Abstract
Stress granules are dynamic, reversible condensates composed of RNA and protein that assemble in eukaryotic cells in response to a variety of stressors and are normally disassembled after stress is removed. The composition and assembly of stress granules is well understood, but little is known about the mechanisms that govern disassembly. Impaired disassembly has been implicated in some diseases including amyotrophic lateral sclerosis, frontotemporal dementia, and multisystem proteinopathy. Using cultured human cells, we found that stress granule disassembly was context-dependent: Specifically in the setting of heat shock, disassembly required ubiquitination of G3BP1, the central protein within the stress granule RNA-protein network. We found that ubiquitinated G3BP1 interacted with the endoplasmic reticulum– associated protein FAF2, which engaged the ubiquitin-dependent segregase p97/VCP (valosin-containing protein). Thus, targeting of G3BP1 weakened the stress granule–specific interaction network, resulting in granule disassembly.
One Sentence Summary:
Ubiquitination of G3BP1 mediates FAF2- and p97/VCP-dependent disassembly of heat-induced stress granules
Main Text:
Biomolecular condensation is a vital strategy of cellular organization that regulates a variety of biological functions. Ribonucleoprotein (RNP) granules are a highly conserved class of biomolecular condensates that govern many aspects of RNA metabolism (1, 2). One prominent type of RNP granule is the stress granule, a dynamic and reversible cytoplasmic assembly formed in eukaryotic cells in response to a variety of stressors. Formation of stress granules typically follows upon inhibited translation initiation and polysome disassembly, which causes a rapid increase in the cytoplasmic concentration of uncoated mRNA. This rise in cytoplasmic mRNA triggers multicomponent liquid-liquid phase separation (LLPS) with RNA-binding proteins, creating a condensed liquid compartment that remains in dynamic equilibrium with, yet distinct from, the surrounding cytosol (3–5). Impaired dynamics of RNP granules such as stress granules are implicated as an important contributor to certain pathological conditions, including neurodegenerative diseases (6).
Recent investigations of stress granule assembly have yielded insight into how multicomponent networks self-organize to form a compartment that is distinct from its surroundings. Stress granules are composed of RNA and protein that interact via protein-protein, protein-RNA, and RNA-RNA interactions, many of which may be weak and transient. However, once the sum of these interactions breaches a particular threshold, known as the percolation threshold (6), the individual molecules form a system-spanning network that separates itself from its milieu – in other words, LLPS ensues (3–5). The concept of a percolation threshold is generalizable to other biomolecular condensates, each of which encodes a system-specific threshold for LLPS.
In U2OS cell stress granules, there are ~36 proteins that, together with RNA, provide the majority of the interactions that set the percolation threshold for RNA-dependent LLPS (3). Whereas each constituent node of this network contributes toward the sum of interactions required to reach the percolation threshold, a small subset of constituents of high centrality within the interaction network predominately establish this threshold (3, 7). The most important proteins in the stress granule interaction network are G3BP1 and G3BP2, which provide the largest contribution to establishing the percolation threshold for stress granule assembly (3–5). Indeed, elimination of G3BP1/2 proteins can preclude stress granule assembly (3, 8) whereas their enforced activation by optogenetic induction of dimerization can initiate stress granule assembly (3, 9). Higher order regulation over stress granule assembly may also be afforded by post-translational modifications of stress granule proteins that impact the interaction network (10).
In healthy cells, stress granules are transient, dynamic structures whose presence generally corresponds to the duration of time during which a stress is applied. However, exactly how stress granules are removed from cells remains unknown. Specifically, the role of autophagy in the elimination of stress granules is unclear: Whereas some studies have shown that stress granule removal is autophagy-dependent (11), autophagy-independent elimination has also been reported (12). Autophagy independence is consistent with evidence suggesting that stress granule constituents are recycled, with mRNPs reentering the translational pool after removal of stress (13).
Insight into reconciling these apparent contradictions may come from studies examining the role of context in defining the mechanisms of stress granule assembly and disassembly. As shown in our companion paper (14), stress granules formed by heat stress are disassembled via ubiquitin-dependent mechanisms, whereas those formed in response to arsenite are not. Thus, the key nodes and cellular processes engaged in disassembly of stress granules are specific to the context of each stress. This finding may explain why ubiquitination is dispensable for arsenite-induced stress granule dynamics (15), whereas the ubiquitin-binding adaptor protein ZFAND1 is required (16). Here we wanted to ascertain the extent to which the mechanisms involved in stress granule elimination depend on the nature of the initiating stress.
G3BP1 undergoes K63-linked ubiquitination upon heat shock
In our companion paper, we found that G3BP1 is ubiquitinated when stress granules are assembled in response to heat shock, but not when cells are exposed to other types of granuleinducing stress (14). Furthermore, we found that ubiquitination was not required for heat shock-induced stress granule assembly but was essential for their rapid disassembly after removal of stress. However, the mechanism whereby ubiquitination was required for stress granule disassembly remained unclear (14).
In addition to its essential role in stress granule formation, G3BP1 is required to maintain the assembly of stress granules, as pharmacological disruption of G3BP1 dimers leads to rapid disassembly of stress granules (3). Thus, we hypothesized that ubiquitination and targeted degradation of G3BP1 may have a key role in disassembly.
