate of 10 μl/min.
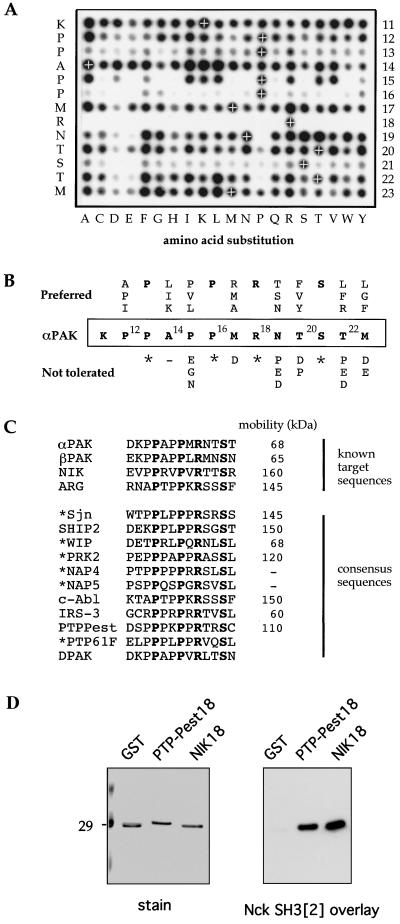
Substitution analysis of PAK(11-23) binding to Nck SH3[2].
All 260 peptides corresponding to the core binding sequence (PAK13) and containing every possible single-point substitution were synthesized on cellulose and assayed by Nck SH3[2] overlay (Fig. 5A). This analysis reveals both preferred residues and nontolerated substitutions and is more informative than alanine scanning substitution. Following a 10-min exposure to the storage screen, spot values were quantified: wild-type peptide sequences yielded (7.2 ± 1.4) × 106 U (n = 13). Residues producing the strongest binding (i.e., preferred) or values of <1 × 106 at a given position are presented in Fig. 5B. The requirement for prolines (particularly P13 and P16) within the sequence is not unexpected, but two novel aspects were noted. First, the conserved R18 is absolutely required for binding (K18 is not accepted). Second, C-terminal to this arginine, neither proline nor acidic residues are tolerated, suggesting that both structure and charge can regulate binding. The fact that serine is clearly preferred by Nck SH3[2] at αPAK21 has already been noted (43). The PAK18-1 (wild-type) sequence is optimal for Nck binding at most positions: stronger binding by NIK18-2 than by PAK18-1 can be attributed to an arginine in the position equivalent to PAK M22 (Fig. 3A). The failure of NIK18-1 (DPCPPSRSEGL) to bind Nck (Fig. 3B) is related to poorly tolerated residues (underlined). Within the αPAK sequence, S21 is known to be autophosphorylated (30); substitution by acidic residues completely blocks binding (also see Fig. 3C).
FIG. 5.
Positional residue preferences for peptide binding to Nck SH3[2]. (A) Synthetic peptides corresponding to the αPAK Nck binding sequence (indicated on the left and marked by white crosses) and containing all possible point substitutions (260 peptides) were synthesized in an array. The filter was blocked with 1% bovine serum albumin and probed with 32P-labeled Nck SH3[2] under standard conditions. A 10-min exposure is shown. (B) From phosphorimager analysis of panel A, preferred (highest counts) or required (boldface) and poorly tolerated (<20% of wild type) peptides are tabulated. Note that the presence of proline or acid residues in the C-terminal half of the peptide drastically reduces binding. (C) Identification of putative binding sequences in known Nck targets. Mammalian proteins identified by PHI-Blast (58) using the PAK 13 sequence and the motif P-X-[AIKLPRTV]-P-X-R-[exclude DEP]-[exclude DEP]-S. Matches were found in all targets that had previously been shown to directly bind Nck (asterisks), while other proteins with the potential to bind Nck are also shown. The conserved residues are highlighted in boldface. (D) Nck SH3[2] can bind to the proline-rich region of PTP-PEST (328 to 344); as a positive control, the NIK18 peptide (2 μg) was corun on a 10% polyacrylamide gel and analyzed by overlay for binding). Asterisks represent sites where only 1 residue is tolerated.
Nck SH3[2] binding proteins are identified from sequence analysis.
Other than PAKs, only the tyrosine kinase Arg has an Nck binding site (residues 659 to 671), which has been precisely mapped (55) and which we now predict binds via SH3[2]. The motif PXXPXRXXS, but conforming to the PAK peptide-based consensus, was detected in many Nck partners (aligned in Fig. 5C), including synaptojanin 1, Nck-associated protein 4 (NAP4) and NAP5 (34), PRK2 (43), and WASP-interacting protein (WIP). Novel candidate binders were detected by PHI-Blast (58), including the phosphatidylinositol polyphosphate-5-phosphatase SHIP2, the p60 IRS-3, the p150 c-Abl, and members of the PTP-PEST tyrosine phosphatase family. Interestingly, a Drosophila protein, dPTP61F, is a potential partner for the Drosophila Nck homologue (10) and contains a consensus binding site for Nck SH3[2]; no direct mammalian dPTP61F homologue is known.
