Abstract
An important component underlying the disparity in HIV risk between race/ethnic groups is the preferential transmission between individuals in the same group. We sought to quantify transmission between different race/ethnicity groups and measure racial assortativity in HIV transmission networks in major metropolitan areas in the United States. We reconstructed HIV molecular transmission networks from viral sequences collected as part of HIV surveillance in New York City, Los Angeles County, and Cook County, Illinois. We calculated assortativity (the tendency for individuals to link to others with similar characteristics) across the network for three candidate characteristics: transmission risk, age at diagnosis, and race/ethnicity. We then compared assortativity between race/ethnicity groups. Finally, for each race/ethnicity pair, we performed network permutations to test whether the number of links observed differed from that expected if individuals were sorting at random. Transmission networks in all three jurisdictions were more assortative by race/ethnicity than by transmission risk or age at diagnosis. Despite the different race/ethnicity proportions in each metropolitan area and lower proportions of clustering among African Americans than other race/ethnicities, African Americans were the group most likely to have transmission partners of the same race/ethnicity. This high level of assortativity should be considered in the design of HIV intervention and prevention strategies.
Keywords: HIV, phylogenetics, network, race, ethnicity, transmission, MSM
Background
The lifetime risk of contracting HIV-1 for African American men is 1 in 20, seven-times higher than the lifetime risk for white men in the United States.1 African Americans represent only 12% of the U.S. population, but 43% of people living with HIV.2
The drivers of this disparity are numerous and a detailed assessment was presented by Adimora et al.3 An important component of this disparity is that differences in HIV risk can be sustained and amplified when transmission preferentially occurs among individuals with similar characteristics to themselves: a phenomenon described as “preferential assortativity.”4 Structural determinants such as residential segregation5 and the ratio of men to women in local populations6 are further amplified and compounded within the African American community as a whole. Therefore, sexual network structure can exacerbate HIV risk among African American populations.3,7
Interviews and surveys have been used to ascertain levels of interracial and intraracial sexual sorting in HIV-infected and at-risk populations,8–12 but these studies are resource-intensive and possible only for small numbers of participants. The data collected from these studies are egocentric, and thus subject to recall bias and possible incorrect identification of partners' race/ethnicity. With some exceptions, these studies have focused on self-identified men who have sex with men (MSM).8–12 Importantly, sexual partnering is not necessarily indicative of risk for transmission, as individuals may adapt their risk behavior based on perception of their partner's risk. For example, a longitudinal study of young MSM found that young nonblack MSM were more likely to use a condom with young black MSM.9
Molecular epidemiology offers an alternative route to understanding transmission patterns within large populations. Individuals whose HIV sequences are closely related can be considered potential transmission partners and the relationships between groups can be ascertained at the population level.13–17 In New York City (NYC) and other locations around the United States, genetic linkage has proven a more reliable indicator of potential transmission partners than partner naming alone.17–19
African Americans may be more likely to have partners of the same/race ethnicity as themselves than other race/ethnic groups. Studies of sexual networks among MSM in the United States and in the United Kingdom have found three times more homophilic links among black MSM than expected by chance.12,20 A nationwide analysis of the molecular transmission network by race/ethnicity among United States MSM found that 78% of the potential transmission partners of African Americans were also African American, compared with 64% of white MSM and 49% of Latinos having potential transmission partners of the same race/ethnicity.21
This single molecular analysis on assortativity by race/ethnicity focused solely on MSM, excluding a sizable fraction of MSM whose transmission risk may not be accurately recorded in surveillance databases,22,23 as well as other risk groups. Here, we extend this analysis to all risk groups and focus on three distinct, large metropolitan areas to investigate the impact of race/ethnicity composition on sorting patterns in molecular transmission networks.
We reconstructed HIV molecular transmission networks for the three most populous American metropolitan areas: NYC (population 8.5 million), Los Angeles County (LAC; population 10.2 million), and Cook County, which includes Chicago (population 5.2 million). Our aim was to measure transmission between different race/ethnicity groups and describe race/ethnicity specific assortativity within each jurisdiction.
Methods
Surveillance data
The University of California San Diego (UC San Diego) Institutional Review Board (IRB) certified the study as not qualifying as human subjects' research according to the Code of Federal Regulations Title 45 part 46 and UC San Diego Standard Operating Policies and Procedures. The study therefore did not require IRB review.
HIV-1 protease and reverse transcriptase (pol) genetic sequences generated for antiretroviral resistance testing have been routinely reported to surveillance since 2005 in NYC and LAC and since 2012 in Illinois. At the time of analysis, sequences for study were available through March 2018 for NYC, December 2016 for LAC, and June 2018 for Illinois. We restricted our analysis of Illinois data to diagnosed individuals residing in Cook County, as this county represents the urban center most comparable to NYC and LAC.