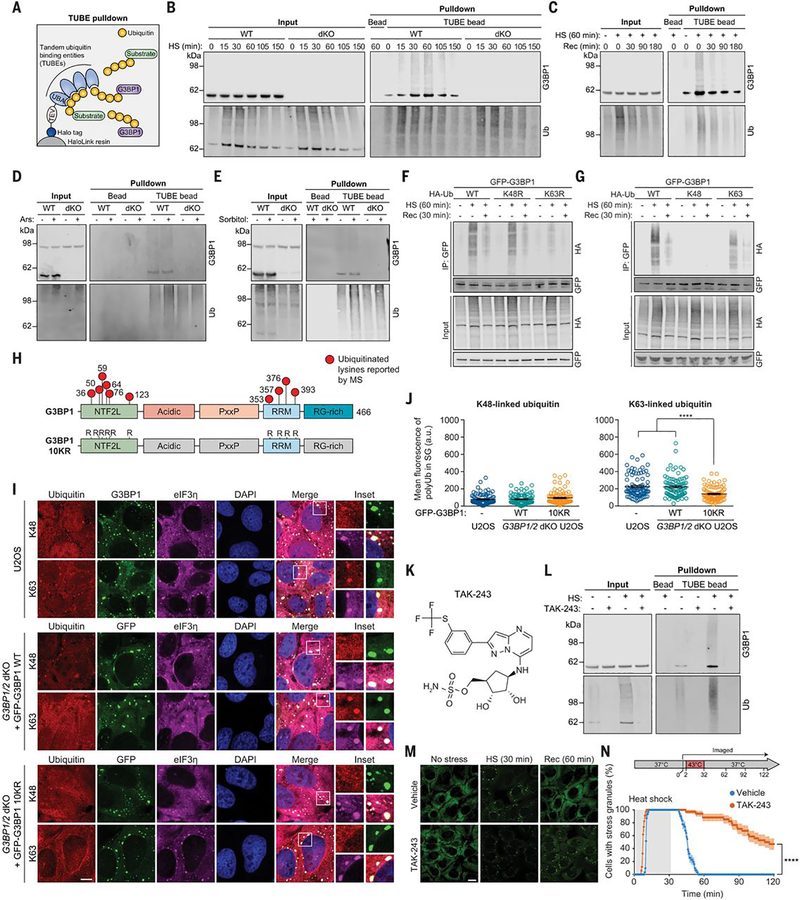
To confirm that G3BP1 is ubiquitinated in response to heat shock, we performed tandem-ubiquitin binding entity (TUBE) pulldown of ubiquitinated proteins from U2OS cells exposed to heat shock (43°C) followed by immunoblotting (Fig. 1A), using G3BP1/2 double knockout (dKO) cells as controls. We detected increased levels of polyubiquitin-conjugated G3BP1 as early as 15 min after exposure to heat stress; with continuing stress, these levels peaked at ~60 min and remained detectable in the soluble fraction for at least 150 min (Fig. 1B). After stress was removed, levels of polyubiquitinated G3BP1 decreased, returning to baseline within 3 hours after a 60-min heat stress (Fig. 1C). Polyubiquitin conjugation of G3BP1 was specific to heat stress: Despite an increase in total polyubiquitin conjugates in response to oxidative or osmotic stress, cells exposed to these stresses did not accumulate polyubiquitinated G3BP1 (Fig. 1, D and E).
Fig. 1. G3BP1 undergoes K63-linked ubiquitination in response to heat stress.
(A) Illustration showing TUBE capture of ubiquitinated G3BP1. TEV, tobacco etch virus protease cleavage site. (B and C) Immunoblots of TUBE-captured cell extracts showing levels of ubiquitinated G3BP1 in U2OS cells after different durations of 43°C heat shock, using U2OS G3BP1/2 dKO cells as controls (B), and during 37°C recovery (C). (D and E) Immunoblots of TUBE-captured cell extracts showing levels of ubiquitinated G3BP1 in response to oxidative stress (0.5 mM NaAsO2, 1 hour) (D) or osmotic stress (0.4 M sorbitol, 1 hour) (E). (F and G) Immunoblots of cell extracts captured with antibody to GFP, showing K63-linked ubiquitination of G3BP1. Transfected HEK293T cells were exposed to heat shock (43°C, 1 hour) and recovery (37°C, 30 min). K48R and K63R prevent the formation of K48-linked (K48R) or K63-linked (K63R) chains; K48 and K63 permit K48-linked or K63-linked chains exclusively. HA, hemagglutinin; IP, immunoprecipitate. (H) G3BP1 domain labeled with lysines on which ubiquitination has been reported. In the G3BP1 10KR mutant, six lysines in NTF2L and four lysines in RRM are mutated to arginine. (I) Immunofluorescent staining of fixed U2OS WT and U2OS G3BP1/2 dKO cells stably expressing GFP-G3BP WT and 10KR. Scale bar, 10 μm. (J) Fluorescence intensities of K48- and K63-linked ubiquitin in eIF3η-positive stress granules from three technical replicates are plotted in (n = 90). Error bars indicate SEM. ****P < 0.0001 (ANOVA with Tukey’s test). (K) Structure of TAK-243. (L) Immunoblot of TUBE-captured cell extracts showing block of heat shock–induced G3BP1 ubiquitination by TAK-243. U2OS cells were treated with DMSO or TAK-243 for 1 hour prior to heat shock. (M and N) Fluorescent imaging of U2OS cells stably expressing GFP-G3BP1 were treated with DMSO or TAK-243 (1 hour) prior to imaging. Representative images are shown in (M). In (N), GFP signals were monitored at 30-s intervals to count cells with two or more stress granules from three technical replicates (vehicle n = 32, TAK-243 n = 35). Scale bar, 20 μm. Error bars indicate SEM. ****P < 0.0001 (Mantel-Cox test).
Polyubiquitin chains can be formed by conjugating ubiquitin to one of seven lysine (K) residues of another ubiquitin molecule, giving rise to K6-, K11-, K27-, K29-, K33-, K48-, or K63-linked polyubiquitin chains (17). Among these, K48- and K63-linked chains are the most abundant and functionally well-characterized linkage types (18). To determine which of these linkage types were present in polyubiquitinated G3BP1, we used ubiquitin mutants that prevent the formation of K48-linked (K48R) or K63-linked (K63R) chains or that permit K48-linked or K63-linked chains exclusively (K48 or K63 being the only available lysines). Expression of K63R inhibited accumulation of polyubiquitin-conjugated G3BP1 upon heat shock, whereas expression of K48R had no impact (Fig. 1F). Conversely, cells expressing K63 ubiquitin (in which the only available lysine is K63) accumulated polyubiquitin-conjugated G3BP1, whereas cells expressing K48 ubiquitin did not (Fig. 1G), which suggests that heat shock promotes K63-linked polyubiquitination of G3BP1. The levels of K63-linked polyubiquitinated G3BP1 were lower than the levels of polyubiquitinated G3BP1 using wild-type ubiquitin; therefor, other linkage types may also contribute to this polyubiquitination.
Heat shock-induced stress granules accumulate K63-linked polyubiquitin chains that are required for stress granule disassembly
Examination of public databases suggests 10 lysine residues in G3BP1 that are potentially subject to ubiquitination (19). Of these, six residues are located within the N-terminal NTF2-like (NTF2L) domain and four in the C-terminal RNA-recognition motif (RRM) (Fig. 1H). When we mutated these 10 residues (G3BP1 10KR) and expressed this construct in G3BP1/2 dKO cells, we found no ubiquitination of G3BP1 in response to heat shock, which suggests that G3BP1 10KR functions as a ubiquitination null mutant (fig. S1A).