We were interested in the possibility that p110 (Fig. 1; also see Fig. 7) might correspond to proteins of the PTP-PEST family which associate with FCs through paxillin (51). When tested, the proline-rich sequence derived from PTP-PEST indeed bound Nck SH3[2] as avidly as did NIK18 (Fig. 5D). Thus, it is likely that the proteins listed in Fig. 5C indeed have the potential to bind Nck.
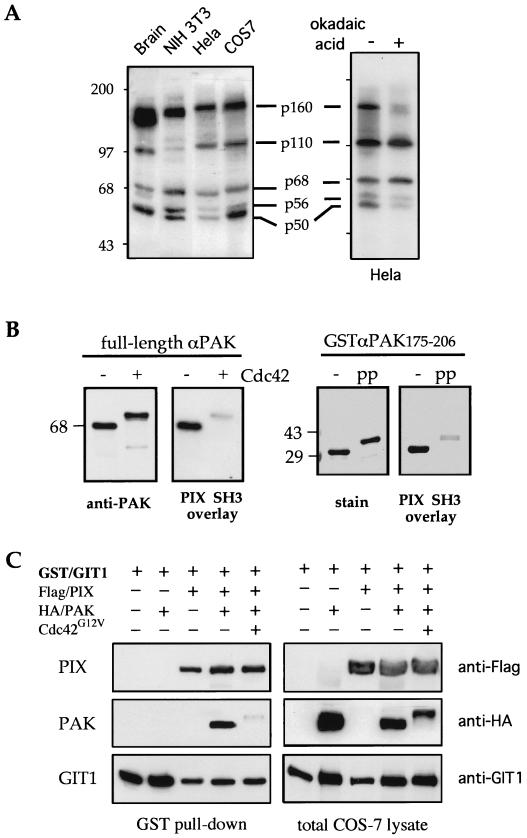
FIG. 7.
Identifying Nck and PAK interactions that are negatively regulated by phosphorylation. (A) Nck SH3[2] binding proteins detected in total proteins (80 μg/lane) from cultured cells are compared to those in brain extract. The sizes of the various targets are indicated; of these, p160 is suggested to represent NIK and p68 is suggested to represent αPAK. The identities of the relatively abundant p110 and p50 species are unclear. The right gel shows reduced Nck SH3[2] binding to HeLa cell extracts following 100 nM okadaic acid treatment (30 min) to induce general phosphorylation. Proteins with mobilities corresponding to 110 and 68 kDa showed no change; however, the p160, p60, and p58 bands showed significant loss of binding. These data are representative of two experiments. (B) Immunoprecipitated αPAK was activated in vitro with GTPγS-Cdc42 and probed with Ras-PIX SH3. Loss of binding correlated with autophosphorylation. The GST-αPAK175-206 PIX-binding peptide (2 μg) was incubated with active recombinant αPAK (0.4 μg) plus 0.5 mM ATP (1 h; 30°C). The complete phosphorylation of S198 and S203 is indicated by its shifted mobility (phosphoprotein [pp]). The PIX SH3 overlay shows a decrease in binding due to this phosphorylation. (C) PAK binding to PIX-GIT1 is negatively regulated by kinase activation. COS-7 cells were transfected with combinations of expression plasmids as shown. The right gels show each of the proteins detected in total lysates (80 μg); the left gels show proteins associating with the expressed GST-GIT1. αPAK activation by Cdc42G12V is accompanied by a mobility shift and loss of binding. +, present; −, absent.
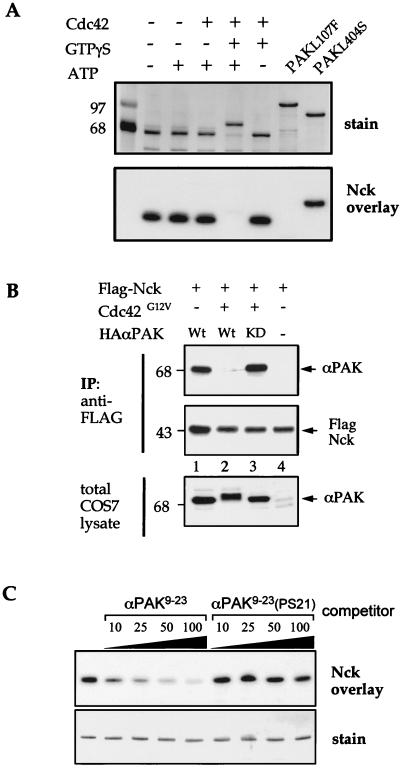
PAK autophosphorylation regulates Nck binding.