For each case reported to the local HIV surveillance system, additional clinical and demographic data are collected in the enhanced HIV/AIDS Reporting System (eHARS).24 Data available in eHARS include race/ethnicity, transmission risk (MSM, people who inject drugs [PWID], MSM/PWID, heterosexual, perinatal, other, unknown), age at diagnosis, and date of diagnosis.
The HIV case report form collects data for ethnicity (Hispanic/Latino, or not) and race (American Indian/Alaska Native, Asian, black/African American, Native Hawaiian/Pacific Islander, white or unknown), separately. These data are consolidated into a single race/ethnicity description for reporting by public health agencies, with those identifying as Hispanic/Latino classified as such, and those not identifying as Hispanic/Latino classified based on their race. More than one race can be selected, and those who do are classified as mixed race. In our analysis, Asians, African Americans, whites, and Latinos were analyzed as separate groups; other groups, including those of mixed race, were combined into “other and unknown.”
We grouped age at diagnosis into decades (13–18, 19–24, 25–29, 30–39, 40–49, 50–59, 60+) for analysis. We did not restrict our analysis based on previous antiretroviral drug exposure or remove drug-resistance associated codons, as our network reconstruction algorithm is robust to their inclusion.17,25
As of 2016, 87,700 persons were estimated to be living with HIV in NYC, 60,000 in LAC, and 29,032 in Cook County. Sequences were available for 69,317 in NYC, with sequence completeness (the proportion of newly diagnosed people with a genotype) of 72.3% in the 5 years prior. LAC HIV surveillance had received HIV-1 genetic sequences from 22,860 individuals, with completeness of 59.6% in the 5 years prior. In Cook County, 7,158 sequences were available and completeness was 42.9% in the 5 years prior. For each eligible person, we included the first reported HIV protease/reverse transcriptase genotype sequence. In all three metropolitan areas, some sequences may originate from people who have since died and are not counted among the number of people living with HIV.
The data analyzed here were collected as part of routine HIV surveillance activities and are protected by local statute. The data cannot be submitted to public databases. These data were shared with investigators under data use agreements with UC San Diego.
Transmission network reconstruction
We reconstructed the genetic network for each jurisdiction using HIV-TRACE.26 The earliest reported pol sequence from each individual was aligned to HXB2 (positions 2253–3749) and TN93 pairwise distances were calculated.27 Individuals are linked to each other in the network if their pairwise genetic distance falls below a preselected threshold. We consider potential transmission partners to be those who link to each other in the network. However, we note that genetic networks comprise far more links than the true transmission network, and so most links in the network do not represent true transmission events. For the primary analyses presented here, we used a genetic distance threshold of ≤0.015 substitutions/site (1.5%), because this distance is within the range observed between viruses within a single individual25 and between named sexual partners considered to be transmission partners.17
For sensitivity analyses, we applied genetic distance thresholds of ≤0.01 (1.0%) and ≤0.005 (0.5%) substitutions/site, because a conservative threshold of ≤0.005 substitutions/site has been demonstrated to be more consistent with transmission events in simulations.28
Statistical analysis
In this analysis, we focused on individuals in the four most numerous race/ethnicity categories: African American/black, Asian, Hispanic/Latino, and white. Other race/ethnicity groups (e.g., Native American, Pacific Islanders, multiracial, and so on) were too small to permit robust analysis and were grouped into a single “other” category. The race/ethnicity compositions of each metropolitan area were compared using a χ2 test. The correlation between race/ethnicity and having at least one transmission partner in the genetic network (i.e., clustering) was assessed using univariate logistic regression in each metropolitan area.
Assortativity is a network metric, which describes, for a given characteristic, (e.g., transmission risk factor) the tendency for individuals in the network (represented by nodes) to link to others with the same trait (e.g., do PWIDs tend to cluster with other PWIDs?).29 Assortativity varies between −1 (completely disassortative) and +1 (completely assortative), and in our analysis, it was calculated using the function available in the R igraph package.30 Assortativity can be measured for the trait as whole (e.g., do individuals in the same age category link to each other?) or broken down for each category of that trait (e.g., which age category is most homophilic?).
First, we calculated assortativity for age at diagnosis, transmission risk, and race/ethnicity for each metropolitan area. For each jurisdiction, we generated a null expectation for assortativity by permuting trait labels across the static network 1,000 times and recalculating assortativity. For race/ethnicity analyses, we calculated assortativity for each of the four predominant race/ethnicity groups: African American, Asian, Latino and white. Again, we generated null distributions within each jurisdiction for calculating statistical significance.
Next, we analyzed transmission between race/ethnicity groups by calculating the proportion of genetic links to other race/ethnic groups in each transmission network. To avoid bias based on degree centrality, as some individuals have far more links than others, each potential transmission partner for a given person was assigned a weight based on k (the number of potential transmission partners associated with the person). We counted links for each individual in the network, assigning a weight 1/k to each of their potential transmission partners, as done previously.21 We then permuted labels 1,000 times to generate null distributions and assess statistical significance of observed patterns.