Stress granules induced by heat stress (but not arsenite stress) exhibit a robust accumulation of polyubiquitin signal (14). To examine the contributions of K48- versus K63-linked chains to this ubiquitin signal within stress granules, we used linkage-specific antibodies. Whereas K48- and K63-linked polyubiquitin signals were both uniformly distributed throughout the cell under basal conditions (fig. S1B), only K63-linked polyubiquitin signals accumulated in stress granules upon heat shock in wild-type U2OS cells (Fig. 1I). Furthermore, K63-linked polyubiquitin signal was significantly diminished in G3BP1/2 dKO cells expressing GFP-G3BP1 10KR fused to green fluorescent protein (GFP-G3BP1 10KR), whereas G3BP1/2 dKO cells expressing wild-type GFP-G3BP1 (GFP-G3BP1 WT) accumulated K63-linked polyubiquitin signal in stress granules at levels comparable to those observed in wild-type U2OS cells (Fig. 1, I and J). Thus, a significant portion of heat shock-induced polyubiquitin signal in stress granules arises from K63-linked polyubiquitination of G3BP1. Polyubiquitin conjugation of G3BP1 was eliminated by treatment with TAK-243, a small-molecule inhibitor of UBA1, an E1 ubiquitinactivating enzyme (Fig. 1, K and L). Consistent with our previous observations (14), blocking ubiquitination by TAK-243 significantly delayed stress granule disassembly during recovery from heat stress (Fig. 1, M and N and movie S1). Thus, ubiquitination is required for stress granule disassembly after heat shock. Similar results were obtained upon small interfering RNA (siRNA)-mediated knockdown of UBA1 (14). We next sought to investigate the relationship between polyubiquitin conjugation of G3BP1 and stress granule dynamics.
Ubiquitination within G3BP1 NTF2L is required for disassembly of heat shock-induced stress granules
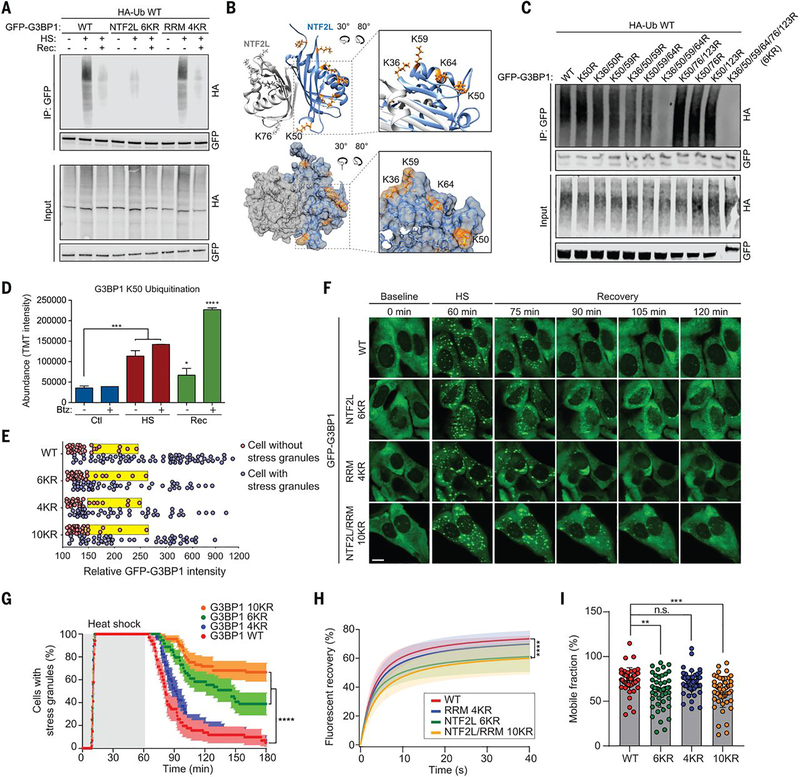
To investigate the effect of G3BP1 ubiquitination on stress granule dynamics, we first sought to identify the heat shock-dependent ubiquitination site(s) within G3BP1. To this end, we generated constructs in which six lysine residues in the NTF2L domain (6KR) or four lysine residues in the RRM domain (4KR) were mutated to arginines (fig. S2A). When expressed in G3BP1/2 dKO cells, G3BP1 6KR showed greatly reduced ubiquitination in response to heat shock, whereas G3BP1 4KR showed no apparent change in polyubiquitin conjugation; this result implies that the NTF2L domain is preferentially ubiquitinated upon heat shock (Fig. 2A). The crystal structure of the NTF2L dimer (PDB: 5FW5) indicates that all six lysine residues are surface-exposed. K36, K50, K59, and K64 are clustered to form a positively charged surface on the lateral sides of homodimeric NTF2L domains (Fig. 2B); K50 and K76 from each monomer lie adjacent to the dimeric interface; and K123 is located near the hydrophobic grove between α helices, where the nsP3 protein of Old World alphaviruses binds to inhibit stress granule assembly (fig. S2B). To gain insight into which lysines in G3BP1 are ubiquitinated, we generated an additional series of Lys → Arg (K-to-R) mutations. Mutation of the four clustered lysine residues (K36/50/59/64R) nearly abolished heat shock-dependent ubiquitination of G3BP1 in G3BP1/2 dKO cells, whereas other combinations of mutations to NTF2L lysines had no strong effect (Fig. 2C). However, ubiquitination levels further decreased in the 6KR mutant, and we therefore used G3BP1 6KR as a ubiquitination-deficient mutant. Ubiquitination of G3BP1 in this region is consistent with results from an independent approach (14). Specifically, paired di-Gly and tandem mass tag (TMT) analysis revealed enrichment of ubiquitinated G3BP1 peptide containing K50 [K.NSSYVHGGLDSNGK50PADAVYGQK.E (20)] upon heat shock and subsequent decrease in this ubiquitinated species during recovery from stress (Fig. 2D). The decrease in ubiquitinated G3BP1 during recovery appears to reflect proteasome-dependent degradation, because treatment with the proteasome inhibitor bortezomib (Btz) led to accumulation of ubiquitinated G3BP1 (Fig. 2D).
Fig. 2. Ubiquitination of the NTF2L domain of G3BP1 is required for the disassembly of stress granules.