Since certain substitutions by acid residues (Fig. 5A) were deleterious to PAK binding, this suggested that phosphorylation within these sequences might provide a mechanism to regulate such binding. Indeed, purified COS-7-derived recombinant αPAK loses its ability to bind Nck SH3[2] upon activation by GTPγS-Cdc42 in an ATP-dependent manner (Fig. 6A). Similarly, recombinant (autophosphorylated) GST-PAKL107F did not bind to Nck. The disruptive effect of phosphorylation on the PAK-Nck complex was also borne out by in vivo analysis. Complexes between PAK and Nck occur in cultured cells (14), and also when they are coexpressed (Fig. 6B). The expression of Cdc42G12V with PAK (and Nck) leads to kinase activation (as detected by an upward shift in mobility), but Nck immunoprecipitates no longer contain PAK (Fig. 6B, lane 2). Cdc42G12V does not promote dissociation of the PAK-Nck complex with the catalytically inactive αPAKK298A (lane 3), showing that the effect is indeed mediated through autophosphorylation. Thus, within cells it seems likely that following recruitment by GTP-Cdc42 or GTP-Rac1, PAK-Nck complexes dissociate upon PAK activation.
FIG. 6.
Phosphorylation of PAK regulates Nck binding. (A) Effect of Cdc42-mediated activation of PAK on Nck SH3[2] binding. The top gel shows Coomassie blue-stained COS-7-expressed Flag-αPAK immunoprecipitated and incubated (30 min; 30°C) in the presence (+) of 2 μg of recombinant Cdc42 with (+) 0.1 mM GTPγS or GDP and/or 0.2 mM ATP as indicated. Neither Cdc42-activated αPAK nor intrinsically active recombinant GST-αPAKL107F binds Nck. As a control, the unphosphorylated GST-αPAKL404S (1 μg) bound normally. (B) PAK activation in vivo leads to dissociation of αPAK and Nck. COS-7 cells were transfected with 3 μg of the constructs per 100-mm-diameter plate as shown. The expression of αPAK is shown in the lower gel. Flag-Nck immunoprecipitates contain αPAK as expected, but not when the kinase is activated. As a control, kinase-dead (KD) αPAK(K298A) was found to be equally associated with Nck even with Cdc42G12V present. Wt, wild type. (C) Phosphorylation of S21 blocks Nck binding. Increasing concentrations (micromolar) of synthetic peptides corresponding to PAK9-23 or its phospho-S21 counterpart were coincubated with the Nck-SH3[2] probe. Each lane of the upper gel corresponds to a strip of filter incubated separately with the probe. The phosphorylated variant was unable to compete for binding to the GST-PAK1-250 target (0.5 μg per lane).
In peptides derived from the N terminus of αPAK, S21 is not phosphorylated by exogenous active PAK (data not shown), but this represents an intramolecular phosphorylation site in αPAK (30). We therefore tested the notion that S21 phosphorylation plays a key role by using a synthetic PAK9-23 peptide and its S21-phosphorylated counterpart to compete with Nck SH3[2] probe for binding to immobilized GST-PAK1-250 (Fig. 6C). Over the concentration range tested (10 to 100 μM), synthetic PAK9-23 peptide, but not the phosphorylated version of the peptide, was effective as a competitor.
Is phosphorylation widely used as a regulator of SH3 interactions?
Since all of the Nck binding sites we identified (Fig. 5C) have the potential for regulation by phosphorylation, we were interested in determining which of them might exhibit such an effect in vivo. Figure 7A, left, shows that similar Nck targets exist in cultured cells. We therefore briefly treated HeLa cells with okadaic acid (inhibiting serine-threonine phosphatase activity) and tested total lysates by overlay for Nck binding. The p50 and p160 (NIK) bands were clearly reduced by such treatment (a representative result is shown in Fig. 7A). The p68 band was neither reduced nor shifted, indicating that PAK was not activated under these conditions. This method has the potential to rapidly detect regulatable SH3 interactions but does not identify all such events.
Interestingly, activation of αPAK also substantially reduces PIX SH3 binding (Fig. 7B). Since the PIX-binding peptide αPAK175-206 is phosphorylated by exogenous PAK, we could demonstrate that it is the phosphorylation of GST-PAK175-206 (as monitored by mobility shift after incubation with the kinase) that is responsible for the loss of PIX binding, which was similar to autophosphorylated full-length αPAK (Fig. 7B, left). Within the peptide, the only two serines (S198 and S203) are known PAK autophosphorylation sites (30). We then investigated the interaction of PAK and PIX during kinase activation in vivo. To more faithfully replicate the cellular context, we also introduced GIT1 (4, 42), which we find to be constitutively bound to βPIX and phosphorylated by PAK (data not shown). αPAK associated with GIT1 in a βPIX-dependent manner when relevant combinations were expressed in COS-7 cells (Fig. 7C). However, in cells expressing Cdc42G12V (lane 5), only a small fraction of the autophosphorylated PAK was found associated with the complex, though there was no change in the binding of PIX to GIT1.