Finally, for the LAC network, we repeated our assortativity and mixing analyses on networks stratified by risk group (MSM, heterosexual, PWID, MSM/PWID), year of diagnosis (2012–2016), and age category (18–24, 25–29, 30–39, 40–49, 50+).
Undersampling bias
Sampling bias can bias assortativity measurements in networks,31 and the lower clustering frequency among African Americans may reflect a pattern of underdiagnosis or lower rates of genotyping in this group.2,32–34 To investigate the effect of potential undersampling of African Americans, we carried out a sensitivity analysis in our midsize network, LAC, by repeatedly (1,000 times) downsampling a proportion (25%, 50% and 75%) of African Americans and recalculating assortativity each time.
Results
The race/ethnicity compositions of NYC, LAC, and Cook County differ
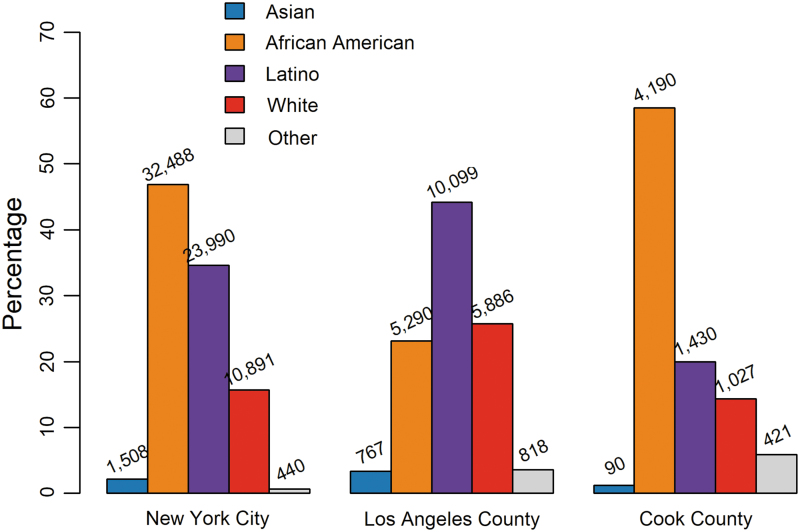
The race/ethnic distribution of the HIV-infected population with reported HIV sequences varied across the three metropolitan areas (Fig. 1; χ2; p < .001). The majority of people living with HIV in LAC with a reported genotype were Latino, whereas in NYC and Cook County the majority of people were African American. The proportion of Asians with a genotype in each jurisdiction was low, ranging between 1.3% and 3.4%. The breakdown of each population by sex, risk group, and age category is shown in Supplementary Table S1; MSM was the predominant reported transmission risk in all three populations.
FIG. 1.
Race/ethnicity composition of people living with HIV with a reported HIV genotype in NYC (2005–2018), LAC (2005–2016), and Cook County (2012–2018). The column heights indicate proportional representation and the number of individuals of each race/ethnicity is displayed above each column. LAC, Los Angeles County; NYC, New York City. Color images are available online.
African Americans are less likely to cluster
The proportion of individuals clustering at the 0.015 substitutions/site distance threshold in LAC and Cook County was 35.6% and 33.2%, respectively, substantially higher than the 22.2% clustering proportion in NYC (Table 1). In NYC, Asians, Latinos, and whites all clustered at higher frequencies than African Americans (p < .001; Table 1; Supplementary Tables S2 and S3). In LAC, Asians and Latinos clustered more frequently than African Americans (p < .001), but there was no difference between African Americans and whites. In Cook County, African Americans clustered less frequently than Latinos (p < .001), but there was no difference with other race/ethnicities.
Table 1.
Proportion of Individuals Clustering and Odds Ratios for Clustering for Each Race/Ethnicity and Each Analyzed Jurisdiction (0.015 Substitutions/Site)
| Race/ethnicity | New York City |
Los Angeles County |
Cook County |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Clustered (%) | OR (95% CI) | Total | Clustered (%) | OR (95% CI) | Total | Clustered (%) | OR (95% CI) | |
| Asian | 1,508 | 513 (34.0%) | 2.4 (2.1–2.6)*** | 767 | 308 (40.2%) | 1.5 (1.3–1.8)*** | 90 | 27 (30.0%) | 0.9 (0.6–1.4) |
| African American | 32,488 | 5,828 (17.9%) | REF | 5,290 | 1,607 (30.4%) | REF | 4,190 | 1,379 (32.9%) | REF |
| Latino | 23,990 | 5,601 (23.3%) | 1.4 (1.3–1.5)*** | 10,099 | 4,074 (40.3%) | 1.5 (1.4–1.7)*** | 1,430 | 539 (37.7%) | 1.2 (1.1–1.4)** |
| White | 10,891 | 3,295 (30.3%) | 2.0 (1.9–2.1)*** | 5,886 | 1,868 (31.7%) | 1.1 (1.0–1.2) | 1,027 | 332 (32.3%) | 1.0 (0.8–1.1) |
| Other | 440 | 118 (26.8%) | 1.7 (1.4–2.1)*** | 818 | 276 (33.7%) | 1.2 (1.0–1.4) | 421 | 103 (24.5%) | 0.7 (0.5–0.8)** |
| Total | 69,317 | 15,355 (22.2%) | – | 22,860 | 8,133 (35.6%) | – | 7,158 | 2,380 (33.2%) | – |
p < .01; ***p < .001.