(A) Immunoblot of transfected HEK293T cell extracts captured with antibody to GFP after exposure to no stress, heat shock for 1 hour, or 37°C recovery for 30 min. (B) Dimeric structure of G3BP1 NTF2L domain. K36, K50, K59, and K64 are highlighted in enlarged images (orange). (C) Immunoblot of transfected HEK293T cell extract captured with GFP antibody after exposure to heat shock for 1 hour. (D) TMT intensity of ubiquitinated G3BP1 peptides during heat shock and recovery with or without bortezomib (Btz). Error bars indicate SD. *P < 0.05, ***P < 0.001, ****P < 0.0001 (ANOVA with Dunnett’s test). (E) Percolation threshold of GFP-G3BP1 WT, 6KR, 4KR, and 10KR exposed to heat shock for 1 hour. U2OS G3BP1/2 dKO cells without stress granules (red) and with stress granules (blue) are plotted from three biological replicates according to GFP intensities (WT n = 109, 6KR n = 86, 4KR n = 78, 10KR n = 85). Yellow boxes indicate the cells with 25% highest levels of GFP within the stress granule–null group. (F) Stress granule disassembly in U2OS G3BP1/2 dKO cells expressing G3BP1 WT, 6KR, 4KR, or 10KR. Scale bar, 20 μm. (G) GFP signals were monitored at 30-s intervals at 37°C for 2 min, 43°C for 60 min, and 37°C for 118 min to count cells with two or more stress granules from three biological replicates (WT n = 38, 6KR n = 36, 4KR n = 39, 10KR n = 39). Error bars indicate SEM. ****P < 0.0001 (Mantel-Cox test). (H) FRAP of GFP-positive puncta in U2OS G3BP1/2 dKO cells transfected with GFP-G3BP1 WT, 6KR, 4KR, or 10KR and exposed to heat shock (1 hour) followed by recovery (10 min) from three biological replicates is plotted as mean ± SEM (WT n = 48, 6KR n = 48, 4KR n = 45, 10KR n = 49). ****P < 0.0001 (ANOVA with Tukey’s test). (I) Quantification of mobile fraction at 40 s of recovery from three biological replicates (WT n = 48, 6KR n = 48, 4KR n = 45, 10KR n = 49). Error bars indicate SD. **P < 0.01, ***P < 0.001 (ANOVA with Dunnett’s test); n.s., not significant.
We next sought to determine the role of G3BP1 ubiquitination in stress granule dynamics, including assembly, disassembly, and dynamic exchange between the stress granule and surrounding cytoplasm. To this end, we first generated stable cell lines in which G3BP1 6KR, 4KR, or 10KR were re-introduced into G3BP1/2 dKO cells at levels comparable to endogenous G3BP1 levels in wild-type cells (fig. S2, C and D). To rule out the possibility that the K-to-R mutations impair intrinsic properties of G3BP1, we performed a series of control experiments. First, we assessed the impact of K-to-R mutations on G3BP1 dimerization and binding to interaction partners. By co-immunoprecipitation, we confirmed that GFP-G3BP1 WT, 6KR, 4KR, and 10KR mutants formed dimers with endogenous WT G3BP1 (fig. S2E). Moreover, 6KR mutations in the NTF2L domain did not alter G3BP1’s ability to interact with caprin 1 and USP10 through this domain (fig. S2F), nor did 4KR mutations in the RRM domain alter its ability to bind RNA (fig. S2G), which suggested that intrinsic properties of G3BP1 were not compromised by K-to-R substitution.
The most sensitive test of perturbation to G3BP1 function is the concentration threshold needed to trigger stress granule assembly in cells – the percolation threshold. To test this, we transiently transfected G3BP1 mutants into G3BP1/2 dKO cells and measured the G3BP1 concentration threshold at which stress granule assembly was initiated (3). The concentration threshold for stress granule assembly for the 6KR, 4KR, and 10KR mutants was unchanged from G3BP1 WT (Fig. 2E). Thus, the K-to-R mutants block G3BP1 ubiquitination but do not otherwise impair intrinsic phase separation properties or interactions that support stress granule assembly.
We next assessed how preventing ubiquitination of G3BP1 impacted stress granule dynamics. Upon heat shock, all three mutant forms of G3BP1 assembled stress granules with kinetics identical to G3BP1 WT (Fig. 2, F and G and movie S2), indicating that assembly of stress granules is independent of ubiquitination of G3BP1. This result is consistent with our findings that global inhibition of ubiquitination with TAK-243 did not alter the dynamics of stress granule assembly in response to heat shock (Fig. 1, M and N) (14). However, 6KR and 10KR mutations resulted in significantly prolonged stress granule disassembly (Fig. 2, F and G and movie S2). G3BP1 10KR (KR mutations in NTF2L and RRM domains) had a more severe impact on stress granule disassembly than did G3BP1 6KR (KR mutations in NTF2L only); hence, the four lysine residues in the RRM domain may also contribute to stress granule disassembly, although the levels of polyubiquitination in the RRM domain were below the detection limit by immunoblotting. When we assessed G3BP1 dynamics within stress granules during heat shock by fluorescent recovery after photobleaching (FRAP) analysis, when stress granules were fully assembled, all three mutant forms of G3BP1 showed fluorescence recovery at a rate identical to that of WT G3BP1 (fig. S2H). In contrast, after heat stress was removed and the disassembly phase had begun, the G3BP1 6KR and 10KR mutants showed significantly slower recovery rates and greater immobile fractions relative to wild-type and 4KR proteins (Fig. 2, H and I). Thus, mutations impeding ubiquitination of NTF2L caused a relative reduction in G3BP1 mobility that was specific to the disassembly phase.
Heat shock induces interaction of VCP with ubiquitinated G3BP1
Depletion of cellular adenosine triphosphate (ATP) by the addition of 2-deoxyglucose (2-DG), an inhibitor of the glycolytic pathway, impairs stress granule assembly and subsequent dynamic behavior, implicating energy-dependent processes in assembly dynamics (21). However, the requirement of cellular ATP for the disassembly of stress granules has not been explored. Thus, we examined the contribution of ATP to stress granule disassembly. Under basal conditions, addition of 2-DG led to a ~40% reduction in cellular ATP levels after 30 min, as assessed by a luminescent signal-based ATP sensor (fig. S3A). In contrast, addition of 2-DG after 60 min of heat shock and immediately before recovery led to a ~70% reduction in cellular ATP levels within 10 min (fig. S3A). Concurrently, we observed defects in disassembly of stress granules induced by heat shock or arsenite stresses (fig. S3, B to E, movie S3), which suggests that ATP is hydrolyzed during the recovery phase and that stress granules are disassembled via energy-dependent processes irrespective of stress type (i.e., heat shock or arsenite).