FC targeting of PAK is under dynamic regulation.
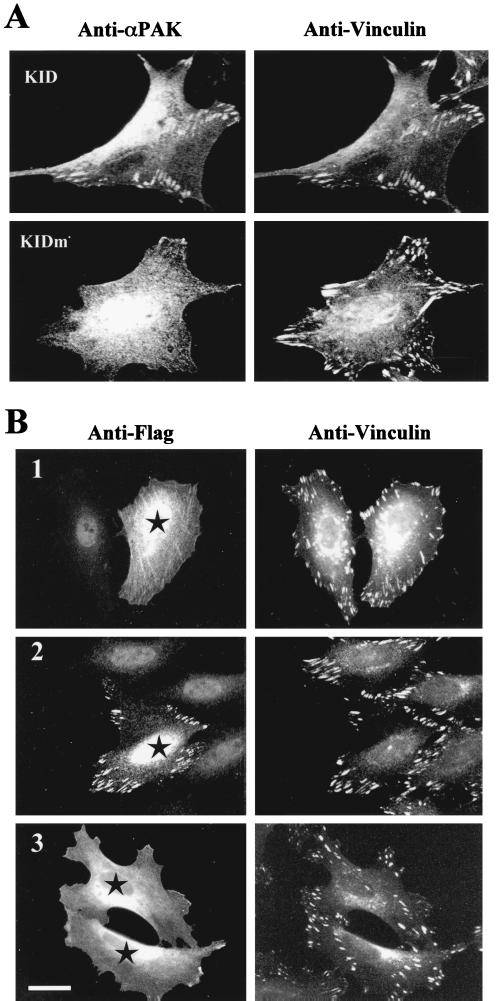
We previously demonstrated that PAK binding to PIX is necessary but not sufficient for localization to FCs (32). Taking this together with our observations that both Nck and PIX binding are negatively regulated by phosphorylation, we considered a model in which activation of PAK prevents its constitutive association with FCs, resulting in dynamic equilibrium between the bound and cytosolic states (Fig. 8). Since PAK associates poorly with the RhoA-type FCs found in resting cells, we tested this notion by transfecting a PAK kinase inhibitor domain, KID (PAK83-149), into NIH3T3 cells and then localized the endogenous αPAK, which indeed became associated with FCs (Fig. 9A). By contrast, the inactive mutant KIDm(L107F)-expressing cells showed normal perinuclear PAK staining. Similarly, when we introduced Flag-αPAK into HeLa cells (Fig. 9B, image 1), the kinase inhibitor caused a shift in equilibrium towards FCs (image 2). Interestingly Flag-ΔN22αPAK, lacking the Nck binding sequence, failed to efficiently bind FCs when cotransfected with KID (image 3). Taken together, these results explain previous observations that N-terminal PAK1-250, but not the wild-type kinase, is targeted to RhoA-type FCs (30).
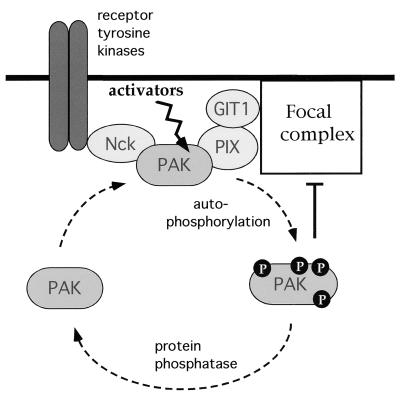
FIG. 8.
PAK undergoes dynamic regulation of its interactions. PAK is capable of forming complexes with Nck and PIX through distinct proline-rich sequences. The kinase can be recruited to juxtamembrane sites either through the interaction of Nck with various tyrosine-phosphorylated receptors or by the interaction of PIX with elements of FCs. Activation of PAK requires autophosphorylation and causes FC disassembly; however, the kinase also loses affinity for its partners, resulting in PAK cycling back to the cytosol.
FIG. 9.
Regulation of PAK association with FCs. (A) Inhibition of PAK leads to an accumulation of PAK in focal adhesions. NIH 3T3 cells were transfected with KID PAK83-149 and stained with anti-αPAK and anti-vinculin. Only in cells expressing the inhibitor was there clear colocalization of PAK with the underlying focal adhesions. The inactive KID(L107F) mutant (KIDm) was used as a control. (B) HeLa cells were transfected (stars) as follows: Flag-αPAK (image 1), Flag-αPAK plus GST-KID (59) (image 2), and Flag-ΔN22αPAK plus GST-KID (image 3). Immunofluorescent detection of rabbit anti-Flag (Upstate Biotechnology Inc.) and mouse anti-vinculin was then performed. With inhibitor, the αPAK robustly stained vinculin-rich FCs, but PAK lacking the first 22 residues (i.e., the Nck binding site) was poorly localized to these structures. Bar, 10 μm.
