Abstract
Single-cell RNA sequencing (scRNA-seq) revolutionized our understanding of disease biology. The promise it presents to also transform translational research requires highly standardized and robust software workflows. Here, we present the toolkit Besca, which streamlines scRNA-seq analyses and their use to deconvolute bulk RNA-seq data according to current best practices. Beyond a standard workflow covering quality control, filtering, and clustering, two complementary Besca modules, utilizing hierarchical cell signatures and supervised machine learning, automate cell annotation and provide harmonized nomenclatures. Subsequently, the gene expression profiles can be employed to estimate cell type proportions in bulk transcriptomics data. Using multiple, diverse scRNA-seq datasets, some stemming from highly heterogeneous tumor tissue, we show how Besca aids acceleration, interoperability, reusability and interpretability of scRNA-seq data analyses, meeting crucial demands in translational research and beyond.
INTRODUCTION
Major breakthroughs in our understanding of rare cell types, tissue heterogeneity, cell differentiation and transcriptional regulation have been enabled by the increased resolution in detecting gene expression provided by single-cell RNA-sequencing (scRNA-seq). Encouraged by early successes, pharmaceutical research has also embraced the technology – to accelerate drug discovery. In this context, scRNA-seq is used to better understand disease phenotypes (1), to assess drug targets (2), to characterize microphysiological systems (3) and to measure cell-type-specific pharmacology and toxicity of drug candidates (4). In addition, scRNA-seq assists the characterization of in vitro and in vivo disease- and safety models by offering insights into cell-to-cell communication (5), cell activation (6) or differentiation trajectories (7).
Current challenges in the analysis of single-cell transcriptomics data are predominantly related to the biological interpretation of the analysis results rather than to the computation thereof (8). Whereas the computational part can be automated, biological interpretation still requires manual user interaction and expert knowledge, often leading to hurdles in translational research. Therefore, we automated and standardized multiple analysis steps, in line with current best practices and benchmarks (9–14), focused on streamlining a major bottleneck (15)—the cell type annotation process—as well as on facilitating results usage in downstream applications such as bulk RNA-seq deconvolution. Our toolkit Besca (Figure 1) will thus allow translational researchers (and not only) to take full advantage of the rapidly growing amount, size, and scope of single cell data generated (16,17) and will facilitate consistent biological investigation.
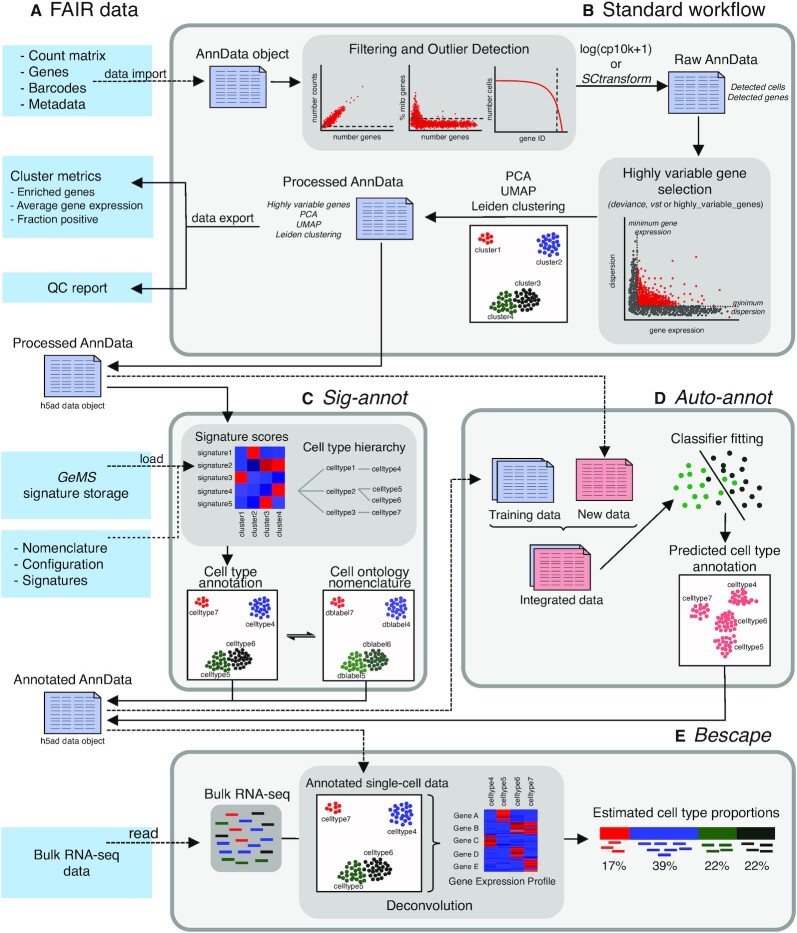
Figure 1.
Besca provides streamlined single-cell transcriptomics data analysis modules and exchange file formats. (A) Well-defined interoperable input and output file formats, cluster metrics, a quality control report and a signature storage ensure reusability of data. (B) The standard workflow internalizes a raw count matrix and generates a quality control report as well as a processed dataset post filtering, normalization, highly variable gene selection, batch correction, and clustering. (C, D) Clusters identified from the standard workflow are annotated using either signature-based hierarchical cell annotation (Sig-annot module, C) or a supervised machine learning-based algorithm trained on previously annotated datasets (Auto-annot module, D). (E) The annotated datasets can be used to deconvolute bulk RNA-seq data based on gene expression profiles generated from annotated single-cell datasets utilizing the Bescape module.
Besca is an open-source Python library that is compatible with and extends Scanpy (18), one of the most established and up-to-date single-cell analysis toolkits. It uniquely streamlines scRNA-seq data processing beyond the clustering step, by providing a selection of robust analysis methods, cell annotation approaches and underlying nomenclature, ensuring interoperability and reusability of analysis, quality controls and results. Importantly, it remains fully customizable beyond the standards introduced here. For instance, we have expanded Besca to support the analysis of datasets generated by the recently developed CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) (19) method.
Further, the Besca proportions estimate (Bescape) module integrates SCDC (20) and MuSiC (21), allowing to directly apply Besca-generated results and cell annotations to deconvolute bulk transcriptomics data (Figure 1E). This enhances currently available bulk RNA-seq data, for instance in the case of larger clinical settings that do not have the capacity to perform scRNA-seq and where signals are often confounded by heterogeneity related to distinct cell type composition (22). The resulting estimated cell compositions can be used directly as biomarkers or as covariates towards getting more robust differential gene expression results facilitating the understanding of disease biology or treatment responses.
Besca targets scientists in translational research, helping bioinformaticians streamline scRNA-seq analyses and increasing comparability between studies, while at the same time offering a low hurdle entry point into such analyses for wet lab scientists with limited programming skills. The toolkit is especially relevant for research groups that deal with large amounts of internal data as well as re-analyse public data and need to compare and reuse results or provide them as a basis for downstream applications. For instance, cell type gene signatures and annotation schemas provided with Besca were tested and applied across multiple studies, tissues and species, ranging from healthy PBMCs to highly heterogeneous tumor tissue. They represent one of the most comprehensive hand-curated publicly available resources that can be employed out of the box, with minimal adjustments for novel datasets. Cell type annotations were harmonized and can be reused across studies, also allowing to cross-validate the discovery of new cell types from multiple studies such as inflammatory fibroblasts in colitis described below. Finally, results can be seamlessly reused from one application to another, for instance by using single-cell gene expression profiles for tissue-specific cell deconvolution of bulk RNA-seq.
MATERIALS AND METHODS
Example data
The following publicly available single-cell datasets from ten studies were reprocessed (see also Table 1). Besca allows to load the unprocessed as well as the analyzed datasets with a single function call. All studies utilized the microfluidics platform from 10X Genomics, today's most used platform for disease understanding and drug discovery, and therefore Besca’s workflows are optimized for this platform. In addition, Besca was applied to more than a hundred internal and public datasets including additional platforms (data not shown).
Table 1.
Dataset overview. Datasets include hematopoietic cells of peripheral blood and bone marrow, intestine and pancreas in health and disease. Besca’s functionality is exemplified on these datasets utilizing Jupyter notebooks (N), R markdowns (R), this manuscript (M) and its supplementals (S)
| Tissue | Area of interest | Dataset | Original data accession | Processed data DOI | CITE-seq | Standard workflow | Sig-annot, signature-based cell annotation | Auto-annot, supervised cell annotation | Bescape, bulk RNA-seq deconvolution |
|---|---|---|---|---|---|---|---|---|---|
| Bone marrow and peripheral blood | Healthy | PBMC3k | 10xgenomics.com | 10.5281/zenodo.3948150 | No | S, N | N | M4, S, N | - |
|
|
Mixed-phenotype acute leukemia | Granja2019 | GSE139369 | 10.5281/zenodo.3944753 | Yes | N | M2, N | M4, S, N | - |
|
|
Vaccine responsivenes | Kotliarov2020 | 10.35092/yhjc.c.4753772 | 10.5281/zenodo.3938290 | Yes | N | M3, N | M4, S, N | M6, R |
| Intestine | Ulcerative colitis | Smillie2019 | SCP259 | 10.5281/zenodo.3960617 | No | N | N | M5, S, N | - |
|
|
Crohn's disease | Martin2019 | GSE134809 | 10.5281/zenodo.3862132 | No | N | N | M5, S, N | - |
|
|
Mouse | Haber2017 | GSE92332 | 10.5281/zenodo.3935782 | No | N | N | M5, S, N | - |
|
|
Colorectal cancer | Lee2020 | GSE132465 | 10.5281/zenodo.3967538 | No | N | S, N | - | - |
| Pancreas | Type II Diabetes | Segerstolpe2016 | E-MTAB-5061 | 10.5281/zenodo.3928276 | No | N | N | S, N | M6, R |
|
|
Pancreatic ductal adenocarcinoma | Peng2019 | PRJCA001063 | 10.5281/zenodo.3969339 | No | N | S, N | S, N | - |
|
|
Healthy | Baron2016 | GSE84133 | 10.5281/zenodo.3968315 | No | N | N | S, N | - |
M = main manuscript; S = supplementary material; N = notebook on GitHub (https://github.com/bedapub/besca_publication_results); R = R Markdown on GitHub (https://github.com/bedapub/bescape)
2, Fig.2; 3, Fig. 3; 4, Fig. 4; 5, Fig. 5; 6, Fig. 6.
Three of the datasets shown in this manuscript cover blood- and bone-marrow-derived hematopoietic cells:
PBMC3k (https://doi.org/10.5281/zenodo.4441679) includes healthy peripheral blood mononuclear cell (PBMC) samples from one donor, a reference dataset often used in single-cell tutorials based on 10X Genomics data (https://www.10xgenomics.com/).
Granja2019 (https://doi.org/10.5281/zenodo.4419527) includes bone marrow mononuclear cell (BMMCs) and PBMC samples from healthy donors (23). In addition to scRNA-seq, several protein markers were also probed by CITE-seq.
Kotliarov2020 (https://doi.org/10.5281/zenodo.4350119) includes baseline PBMC samples from healthy donors, who were high and low responders to influenza vaccines (24). In addition to scRNA-seq, a high number of protein markers were also probed by CITE-seq.
Four datasets reveal the cell composition in intestinal tissue:
Smillie2019 (https://doi.org/10.5281/zenodo.3960617) includes colon epithelium and lamina propria samples from healthy donors and ulcerative colitis patients (25).
Martin2019 (https://doi.org/10.5281/zenodo.3862132) includes ileal lamina propria samples from Crohn's disease patients (26).
Haber2017 (https://doi.org/10.5281/zenodo.4447233) includes murine small intestine samples (27).
Lee2020 (https://doi.org/10.5281/zenodo.3967538) includes tumor and non-malignant colon samples from colorectal cancer (CRC) patients (28).
Three datasets are pancreas-derived:
Segerstolpe2016 (https://doi.org/10.5281/zenodo.3928276) includes pancreatic islet cells from healthy donors and type 2 diabetic patients (29).
Peng2019 (https://doi.org/10.5281/zenodo.3969339) includes tumor and non-malignant pancreatic samples from pancreatic ductal adenocarcinoma (PDAC) and non-pancreatic tumor patients (30).
Baron2016 (https://doi.org/10.5281/zenodo.3968315) includes pancreatic samples from healthy donors (31).
Methods, implemented tools or functions and parameter choices
For all Besca modules, the methods, the implemented tools or functions, and the parameter choices are summarized in Supplementary Table S1 together with motivations and remarks regarding their choice.
Besca’s standard workflow
Besca’s standard workflow starts with loading the count matrix obtained from a preprocessing pipeline (demultiplexing, read alignment, feature counting), and the annotation of the matrix, including barcodes, genes and, if available metadata associated to the datasets, including biological (e.g. donor, experimental condition) and technical (e.g. batches, protocols differences) variables. Before proceeding with analysis, quality control (QC) is performed. This includes visualizing drop-outs and sequencing saturation as well as performing cell and gene filtering.