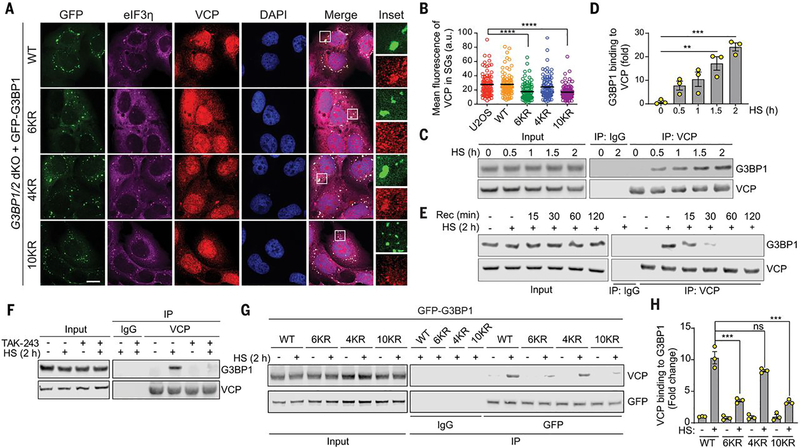
The stress granule proteome contains a large group of proteins with adenosine triphosphatase (ATPase) activities (21, 22), and the ATPase VCP is recruited to stress granules and contributes to their clearance (11, 16). VCP is a ubiquitin-dependent protein segregase coupled to both proteasome-dependent and autophagy-dependent degradation (11, 12, 16). Because disassembly of heat shock-induced stress granules depends on the ubiquitination of G3BP1, we hypothesized that VCP acts on ubiquitinated G3BP1 to promote disassembly of stress granules. Thus, we first tested whether recruitment of VCP to stress granules is contingent upon ubiquitination of G3BP1. We first confirmed that VCP was recruited to stress granules upon heat shock in wild-type U2OS cells (fig. S4A). Although G3BP1/2 dKO cells do not form stress granules (3, 8), reintroduction of exogenous GFP-G3BP1 WT or 4KR in these cells restored VCP recruitment to stress granules to levels similar to those observed in WT U2OS cells (Fig. 3, A and B, fig. S4A). However, VCP recruitment to stress granules was significantly reduced in G3BP1/2 dKO cells stably expressing GFP-G3BP1 6KR or 10KR (Fig. 3, A and B). Thus, ubiquitination of G3BP1 contributes to the recruitment of VCP to stress granules. Although G3BP1 is not the only stress granule protein that is ubiquitinated (14), this data are consistent with the pronounced abundance of G3BP1 among the protein constituents of stress granules (21).
Fig. 3. VCP interacts with ubiquitinated G3BP1 in response to heat shock.
(A) Fluorescent imaging of U2OS G3BP1/2 dKO cells stably expressing GFP-G3BP WT, 6KR, 4KR or 10KR exposed to heat shock for 1 hour. Scale bar, 20 μm. (B) Fluorescence intensities of VCP in eIF3η-positive stress granules from three technical replicates (n = 90). Error bars indicate SEM. ****P < 0.0001 (ANOVA with Tukey’s test). (C) Immunoblot of U2OS cell extracts exposed to heat shock for the indicated times. Cell extracts were captured with magnetic beads conjugated with VCP antibody for IP, and resulting beads were analyzed by immunoblot. (D) Quantification of immunoblots from three biological replicates. Error bars indicate SEM. **P < 0.01, ***P < 0.001 (ANOVA with Tukey’s test). (E) Immunoblot showing G3BP1-VCP interaction in U2OS cells based on duration of recovery after heat shock for 2 hours. Cell extracts were captured with magnetic beads conjugated with antibody to VCP for IP, and the resulting beads were analyzed by immunoblot. (F) Immunoblot of U2OS cells treated with DMSO or TAK-243 (60 min) and exposed to heat shock for 2 hours. Cell extracts were captured with magnetic beads conjugated with antibody to VCP for IP. Blots show an attenuated G3BP1-VCP interaction with inhibition of ubiquitination. (G) Immunoblot of U2OS G3BP1/2 dKO cells stably expressing G3BP1 WT, 6KR, 4KR, or 10KR and exposed to heat shock for 2 hours. Cell extracts were captured with magnetic beads conjugated with antibody to GFP for IP. (H) Quantification of immunoblots from three biological replicates. Error bars indicate SEM. ***P < 0.001 (ANOVA with Tukey’s test).
Consistent with these findings, endogenous G3BP1 co-immunoprecipitated with VCP upon heat shock (Fig. 3, C and D). This interaction gradually increased during a 2-hour heat shock period and decreased to baseline levels within 1 hour of recovery (Fig. 3, C to E). TAK-243 treatment abolished the VCP-G3BP1 interaction upon heat shock (Fig. 3F), confirming that the interaction is ubiquitin-dependent. To further assess the role of G3BP1 ubiquitination in this interaction, we expressed WT, 6KR, 4KR, and 10KR mutants in G3BP1/2 dKO cells and assessed their binding to VCP. 6KR and 10KR mutants showed significantly reduced binding to VCP compared to G3BP1 WT upon heat shock, whereas VCP interaction was not significantly affected in the 4KR mutant (Fig. 3, G and H). This observation is consistent with ubiquitination of the G3BP1 NTF2L domain, but not that of the RRM domain, being primarily responsible for interaction with VCP. Both 6KR and 10KR mutants still showed modest interaction with VCP upon heat shock, although the levels were substantially lower than for G3BP1 WT. Because we did not observe VCP-G3BP1 interaction in cells treated with TAK-243, this finding may be attributable to indirect interaction of VCP with other ubiquitinated proteins bound to G3BP1. Finally, the levels of ubiquitinated G3BP1 were increased by the addition of a VCP inhibitor (CB-5083) and stabilized by the addition of the proteasome inhibitor bortezomib; these findings suggest that ubiquitinated G3BP1 is targeted by VCP for proteasomal degradation during stress granule disassembly (fig. S4, B and C).