DISCUSSION
The molecular basis of SH3 binding interactions.
SH3 domains occur in at least 87 distinct human proteins (48). Since the report of a 3BP-1 proline-rich sequence as the target for the Abl-SH3 domain (9), numerous proline-rich binding partners for SH3 domains have been described, all containing at their cores the binding motif PXXP (41). The only known naturally occurring exception is the central PIX SH3 binding sequence (PPPVIAPRPEHTKS) in αPAK and βPAK (3, 32). On the C-terminal site of this motif, the S198 is an autophosphorylation site that in this study we show to negatively regulate binding.
The structures of several SH3-peptide complexes have been available for some time (56, 57), but the role of determinants outside of the core sequence is less well understood. Src, Fyn, and Yes SH3 domain-selected type II peptides display absolute specificity for asparagine at the +1 position adjacent to the conserved arginine (46). For Hck-SH3 binding to human immunodeficiency virus type 1 Nef, important contacts are provided by the RT loop of the SH3 domain, increasing both the specificity and affinity of binding (23). The ability of Nef to bind Hck-SH3, but not the related Fyn-SH3, is apparently determined by a single isoleucine within this loop (22). A more recent study indicates that residues within the RT loop reorient to form an “induced fit” with the PXXP flanking a helical region of Nef (1). This may be the basis for the slow kinetics of tight binding that we observe with Nck SH3[2]. It is also possible that the S- or T-rich C-terminal region in Nck SH3[2] binding sequences (cf. PAK18) needs to adopt a helical conformation, which would explain why proline substitution blocks binding (Fig. 5A).
The Nck SH3[2] domain engages selectively.
The adaptor proteins of the Grb2, Crk, and Nck families, which contain only SH2 and SH3 domains, play pivotal roles in many signal transduction cascades (41) and perhaps for this reason can interact with multiple SH3 targets. The best studied of these is Grb2, which was first identified as associating with Sos1 (and -2) via interactions of both SH3 domains, although the N-terminal domain contribution is more important. Affinity may not be the primary issue, since it has been demonstrated that small peptides containing N-substituted residues at positions normally occupied by prolines can yield Grb2 SH3 ligands with much higher affinities (Kd up to 40 nM, some 100 times that of the natural Sos-derived 12-mer peptide) (38).
The Nck SH3[2] domain achieves selectivity in its choice of targets through particular interactions C-terminal to the PXXP motif. Because neither proline or acidic residues are tolerated in these C-terminal positions, this serves to seriously limit the availability of targets. Many “proline-rich” sequences in the database containing PXXPXRXXS motifs also contain proline or acidic residues at inappropriate positions. By contrast, the conserved sites in the PTP-PEST family phosphatases which are recognized by the Csk SH3 domain include acidic and proline residues (underlined) in corresponding positions (PPLPERTPESFIV) that facilitate binding (17).
Identifying biologically relevant Nck SH3 partners.
The prototype target for the Abl SH3 domain (9) was identified by ligand overlay expression screening, which has been used successfully in many subsequent studies (33, 36, 43). One drawback is that many SH3 domains bind to the same proteins in vitro, for example, SH3 domains from p85α, PLCγ, c-Src, fgr, Grb2, and fyn (∼50% of those tested) all bind dynamin robustly (16), although it is unlikely that they all regulate dynamin function. One approach is to consider the strength of interaction: it is likely that there is in vivo competition for SH3 binding among a variety of potential partners. Synthetic libraries have therefore been screened to define the optimal binding sequence with a view to identifying these sequences in target proteins. For example, using the cortactin SH3 domain, a PPXPXKP consensus was derived (52). This motif is indeed present in known cortactin targets, 180-kDa CortBP1 (12) and the recently described CBP90 (40), which probably corresponds to 80- and 85-kDa binding proteins detected in brain extracts (Fig. 1B).
Partners for Nck experimentally identified in our study include NIK, synaptojanin-1, PTP-PEST, αPAK, αPAK, and hnRNP-K. Of these, synaptojanin, PTP-PEST, and hnRNP-K have not previously been reported to bind Nck. Synaptojanin is an inositol 5′-phosphatase present in nerve terminals (36), which is interesting in view of the roles of Drosophila Nck and Dock in axonal guidance. The more ubiquitous synaptojanin-2 does not contain a Nck binding sequence but instead interacts with Grb2 (37). PTP-PEST probably corresponds to the p110 protein seen in testis, spleen, and thymus, as this protein, tyrosine phosphatase, is enriched in the immune system. The other known Nck binding proteins, WIP, PRK2, NAP-4, and NAP-5, contain suitable target sequences (Fig. 5) but are probably lower-abundance proteins. Except in the case of PAK, we have yet to establish whether these proteins are in vivo Nck targets; the availability of Nck-deficient cells may help to resolve this issue in the future.