CI, confidence interval; OR, odds ratio.
Race/ethnicity is the most assortative factor in the transmission network
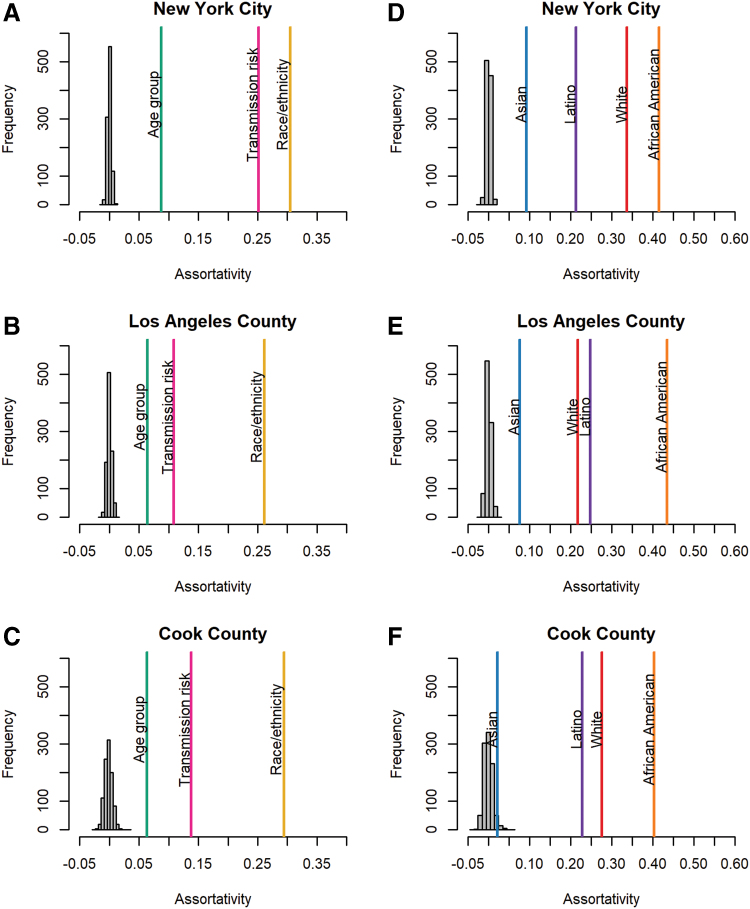
In all three metropolitan areas and at all distance thresholds, potential transmission partners were significantly assortative by age group at diagnosis, transmission risk, and race/ethnicity (p < .001; Fig. 2A–C; Supplementary Fig. S1); observed values exceeded the null expectation for assortativity values. Of all the attributes analyzed, assortativity by race/ethnicity was consistently the strongest, followed by transmission risk and finally by age at diagnosis. These findings indicate that genetic transmission partners were more likely to be of the same race/ethnicity than they were to be of the same age or transmission risk group. This pattern was robust across metropolitan areas and genetic distance thresholds.
FIG. 2.
Assortativity by diagnosis age, transmission risk, and race/ethnicity in (A) NYC, (B) LAC, and (C) Cook County; and assortativity for each of the predominant race/ethnicities: African American, Asian, Latino, and white in (D) NYC, (E) LAC, and (F) Cook County. Individuals were linked to each other in the network if they were ≤0.015 substitutions/site divergent. The null expectation for assortativity values based on 1,000 permutations is shown in gray. Color images are available online.
African Americans are the most assortative race/ethnic group
We next compared assortativity among the predominant race/ethnic groups. In all three metropolitan areas at various genetic distance thresholds, Latinos, African Americans, and whites were each assortative by race/ethnicity (p < .001; Fig. 2D–F; Supplementary Fig. S2). Asians were assortative in NYC and LAC at all distance thresholds evaluated, but they were only significantly assortative in Cook County at the most conservative distance threshold, 0.005 substitutions/site. African Americans were consistently the most assortative group in the transmission network, even in LAC where they were not the predominant race/ethnicity.