During cell filtering all barcodes that likely do not correspond to viable cells are removed. Cell filtering is performed on the basis of three QC covariates: the number of counts per barcode, the number of genes per barcode, and the relative contribution of mitochondrial genes per barcode. Each of the covariates are examined for outliers by thresholding as described in (10). During gene filtering, transcripts which are only expressed in a few cells are removed to reduce dataset dimensionality. As recommended by Luecken and Theis (10), the filtering threshold for genes should be set to the minimum cell cluster size that is of interest. As QC filtering is highly dependent on the dataset, the filtering thresholds need to be defined by the user before running the workflow. To guide the user, Besca offers a wrapper function based on Scater's isOutlier (32) which estimates the outlier cells and genes based on the number of median absolute deviations for a given QC. Correctly chosen thresholds are verified through knee-plot graphics within the pipeline.
After QC, the expression values are normalized. Normalization is performed using count depth scaling and count values are log(x+1)-transformed. Alternatively, the variance stabilizing transformation approach of SCtransform can be applied, which returns the Pearson residuals from a regularized negative binomial regression model applied on UMI count data (33). To reduce dataset dimensionality before clustering, the highly variable genes within the dataset are selected. By default, genes are defined as being highly variable when they have a minimum mean expression of 0.0125, a maximum mean expression of 3 and a minimum dispersion of 0.5.
Technical variance is removed by regressing out the effects of count depth and mitochondrial gene content and the gene expression values are scaled to a mean of 0 and variance of 1 with a maximum value of 10. It needs to be mentioned here that correction of mitochondrial gene content might not be considered a technical variance correction but removal of biological variability. If this correction is not desired, the threshold for mitochondrial gene content correction can be set to 1. Based on the best practices suggested by Luecken and Theis, technical variance should be corrected before selection of highly variable genes. In Besca’s standard workflow this order is reversed, due to regress-out being a very time-consuming computational process which can be significantly sped up by only calculating corrected values for the previously selected highly variable genes. Based on recent benchmarking (12), developments in the community, and our own comparison (Supplementary Table S2), we recommend omitting the regress-out step in case of doubt, especially if SCtransform is used for normalization.
Finally, dimensionality reduction and clustering is performed. The first 50 principal components are calculated and used as input for calculation of the 10 nearest neighbors. The intrinsic dimensionality of the data can also be estimated with a function based on the intrinsicDimension R package (34). This method estimates the intrinsic number of dimensions using a translated Poisson mixture model and was shown to provide a better separation of cell subpopulations after clustering (12). BBKNN can be used for batch correction (35). The neighbourhood graph is then embedded into 2D space using the UMAP (Uniform Manifold Approximation and Projection) algorithm (36). Cell communities are detected using the Leiden algorithm (37) at a resolution of 1 by default.
For CITE-seq data, the protein marker abundance values are loaded separately to the gene expression values and stored in its own data object. Previously determined cell barcode filtering to identify viable cells on the basis of gene expression values is applied to the CITE-seq data. Unlike gene expression counts, protein marker counts are normalized using centred log ratios, a per-ADT transformation that divides each count by the geometric mean of that ADT counts (19). Alternatively, the DSB method can be selected to normalize the protein counts, which denoises the data by subtracting the background noise of empty droplets (38). If less than 50 markers were measured, the entire count matrix is used as input for the nearest neighbour calculation. Otherwise, as in the gene expression data, the first 50 principal components are calculated. The rest of the CITE-seq pipeline is analogous to the gene expression pipeline. At the end of the workflow the results are homogenized into one data object which contains clustering and visualization results of both gene expression and protein abundance from CITE-seq data. The subsequent cell annotation can then either be performed on the RNA-based clusters or on the ADT-based clusters (following CLR or DSB normalization).
Analysis results are exported into interoperable file formats to allow FAIR data management of analysis results. This includes the Matrix Market exchange format (https://math.nist.gov/MatrixMarket/formats.html) for sparse count matrices, GCT (https://software.broadinstitute.org/software/igv/GCT) for dense count matrices, and simple tab-separated or comma-separated values formats for metadata and as interface for the cell deconvolution package Bescape, respectively. Clustering results or cell type labeling are exported including pre-computed average expression and ranked marker gene lists per cluster or cell type.
Annotation of cell types based on CITE-seq data
A fine-grained annotation of the cells contained within the Kotliarov2020 dataset (24) was generated on the basis of the labelled protein antibody counts from CITE-seq. The normalized protein counts were exported to FCS files using the R package flowCore (39,40) (R package version 2.0.1) and loaded into FlowJo™ Software (FlowJo™ Software Mac Version 10.6.2. Ashland, OR: Becton, Dickinson and Company; 2019). The gating strategy used to identify individual cell populations is outlined in Supplementary Figure S1. Gating of individual cell populations was based on the gating strategy utilized in (41). Barcodes from identified cell populations were exported from FlowJo™ Software to csv files and loaded into Besca for visualization.
Sig-annot, signature-based automated cell type annotation
The annotation process has three components:
a nomenclature table with long and short names, according to Cell Ontology (42), see Supplementary Table S3 and https://github.com/bedapub/besca/blob/master/besca/datasets/nomenclature/CellTypes_v1.tsv
a configuration file including all the cell types to be considered, their parent (or ‘none’), a factor to be multiplied with the cut-off for scoring a cluster positive or negative for the signature based on the Mann–Whitney test and the order in which to consider the signatures (only first positive one matching a cluster will be taken into account). Two distinct default configuration files are provided with Besca, covering mouse and human. Users are free to adjust the parameters in the files, and tailor these according to tissues or dataset. Human: Supplementary Table S4 and https://github.com/bedapub/besca/blob/master/besca/datasets/genesets/CellNames_scseqCMs6_config.tsv Mouse: Supplementary Table S5 and https://github.com/bedapub/besca/blob/master/besca/datasets/genesets/CellNames_scseqCMs6_config.mouse.tsv
a GMT file with the signatures, in line with the nomenclature table, see Supplementary Table S6 and https://github.com/bedapub/besca/blob/master/besca/datasets/genesets/CellNames_scseqCMs6_sigs.gmt
Sig-annot mimics and automates the classical manual marker-based cell annotation process—for a given set of clusters, signatures and cell types, it determines the best match given previous knowledge of a hierarchy between the cell types. As exemplified in the accompanying cell annotation workbook, the hierarchy provided in the configuration file (component 2) can be visualized as a graph and is then used to call cell types at distinct levels. Level 1 would thus contain major cell types such as epithelial, endothelial, hematopoietic cells. Once clusters will be attributed to these cell types (or to ‘animal cell’ in case none of the corresponding signatures score above the given cutoff), the next level will be attributed. For instance, for hematopoietic cells, this corresponds to lymphocytes of B lineage, myeloid leukocytes, T cells and innate lymphoid cells. Only cells called as hematopoietic in level 1 will be considered for these hematopoietic subtypes, thus reducing the requirement for highly specific markers with regards to the cell population and enabling fine-grained classification. Currently, up to four levels are supported in the nomenclature file, however this can be easily extended should the data and cell type knowledge allow or require it.
For simplicity, we maintained one single large GMT file containing all signatures, however irrelevant cell types that may perturb the annotation can be easily excluded by providing them as an additional parameter. Cutoffs for cluster attribution per signature are set in the configuration files, but can also be manually modified in the annotation notebook after data inspection, to ensure flexibility and best fits with the data at hand. They represent multiplication factors rather than absolute values, as a base cutoff is always determined relative to how a set of ubiquitously expressed genes scores, thus ensuring better translatability across studies.
Finally, the order parameter ensures that more specific signatures are always considered first in the attribution, thus allowing more stringent cutoffs. For instance – epithelial cells, endothelial cells and fibroblasts typically have a more specific transcriptional signal compared to hematopoietic ones, which are mainly characterized by strong expression of PTPRC, which is relatively lowly expressed in plasma cells and myeloids. Thus, in this case, clusters are first attributed to the above-mentioned cell types with a stringent cutoff such that one can be more lenient on the cutoff for calling hematopoietic cells, ensuring inclusion of plasma and myeloid cells. This concept is then applied to all levels and signatures as ranks are pre-specified in the configuration file.
We note that in our experience, while new datasets of distinct types (tissue, technology) may require some parameter adjustments, modifications of the analysis on the same or a highly similar dataset (as is typically required when including/excluding additional samples or trying multiple clustering parameters for instance) can then be rerun with the exact settings, thus saving all the time one would require for the manual attribution of clusters. For large datasets covering tens of types of cells this is a substantial gain. Finally, the obtained annotation will all be in line with the nomenclature table (component 1) and thus the Cell Ontology (42), greatly facilitating comparisons between studies and analyses, as also exemplified here.
Auto-annot, supervised automated cell type annotation
Besca’s Auto-annot module, a supervised machine learning workflow, can be run independently from the standard workflow and works as follows:
Initially the training datasets are merged to form a combined training dataset using Scanorama (43), in the case where multiple training datasets are available, and complemented with the testing dataset. Scanorama is one of the most robust tools for the integration task and performs well on complex real data (44,45), especially on data from the 10X Genomics platform (13). A parameter specifies if the resulting integrated gene expression matrix contains the intersection of all genes, the intersection of previously selected highly variable genes, or genes of a previously defined signature.
Secondly, the Python package scikit-learn (https://scikit-learn.org) is used to train a classifier based on the merged training datasets. Two classification approaches are implemented, SVM and logistic regression. For SVM, one can choose between SVM with linear kernel (linear); SVM with linear kernel using stochastic gradient descent (sgd); SVM with radial basis function kernel (rbf), which should be used on small datasets only due to longer runtime. For logistic regression, the options are multinomial loss (logistic_regression); logistic regression with one versus rest classification, without normalised probability scores (logistic_regression_ovr); logistic regression with elastic loss, cross validated among multiple l1 ratio (logistic_regression_elastic). In our evaluations, logistic regression and SVM generally provide very similar results. We thus recommend to use logistic_regression as the default option as its runtime is superior to the SVM, especially for larger datasets, reducing the runtime from hours to minutes, depending on resources. In addition, the threshold functionality with logistic regression allows for more informative results and can also act as a sanity check. Since the different logistic regression tools usually provide almost identical results, we recommend the standard implementation to avoid unnecessary complexity.
Finally, the fitted model is used to predict cell types in the test dataset and predictions are added to the metadata. A probability threshold can be defined for logistic regression classifiers, to classify only cells reaching the defined threshold. In order to compare the predicted cell types to a ground truth already annotated in the test datasets, a report can be generated including precision, recall, and F1 metrics as well as confusion matrix and automatically annotated UMAP plots. These scores should be interpreted with care for the following three reasons: (i) most scoring methods do not take into account the class imbalance prevalent in scRNA-seq datasets; (ii) when the training and testing set do not contain the same set of cell types, scores are not always defined; (iii) in scoring, misclassifications into very similar subtypes are not treated differently to completely incorrect annotations. Such biases are easily avoided by basing one's interpretation on UMAP visualizations instead of summary scoring functions.
Bescape, cell deconvolution
At the core of the cell deconvolution algorithm is a regression-based problem. The concept is not novel, as it has already been investigated for microarray data (46). The combination of how newly derived cell specific GEP from scRNA-seq data can be used is the key factor that has evolved considerably over time. At a broad level, there are two categories of cell deconvolution, it is either a full deconvolution where neither the source nor the mixing process is known or a partial deconvolution where there is priori knowledge of the sources or the mixing process. Although a completely unsupervised approach can be taken, where the non-negative matrix factorization is suitable, it has been proven to show low accuracy and difficulty in handling the collinearity of the genes (20). The research focus is thus currently placed on partial deconvolution with known signatures used as bases to estimate the proportions in the bulk tissue. Such approaches have been developed using constrained least squares regression (EPIC) (47) and ν-support vector regression (CIBERSORT) (48). These methods either use microarray or a mixture of bulk RNA and scRNA-seq data to build a single GEP as a basis vector.
To facilitate direct incorporation of reference scRNA-seq datasets and at the same time address outstanding methodological shortcomings, we included two recent cell deconvolution methods in Bescape. MuSiC (21) uses a constrained least square regression but factors in gene weights to reduce the impact of genes with low cell type specificity. Thus, it eliminates the need for preselection of genes. Importantly, it also addresses the hierarchical nature of cell lineages with a recursive tree guided search, similar to the gating strategy in FACS, by first grouping similar cell types into the same cluster and estimating cluster proportions, then recursively repeating the previous step within each cluster identified. At each recursion stage, the focus is only on differentially expressed genes across cell types within the cluster, avoiding signal dilution from unspecific genes.
The second method included in the Bescape module is SCDC (20), an ensemble approach allowing for multiple scRNA-seq reference datasets. In short, similar to MuSiC, a weighted non-negative least square regression is adopted but differs slightly on how the weights are assigned to the genes. The salient point of the method is an additional layer of abstraction being introduced by assigning different weights for each reference scRNA-seq dataset. Higher weights are attributed to reference datasets that can fit the gene expression profiles of bulk RNA-seq samples better based on defined performance metric.