The mechanism of stress granule clearance shifts from disassembly to autophagy-dependent degradation during prolonged stress
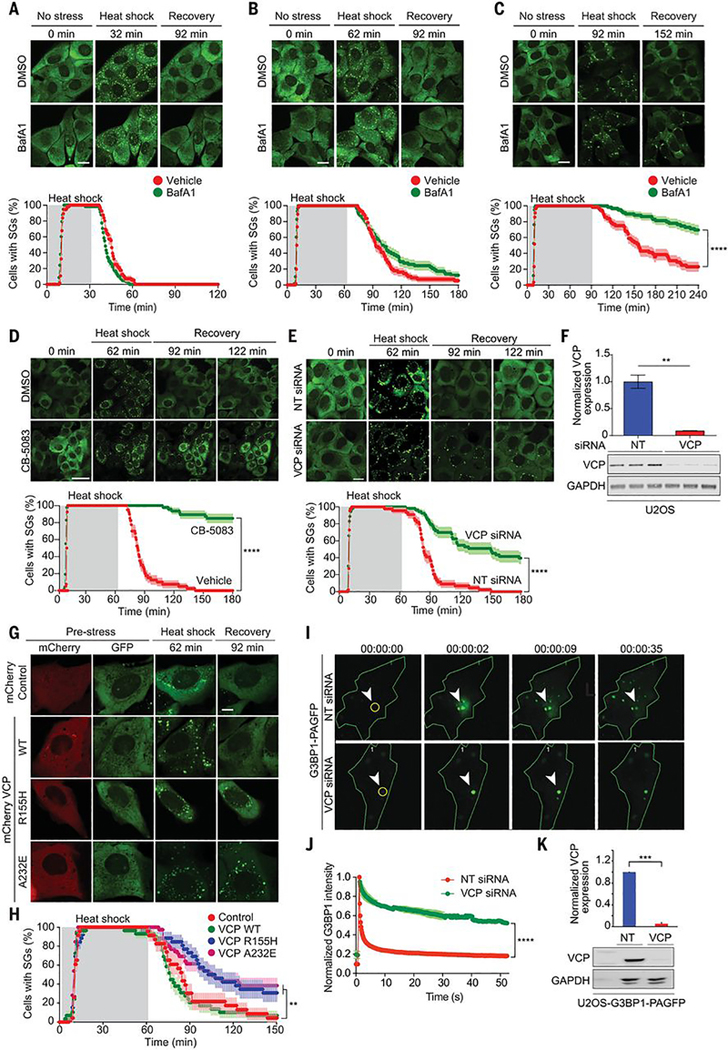
VCP is essential for autophagy-dependent clearance of persistent stress granules, such as those arising from prolonged stress or disease mutations (11, 23–25). A recent report corroborated the importance of VCP in stress granule clearance but found the process to be autophagy-independent (12). This latter report is consistent with evidence indicating that stress granule constituents are often recycled, with mRNPs reentering the translational pool after removal of stress; however, it was unclear how to reconcile these results with the earlier reports (13). We reasoned that the mechanism of stress granule clearance might be influenced by the chronicity of the initiating stress. To test this hypothesis, we compared the kinetics of stress granule assembly and disassembly in the presence and absence of bafilomycin A1 (BafA1), a vacuolar H+-ATPase inhibitor that blocks autophagy (26). With a short (30-min) exposure to heat shock, stress granules rapidly assembled and then completely disassembled, with all stress granules resolved by 30 min after removal from heat shock (Fig. 4A, movie S4). With an intermediate (60-min) exposure to heat shock, stress granules also rapidly assembled and completely disassembled, although the disassembly phase was protracted to 60 min in proportion to the longer duration of stress (Fig. 4B, movie S5). BafA1 treatment did not have a significant impact on the kinetics of disassembly after a 30- or 60-min heat shock, indicating that disassembly was taking place independent of any autophagic degradation. With more prolonged heat shock (90 min), the kinetics of stress granule assembly were again unchanged, but disassembly became even more protracted and highly sensitive to BafA1 treatment, indicating a transition to autophagy-dependent clearance of stress granules (Fig. 4C, movie S6). These results are consistent with prior observations of autophagy-dependent clearance of persistent stress granules such as those initiated by disease mutations (11). In such instances, some stress granule disassembly is observed, but autophagy is required for complete removal of stress granules.
Fig. 4. VCP regulates disassembly of stress granules.
(A to C) Top: U2OS/GFP-G3BP1 were treated with DMSO or bafilomycin A1 (BafA1) for 18 hours before live imaging. Scale bar, 20 μm. Bottom: GFP signals were monitored at 30-s intervals at 37°C for 2 min, 43°C for 30 min (A), 60 min (B), or 90 min (C), and 37°C for 88 min (A), 118 min (B), or 148 min (C) to count cells with two or more stress granules from three technical replicates [DMSO n = 56, BafA1 n = 58 in (A); DMSO n = 56, BafA1 n = 58 in (B); DMSO n = 52, BafA1 n = 59 in (C)]. Error bars indicate SEM. ****P < 0.0001 (Mantel-Cox test). (D and E) U2OS/GFP-G3BP1 cells were treated with DMSO or CB-5083 (1 hour) (D) or transfected with nontargeting (NT) or VCP siRNA (E) before live imaging. GFP signals were monitored at 30-s intervals at 37°C for 2 min, 43°C for 60 min, and 37°C for 118 min to count the cells with two or more stress granules [DMSO n = 40, CB-5083 n = 47 in (D); NT siRNA n = 44, VCP siRNA n = 53 in (E)]. Scale bars, 50 μm (D), 20 μm (E). Error bars indicate SEM. ****P < 0.0001 (Mantel-Cox test). (F) Immunoblot of U2OS cells transfected with NT or VCP siRNA from three biological replicates. Error bars indicate SEM. **P < 0.01 (Student’s t test). (G) U2OS/GFP-G3BP1 cells were transfected with pmCherry-N1, VCP WT-mCherry, VCP R155H-mCherry, or VCP A232EmCherry. Scale bar, 10 μm. (H) GFP signals of mCherry-positive cells were monitored at 60-s intervals at 37°C for 2 min, 43°C for 60 min, and 37°C for 88 min to count cells with two or more stress granules from three biological replicates (control n = 23, VCP WT n = 29, VCP R155H n = 26, VCP A232E n = 39). Error bars indicate SEM. **P < 0.01 (Mantel-Cox test). (I) U2OS cells expressing photoactivatable G3BP1 (G3BP1-PAGFP) were transfected with NT or VCP siRNA. (J) Intensities of GFP signals within activated ROIs were monitored at 200-ms intervals from three technical replicates. ****P < 0.0001 (ANOVA with Sidak’s test). (K) Immunoblots of U2OS cells expressing G3BP1-PAGFP transfected with NT or VCP siRNA from three biological replicates. ***P< 0.001 (Student’s t-test).