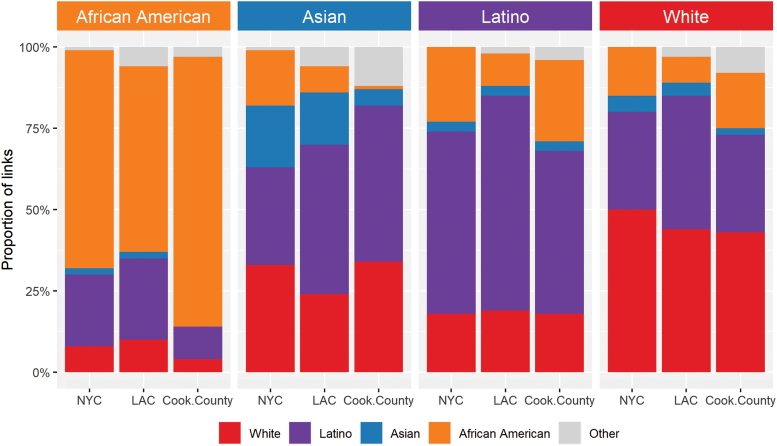
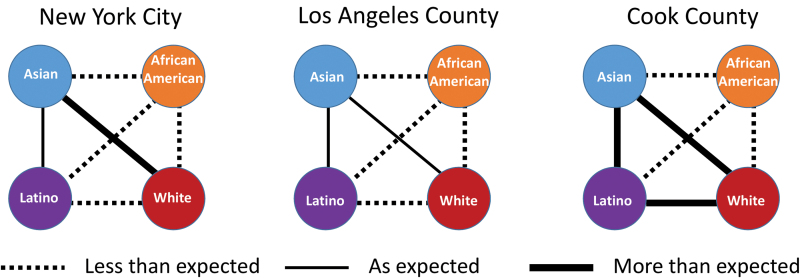
African Americans link less than expected to all other groups
We then calculated the proportion of linked partners of each pair of race/ethnicities (Fig. 3) and evaluated how observed sorting among race/ethnicity groups differed from our expectation if potential transmission partners were mixed at random in the network (Fig. 4; Supplementary Figs. S3–S5; Supplementary Tables S4–S7). In agreement with the assortativity findings, African Americans were the group who most consistently had potential transmission partners of the same race/ethnicity. In Cook County, 83% of their links were to other African Americans; in NYC and LAC, the proportions were 67% and 57%, respectively (Fig. 3). In all jurisdictions, African Americans were less frequently linked to other race/ethnicity groups than expected (Fig. 4). Asians were the only group to link as expected or more than expected to other groups. In NYC, Asians mixed more than expected with whites, and in Cook County they mixed more than expected with all groups, consistent with the observation that they were not assortative in Cook County.
FIG. 3.
Estimated proportion of potential partners from each race/ethnic group in NYC, LAC, and Cook County. Note that the number of individuals in each group varies widely and thus the statistical significance of differences cannot be assessed from these distributions. Color images are available online.
FIG. 4.
Expected/observed number of links between race/ethnicities (African American, Asian, Latino, and white) in each of the three metropolitan areas. Lines between race/ethnic groups indicate whether the numbers observed are lower than expected, as expected, or higher than expected (p < .01). Detailed distributions are available in Supplementary Figures S3–S5. Color images are available online.
Latinos also linked more to other Latinos than to other race/ethnicities: 56%, 66%, and 50% in NYC, LAC, and Cook County, respectively (Fig. 3). Latinos and whites were linked to each other less than expected in NYC and LAC but more than expected in Cook County (Fig. 4). Both Latinos and African Americans formed homophilic links twice as often as expected by chance, consistent across metropolitan areas and genetic distance thresholds (Supplementary Tables S4–S6). When we focused our analysis solely on the network among MSM (in the Los Angeles network only), we found that African Americans formed homophilic links 3.6 times more than expected (Supplementary Table S7). Assortativity and mixing patterns were consistent when we stratified the network by year of diagnosis.
Differences in assortativity are robust to undersampling of African Americans
In our sensitivity analysis in LAC, assortativity by race/ethnicity dropped as an increasing proportion of African Americans were removed from the network (Supplementary Fig. S6), suggesting that if African Americans are indeed undersampled, observed assortativity by race is likely to be underestimated in our analyses. Similarly, the assortativity of African Americans decreased as a higher proportion of African Americans were dropped from the network (Supplementary Fig. S7). Again, this result would indicate that assortativity of African Americans is likely underestimated and our estimates would increase if sampling of African Americans were higher.
Discussion
We examined assortativity patterns in HIV-1 molecular transmission networks in three large metropolitan areas in the United States (NYC, LAC, and Cook County) and found that race/ethnicity was more assortative than either reported transmission risk or age at diagnosis. Despite distinct racial/ethnic compositions in each of these metropolitan areas, African Americans were the group most likely to have potential transmission partners of the same race/ethnicity: their networks were most assortative and they were the least likely to have viruses genetically linked to individuals of any other race/ethnicity.