The selection of MuSiC and SCDC have been included in the Bescape module to allow for the deconvolution feature to use GEPs derived after scRNA-seq analysis with our standard workflow, while taking full advantage of the aforementioned improvements made by these methods. The long term goal is to keep this module open for novel methods to be evaluated and added by the user community. In addition, although benchmarking of the different methods is out of scope for this current work (see e.g. (49)), we have included the results of running CIBERSORTx (50) on the two publicly available datasets in the supplementals for reference (Supplementary Figure S2 and S3).
Generating simulated bulk
Simulated bulk RNA-seq was generated to evaluate the estimated proportions of the selected cell types with ground truth from a known in silico mixture. The annotated scRNA-seq data can be used directly by SCDC and MuSiC where no user specified feature selection based on marker genes is needed, instead a higher weight is assigned to features showing high variability across annotated cell types and low variability across samples (20,21). The simulated bulk is based on linear regressions where the cell fractions (weights) are taken from a uniform distribution, thus without factoring in any prior knowledge of the range of cell proportions of the different cell types, and scaled for the total to add up to 1. The GEPs of the cell types constitute the basis matrix needed to construct the bulk RNA-seq vector. This step is repeated for several instances representing different subjects’ bulk RNA-seq data.
RESULTS
To demonstrate the broad applicability of Besca, we reprocessed publicly available single-cell data from ten studies, across four tissues, seven disease states, including diabetes, inflammatory bowel disease, colorectal and pancreatic cancer (see Table 1 and Materials and Methods). We show how our proposed toolkit can be used to quickly obtain biological insights and generate reusable results from these highly diverse datasets. Further functionalities of Besca and more examples can be found in the supplementary material, example workbooks on GitHub (https://github.com/bedapub/besca_publication_results), and in the tutorials available from the documentation (https://bedapub.github.io/besca/).
A standard workflow streamlining scRNA-seq and CITE-seq analyses
The Besca standard workflow offers a streamlined series of steps, starting from a gene-by-cell count matrix (Figure 1A) and ending with cell clustering (Figure 1B). Based on Scanpy (18), it facilitates performing analysis of single-cell transcriptomics data in a reproducible and comparable manner. Good practices and FAIR (findability, accessibility, interoperability, reusability) principles (51) enable comparisons between all datasets analysed with Besca. The standard workflow detailed in the Methods broadly follows the steps of single-cell analysis described at length by Luecken and Theis (10) and also allows for the processing of CITE-seq (19) data. Several of the top-performing analysis methods according to multiple benchmarking studies and best practice recommendations (9–14) are integrated, ensuring robustness and flexibility (Supplementary Table S1).
The standard workflow generates a quality control (QC) report and a log file which summarize the performed analysis (Figure 1A, B). For future reuse, all of the analysis results are written to files in interoperable exchange data formats (see Materials and Methods) including output files of precomputed metrics, such as average gene expression or marker gene rankings (Figure 1A). These features are important for reusability and result reporting in collaborative efforts. They distinguish Besca from other analysis toolkits such as Seurat (33), Scanpy (18), Scater (32) and scvi-tools (52).
Additional downstream analyses such as automated cell type annotation can be run directly on the output of the standard workflow. The cell type annotation of the clusters can be performed using the Sig-annot (Figure 1C) or Auto-annot (Figure 1D) methods described thereafter. A re-clustering framework is available to focus on specific cell populations or clusters. This procedure re-initiates the highly variable gene selection on a subset of selected cells and reapplies the chosen clustering algorithm to better decipher finer grained cell subtypes. In addition, plotting functions for frequently used visualizations are implemented to illustrate gene expression variation under certain conditions (e.g. treatment effect) or to show the cell type composition found in the analyzed dataset. The standard workflow and subsequent marker-based cell annotation are exemplified in Supplementary Figure S4 utilizing the PBMC3k dataset (see Table 1 and Materials and Methods).
A gene signature management system
The integration of multiple scRNA-seq datasets allows for the accumulation of knowledge and insights about biological tissues, cells, cell states and diseases. As the development of suitable scRNA-seq integration data increases, a key challenge in single-cell data analysis workflows is the accurate dissemination of this knowledge and the appropriate reuse of the information gathered. In particular, it is of utmost importance to be able to re-apply gene signatures extracted from individual studies across studies and within analyses.
In contrast to other single cell analysis toolkits (18,32,33,52), Besca is focusing on the standardization and reusability of analyses, and therefore we connected Besca to the Geneset Management System (GeMS) (https://github.com/bedapub/GeMS). GeMS is a light web-based platform that enables the centralized management of genesets using structured formats and a local application programming interface for geneset information retrieval and organization. The application is built on top of the Flask micro-framework (https://flask.palletsprojects.com) using MongoDB (http://www.monogdb.com), an open-source, document-based database as its backend.
Once GeMS is deployed, Besca allows the export of gene signatures to the GeMS database (for example a geneset of marker genes from distinct populations) and the retrieval of user-defined signatures (Figure 1A). It is also possible to check for geneset similarity to avoid redundancy within the database and check for signature specificity. GeMS is distributed with initial public genesets extracted from Reactome (53), CREEDS (54), CellMarker (55) and MSigDB (56,57) and can be filled with new genesets.
In addition to these public resources, Besca is also distributed with over 100 hand-curated signatures related to cells of different tissues, including hematopoietic, intestinal and pancreatic cell types, which can be used for cell type annotation, as discussed below. We assessed the quality of these signatures by comparing them with genes that are preferentially expressed in tissues and cell types, which we identified previously from bulk expression studies (58,59), and by querying their expression in the gene expression compendia Human Protein Atlas (60), which integrates newly generated data with data from GTEx (61) and FANTOM5 (62). Both approaches confirmed the quality of the Besca signatures by revealing the consistency between signatures identified from single-cell techniques and their expression profile in bulk studies (Supplementary Document 1, Supplementary Figures S5 and S6). Our toolkit is made for direct usage of these genesets for signature enrichment analysis and can compute bi-directional scores combining up and down-regulated genes into one metric, highly relevant for treatment-specific signatures, for instance.
Automated and harmonized cell type annotation
Cell type identification in scRNA-seq poses great challenges, mainly related to the lack of a biological consensus of what a cell type actually represents and a patchy overview of existing cell types and their identity footprints on the transcriptomic level (63,8). During recent years, a large number of approaches and computational methods have been developed to address the attribution of cells to discrete types, however a one-fit-for-all approach is still lacking and the identification of highly specific, very similar or novel cell types remains challenging (64–66). At the most basic level, cell types are attributed iteratively to individual clusters after manual inspection of the expression of a handful of markers according to expert biological knowledge. The vast majority of scRNA-seq-based publications have taken this approach in the past (see e.g. (23–31)), in line with the above-mentioned limitations of specialized tools. However, such an approach is highly dependent on the availability of expert knowledge, does not scale to processing a large number of samples, and is poorly reproducible across individual studies.
In order to standardize this process, while maintaining the flexibility of adjusting marker genes and expression cut-offs across studies according to prior knowledge, we developed Sig-annot (Figure 1C), a Besca module that provides a hierarchical signature-enrichment based approach for cell type annotation (see next paragraph). To guarantee consistency across studies and communities, beyond scRNA-seq, the proposed cell type annotation schemas are based on the Cell Ontology (42), which is accessible via the Experimental Factor Ontology (67). The controlled vocabularies at different cell type hierarchies are summarized in a nomenclature sheet (Supplementary Table S3) and can be easily extended with further cell types. Newly generated cell type annotations in this manuscript provide the most fine-grained annotation as DBlabel assignment, which follows the Cell Ontology whenever possible, as well as higher level annotations according to the nomenclature sheet distributed with Besca.
Sig-annot, Besca’s signature-based hierarchical cell annotation schema
Sig-annot is Besca’s streamlined version of the manual process of cluster attribution based on marker gene enrichment, including ready-to-go annotation schemas for a broad range of cell types, with a particular focus on immune cells. The flexible, multi-level identification schemas are based on a nomenclature file (Supplementary Table S3) containing over 300 cell types and their hierarchical relations as well as the corresponding cell type signatures (see Materials and Methods). Depending on signatures and employed cutoffs, novel cell types not yet covered will be assigned as ‘animal cell’, awaiting inclusion in the schemas. As the scoring is not dependent on a sample's heterogeneity, the annotation result is identical no matter if cells of a single type are present (e.g. only naive B cells) or if a mix of diverse types is enquired (e.g. PBMCs or tumor digest), ensuring consistency and broad applicability. Default configuration files for human and mouse are provided, covering a large range of tissues and over 100 cell types (human: Supplementary Table S4, mouse: Supplementary Table S5). These files are easily customizable and users are free to provide additional schemas or annotations. The corresponding cell type signatures provided with Besca (Supplementary Table S6) are derived and adapted from various scRNA-seq experiments and publications, with subsequent manual curation, providing a comprehensive resource of harmonized cell type markers to be used out-of-the-box in novel experiments (see also Supplementary Document 1).
As demonstrated here, the signatures can be applied across tissues and potentially even species (with some dataset-specific adjustments) and represent a fast and consistent way of determining the most likely cell type composition in complex, large-scale scRNA-seq experiments. Other available marker-based cell annotation tools, like CellAssign (68), scCATCH (69), SCINA (70) and SCSA (71), focus on the underlying attribution method and only provide very limited (typically less than ten) and often tissue-specific gene sets. Alternatively, they provide comprehensive but highly redundant or even inconsistent sets. In contrast, we deliberately facilitate sharing of state-of-the-art signatures and validate their quality based on bulk RNA-seq data (Supplementary Document 1, Supplementary Figures S5 and S6). Thereby we aim to enhance the speed and reproducibility of the annotation process, in particular for analysts that may not be experts in the cell type composition of the sample they are analyzing.
For convenience, we have implemented various functions to guide the annotation based on the Sig-annot framework, and also provide visualization at individual steps in a dedicated cell annotation notebook. For instance, one can visualize the relation between the individual cell types as a graph (Figure 2A), plot the enrichment of individual signatures across all clusters in the dataset as a heatmap (Figure 2B), directly generate annotations at distinct levels in the cell hierarchy and add these in bulk to the AnnData (https://anndata.readthedocs.io) metadata.
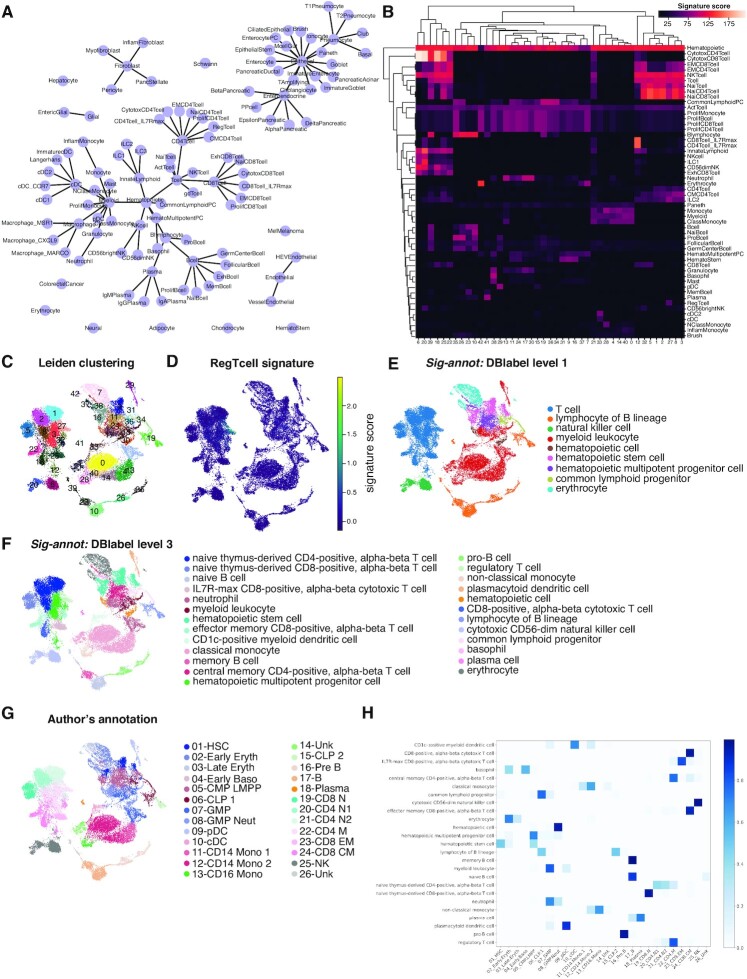
Figure 2.