VCP is required for disassembly of stress granules
Heat shock-dependent ubiquitination of G3BP1 and subsequent interaction with VCP prompted us to examine whether VCP activity is also required for disassembly of more typical heat shock-induced stress granules. Chemical inhibition of VCP using two different inhibitors (Fig. 4D, fig. S4D, and movie S7) and siRNA knockdown of VCP (Fig. 4, E and F, and movie S8) each led to significant delay in the disassembly of heat shock-induced stress granules. Expression of disease-causing mutant forms of VCP (A232E and R155H) causes the accumulation of poorly dynamic stress granules (11). In this prior study it was unclear whether VCP mutations generally impaired autophagy-dependent clearance, including stress granules, or whether VCP played a more direct role in controlling some aspect of stress granule dynamics that was also impaired by disease mutations. The appreciation that VCP is important for stress granule disassembly afforded the opportunity to determine whether this activity of VCP is also impaired by disease mutations. Thus, we tested the impact of mutant VCP on stress granule dynamics. Whereas exogenous expression of wild-type VCP did not alter rates of stress granule assembly or disassembly, expression of VCP A232E or R155H significantly delayed disassembly of stress granules upon removal of heat stress (Fig. 4, G and H and movie S9). Thus, mutant VCP proteins have a dominant negative effect on stress granule disassembly similar to that observed in the presence of VCP inhibitors or VCP knockdown.
To further assess the impact of VCP on G3BP1 dynamics in stress granules, we generated a stable U2OS cell line expressing G3BP1 conjugated to photoactivatable GFP (G3BP1-PAGFP) (27) that allowed us to explore intracellular G3BP1 dynamics by tracking photoactivated molecules. Regions of interest (ROIs) within cells expressing G3BP1-PAGFP were excited briefly with a ~400-nm laser to activate selected pools of G3BP1, followed by time-lapse 488-nm imaging. Stress granule-localized G3BP1 proteins were successfully visualized after PAGFP photoconversion, which enabled monitoring their mobility (Fig. 4I). In control cells transfected with nontargeting siRNA, the fluorescence intensity of stress granules diminished over time within ROIs and redistributed to nearby stress granules (Fig. 4, I to K). Photoactivated G3BP1-PAGFP in cells depleted of VCP displayed significantly longer residence time in stress granules and limited redistribution of signal to neighboring stress granules; this suggests that VCP is needed to facilitate G3BP1 exit from stress granules (Fig. 4, I to K, and movies S10 and S11).
FAF2, an endoplasmic reticulum-associated VCP adaptor, recruits G3BP1 to promote disassembly of stress granules
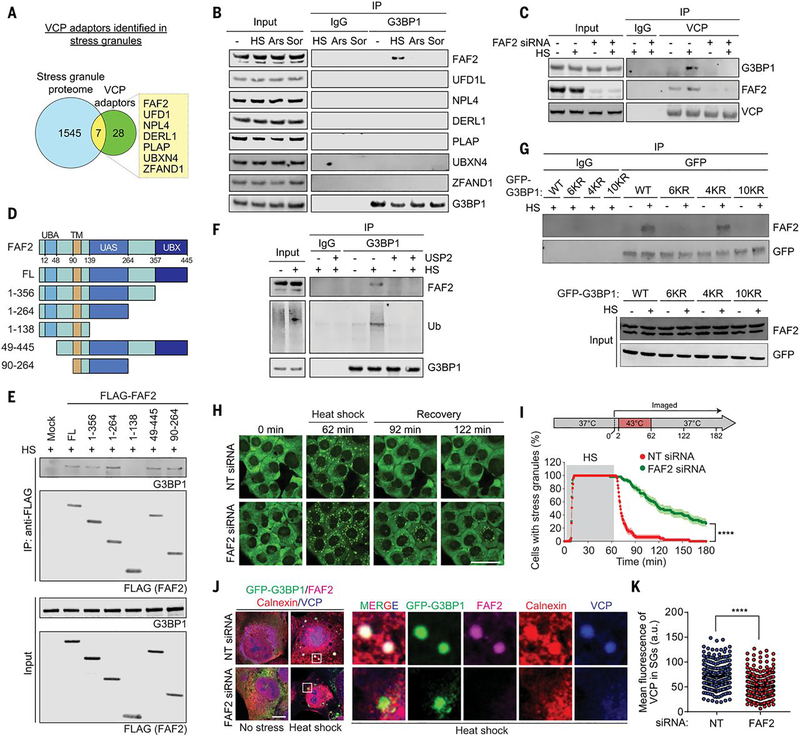
VCP associates with a large number of interacting partners and cofactors that regulate its activities in a variety of cellular pathways (28–31). VCP binds to ubiquitinated substrates largely through complexes that include cofactor proteins with ubiquitin-binding domains that function as ubiquitin adaptors for VCP (31). Approximately 35 VCP cofactor proteins have been identified in mammalian cells (16, 31, 33, 34), although in most cases the pathways in which these cofactors function, and the substrates that they recognize, remain poorly defined. Thus, we sought to identify the cofactor(s) that link VCP to stress granules by binding ubiquitinated G3BP1 and thereby influencing disassembly of heat shock-induced stress granules.