The insularity of African Americans in the genetic transmission network was seen in LAC, where African Americans were the minority of HIV cases, and in Cook County, where African Americans account for the majority of HIV cases. The number of homophilic links for African Americans was twice as high as expected by chance, and among African American MSM in the LAC network, it was 3.6 times higher than expected, in alignment with surveys of black MSM sexual networks in the United Kingdom and the United States.12,20
This racial assortativity in the HIV genetic transmission network could be explained by a number of factors. African Americans may be more likely to select other African Americans as their sexual partners, because of sexual preference, geographical proximity, culture, or life circumstances,35 or they may be less likely to be selected by other groups for these same reasons. Alternatively, as we are reconstructing the transmission network and not the contact network, linkage may be indicative of risk behavior; specifically, other groups may reduce risk taking behavior with African American partners due to perceived increased risk.
The finding that African Americans were the race/ethnicity most likely to have genetic partners from the same race/ethnicity was consistent with previous analysis21 and with contact networks obtained through interviews.35
In NYC and Cook County, Latinos were less assortative than whites, as seen in both those analyses, but Latinos in LAC (where they are more numerous) were more assortative than whites. In all three metropolitan areas African Americans nonetheless were linked with Latinos at high rates. Therefore, we tested the significance of these observed patterns compared to what we would expect if individuals were sorting at random and found that in fact African Americans, Latinos, and whites all mixed assortatively. In contrast to the previous analysis, which aggregated data from across the United States,21 we analyzed well-delineated metropolitan areas, providing regional nuance to sorting patterns. Nonetheless, patterns pertaining to African Americans were consistent across all three jurisdictions.
To better understand transmission patterns within and across race/ethnicities, we opted to analyze all transmission risk groups, not only MSM, because eliminating a portion of sequences will fragment the network. In addition, some MSM do not disclose their risk behavior,23 and their exclusion from the network could bias assortativity inference. Further, many people who reported injection drug use as their primary risk factor acquire HIV sexually, as has been seen in NYC.17,36,37 Nevertheless, over 70% of the persons in our analysis were MSM, and MSM are more likely to cluster than other transmission risk groups.17,37 Therefore, we acknowledge that the patterns observed here are dominated by the transmission dynamics of MSM and their networks.
HIV transmission network reconstruction from genetic data has been criticized for suggesting misleading associations between clustering and transmission covariates (e.g., age28 and recency of infection38), and for its sensitivity to missing data39 and to clustering method.40 Measures of assortativity are also affected by missing data.31 Because other clustering methods rely on the construction of phylogenies, they are hard to apply to datasets as large as these41,42; reassuringly, for small genetic distances (<0.015 substitutions/site), linkages are highly conserved across clustering methods.42,43 The lower proportion of clustering in NYC has been previously noted and is likely due to the age of the epidemic and the inclusion of older, unlinked infections within that dataset.44 Nonetheless, our findings were robust to different genetic distance thresholds and to downsampling of our data. In fact, when we lowered our linkage threshold to 0.5%, which increased our probability of capturing true transmission links, assortativity by race/ethnicity increased.
Because age classification is a dynamic, continuous label, it is sensitive to time and to missing data.45 In contrast, estimates of mixing between race/ethnic groups will not be significantly affected by missing data, because an unsampled intermediary will not modify the network count of assortative and disassortative links.45 Nonetheless, as many of our findings pertained to the lower connectivity of African Americans in the network, we considered that our results could be the result of lower rates of diagnosis and engagement in care among this group.32–34 In NYC and LAC, African Americans diagnosed with HIV were less likely to be genotyped, but this was not the case in Cook County. However, when we downsampled African Americans, assortativity by race/ethnicity and the assortativity of African Americans decreased, indicating that if underdiagnosis and underreporting of HIV in African Americans is present in these jurisdictions, the degree to which African Americans are assortative in the transmission networks would have been underestimated in our analyses.
Although the disparity in lifetime risk of acquiring HIV between African American and white MSM cannot be explained by assortativity alone,3,46,47 our findings highlight the need for prevention and intervention activities that specifically serve African Americans, for example, improving access to antiretroviral therapy and increasing awareness of preexposure prophylaxis (PrEP). Uptake of PrEP has been lower among African Americans than other race/ethnic groups.48,49
We found that transmission networks were more assortative by race/ethnicity than by any other factor we explored, and that African Americans were most likely to be the genetic potential transmission partners of African Americans. Sexual contact networks are likely to display this same pattern. Interventions specifically focused on highly assortative communities, such as African Americans, have the potential to reverberate through the sexual network to magnify the effect of that intervention and have a greater impact on decreasing HIV transmission.
Supplementary Material
Acknowledgment
We wish to thank Andrew Frick for help with data processing.