Besca’s Sig-annot module applied. (A) Overview of the cell type hierarchy provided with Besca’s Sig-annot module and employed for annotating the datasets in the current manuscript. (B–E) Granja2019 data containing hematopoietic cells of multiple healthy donors from blood and bone marrow, probed by CITE-seq. (B) Hierarchically clustered heatmap showing enrichment of main signatures employed in the annotation across Leiden clusters, facilitating the evaluation of cluster attribution. (C) Overview of clustering in 2D UMAP space. (D) Overview of one of the signatures employed in cell annotation; regulatory T cells are typically rare in scRNA-seq experiments and often missed in annotation processes, but can be clearly detected in the Granja2019 dataset based on the Besca included signature. (E) Sig-annot cell type attribution at level 1, consisting of major cell types such as T cells and myeloid cells. All detected populations are broadly consistent with the original annotation (G). (F) Sig-annot cell type attribution at level 3, the highest resolution provided in Besca’s cell annotation schema. The detected populations are consistent with the original Granja annotation (G), cover T cell subsets with higher granularity (e.g. including regulatory T cells) and attribute the previously unknown (‘14_Unk’ and ‘26_Unk’) clusters as well. (G) Original cell type attribution as obtained from Granja et al. Annotated cell populations are highly consistent with clusters obtained from the reanalysis of the original data following the Besca standard workflow. (H) Confusion matrix showing overlap proportions between the original author's annotation of the dataset and Sig-annot cell type attribution at level 3. Further comparisons are summarized in Supplementary Table S7.
To exemplify this approach and its utility across samples of various origins and characteristics, we apply it to recent publicly available datasets covering most known hematopoietic cell types (72) in blood and tissue, showing that we are able to reproduce and enhance the original expert-driven annotations (23,24) (Figure 2 and 3). For instance, while regulatory T cells were annotated as group ‘22-CD4 M’ in the Granja2019 data, they now appear as a stand-alone cell group (Figure 2C–H and Supplementary Table S7). As one of the datasets also contains information on the expression of a large number of surface protein markers, we can confirm that our cell type attribution (Figure 3A and C) is in line with our current protein-level understanding of hematopoietic cell biology (Figure 3B and D, Supplementary Figure S1 and Supplementary Table S7). As described in detail in the Supplementary Document 2, this result is also consistent with annotations automatically obtained by SingleR (73), scANVI (52) and to some extent CellAssign (68) (but only at a limited granularity and ignoring the large fraction of unassigned cells), three broadly used and well-performing annotation tools using highly distinct underlying algorithms (64–66).
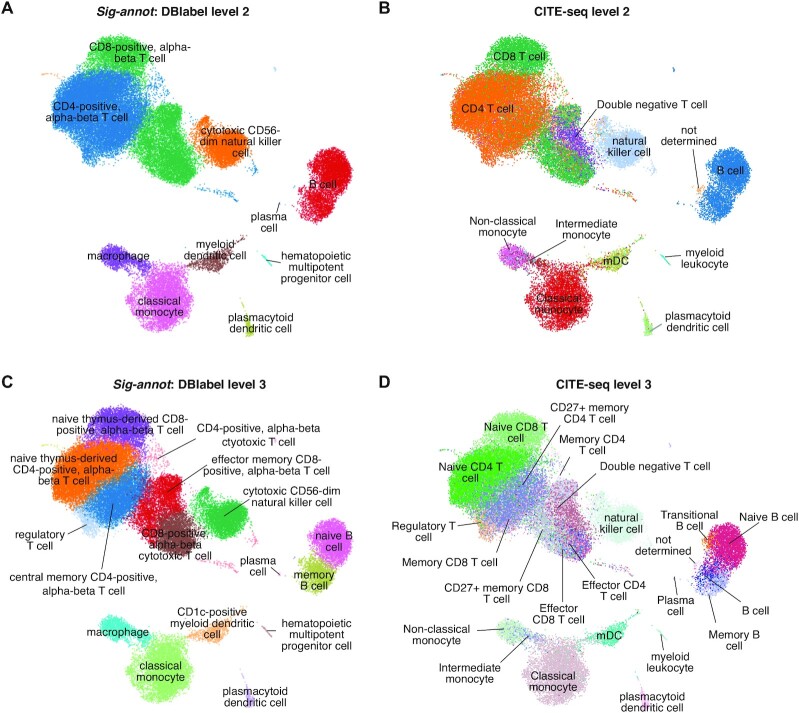
Figure 3.
Sig-annot applied to Kotliarov2020 data containing hematopoietic cells of multiple healthy donors from blood, probed by CITE-seq. (A) RNA signature-based cell type attribution at level 2, consisting of cell subtypes such as CD4+ T cells and classical monocytes. (B) Protein-marker based annotation using a gating method of classical FACS markers at a similar hierarchical depth as described in (A). Cell attribution is highly consistent with the automated RNA based results. (C, D) RNA signature-based (C) and protein-based (D) cell type attribution at the most fine-grained level 3. Even immune cell subtypes such as memory versus naive B cells or rare populations such as regulatory T cells and plasmacytoid dendritic cells are correctly attributed.
Importantly, we demonstrate that our approach is also applicable to more complex settings such as heterogeneous tumor samples, as exemplified by the annotation of publicly available colorectal and pancreatic cancer data (see Supplementary Figures S7, S8 and Supplementary Table S7). Specifically, when we re-analyzed 23 samples of tumor and non-malignant colon samples from colorectal cancer (CRC) patients (28) using Besca and Sig-annot, we found high correspondence with the original annotation not only at basic annotation levels such as main cell types (hematopoietic cells, fibroblasts, epithelial cells, tumor cells) (Supplementary Figure S7d), but also at the highest resolution (e.g. regulatory T cells, myeloid dendritic cells) (Supplementary Figure S7h–i). We made similar observations when we reanalyzed 11 tumor and non-malignant pancreatic samples from pancreatic ductal adenocarcinoma (PDAC) and non-pancreatic tumor patients (30), additionally obtaining finer-grained resolution compared to the original annotation (Supplementary Figure S8).
It remains difficult to compare annotations from different authors using different nomenclatures. To demonstrate the validity of Besca’s standard workflow followed by the Sig-annot approach, we evaluated the silhouette score (74), which is independent from a ground truth or cell type nomenclatures. The silhouette score helps determine if the granularity of cell types do reflect the variance between cell types compared to the intra-cluster variance. For all ten datasets, we assessed the cell type annotations from the original authors, the Besca-derived fine-grained DBlabel annotation, a coarse grained DBlabel annotation at level 2, and a random assignment (Supplementary Document 3). For example, in the case of the PBMC3k dataset, the silhouette score is 0.25 for the high granularity annotation, which was derived after reclustering (see Supplemental Figure S4), whereas the coarser resolution leads to a much higher silhouette score of 0.67. In contrast, the Lee2020 dataset shows 0.04 for the author's annotation, 0.20 for the fine-grained and 0.19 for the coarse-grained Sig-annot annotation. Here, colorectal cancer cells are numerous and divided into many clusters (Supplementary Figure S7a), but the cell type annotations (Supplementary Figure S7d–h) do not capture their diverse functions rendered in transcriptomic space, leading to overall low silhouette scores. For the other datasets we calculated the following average silhouette scores: Granja2019: 0.23 (author's annotation), 0.21 (fine-grained Sig-annot), 0.25 (coarse-grained Sig-annot); Kotliarov2020: 0.28, 0.29, 0.55; Smillie2019: 0.13, 0.14, 0.22; Martin2019: 0.30, 0.37, 0.32; Haber2017: 0.16, 0.25, 0.19; Segerstolpe2016: 0.06, 0.44, 0.19; Peng2019: 0.39, 0.12, 0.16; Baron2016: 0.43, 0.50, 0.14. Overall, the silhouette scores show that the Sig-annot annotations do consistently capture the variability of cells present as well as the author's annotations (see Supplementary Document 3 for more details).
We investigated the clustering and annotation results with additional quantitative metrics, including the adjusted mutual information (AMI) and adjusted Rand index (ARI), which compare two clusterings independent of their label names, and the accuracy and F1 score, which directly compare two annotations, but can only be applied in case of overlapping label names. To be able to compare to the author's annotation, we translated their cell type names to the DBlabel nomenclature (Supplementary Table S3) at different hierarchical levels. We compared the Sig-annot annotation to the translated author's annotation for the Granja2019 data (Figure 2), the Lee2020 data (Supplementary Figure S7), and the Peng2019 data (Supplementary Figure S8). The accuracy (Acc) and F1 scores are generally higher for the coarse-grained annotations and decrease with more fine-grained levels: Granja2019 Acc: 0.82 (level 1), 0.73 (level 2), 0.52 (level 3); Lee2020 Acc: 0.65, 0.76, 0.48; Peng2019 Acc: 0.81 (level 1), 0.69 (level 3); whereas the AMI and ARI scores remain stable across these levels: Granja2019 AMI: 0.80, 0.82, 0.79; Lee2020 AMI: 0.93, 0.83, 0.80; Peng2019 AMI: 0.92, 0.89. Additional scores for all datasets tested and visualized in the main or supplementary figures are summarized in Supplementary Table S7.
We note that we employed the same set of signatures and highly similar configuration files across datasets, successfully obtaining consistent annotations of hematopoietic cells derived from independent experiments, each with distinct levels of resolution and cell type frequency and representation, covering human blood, bone marrow, tumor and non-malignant pancreas and intestine. Importantly, our approach is automated, in the sense that only minimal changes (if any) are required for re-annotating each dataset should e.g. filtering/clustering be modified. It is also fully reproducible if the signature matrix and configuration files are stored for each annotation event. The distinct levels provide flexibility in terms of the annotation depth—one can easily choose to inspect differences between myeloid cells and T cells, or alternatively examine myeloid cell subsets, as each cell is attributed all hierarchical annotation levels present in the configuration file.
Auto-annot, Besca’s supervised machine learning module for cell type annotation
In addition to the signature-based annotation approach, Besca provides the Auto-annot module (Figure 1D), a supervised machine learning workflow for automated cell type annotation based on well annotated training datasets. Recently, supervised machine learning has become a popular alternative to signature-based cell type annotation (68,75–77). Benchmarking studies of such methods revealed that tailored single-cell classifiers or deep learning algorithms do not perform significantly better than conventional general purpose machine-learning methods (64,78). Therefore, we implemented methods for supervised machine learning based on the classical approaches and robust libraries for support vector machines (SVMs) or logistic regression utilizing scikit-learn (https://scikit-learn.org). One or multiple annotated reference datasets can be used to train a classifier for the annotation of a test dataset. Further details of the implementation are described in the Methods. Additionally, a semi-supervised generative model implemented by scANVI (52) can be utilized directly from Besca’s functions.
We demonstrate the application of Auto-annot on scRNA-seq data from healthy PBMCs. The datasets Kotliarov2020 and Granja2019 (Table 1, (23,24)) were used to train a logistic regression model, which was then tested on the PBMC3k dataset (Table 1, https://www.10xgenomics.com/). An initial annotation of the PBMC3k dataset was performed using Besca’s workflow and the Sig-annot procedure with a reclustering on NK and T cells (Supplementary Figure S4). We note that the training data includes far more cells and is annotated more fine-grained, a scenario we expect for predicting cell identities in newly sequenced datasets, while training on deeply annotated reference datasets derived from larger cell atlases.
The reference annotation (Figure 4A) is broadly reproduced by the predicted annotation (Figure 4B, Acc 0.64, AMI 0.62, see Supplementary Table S7 for additional measures), which also highly overlaps with the unsupervised Leiden clustering from Besca’s standard workflow (Figure 4C). For B cells, it provides even higher resolution than the reference annotation, correctly separating them into memory and naive B cells (Figure 4B), as independently confirmed by the according signatures (Figure 4D, E). The automated annotation for T cells shows some ambiguity, which reveals the limitations of the method (Figure 4A, F). Still, the specific IL7R-max CD8 T cells were correctly identified (Figure 4F) showing that accurate subdivisions within T cells are possible.
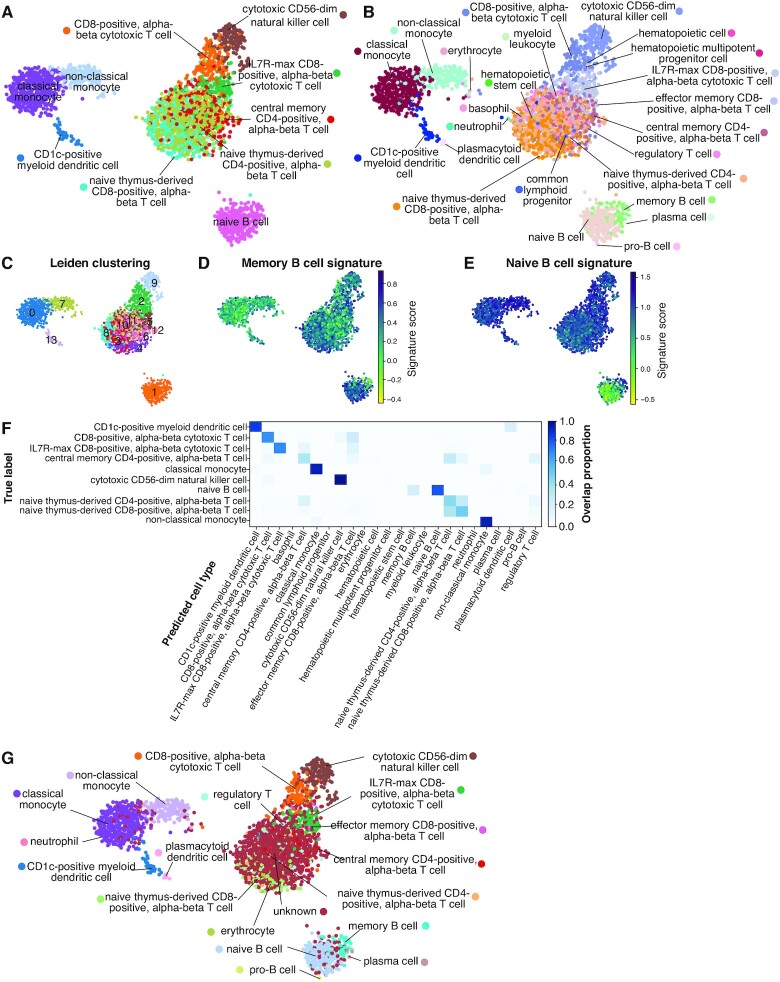
Figure 4.