The SH3[2] domain and Nck function in vivo.
The importance of the SH3[2] domain is exemplified in Drosophila, where photoreceptor cell projection to the optic ganglia requires the Nck homologue Dock. In a dock-null background, expression of Dock-containing mutations in SH3[1], SH3[3], or SH2 restores the projection, but mutants of the SH3[2] domain do not (44). Myristoylated PAK can rescue the loss of Dock, indicating that PAK is a key downstream component (19). This rescue requires the function of both the p21-binding and kinase domains of PAK. Dock can interact with the protein tyrosine phosphatase dPTP61F (10). A putative Nck SH3[2] consensus binding sequence PPPLPPRVQSLN335 is present in dPTP61F. Likewise, the members of the mammalian PTP-PEST family contain similar Nck binding sequences.
The ability of membrane-targeted Nck to promote FC disassembly in cultured mammalian cells through its SH3[2] PAK-binding domain (data not shown) could be relevant to the function of Nck in the nervous system. R-cell projection patterns are abnormal in Drosophila Pak mutants, with axons forming unusually thick bundles (19). PC12 cells expressing kinase-inactive βPAK exhibit stunted neurite outgrowth patterns associated with an increase in cellular FCs, whereas overexpression of wild-type βPAK leads to increased neurite outgrowth (39). Growth factors that stimulate neurite outgrowth (which is a Cdc42- and Rac1-dependent process) probably thereby promote FC turnover through PAK.
Regulation of PAK-SH3 interactions.
The affinity of the well-studied Grb2 for SOS is decreased upon phosphorylation of four sites within the proline-rich C-terminal end of SOS by mitogen-activated protein kinases. This provides a means of controlling Ras activation (11). Extracellular signals can also stimulate specific dephosphorylation events that regulate SH3 interactions: a recent example involves β-arrestin recruitment by G-coupled receptors, leading to dephosphorylation of the phospho-S412, thereby allowing interaction with the Src SH3 domain (29).
In the case of Nck, it seems likely that alternative partners are recruited under different conditions. A prevalence of S and T residues C-terminal to the conserved arginine (PXXPXR) in many Nck targets suggests that these are negative regulatory sites. The Nck-PAK-PIX complex will remain stable at its site of action (for example, FCs) only as long as PAK remains in a basal or partially active state (Fig. 8). It appears that activation of Nck-recruited PAK occurs through both Cdc42-dependent and -independent mechanisms (28) and will be affected by local concentrations of GTP-Cdc42 or other factors. Autophosphorylated PAK loses its affinity for PIX, involving phosphorylation of αPAK S198 and S203 (30), which flank the C-terminal side of the PIX SH3 binding site as well as Nck (or perhaps related SH3s). Thus, wild-type kinase cycles between cytosol and PIX-containing FC sites, but when the catalytic activity of PAK is inhibited, it forms a stable association with FCs (Fig. 9). The role of Nck binding for FC localization is presently not clear. PAK therefore plays a role in RhoA-type FCs (because it is continuously cycling through these structures), although at steady state we do not see it concentrated at these sites. Future studies looking at real-time FC dynamics will be able to address this issue. The down-regulation in binding will generally serve to limit PAK's disassembling action on FCs. At present, it is not clear why PAK is observed in Cdc42- and Rac-induced FCs, where the kinase is expected to be most active. Since in their active state these p21s themselves localize to FCs (30), they might be responsible for shifting the PAK distribution.
PAK dissociation also limits the effects of PIX, whose interaction with its unique partner, PAK, is required for GEF activity (39) and thus cytoskeletal effects, e.g., lamellipodium formation (32). Because formation of GTP-Rac1 itself leads to further recruitment of PAK and associated PIX to the membrane, a mechanism to break this positive-feedback cycle is conferred by the loss of PIX binding upon PAK activation. In support of this, we and others have observed that kinase-dead PAK is more efficient in driving Rac-dependent lamellipodium formation through PIX (39, 49) as a consequence of the stabilization of the PIX-PAK complex. That these PAK effects might be independent of Rac1 (50) seems unlikely.
In conclusion, the SH3[2] domain of the adapter protein Nck is capable of forming complexes with a number of target proteins through a preferred motif. The presence of a phosphorylatable serine in the motif may represent a general mechanism to regulate binding. We observe that other SH3 domains exhibit distinct subsets of targets (Fig. 1), probably based on different motifs flanking the PXXP core, which may be dissected using this peptide approach. Among Nck binding proteins, the phosphorylation states of target sequences in vivo are clearly key factors in regulating complex formation. It will be of interest to see if this mechanism is widely utilized.
ACKNOWLEDGMENT
This work is funded by the Glaxo Singapore Research Fund.