Authors' Contributions
M.R.-C., J.O.W., and N.B. conceived of and designed the study. N.B., C.H., K.P., F.M., L.A.F., Z.S., and L.V.T. generated, organized, and contributed data. M.R.-C., J.O.W., and C.H. conducted the analysis. M.R.-C., N.B., and J.O.W. interpreted the data. M.R.-C. and J.O.W. drafted the article and all authors revised and approved the final version of the article.
Author Disclosure Statement
J.O.W. received funding from Gilead Sciences, Inc. as grants paid to UC San Diego. The other authors declare no conflicts of interest.
Funding Information
This work was supported by a California HIV/AIDS Research Program IDEA Award (ID15-SD-052), an NIH-NIAID K01 Career Development Award (K01AI110181), an NIH-NIAID R01 (AI135992), and support from the Third Coast Center for AIDS Research (CFAR) (P30 AI117943). M.R.-C. is jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement and this award is part of the EDCTP2 program supported by the European Union (MR/R015600/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Supplementary Material
References
- 1. Hess KL, Hu X, Lansky A, Mermin J, Hall HI: Lifetime risk of a diagnosis of HIV infection in the United States. Ann Epidemiol 2017;27:238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention. HIV Surveillance Report, Diagnoses of HIV Infection in the United States and Dependent Areas, Atlanta, GA, 2016, 2017.
- 3. Adimora AA, Schoenbach VJ, Floris-Moore MA: Ending the epidemic of heterosexual HIV transmission among African Americans. Am J Prev Med 2009;37:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aral SO: Patterns of sexual mixing: Mechanisms for or limits to the spread of STIs? Sex Transm Infect 2000;76:415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ibragimov U, Beane S, Adimora AA, et al. : Relationship of racial residential segregation to newly diagnosed cases of HIV among Black heterosexuals in US Metropolitan Areas, 2008–2015. J Urban Health 2019;96:856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adimora AA, Schoenbach VJ, Taylor EM, Khan MR, Schwartz RJ, Miller WC: Sex ratio, poverty, and concurrent partnerships among men and women in the United States: A multilevel analysis. Ann Epidemiol 2013;23:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aral SO, Adimora AA, Fenton KA: Understanding and responding to disparities in HIV and other sexually transmitted infections in African Americans. Lancet 2008;372:337–340. [DOI] [PubMed] [Google Scholar]
- 8. Barlow D, Daker-White G, Band B: Assortative sexual mixing in a heterosexual clinic population—A limiting factor in HIV spread? AIDS 1997;11:1039–1044. [DOI] [PubMed] [Google Scholar]
- 9. Clerkin EM, Newcomb ME, Mustanski B: Unpacking the racial disparity in HIV rates: The effect of race on risky sexual behavior among Black young men who have sex with men (YMSM). J Behav Med 2011;34:237–243. [DOI] [PubMed] [Google Scholar]
- 10. Fujimoto K, Williams ML: Racial/ethnic differences in sexual network mixing: A log-linear analysis of HIV status by partnership and sexual behavior among most at-risk MSM. AIDS Behav 2015;19:996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newcomb ME, Mustanski B: Racial differences in same-race partnering and the effects of sexual partnership characteristics on HIV Risk in MSM: A prospective sexual diary study. J Acquir Immune Defic Syndr 2013;62:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raymond HF, McFarland W: Racial mixing and HIV risk among men who have sex with men. AIDS Behav 2009;13:630–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kouyos RD, von Wyl V, Yerly S, et al. : Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis 2010;201:1488–1497. [DOI] [PubMed] [Google Scholar]
- 14. Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ: Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med 2008;5:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hue S, Pillay D, Clewley JP, Pybus OG: Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci U S A 2005;102:4425–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ragonnet-Cronin M, Lycett SJ, Hodcroft EB, et al. : Transmission of non-B HIV subtypes in the United Kingdom is increasingly driven by large non-heterosexual transmission clusters. J Infect Dis 2016;213:1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. : Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog 2017;13:e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dennis AM, Pasquale DK, Billock R, et al. : Integration of contact tracing and phylogenetics in an investigation of acute HIV infection. Sex Transm Dis 2018;45:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith DM, May SJ, Tweeten S, et al. : A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS 2009;23:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doerner R, McKeown E, Nelson S, Anderson J, Low N, Elford J: Sexual mixing and HIV risk among ethnic minority MSM in Britain. AIDS Behav 2012;16:2033–2041. [DOI] [PubMed] [Google Scholar]
- 21. Whiteside YO, Song R, Wertheim JO, Oster AM: Molecular analysis allows inference into HIV transmission among young men who have sex with men in the United States. AIDS 2015;29:2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hue S, Brown AE, Ragonnet-Cronin M, et al. : Phylogenetic analyses reveal HIV-1 infections between men misclassified as heterosexual transmissions. AIDS 2014;28:1967–1975. [DOI] [PubMed] [Google Scholar]