Auto-annot applied to PBMCs using a logistic regression model trained on the Kotliarov2020 and Granja2019 datasets and tested in the PBMC3k dataset. (A) Overview of DBlabel annotations in 2D UMAP space for the PBMC3k test dataset. (B) Auto-annot largely recovers the original cell types. Finer divisions are uncovered in B cells, but resolution is lost for some T cell subtypes. (C) Overview of Leiden clustering in 2D UMAP space shows high overlap with predictions and illustrates the difficulty of finding subclusters in overlapping T cell communities. (D) The memory B cell signature supports the separation of the B cell cluster in (B). (E) Idem for the naive B cell signature. (F) The confusion matrix shows that misclassifications, if they do occur, generally misannotate very similar cell types. (G) Overview of Auto-annot labels with threshold. Ambiguity in some T cell subtypes leads to classification as unknown, all other cell types remain identified. The corresponding confusion matrix can be found in Supplementary Figure S9. Further comparisons are summarized in Supplementary Table S7.
In order to avoid false positive annotations it is possible to set a threshold for cells with low annotation scores. The threshold approach labels most of the ambiguous T cells as unknowns (Figure 4G and Supplementary Figure S9), removing almost all misclassifications at the cost of some cell types. As a result, central memory CD4 T cells remain virtually undetected, resulting in lower measures: Acc 0.44, AMI 0.58 (see also Supplementary Table S7). However, little changes occur when it comes to other cell types, including IL7R-max CD8 T cells, suggesting that this approach mainly flags out ambiguous attributions.
For comparison, we also trained a SVM model in the same scenario (Supplementary Figure S10). Compared to the logistic regression approach it labels most of the T cells into one larger naive CD4 T cell cluster resulting in higher accuracy scores (Acc 0.71, AMI 0.69, Supplementary Table S7), but neglecting the subdivision of these T cells compared to the logistic regression model.
It is notable how accurate the supervised approach works with a fine-grained training annotation. Still, an automated annotation based on less fine-grained cell types leads to even clearer results and higher accuracy: Acc 0.81, AMI 0.7; Multiple different cell types being co-located in the same broad cell type class from the reference annotation does not occur when we applied it to broader cell types (see Supplementary Figure S11 for Optimised Classes and Supplementary Table S7).
We performed additional cross-validation of the supervised Auto-annot approach on hematopoietic cells using the Granja2019 and Kotliarov2020 datasets on their own (see Supplementary Figures S12, S13 and Supplementary Table S7) and on pancreatic cells utilizing the Segerstolpe2016, Peng2019 and Baron2016 annotations in three different combinations (see Supplementary Figures S14, S15, S16, and Supplementary Table S7). In addition, we compared the results of the Auto-annot module to the results from SingleR (73), scANVI (52) and CellAssign (68) in the PBCM3k dataset (see Supplementary Document 2). In short, the comparison revealed a similar performance of the logistic regression in Auto-annot compared to scANVI and SingleR and the accuracy and F1 measures for the PBMC3k data (see above) are comparable to the scANVI (Acc 0.67) and SingleR (Acc 0.68) results (see also Supplementary Table S7).
Together, our results show that these approaches work best when the training set contains all cell types present in the test set, and when transcriptional differences between cell types are large and stable. These observations were also made in a recent benchmarking exercise of automated cell annotation tools (79). Benchmarking such methods, especially for the needed case of inter-study predictions and difficult to separate cell types remain difficult to assess due to the lack of a reliable ground truth (66). We note that benchmarking studies revealed larger differences in performance between evaluated tissues or datasets than between the different methods assessed (64–66), highlighting the challenge.
Auto-annot facilitates a cohesive understanding of intestinal cell types
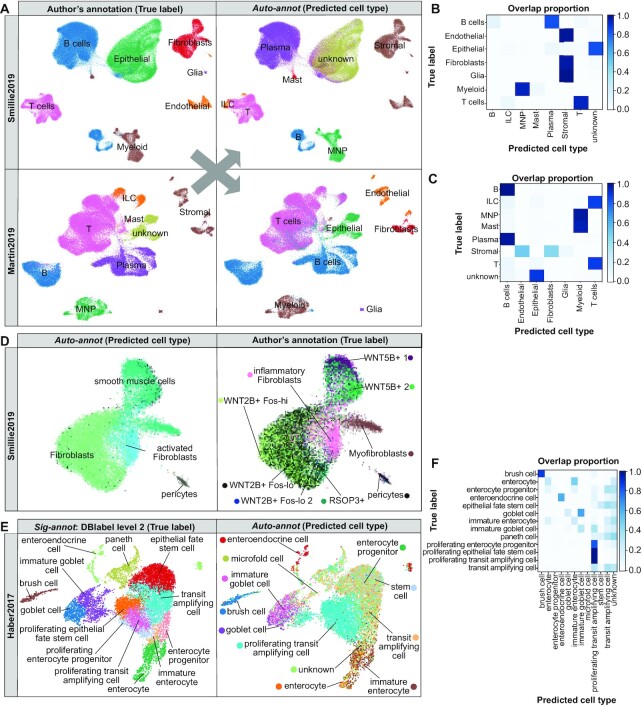
Recent studies revealed the intestinal cell type composition utilizing single-cell transcriptomics of intestinal biopsies taken from inflammatory bowel disease (IBD) patients (including ulcerative colitis and Crohn's disease), healthy donors or mice (25–27) as reviewed in (80). The utilized cell type nomenclatures are inconsistent between these studies and while various novel cell types were discovered, a consolidated understanding of all intestinal cell types is still missing (80,81). Here, we exemplify the power of Besca’s supervised machine learning method Auto-annot in resolving such inconsistencies by cross-validating disparate cell type annotations. We focus on two major studies: Smillie2019 (human colon epithelium and lamina propria during ulcerative colitis) (25) and Martin2019 (human ileum lamina propria during Crohn's disease) (26). In addition, we perform cell type annotation across species using the Haber2017 dataset (mouse small intestine epithelium) (27).
Firstly, we use the Smillie2019 and Martin2019 datasets to train a model with one dataset and apply it to the other, respectively (Figure 5A–D, Supplementary Figures S17, S18 and Supplementary Table S7). Both datasets were processed with Besca’s standard workflow and cell type annotations were adopted from the respective publications, including a coarse (Figure 5A, left and Supplementary Figure S17) as well as a fine grained cell type annotation (see Supplementary Figure S18 and for Smillie2019 fine-grained fibroblasts Figure 5D, left). Epithelial cell annotations are missing from the Martin2019 author's annotation, because those cells were excluded from the analysis in the original study. Therefore, they are labelled as ‘unknown’ in our comparison. The Auto-annot module identifies the corresponding cell types in the unseen dataset, respectively, revealing differences of the individual coarse annotations. For instance, while B and plasma cells were subclassified in Martin2019, only Smilie2019 separated the stromal compartment into fibroblasts, glial and endothelial cells. Taken together, confidence in such cell communities can easily be obtained by Auto-annot.
Figure 5.
Supervised machine learning to compare intestinal cell type annotations in scRNA-seq data. (A) UMAP representations of the coarse-grained cell types annotated in the Smillie2019 and Martin2019 datasets based on author's annotations (left) and predictions based on Besca’s Auto-annot module (right). (B, C) Confusion matrices comparing the true labels from the author's annotation and the cell types predicted in the Smillie2019 dataset from the Martin2019 annotation (B) and in the Martin2019 dataset predicted from Smillie2019 (C). (D) Discovery of inflammatory or activated fibroblasts point to the same cell community in both studies as exemplified in the Smillie2019 dataset by the author's annotation (left) and prediction from Martin2019 (right). (E) UMAP representations of the mouse small intestinal epithelial cells from Haber2017 showing the reference DBlabel cell type annotation (left) and cell types predicted from Smillie2019 human colon (right), and (F) the corresponding confusion matrix. Further comparisons are summarized in Supplementary Table S7.
Most interestingly, in both studies a new type of disease-relevant fibroblasts was discovered and named inflammatory fibroblasts in ulcerative colitis (25) or activated fibroblasts in Crohn's disease (26). Here, we show as an example to compare fine-grained cell types, how our machine learning approach could clearly confirm that these two fibroblast communities correspond to the same cell type (Figure 5D and Supplementary Figure S18), unified in our DBlabel nomenclature as an ‘inflammatory fibroblast’, with a specific signature.
The fine-grained comparison revealed further differences in the cell type annotation, e.g. a cell community from the enteric nervous systems, which is named glial cells in Smillie2019 and enteric neurons in Martin2019 (Supplementary Figure S18). Another example are B cells, which are separated into follicular, germ center, and cycling B cells in Smillie2019, whereas Martin2019 provides a separation into naive and memory B cells (Supplementary Figure S18). Those differences in annotation could be driven by biological differences, experimental differences, or simply different cell type nomenclatures used. The results show that our approach can be used to match cell type identities across studies and obtain a more cohesive picture of a tissue's cell type composition.
Finally, we performed a cross-species comparison. The Haber2017 small intestine mouse dataset includes only epithelial cells and was used as a test dataset. As the training dataset we chose the Smillie2019 human colon dataset and trained the machine learning model on the epithelial cells only leading to accuracy scores between 0.23 (fine-grained DBlabel) and 0.66 (coarse-grained DBlabel level 2). This approach clearly identified enterocytes, enteroendocrine cells, goblet cells and brush (tuft) cells (Figure 5E, F). The overall gradient from stem and transit amplifying cells to precursor and fully differentiated cells was mainly reproduced, but with less accuracy than the aforementioned discrete cell types (Figure 5E, F). Paneth cells are highly abundant in the mouse small intestine (27), but mainly absent in colon and not annotated in the human colon training data (25). They were wrongly identified as stem cells or transit amplifying cells, which are their neighbouring cells in the intestinal stem cell niche. Still, most Paneth cells did not get assigned to any known cell type (Figure 5F), due to a threshold for cells with low annotation score in the Auto-annot method.
Similar results were achieved by using the fine-grained author's annotation from Smillie2019 and by the reverse prediction from mouse to human (Acc 0.18–0.60, see also Supplementary Table S7). The results show that a cross-species prediction is generally possible and Auto-annot can be applied to provide fast insights for translational research (see Supplementary Figure S19).
The quantitative assessment of all these comparisons between Smillie2019, Martin2019, and Haber2017 summarized in Supplementary Table S7 reveals high similarity between the annotations at coarse-grained levels (DBlabel level 1 and 2, Acc 0.82–0.96) and a significant drop in the comparability with the more fine-grained levels (Acc 0.08–0.71), which suggests a need for harmonizing such annotations across datasets. Precision, recall and F1 metrics per cell type are provided in the Supplementary Reports ZIP file for each comparison.
scRNA-seq-informed cell deconvolution through Bescape
Cell deconvolution aims to estimate cell type proportions from bulk transcriptomic data based on cell type specific gene expression profiles (GEPs). Derivation of GEPs relevant for different bulk RNA-seq experiments has remained a challenge. As scRNA-seq data is being collected and annotated at an unprecedented rate, this offers the potential to leverage on the newly gathered knowledge (82). As the deconvolution algorithms have made significant progress over the past years (48,83), the focus is now being placed on the specificity of the GEPs that are used as basis vectors to estimate the cell composition addressing platform, tissue and indication variability (84). This is where Besca’s cell type annotations from scRNA-seq have a direct impact.
Besca’s deconvolution framework Bescape facilitates the usage of established deconvolution methods directly on any scRNA-seq data of choice (Figure 1E). This is in contrast to most available tools, which do not offer the flexibility to introduce user defined cell specific GEPs, instead relying solely on the authors’ carefully curated ones. The application and performance of the cell deconvolution results are then limited to the scope of the tissue and cell types embedded in the curated set. For example, GEPs derived from microarray data from hematological malignancies will have a limited scope of application in deconvoluting cell proportions from bulk RNA-seq sequenced from solid tumor biopsies.
In order to allow for simple incorporation of reference scRNA-seq datasets to generate GEPs for cell types of interest and addressing challenges such as collinearity of closely related cell types, Bescape includes two cell deconvolution tools, SCDC (20) and MuSiC (21) (see Materials and Methods), which performed among the best in a recent benchmark (22). As most deconvolution methods are implemented in R, several steps are required which have been implemented to run seamlessly in the background within Bescape in a containerized environment (see Supplementary Figure S20).