REFERENCES
- 1.Arold S, O'Brien R, Franken P, Strub M P, Hoh F, Dumas C, Ladbury J E. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37:14683–14691. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- 2.Bagrodia S, Taylor S J, Creasy C L, Chernoff J, Cerione R A. Identification of a mouse p21Cdc42/Rac activated kinase. J Biol Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 3.Bagrodia S, Taylor S J, Jordon K A, Van Aelst L, Cerione R A. A novel regulator of p21-activated kinases. J Biol Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- 4.Bagrodia S, Bailey D, Lenard Z, Hart M, Guan J L, Premont R T, Taylor S J, Cerione R A. A tyrosine-phosphorylated protein that binds to an important regulatory region on the cool family of p21-activated kinase-binding proteins. J Biol Chem. 1999;274:22393–22400. doi: 10.1074/jbc.274.32.22393. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 6.Booker G W, Gout I, Downing A K, Driscoll P C, Boyd J, Waterfield M D, Campbell I D. Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase. Cell. 1993;73:813–822. doi: 10.1016/0092-8674(93)90259-s. [DOI] [PubMed] [Google Scholar]
- 7.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 8.Chou M M, Fajardo J E, Hanafusa H. The SH2- and SH3-containing Nck protein transforms mammalian fibroblasts in the absence of elevated phosphotyrosine levels. Mol Cell Biol. 1992;12:5834–5842. doi: 10.1128/mcb.12.12.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cicchetti P, Mayer B J, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- 10.Clemens J C, Ursuliak Z, Clemens K K, Price J V, Dixon J E. A Drosophila protein-tyrosine phosphatase associates with an adapter protein required for axonal guidance. J Biol Chem. 1996;271:17002–17005. doi: 10.1074/jbc.271.29.17002. [DOI] [PubMed] [Google Scholar]
- 11.Corbalan-Garcia S, Yang S S, Degenhardt K R, Bar-Sagi D. Identification of the mitogen-activated protein kinase phosphorylation sites on human Sos1 that regulate interaction with Grb2. Mol Cell Biol. 1996;16:5674–5682. doi: 10.1128/mcb.16.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Y, Weed S A, Xiong W C, Marshall T D, Parsons J T. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol Cell Biol. 1998;18:5838–5851. doi: 10.1128/mcb.18.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 14.Galisteo M L, Chernoff J, Su Y C, Skolnik E Y, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 15.Garrity P A, Rao Y, Salecker L, McGlade J, Pawson T, Zipursky S L. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- 16.Gout I, Dhand R, Hiles L D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, Campbell I D, Waterfield M D. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 17.Gregorieff A, Cloutier J F, Veillette A. Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50(csk) J Biol Chem. 1998;273:13217–13222. doi: 10.1074/jbc.273.21.13217. [DOI] [PubMed] [Google Scholar]
- 18.Harden N, Lee J, Loh H Y, Ong Y M, Tan I, Leung T, Manser E, Lim L. A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol Cell Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hing H, Xiao J, Harden N, Lim L, Zipursky S L. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 20.Koyama S, Yu H, Dalgarno D C, Shin T B, Zydowsky L D, Schreiber S L. Structure of the PI3K SH3 domain and analysis of the SH3 family. Cell. 1993;72:945–952. doi: 10.1016/0092-8674(93)90582-b. [DOI] [PubMed] [Google Scholar]
- 21.Ladbury J E, Lemmon M A, Zhou M, Green J, Botfield M C, Schlessinger J. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc Natl Acad Sci USA. 1995;92:3199–3203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C H, Leung B, Lemmon M A, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 1995;14:5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C H, Saksela K, Mirza U A, Chait B T, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 24.Lemmon M A, Ladbury J E, Mandiyan V, Zhou M, Schlessinger J. Independent binding of peptide ligands to the SH2 and SH3 domains of Grb2. J Biol Chem. 1994;269:31653–31658. [PubMed] [Google Scholar]
- 25.Li W, Hu P, Skolnik E Y, Ullrich A, Schlessinger J. The SH2 and SH3 domain-containing Nck protein is oncogenic and a common target for phosphorylation by different surface receptors. Mol Cell Biol. 1992;12:5824–5833. doi: 10.1128/mcb.12.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim W A, Richards F M, Fox R O. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 27.Lu W, Katz S, Gupta R, Mayer B J. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 28.Lu W, Mayer B J. Mechanism of activation of Pak1 kinase by membrane localization. Oncogene. 1999;18:797–806. doi: 10.1038/sj.onc.1202361. [DOI] [PubMed] [Google Scholar]