- 23. Ragonnet-Cronin M, Hue S, Hodcroft EB, et al. : Non-disclosed men who have sex with men in UK HIV transmission networks: Phylogenetic analysis of surveillance data. Lancet HIV 2018;5:e309–e316. [DOI] [PubMed] [Google Scholar]
- 24. Cohen SM, Gray KM, Ocfemia MC, Johnson AS, Hall HI: The status of the National HIV Surveillance System, United States, 2013. Public Health Rep 2014;129:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hue S, Clewley JP, Cane PA, Pillay D: HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 2004;18:719–728. [DOI] [PubMed] [Google Scholar]
- 26. Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO: HIV-TRACE (Transmission Cluster Engine): A tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol 2018;35:1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura K, Nei M: Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512–526. [DOI] [PubMed] [Google Scholar]
- 28. Le Vu S, Ratmann O, Delpech V, et al. : Comparison of cluster-based and source-attribution methods for estimating transmission risk using large HIV sequence databases. Epidemics 2018;23:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newman ME: Mixing patterns in networks. Phys Rev E Stat Nonlin Soft Matter Phys 2003;67(2 Pt 2):026126. [DOI] [PubMed] [Google Scholar]
- 30. Csardi G, Nepusz T: The igraph software package for complex network research. InterJournal 2006; 2006;Complex Systems:1695. [Google Scholar]
- 31. Smith JA, Moody J: Structural effects of network sampling coverage I: Nodes missing at random. Soc Networks 2013;35:652–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nunn A, Zaller N, Cornwall A, et al. : Low perceived risk and high HIV prevalence among a predominantly African American population participating in Philadelphia's Rapid HIV testing program. AIDS Patient Care STDS 2011;25:229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reilly KH, Neaigus A, Jenness SM, Wendel T, Marshall DMt, Hagan H: Factors associated with recent HIV testing among men who have sex with men in New York City. AIDS Behav 2014;18 Suppl 3:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holmes L Jr, Monjok E, Ward D, et al. : Racial variance in rationale for HIV testing in community-based setting in the United States: Evidence from the National Health Interview Survey. J Int Assoc Physicians AIDS Care (Chic) 2008;7:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mustanski B, Birkett M, Kuhns LM, Latkin CA, Muth SQ: The role of geographic and network factors in racial disparities in HIV among young men who have sex with men: An Egocentric Network Study. AIDS Behav 2015;19:1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Des Jarlais DC, Arasteh K, Perlis T, et al. : Convergence of HIV seroprevalence among injecting and non-injecting drug users in New York City. AIDS 2007;21:231–235. [DOI] [PubMed] [Google Scholar]
- 37. Ragonnet-Cronin M, Hu YW, Morris SR, Sheng Z, Poortinga K, Wertheim JO: HIV transmission networks among transgender women in Los Angeles County, CA, USA: A phylogenetic analysis of surveillance data. Lancet HIV 2019;6:e164–e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volz EM, Koopman JS, Ward MJ, Brown AL, Frost SD: Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol 2012 2012;8:e1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Novitsky V, Moyo S, Lei Q, DeGruttola V, Essex M: Impact of sampling density on the extent of HIV clustering. AIDS Res Hum Retroviruses 2014;30:1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Poon AF: Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks. Virus Evol 2016;2:vew031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prosperi MC, Ciccozzi M, Fanti I, et al. : A novel methodology for large-scale phylogeny partition. Nat Commun 2011;2:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ragonnet-Cronin M, Hodcroft E, Hue S, et al. : Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013;14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rose R, Lamers SL, Dollar JJ, et al. : Identifying transmission clusters with cluster picker and HIV-TRACE. AIDS Res Hum Retroviruses 2017;33:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wertheim JO, Murrell B, Mehta SR, et al. : Growth of HIV-1 molecular transmission clusters in New York City. J Infect Dis 2018;218:1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ragonnet-Cronin M, Hodcroft EB, Wertheim JO: Understanding disclosed and cryptic HIV transmission risk via genetic analysis: What are we missing and when does it matter? Curr Opin HIV AIDS 2019;14:205–212. [DOI] [PubMed] [Google Scholar]
- 46. Goodreau SM, Rosenberg ES, Jenness SM, et al. : Sources of racial disparities in HIV prevalence in men who have sex with men in Atlanta, GA, USA: A modelling study. Lancet HIV 2017;4:e311–e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morris M, Kurth AE, Hamilton DT, Moody J, Wakefield S: Concurrent partnerships and HIV prevalence disparities by race: Linking science and public health practice. Am J Public Health 2009;99:1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoots BE, Finlayson T, Nerlander L, Paz-Bailey G, National HIVBSSG: Willingness to take, use of, and indications for pre-exposure prophylaxis among men who have sex with men-20 US Cities, 2014. Clin Infect Dis 2016;63:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lelutiu-Weinberger C, Golub SA: Enhancing PrEP access for Black and Latino men who have sex with men. J Acquir Immune Defic Syndr 2016;73:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.