To extract the information from a reference scRNA-seq dataset, two sets of GEPs are generated from the Besca workflow immediately following the cell type annotation step: (i) using all genes from the scRNA-seq reference dataset without performing any feature selection, through the functionality provided by SCDC and MuSiC or (ii) based on the subset of highly variable genes defined in the standard workflow. The first set of GEPs is suitable for use by MuSiC and SCDC where subsequent weighing of the different genes is performed. The second set of GEPs can be used as input basis matrix for a multitude of cell deconvolution tools such as EPIC (47) and CIBERSORT (48). The resulting GEPs derived from the Segerstolpe2016 and Kotliarov2020 datasets (Table 1, (24,29)) utilizing both strategies are shown for comparison in Supplementary Figures S21-S24.
Here, we focus on the first strategy, applied to both datasets, utilizing SCDC by example, as a representative demonstration of Bescape’s functionality. One of the advantage of SCDC, which follows an ensemble approach, is that it allows for multiple scRNA-seq reference datasets. Bulk RNA-seq was simulated from the pancreatic islets (29) and hematopoietic CITE-seq (24) datasets using the GEPs across all genes from the raw count. The use of simulated bulk RNA-seq, where the ground truth of the in silico ad-mixture is known, allows validation of the estimated cell proportions (see Materials and Methods).
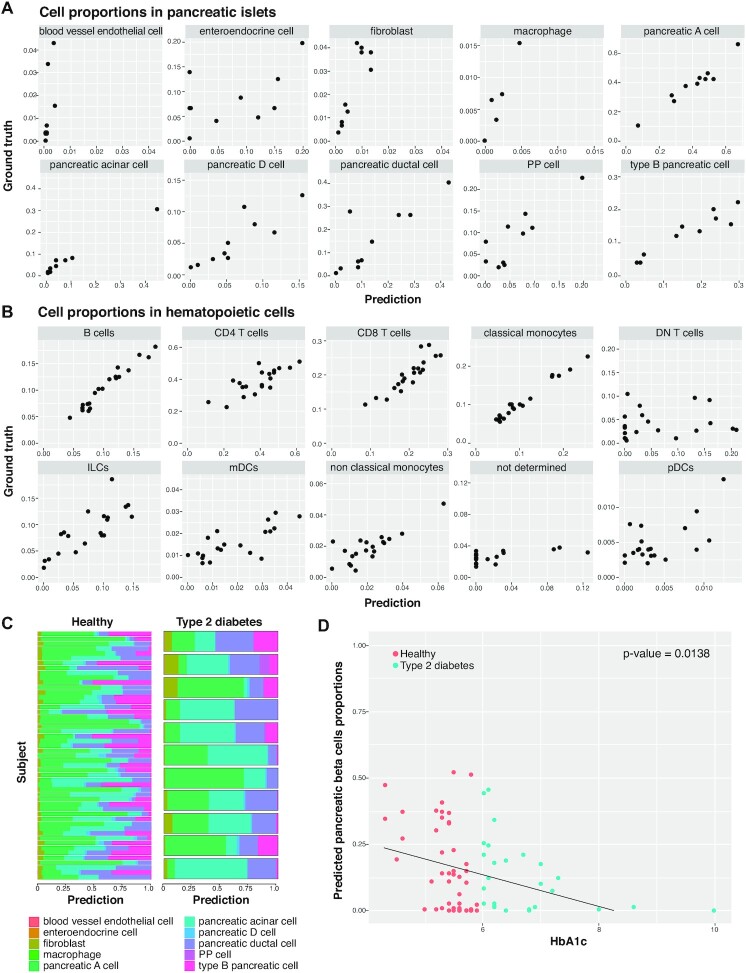
The estimated proportions from these simulated data using SCDC correlate highly with the ground truth across samples for both datasets (Figure 6A,B).The estimated proportions show high Pearson correlation with the ground truth, and corresponding low root mean square deviation (RMSD) and mean absolute deviation (mAD), in both tissues for all the cell types that were annotated from the Besca workflow (Tables 2 and 3). There are a few exceptions where the cell type proportions are of low abundances in the reference scRNA-seq dataset (see Tables 2 and 3) and where the GEPs are less well defined (see Supplementary Figures S21 and S23). More specifically, the deconvolution result is shown to be less performant for blood vessel endothelial cells and enteroendocrine cell in the simulated pancreatic islets dataset and for DN T cells, not determined, pDCs and mDCs labelled cell types in the simulated hematopoietic CITE-seq dataset.
Figure 6.
Cell deconvolution using Bescape. (A) Measured versus predicted cell proportions in pancreatic islets bulk RNA-seq simulated from Segerstolpe2016 (please refer to Table 2 for performance metrics). (B) Measured versus predicted cell proportions in hematopoietic bulk RNA-seq simulated from Kotliarov2020 (please refer to Table 3 for performance metrics). (C) Estimated cell proportions for real pancreatic islets bulk RNA-seq between type 2 diabetes patients and healthy controls. (D) Estimated pancreatic beta cells proportions for real pancreatic islets bulk RNA-seq between type 2 diabetes patients and healthy controls showing a significant negative association with glycated hemoglobin levels (HbA1c) to reject H0: slope = 0 (P-value = 1.38E–3) similar to the analysis performed in MuSiC.
Table 2.
SCDC deconvolution results based on simulated bulk RNA-seq from SCDC GEPs on pancreatic islets reference scRNA-seq from Segerstolpe2016
| Cell type | Pearson correlation | RMSD | mAD |
|---|---|---|---|
| blood vessel endothelial cell | 0.68 | 0.017 | 0.010 |
| enteroendocrine cell | 0.52 | 0.064 | 0.047 |
| fibroblast | 0.87 | 0.020 | 0.017 |
| macrophage | 0.98 | 0.004 | 0.002 |
| pancreatic A cell | 0.97 | 0.043 | 0.033 |
| pancreatic acinar cell | 0.99 | 0.049 | 0.024 |
| pancreatic D cell | 0.88 | 0.023 | 0.018 |
| pancreatic ductal cell | 0.83 | 0.075 | 0.043 |
| PP cell | 0.87 | 0.041 | 0.033 |
| type B pancreatic cell | 0.94 | 0.055 | 0.040 |
Table 3.
SCDC deconvolution results based on simulated bulk RNA-seq from SCDC GEPs on hematopoietic reference CITE-seq data from Kotliarov2020
| Cell type | Pearson correlation | RMSD | mAD |
|---|---|---|---|
| B cells | 0.98 | 0.009 | 0.007 |
| CD4 T cells | 0.79 | 0.075 | 0.062 |
| CD8 T cells | 0.90 | 0.021 | 0.016 |
| classical monocytes | 0.99 | 0.014 | 0.011 |
| DN T cells | 0.13 | 0.078 | 0.058 |
| ILCs | 0.81 | 0.031 | 0.024 |
| mDCs | 0.75 | 0.009 | 0.008 |
| non-classical monocytes | 0.81 | 0.009 | 0.007 |
| not determined | 0.56 | 0.033 | 0.026 |
| pDCs | 0.61 | 0.003 | 0.002 |
In contrast to evaluations on simulated data, it is important to note that the success of cell deconvolution on real datasets can be measured based on two merits. First, on the accuracy to known proportions estimated based on a known or proxy ground truth from matched samples measured with more traditional single-cell means (e.g. immunohistochemistry or flow cytometry). Although this is the preferred measure of success, validating the results compared to a ground truth obtained using known cell types can be difficult, as these more traditional methods for studying cell heterogeneity rely on a limited repertoire of markers of known cell types. Secondly, the success can also be measured based on the results obtained from embedding estimated cell proportions as covariates in prognostic and predictive models. Following the SCDC manuscript (20), we utilized this strategy as described in the next paragraph.
In a recent study of type 2 diabetes (85), the difference in the estimated beta pancreatic cell proportions between type 2 diabetic patients and healthy controls provides an opportunity to test the performance of deconvolution results. Estimated proportions obtained from the real bulk RNA-seq for all ten cell types using Besca cell annotation is shown in Figure 6C. The estimated pancreatic beta cells show the expected lower cell proportions in the type 2 diabetes patients as compared to healthy subjects as shown in Figure 6D.
In sum, both evaluations on simulated bulk RNA-seq data and the ability to detect a downstream biological effect suggest that the GEPs derived after Besca analysis, in combination with the implementation of the SCDC deconvolution algorithm facilitate the accurate deconvolution of bulk RNA-seq samples. Future extension of the Bescape module aims to incorporate additional cell deconvolution methods and further downstream analysis tools to help validate the estimated proportions based on feedback of the user community.
DISCUSSION
No two cells are identical; neither are two scRNA-seq experiments. Cells are extracted from different tissues, treated according to lab-specific protocols, and sequenced with a variety of technologies. Still, the vast amount of available scRNA-seq studies provokes the ambition to reuse the valuable experimental data and to re-assess them by comparing between studies. Streamlined and standardized workflows, such as those presented here, strive to find balance between automation and flexibility, as automation brings efficiency, reusability, and reproducibility of data processing. They strive to bring scRNA-seq results to a level that allows for cross-study comparisons, integration into larger cell atlases and continuous improvement of our general understanding of cell types and their characteristics.
As Besca builds upon and extends concepts and functions from Scanpy, each analysis step remains customizable and it seamlessly integrates with other toolkits such as scvi-tools (52) or specialized analysis tools such as scVelo (86) and CellRank (http://cellrank.org/) for cellular trajectory and fate (87) analysis or Scirpy (88) for T-cell receptor analysis. Besca is however not limited to Python modules and also includes optional R-based methods (e.g. DSB and SCtransform), added through the rpy2 module. This makes Besca very flexible, as it can integrate cutting-edge methods from both the Python and the R community. Still, its strength is to free the user from having to navigate individually through each method and parameter from the vast number of available options (89) by providing robust defaults that work in most cases. This abstraction has the drawback that some steps or parameters are hidden in wrapper functions not visible to the user.
Translational research is utilizing multiple modalities of single cell assays (90) and the integration of these multi-modal data is gaining importance in translational research. Besca’s workflow allows to process complementary RNA expression and protein abundance from CITE-seq experiments simultaneously and is prepared to adapt multi-modal analysis approaches (91) once they mature.
Besca’s Sig-annot module greatly facilitates streamlined and reusable scRNA-seq analyses by automating the annotation of the cell groups obtained by unsupervised clustering. It mirrors the still most widespread and accurate manual annotation approach, but provides a harmonized annotation schema and hence guarantees comparability between studies. It also captures the knowledge of cell type markers that is gained in this process in explicit gene signatures that can be easily shared, applied with other public marker-based annotation methods (68,70), re-assessed and improved across different conditions, studies and technologies.
This signature-based approach is valuable for specific tissues and disease phenotypes as an approach to harmonize annotations across various cell atlases, which is critical for holistic disease understanding (see e.g. (1,80,92–94)). Still, as each tissue and fine-grained cell type needs to be incorporated and optimized for in the annotation schema, despite our inclusion of over 100 cell type markers – one of the largest hand-curated resource currently available—many biological systems are not yet covered. Other recent efforts have pursued streamlining the manual maker-based annotation, including CellAssign (68), scCATCH (69), SCINA (70) and SCSA (71), and utilizing unified cell type nomenclatures and hierarchical relations such as OnClass (95) and scMatch (96).
However, in practice, various limitations remain in their direct and systematic applicability across datasets. For instance, some methods rely on differential expression between the clusters to derive marker genes that are then overlapped with marker gene databases, making them not suitable for samples where a limited number of cells are enquired. The markers provided are either very restricted (less than ten cell types) or at the other extreme, exhaustive, covering entire databases (e.g. CellMarker (55), CancerSEA (97), PanglaoDB (98)), but suffering from redundancy, inconsistency or lack of specificity. Most do not consider hierarchical relations between cell types, or only limited ones (two levels), requiring the user to rerun the annotation or to modify the granularity of the analysis (e.g. naive B cells, B cells or hematopoietic cells).
On the other end of the spectrum, the recently proposed OnClass (95) approach relies on the Cell Ontology, as we do here, building exhaustive hierarchical relations across all available cells. This introduces additional complexity in interpretation, given the very large number of intermediate cell types and relations thereof contained in the ontology. In contrast, in our approach, we chose to simplify this relation and also to ensure flexibility by allowing the user to determine the relations in a practical sense in the configuration sheet, as often many of the intermediate levels are not required or of interest at the single-cell analysis level.
When it comes to automating cell annotation, supervised approaches overcome certain challenges faced by unsupervised clustering (99) and therefore generalize better. Importantly, they allow for the utilization of curated high-quality annotations by transferring them to new studies more efficiently than a marker-based process. Such approaches not only allow for the fast comparison of cell annotations between studies, but even across species. They depend on well annotated reference datasets containing harmonized labelings. We expect cell atlas projects (see e.g. (93,100–104)) to provide such annotations in the near future for all major tissues, which would allow for a wide applicability of supervised approaches. With more data, advanced machine learning methods will dominate including new approaches to transfer annotations between atlases, species, and disease states (105), or to even predict cell types missing from the training data (95,106). Besca is primed to integrate such methods in its framework in the future.