- 29.Luttrell L M, Ferguson S S, Daaka Y, Miller W E, Maudsley S, Della Rocca G J, Lin F, Kawakatsu H, Owada K, Luttrell D K, Caron M G, Lefkowitz R J. Beta-arrestin-dependent formation of β2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 30.Manser E, Huang H-Y, Loo T-H, Chen X-Q, Dong J-M, Leung T, Lim L. Expression of constitutively active αPAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manser E, Leung T, Salihuddin H, Zhao Z-S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 32.Manser E, Loo T-H, Koh C-G, Zhao Z-S, Chen X Q, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda M, Ota S, Tanimura R, Nakamura H, Matuoka K, Takenawa T, Nagashima K, Kurata T. Interaction between the amino-terminal SH3 domain of CRK and its natural target proteins. J Biol Chem. 1996;271:14468–14472. doi: 10.1074/jbc.271.24.14468. [DOI] [PubMed] [Google Scholar]
- 34.Matuoka K, Miki H, Takahashi K, Takenawa T. A novel ligand for an SH3 domain of the adaptor protein Nck bears an SH2 domain and nuclear signaling motifs. Biochem Biophys Res Commun. 1997;239:488–492. doi: 10.1006/bbrc.1997.7492. [DOI] [PubMed] [Google Scholar]
- 35.Mayer B J. Signal transduction: clamping down on Src activity. Curr Biol. 1997;7:295–298. doi: 10.1016/s0960-9822(06)00141-2. [DOI] [PubMed] [Google Scholar]
- 36.McPherson P S, Czernik A J, Chilcote T J, Onofri F, Benfenati F, Greengard P, Schlessinger J, De Camilli P. Interaction of Grb2 via its Src homology 3 domains with synaptic proteins including synapsin I. Proc Natl Acad Sci USA. 1994;91:6486–6490. doi: 10.1073/pnas.91.14.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemoto Y, Arribas M, Haffner C, DeCamilli P. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J Biol Chem. 1997;272:30817–30821. doi: 10.1074/jbc.272.49.30817. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen J T, Turck C W, Cohen F E, Zuckermann R N, Lim W A. Exploiting the basis of proline-recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 39.Obermeier A, Ahmed S, Manser E, Yen S C, Hall C, Lim L. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 1998;17:4328–4339. doi: 10.1093/emboj/17.15.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohoka Y, Takai Y. Isolation and characterization of cortactin isoforms and a novel cortactin-binding protein, CBP90. Genes Cells. 1998;3:603–612. doi: 10.1046/j.1365-2443.1998.00216.x. [DOI] [PubMed] [Google Scholar]
- 41.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 42.Premont R T, Claing A, Vitale N, Freeman J L, Pitcher J A, Patton W A, Moss J, Vaughan M, Lefkowitz R J. β2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quilliam L A, Lambert Q T, Mickelson-Young L A, Westwick J K, Sparks A B, Kay B K, Jenkins N A, Gilbert D J, Copeland N G, Der C J. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem. 1996;271:28772–28776. doi: 10.1074/jbc.271.46.28772. [DOI] [PubMed] [Google Scholar]
- 44.Rao Y, Zipursky S L. Domain requirements for the Dock adapter protein in growth-cone signaling. Proc Natl Acad Sci USA. 1998;95:2077–2082. doi: 10.1073/pnas.95.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 46.Rickles R J, Botfield M C, Zhou X-M, Henrey P A, Brugge J S, Zoller M J. Phage display selection of ligand residues important for Src homology domain binding specificity. Proc Natl Acad Sci USA. 1995;92:10909–10913. doi: 10.1073/pnas.92.24.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sastry L, Lin W, Wong W T, Di Fiore P P, Scoppa C A, King C R. Quantitative analysis of Grb2-Sos1 interaction: the N-terminal SH3 domain of Grb2 mediates affinity. Oncogene. 1995;11:1107–1112. [PubMed] [Google Scholar]
- 48.Schultz J, Milpetz F, Bork P, Ponting C P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 50.Sells M A, Boyd J T, Chernoff J. p21-activated kinase 1 (Pak1) regulates cell motility in mammalian fibroblasts. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen Y, Schneider G, Cloutier J F, Veillette A, Schaller M D. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J Biol Chem. 1998;273:6474–6481. doi: 10.1074/jbc.273.11.6474. [DOI] [PubMed] [Google Scholar]
- 52.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quillam L A, Kay B K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCγ, Crk, and Grb2. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su Y C, Han J, Xu S, Cobb M, Skolnik E Y. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, Matsuda M. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang B, Mysliwiec T, Feller S M, Knudsen B, Hanafusa H, Kruh G D. Proline-rich sequences mediate the interaction of the Arg protein tyrosine kinase with Crk. Oncogene. 1996;13:1379–1385. [PubMed] [Google Scholar]
- 56.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 57.Yu H, Rosen M K, Shin T B, Seidel-Dugan C, Brugge J S, Schreiber S L. Solution structure of the SH3 domain of Src and identification of its ligand-binding site. Science. 1992;258:1665–1668. doi: 10.1126/science.1280858. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z, Schaffer A A, Miller W, Madden T L, Lipman D J, Koonin E V, Altschul S F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Z-S, Manser E, Chen X-Q, Chong C, Leung T, Lim L. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]