Hard-to-classify fine-grained cell types and non-overlapping cell types between reference dataset and test dataset are the major challenges for supervised cell type annotation methods. Similar to the challenges in cell deconvolution methods, cell type annotation methods are prone to spillover effects, a term borrowed from flow cytometry, which lead to wrong predictions of the abundance of a certain cell type due to high correlation with the signature of a related cell type (82,107). In addition, low abundant cell types from the reference dataset might not provide reliable information to predict those cell types in the test dataset, especially if a cell type is missing from the reference, which might be solved with most recent methods such as MARS (106). These and several other challenges are being thoroughly investigated in recent cell deconvolution benchmarking papers (22,108). A similar line of development (e.g. recursive fitting for closely related cell types proposed by MuSiC (21)) may help to more accurately perform cell type classification.
In this manuscript, we assessed logistic regression as part of the Auto-annot module as a robust supervised machine learning method. We argue that it provides a straightforward approach to assign ‘unknown’ labels for such challenging predictions, because a probability score for the assignment of each individual cell to all reference cell types is calculated. This probability offers the application of a threshold that needs to be reached to assign a cell type, or ‘unknown’ otherwise. A limitation of this approach is that it does not provide a generalized fixed threshold or an optimization procedure to determine the threshold. The optimization would require a systematic benchmark across different datasets and scenarios. It would be further complicated by the fact that the ideal threshold depends on the type of subsequent analysis, which might require either a large proportion of annotated cells with lower confidence, or few very-high-confidence annotations. The overlap and granularity of cell types is usually not known a priori for the testing dataset and therefore we recommend to use the threshold to reduce the complexity of the predicted annotation and as a sanity check in combination with traditional marker gene assessment.
Various examples in this manuscript and previous studies show that the automation of cell type annotation is feasible to a certain extent, however the vast majority of discovery publications still resort to expert-driven manual annotation or at least adjustments. With Besca, the analyst retains full control, and is provided with robust, standardized and streamlined approaches that complement each other and enable to focus on the biological challenges in cell type annotation. As cells can be grouped by multiple orthogonal criteria such as surface markers, functions, cell cycle states, differentiation stages, or activation levels (109), a clear definition of concrete cell types remains controversial and a more general concept of cell types will be needed in the future (63).
Setting aside the controversy in cell type definition (110), our work already provides tools and best practices to achieve better reference annotations and to share the gene signatures that capture the knowledge about how they were derived, which is novel compared to most current studies. As the human reference genome, which does not ultimately reflect a human genome consensus (111), but serves many practical purposes (112), accelerated genomic research, such reference cell type annotations will accelerate our understanding of biological systems even though they reflect only a subset of a cell's characteristics.
Finally, these cell type definitions help investigate changes in cell composition and differentially expressed genes within certain cell types, which are often postulated as indications of disease progression or response to stimulation and perturbation (113). While scRNA-seq offers the possibility to investigate these hypotheses, the current cost as well as the technical and logistical challenges associated with the technology are preventing large scale studies (114), particularly in a clinical trial setting. Although this is likely to improve over time as the technology matures, large numbers of biological replicates are currently measured using bulk RNA-seq. In these samples, heterogeneity resulting from the distinct cell type composition of the probed material can often confound the signals, making it difficult to interpret results. By leveraging annotated reference scRNAseq datasets in combination with cell deconvolution methods, the cell composition of bulk RNA-seq samples can be robustly estimated. This information can then either be used directly as biomarkers or as covariates towards inferring more robust differential gene expression results.
It is important to point out the difference in algorithms between the different cell deconvolution tools that are being proposed and widely adopted. MuSiC and SCDC are based on weighted nonlinear least squares (W-NNLS) as opposed to CIBERSORT, which is based on support vector regression (SVR) framework. While the Besca workflow allows the user to extract the GEPs and apply the deconvolution method of their choice, we have included MuSiC and SCDC in the Bescape module for their seamless integration of incorporating multiple scRNA-seq reference data and their performance on a recent benchmark (49).
In sum, Besca extends broadly used analysis workflows based on Seurat (33), Scanpy (18), Scater (32) and scvi-tools (52) with additional functionalities: It ensures a standardized output structure of interoperable file formats to enhance reusability of results. It promotes the usage of a cell type nomenclature and provides one of the most comprehensive hand-curated publicly available resources for cell type gene signatures. It provides a framework, the Sig-annot module, to use those signatures for semi-automated cell type annotation. It provides reprocessed and harmonized reference datasets covering multiple tissues and disease states, including distinct cancer types (Table 1), which can directly be used for comparison, cell type prediction using the Auto-annot module, or deconvolution. It provides plotting functions to investigate data quality, to compare cell type annotations, or to explore differentially expressed genes between conditions.
The core benefits of adopting Besca for scRNA-seq data analysis are automation, standardization, and reusability. We expect that Besca, published as an open-source software contribution to the community, will promote interoperability, reusability, and interpretability of scRNA-seq data. Finally, Besca will be part of the many components that pave the way for a reference catalogue of cell types and their reactions to various perturbations. This catalogue will allow a deeper understanding of human diseases and their interventions.
AVAILABILITY OF DATA AND SOURCE CODE
Besca source code and installation
The Besca source code is available from GitHub: https://github.com/bedapub/besca. Release 2.4 is also available from Zenodo: https://doi.org/10.5281/zenodo.4551125.
Besca can be installed through Python's package manager pip with the following command: pip install git+https://github.com/bedapub/besca.git.
The Bescape source code is available from GitHub: https://github.com/bedapub/bescape.
Bescape can be installed through Python's package manager pip with the following command: pip install bescape.
Besca dependencies
Besca depends mainly on Python 3 (https://www.python.org/), Scanpy (https://github.com/theislab/scanpy), anndata (https://anndata.readthedocs.io/), leidenalg (https://github.com/vtraag/leidenalg), bbknn (https://github.com/Teichlab/bbknn), umap-learn (https://umap-learn.readthedocs.io/), Scanorama (https://github.com/brianhie/scanorama), and scikit-learn (https://scikit-learn.org/).
The Bescape container needs Docker (https://docs.docker.com/get-docker/) or Singularity (https://sylabs.io/guides/3.0/user-guide/installation.html) to run.
Bescape depends mainly on R (https://www.r-project.org/), MuSiC (https://github.com/xuranw/MuSiC), and SCDC (https://github.com/meichendong/SCDC).
Datasets
All processed datasets are available from the Besca community in Zenodo: https://zenodo.org/communities/besca/.
All original datasets are available from the respective publications or under the accession numbers given in Table 1.
Results workbooks
Jupyter notebooks (https://jupyter.org/) used to generate results and figures in this manuscript are available from GitHub: https://github.com/bedapub/besca_publication_results.
The cell type gene signature validation analysis is available from GitHub: https://github.com/bedapub/besca_publication_signature_validation.
R markdown files (https://rmarkdown.rstudio.com/) used to generate results and figures in this manuscript are available from GitHub: https://github.com/bedapub/bescape/tree/master/bescape/docker_files.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all providers of publicly available datasets utilized in this manuscript for making those data available, all Besca users for their feedback, and the anonymous reviewers for their helpful comments to improve the manuscript and the toolkit. We thank all developers of tools mentioned in this manuscript, especially those who maintain Scanpy, SCDC and MuSiC. We thank Andreea Ciuprina, Bhavesh Soni and Mariia Bilous for testing Besca. In particular, we thank Alberto Valdeolivas Urbelz for Besca maintenance, Martha Serrano for data pre-processing, Llucia Alberti Servera for testing and signature refinement, and Ramona Schlenker, Marina Bacac, Stephan Gasser, Mi He, Marisa Mariani and Sylvia Herter for fruitful discussions on cell populations and their gene signatures. This publication is part of the Human Cell Atlas: www.humancellatlas.org/publications.
Authors' contributions: S.C.M., A.J.L., R.S., M.E., L.B., T.K.T., P.C.S. and K.H. contributed to the conception and design of the work. S.C.M., A.J.L., L.W., M.P., A.S., E.U., J.D.Z., T.K.T., P.C.S. and K.H. contributed to analysis and interpretation of data. All authors contributed to the development of the software. S.C.M., A.J.L., L.W., M.P., A.S., J.D.Z., T.K.T., P.C.S. and K.H. wrote the manuscript. All authors approved the final version of the manuscript.
Notes
Present address: Sophia Clara Mädler. Department of Proteomics and Signal Transduction, Max Planck Institute of Biochemistry, Martinsried, Germany.
Contributor Information
Sophia Clara Mädler, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Alice Julien-Laferriere, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland; Soladis GmbH, Basel, Switzerland.
Luis Wyss, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland; Department of Biosystems Science and Engineering, ETH Zurich, Basel, Switzerland.
Miroslav Phan, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland; Department of Biosystems Science and Engineering, ETH Zurich, Basel, Switzerland.
Anthony Sonrel, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland; Department of Molecular Life Sciences, University of Zürich, Zürich, Switzerland.
Albert S W Kang, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Eric Ulrich, Roche Pharma Research and Early Development, I2O Disease Translational Area, Roche Innovation Center Basel, Basel, Switzerland.
Roland Schmucki, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Jitao David Zhang, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Martin Ebeling, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Laura Badi, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Tony Kam-Thong, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Petra C Schwalie, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
Klas Hatje, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, Basel, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
F. Hoffmann-La Roche.
Conflict of interest statement. All authors were previously or are currently employed by F. Hoffmann-La Roche Ltd. AJL is employed by Soladis GmbH. The authors declare that they have no other competing interests.
REFERENCES
- 1. Muus C., Luecken M.D., Eraslan G., Waghray A., Heimberg G., Sikkema L., Kobayashi Y., Vaishnav E.D., Subramanian A., Smilie C. et al. Single-cellmeta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat. Med. 2021; 27:546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M. et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020; 181:1016–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim J., Koo B.-K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020; 21:571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Q., Caudle W.M., Pi J., Bhattacharya S., Andersen M.E., Kaminski N.E., Conolly R.B. Embracing systems toxicology at single-cell resolution. Curr. Opin. Toxicol. 2019; 16:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020; 15:1484–1506. [DOI] [PubMed] [Google Scholar]
- 6. Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P. et al. Single-cell transcriptomics of human t cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019; 10:4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019; 37:547–554. [DOI] [PubMed] [Google Scholar]
- 8. Lee J.T.H., Hemberg M. Supervised clustering for single-cell analysis. Nat. Methods. 2019; 16:965–966. [DOI] [PubMed] [Google Scholar]
- 9. Hie B., Peters J., Nyquist S.K., Shalek A.K., Berger B., Bryson B.D. Computational methods for single-cell RNA sequencing. Annu. Rev. Biomed. Data Sci. 2020; 3:339–364. [Google Scholar]
- 10. Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019; 15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stegle O., Teichmann S.A., Marioni J.C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015; 16:133–145. [DOI] [PubMed] [Google Scholar]
- 12. Germain P.-L., Sonrel A., Robinson M.D. pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single cell RNA-seq preprocessing tools. Genome Biol. 2020; 21:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen W., Zhao Y., Chen X., Yang Z., Xu X., Bi Y., Chen V., Li J., Choi H., Ernest B. et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nat. Biotechnol. 2020; 13:1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andrews T.S., Kiselev V.Y., McCarthy D., Hemberg M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat. Protoc. 2021; 16:1–9. [DOI] [PubMed] [Google Scholar]
- 15. Miao Z., Moreno P., Huang N., Papatheodorou I., Brazma A., Teichmann S.A. Putative cell type discovery from single-cell gene expression data. Nat. Methods. 2020; 17:621–628. [DOI] [PubMed] [Google Scholar]
- 16. Angerer P., Simon L., Tritschler S., Wolf F.A., Fischer D., Theis F.J. Single cells make big data: new challenges and opportunities in transcriptomics. Curr. Opin. Syst. Biol. 2017; 4:85–91. [Google Scholar]
- 17. Svensson V., Vento-Tormo R., Teichmann S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018; 13:599–604. [DOI] [PubMed] [Google Scholar]
- 18. Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018; 19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017; 14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dong M., Thennavan A., Urrutia E., Li Y., Perou C.M., Zou F., Jiang Y. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Brief. Bioinform. 2021; 22:427–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 2019; 10:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avila Cobos F., Vandesompele J., Mestdagh P., De Preter K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinforma. Oxf. Engl. 2018; 34:1969–1979. [DOI] [PubMed] [Google Scholar]
- 23. Granja J.M., Klemm S., McGinnis L.M., Kathiria A.S., Mezger A., Corces M.R., Parks B., Gars E., Liedtke M., Zheng G.X.Y. et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 2019; 37:1458–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kotliarov Y., Sparks R., Martins A.J., Mulè M.P., Lu Y., Goswami M., Kardava L., Banchereau R., Pascual V., Biancotto A. et al. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat. Med. 2020; 26:618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J. et al. Intra- and Inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019; 178:714–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L., Nayar S., Greenstein A.J. et al. Single-Cell analysis of crohn's disease lesions identifies a pathogenic cellular module associated with resistance to Anti-TNF therapy. Cell. 2019; 178:1493–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y. et al. A single-cell survey of the small intestinal epithelium. Nature. 2017; 551:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H.-O., Hong Y., Etlioglu H.E., Cho Y.B., Pomella V., Van den Bosch B., Vanhecke J., Verbandt S., Hong H., Min J.-W. et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020; 52:594–603. [DOI] [PubMed] [Google Scholar]
- 29. Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M.K. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016; 24:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng J., Sun B.-F., Chen C.-Y., Zhou J.-Y., Chen Y.-S., Chen H., Liu L., Huang D., Jiang J., Cui G.-S. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019; 29:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016; 3:346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCarthy D.J., Campbell K.R., Lun A.T.L., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinforma. Oxf. Engl. 2017; 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnsson K., Soneson C., Fontes M. Low bias local intrinsic dimension estimation from expected simplex skewness. IEEE Trans. Pattern Anal. Mach. Intell. 2015; 37:196–202. [DOI] [PubMed] [Google Scholar]
- 35. Polański K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., Park J.-E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020; 36:964–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McInnes L., Healy J., Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. 2018; bioRxiv doi:18 September 2020, preprint: not peer reviewedhttps://arxiv.org/abs/1802.03426v3. [Google Scholar]
- 37. Traag V.A., Waltman L., van Eck N.J. From louvain to leiden: guaranteeing well-connected communities. Sci. Rep. 2019; 9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mulè M.P., Martins A.J., Tsang J.S. Normalizing and denoising protein expression data from droplet-based single cell profiling. 2020; bioRxiv doi:28 February 2021, preprint: not peer reviewed 10.1101/2020.02.24.963603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hahne F., LeMeur N., Brinkman R.R., Ellis B., Haaland P., Sarkar D., Spidlen J., Strain E., Gentleman R. flowCore: a bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009; 10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ellis B., Haal P., Hahne F., Meur N.L., Gopalakrishnan N., Spidlen J., Jiang M., Finak G., Granjeaud S. flowCore: flowCore: basic structures for flow cytometry data bioconductor version: release (3.11). 2020; [Google Scholar]
- 41. Waugh K.A., Araya P., Pandey A., Jordan K.R., Smith K.P., Granrath R.E., Khanal S., Butcher E.T., Estrada B.E., Rachubinski A.L. et al. Mass cytometry reveals global immune remodeling with multi-lineage hypersensitivity to type i interferon in down syndrome. Cell Rep. 2019; 29:1893–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diehl A.D., Meehan T.F., Bradford Y.M., Brush M.H., Dahdul W.M., Dougall D.S., He Y., Osumi-Sutherland D., Ruttenberg A., Sarntivijai S. et al. The cell ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semant. 2016; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hie B., Bryson B., Berger B. Efficient integration of heterogeneous single-cell transcriptomes using scanorama. Nat. Biotechnol. 2019; 37:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chazarra-Gil R., van Dongen S., Kiselev V.Y., Hemberg M. Flexible comparison of batch correction methods for single-cell RNA-seq using batchbench. Nucleic Acids Res. 2021; 49:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luecken M.D., Büttner M., Chaichoompu K., Danese A., Interlandi M., Mueller M.F., Strobl D.C., Zappia L., Dugas M., Colomé-Tatché M. et al. Benchmarking atlas-level data integration in single-cell genomics. 2020; bioRxiv doi:27 May 2020, preprint: not peer reviewed. 10.1101/2020.05.22.111161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abbas A.R., Wolslegel K., Seshasayee D., Modrusan Z., Clark H.F. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009; 4:e6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017; 6:e26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015; 12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Avila Cobos F., Alquicira-Hernandez J., Powell J.E., Mestdagh P., De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 2020; 11:5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019; 37:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilkinson M.D., Dumontier M., Aalbersberg Ij.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu C., Lopez R., Mehlman E., Regier J., Jordan M.I., Yosef N. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst. Biol. 2021; 17:e9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020; 48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Z., Monteiro C.D., Jagodnik K.M., Fernandez N.F., Gundersen G.W., Rouillard A.D., Jenkins S.L., Feldmann A.S., Hu K.S., McDermott M.G. et al. Extraction and analysis of signatures from the gene expression omnibus by the crowd. Nat. Commun. 2016; 7:12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019; 47:D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015; 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang J.D., Hatje K., Sturm G., Broger C., Ebeling M., Burtin M., Terzi F., Pomposiello S.I., Badi L. Detect tissue heterogeneity in gene expression data with BioQC. BMC Genomics. 2017; 18:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roudnicky F., Zhang J.D., Kim B.K., Pandya N.J., Lan Y., Sach-Peltason L., Ragelle H., Strassburger P., Gruener S., Lazendic M. et al. Inducers of the endothelial cell barrier identified through chemogenomic screening in genome-edited hPSC-endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:19854–19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uhlen M., Karlsson M.J., Zhong W., Tebani A., Pou C., Mikes J., Lakshmikanth T., Forsström B., Edfors F., Odeberg J. et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019; 366:eaax9198. [DOI] [PubMed] [Google Scholar]
- 61. GTEx Consortium The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020; 369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Forrest A.R.R., Kawaji H., Rehli M., Kenneth Baillie J., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M. et al. A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wagner A., Regev A., Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 2016; 34:1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M.J.T., Mahfouz A. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019; 20:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang Q., Liu Y., Du Y., Garmire L.X. Evaluation of cell type annotation r packages on single cell RNA-seq data. Genomics Proteomics Bioinformatics. 2020; 10.1016/j.gpb.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao X., Wu S., Fang N., Sun X., Fan J. Evaluation of single-cell classifiers for single-cell RNA sequencing data sets. Brief. Bioinform. 2020; 21:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Malone J., Holloway E., Adamusiak T., Kapushesky M., Zheng J., Kolesnikov N., Zhukova A., Brazma A., Parkinson H. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010; 26:1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang A.W., O’Flanagan C., Chavez E.A., Lim J.L.P., Ceglia N., McPherson A., Wiens M., Walters P., Chan T., Hewitson B. et al. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat. Methods. 2019; 16:1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shao X., Liao J., Lu X., Xue R., Ai N., Fan X. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020; 23:100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang Z., Luo D., Zhong X., Choi J.H., Ma Y., Wang S., Mahrt E., Guo W., Stawiski E.W., Modrusan Z. et al. SCINA: a Semi-supervised subtyping algorithm of single cells and bulk samples. Genes. 2019; 10:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cao Y., Wang X., Peng G. SCSA: a cell type annotation tool for single-cell RNA-seq data. Front. Genet. 2020; 11:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y., Gao S., Xia J., Liu F. Hematopoietic hierarchy – an updated roadmap. Trends Cell Biol. 2018; 28:976–986. [DOI] [PubMed] [Google Scholar]
- 73. Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019; 20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987; 20:53–65. [Google Scholar]
- 75. Pliner H.A., Shendure J., Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods. 2019; 16:983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li C., Liu B., Kang B., Liu Z., Liu Y., Chen C., Ren X., Zhang Z. SciBet as a portable and fast single cell type identifier. Nat. Commun. 2020; 11:1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lin Y., Cao Y., Kim H.J., Salim A., Speed T.P., Lin D.M., Yang P., Yang J.Y.H. scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Mol. Syst. Biol. 2020; 16:e9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Köhler N.D., Büttner M., Theis F.J. Deep learning does not outperform classical machine learning for cell-type annotation. 2019; bioRxiv doi:24 June 2021, preprint: not peer reviewed 10.1101/653907. [DOI]
- 79. Huang Q., Liu Y., Du Y., Garmire L.X. Evaluation of cell type annotation r packages on Single-cell RNA-seq data. Genomics Proteomics Bioinformatics. 2020; 10.1016/j.gpb.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bigaeva E., Uniken Venema W.T.C., Weersma R.K., Festen E.A.M. Understanding human gut diseases at single-cell resolution. Hum. Mol. Genet. 2020; 29:R51–R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Corridoni D., Chapman T., Antanaviciute A., Satsangi J., Simmons A. Inflammatory bowel disease through the lens of Single-cell RNA-seq technologies. Inflamm. Bowel Dis. 2020; 26:1658–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sturm G., Finotello F., Petitprez F., Zhang J.D., Baumbach J., Fridman W.H., List M., Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019; 35:i436–i445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gaujoux R., Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013; 29:2211–2212. [DOI] [PubMed] [Google Scholar]
- 84. Schelker M., Feau S., Du J., Ranu N., Klipp E., MacBeath G., Schoeberl B., Raue A. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat. Commun. 2017; 8:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L., Taneera J., Storm P., Osmark P., Ladenvall C., Prasad R.B. et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13924–13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020; 38:1408–1414. [DOI] [PubMed] [Google Scholar]
- 87. Setty M., Kiseliovas V., Levine J., Gayoso A., Mazutis L., Pe’er D. Characterization of cell fate probabilities in single-cell data with palantir. Nat. Biotechnol. 2019; 37:451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sturm G., Szabo T., Fotakis G., Haider M., Rieder D., Trajanoski Z., Finotello F. Scirpy: a scanpy extension for analyzing single-cell T-cell receptor sequencing data. Bioinforma. Oxf. Engl. 2020; 36:4817–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zappia L., Phipson B., Oshlack A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 2018; 14:e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhu C., Preissl S., Ren B. Single-cell multimodal omics: the power of many. Nat. Methods. 2020; 17:11–14. [DOI] [PubMed] [Google Scholar]
- 91. Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M. et al. Integrated analysis of multimodal single-cell data. Cell. 2021; 184:3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Breschi A., Muñoz-Aguirre M., Wucher V., Davis C.A., Garrido-Martín D., Djebali S., Gillis J., Pervouchine D.D., Vlasova A., Dobin A. et al. A limited set of transcriptional programs define major cell types. Genome Res. 2020; 30:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ecker J.R., Geschwind D.H., Kriegstein A.R., Ngai J., Osten P., Polioudakis D., Regev A., Sestan N., Wickersham I.R., Zeng H. The BRAIN initiative cell census consortium: lessons learned toward generating a comprehensive brain cell atlas. Neuron. 2017; 96:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ponting C.P. The human cell atlas: making ‘cell space’ for disease. Dis. Model. Mech. 2019; 12:dmm037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang S., Pisco A.O., McGeever A., Brbic M., Zitnik M., Darmanis S., Leskovec J., Karkanias J., Altman R.B. Unifying single-cell annotations based on the cell ontology. 2020; bioRxiv doi:4 February 2020, preprint: not peer reviewed 10.1101/810234. [DOI]
- 96. Hou R., Denisenko E., Forrest A.R.R. scMatch: a single-cell gene expression profile annotation tool using reference datasets. Bioinforma. Oxf. Engl. 2019; 35:4688–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z. et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; 47:D900–D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Franzén O., Gan L.-M., Björkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database J. Biol. Databases Curation. 2019; 2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kiselev V.Y., Andrews T.S., Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 2019; 20:273–282. [DOI] [PubMed] [Google Scholar]
- 100. Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M. et al. The human cell atlas. eLife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W. et al. Construction of a human cell landscape at single-cell level. Nature. 2020; 581:303–309. [DOI] [PubMed] [Google Scholar]
- 102. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F. et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018; 172:1091–1107. [DOI] [PubMed] [Google Scholar]
- 103. Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M.B. et al. Single-cell transcriptomics of 20 mouse organs creates a tabula muris. Nature. 2018; 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Snyder M.P., Lin S., Posgai A., Atkinson M., Regev A., Rood J., Rozenblatt-Rosen O., Gaffney L., Hupalowska A., Satija R. et al. The human body at cellular resolution: the NIH human biomolecular atlas program. Nature. 2019; 574:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lotfollahi M., Naghipourfar M., Luecken M.D., Khajavi M., Büttner M., Wagenstetter M., Avsec Ž., Gayoso A., Yosef N., Interlandi M. et al. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol. 2021; 10.1038/s41587-021-01001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Brbić M., Zitnik M., Wang S., Pisco A.O., Altman R.B., Darmanis S., Leskovec J. MARS: discovering novel cell types across heterogeneous single-cell experiments. Nat. Methods. 2020; 17:1200–1206. [DOI] [PubMed] [Google Scholar]
- 107. Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017; 18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jin H., Liu Z. A benchmark for RNA-seq deconvolution analysis under dynamic testing environments. Genome Biol. 2021; 22:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. McKinley K.L., Castillo-Azofeifa D., Klein O.D. Tools and concepts for interrogating and defining cellular identity. Cell Stem Cell. 2020; 26:632–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cell SystemsVoices What Is Your Conceptual Definition of “Cell Type” in the Context of a Mature Organism?. Cell Syst. 2017; 4:255–259. [DOI] [PubMed] [Google Scholar]
- 111. Hatje K., Mühlhausen S., Simm D., Kollmar M. The protein-coding human genome: annotating high-hanging fruits. Bioessays. 2019; 41:1900066. [DOI] [PubMed] [Google Scholar]
- 112. Gibbs R.A. The human genome project changed everything. Nat. Rev. Genet. 2020; 21:575–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang Y., Navin N.E. Advances and applications of single cell sequencing technologies. Mol. Cell. 2015; 58:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018; 36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.