To establish a candidate list of relevant VCP interactors, we integrated a list of 35 VCP cofactors with 1,552 proteins previously identified in stress granules initiated by multiple stressors (Table S1) (3, 21, 35, 36). The stress granule proteome and VCP cofactors had seven proteins in common (Fig. 5A), including ZFAND1, a protein that promotes the clearance of arsenite-induced stress granules by recruiting VCP and the 26S proteasome (16). We confirmed interaction of five of these adaptors with endogenous VCP (fig. S5A). Of the seven proteins in common between the stress granule proteome and known VCP cofactors, only the endoplasmic reticulum (ER)-associated FAF2 (FAS-associated factor 2; also known as UBXD8) demonstrated interaction with G3BP1 upon heat shock (Fig. 5B). FAF2 interacts with VCP via its C-terminal UBX domain (37, 38). Thus, we hypothesized that FAF2 binding to VCP might mediate the interaction between VCP and G3BP1. Indeed, siRNA-mediated knockdown of FAF2 eliminated the VCP-G3BP1 interaction (Fig. 5C and fig. S5B). We next performed systematic deletion analysis of FAF2 to identify the domain needed for interaction with G3BP1. FAF2-G3BP1 interaction did not require the UBA domain, but rather the UAS domain (Fig. 5, D and E). The same strategy of domain interactions of FAF2 (i.e., UBX domain-mediated interaction with VCP and UAS domain-mediated interaction with substrate) is also found in an entirely different context, enabling VCP-dependent degradation of adipose triglyceride lipase (ATGL) in lipid metabolism (37). Nonetheless, we found that formation of a complex between FAF2 and G3BP1 was ubiquitin-dependent, because it was prevented by treatment of G3BP1 with the deubiquitinase USP2 (Fig. 5F) and ubiquitination-deficient G3BP1 6KR and 10KR mutants failed to interact with FAF2 (Fig. 5G). Because FAF2-G3BP1 interaction is ubiquitin-dependent, but does not require the UBA domain of FAF2, these results may suggest the existence of an additional ubiquitin-binding entity within the G3BP1-FAF2-VCP complex.
Fig. 5. FAF2 links ubiquitinated G3BP1 to VCP.
(A) Venn diagram showing overlapping proteins among the stress granule proteome and known VCP adaptors. (B) Immunoblot of U2OS cells extracts capture with antibody to GFP exposed to no stress, heat shock (1.5 hours), oxidative stress (sodium arsenite, 1.5 hours), or osmotic stress (sorbitol, 1.5 hours). (C) Immunoblot of U2OS cells extracts captured with antibody to VCP after transfection with NT or FAF2 siRNA and exposure to heat shock for 1.5 hours. (D) Domain structure of human FAF2 protein and deletion constructs used to investigate the function of individual domains of FAF2. UBA, ubiquitin-associated domain; TM, transmembrane domain; UAS, upstream activation sequence domain; UBX, ubiquitin regulatory X domain. (E) Immunoblot of U2OS cells captured with antibody to FLAG exposed to heat shock for 1.5 hours after transfection of FLAG-FAF2 full length (FL) or deletion mutants. (F) Immunoblot of U2OS cells exposed to no stress or heat shock for 1.5 hours. Cell extracts were incubated with or without purified USP2 and captured with magnetic beads conjugated with antibody to G3BP1 for IP, and the resulting beads were analyzed by immunoblot. (G) Immunoblots of U2OS G3BP1/2 dKO cells stably expressing G3BP1 WT, 6KR, 4KR, or 10KR mutants and exposed to heat shock for 2 hours. Cell extracts were captured with magnetic beads conjugated with antibody to GFP for IP. (H) U2OS/GFP-G3BP1 cells were transfected with NT or FAF2 siRNA. Scale bar, 50 μm. (I) GFP signals were monitored at 30-s intervals at 37°C for 2 min, 43°C for 60 min, and 37°C for 118 min to count cells with two or more stress granules from three technical replicates (NT siRNA n = 47, FAF siRNA n = 59). Error bars indicate SEM. ****P < 0.0001 (Mantel-Cox test). (J) Fluorescent imaging of U2OS/GFP-G3BP1 cells exposed to heat shock for 1 hour. Scale bar, 10 μm. (K) Fluorescence intensities of VCP in stress granules from three biological replicates are plotted as mean ± SEM (NT siRNA n = 272, FAF2 siRNA n = 349). ****P < 0.0001 (Student’s t test).
Disassembly of heat shock-induced stress granules is FAF2-dependent
We next examined the importance of FAF2 in stress granule disassembly. Depletion of FAF2 by siRNA delayed disassembly of heat shock-induced stress granules upon removal of heat stress, phenocopying our earlier observations using chemical inhibitors of VCP (Fig. 5, H and I, and movie S12). In contrast, depletion of six other adaptors, including ZFAND1, did not significantly alter the disassembly of heat shock-induced stress granules upon removal of heat stress (fig. S5C), which suggests that VCP specifically engages FAF2 to regulate the dynamics of stress granules induced by heat shock. Depletion of ZFAND1 delayed disassembly of arsenite-induced stress granules upon removal of arsenite as expected (16), whereas depletion of FAF2 failed to delay disassembly of arsenite-induce stress granules, demonstrating a context-dependent control of stress granule dynamics by ZFAND1 and FAF2 (fig. S5, D to G).
The FAF2-G3BP1-VCP pathway of stress granule disassembly occurs at the ER membrane
FAF2 is an ER membrane-associated protein that contains a stretch of 28 hydrophobic amino acid residues that are inserted into the cytosolic surface of the ER membrane (39). FAF2 is perhaps best known for its role in ubiquitin-dependent degradation of misfolded ER proteins in the context of ER-associated protein degradation (ERAD) (40, 41). FAF2 is also essential for VCP-dependent degradation of ATGL in ER-associated lipid droplet metabolism (37). The VCP-FAF2 complex disassembles mRNPs by promoting the extraction and degradation of ubiquitinated HuR, an RNA-binding protein, from mRNA (41). Thus, there is precedent for FAF2 involvement in ubiquitin-dependent degradation of diverse substrates, including RNP granule constituents, at the ER membrane.
To assess whether the FAF2-G3BP1-VCP pathway of stress granule disassembly occurs at the ER membrane, we began by examining the spatiotemporal relationship among the ER, FAF2, G3BP1, and VCP upon heat shock. At baseline, FAF2 was diffusely distributed with the ER, as indicated by colocalization with the ER marker calnexin (fig. S6A). Upon heat shock, calnexin and FAF2 signals appeared as bulges within ER tubules that colocalized with stress granules marked by GFP-G3BP1 (Fig. 5J). VCP showed a similar colocalization with G3BP1, FAF2, and calnexin upon heat shock (Fig. 5J). In contrast, arsenite stress did not induce appreciable FAF2 localization with stress granules (fig. S6B). VCP accumulation in heat stress-induced stress granules was significantly decreased in cells depleted of FAF2, whereas the more modest VCP accumulation in arsenite-induced stress granules was not influenced by depletion of FAF2 (Fig. 5K and fig. S6, C and D). These findings support our hypothesis that FAF2 functions as a VCP adaptor that connects the ER to heat shock-induced stress granules by interacting with ubiquitinated G3BP1.
Discussion
Whereas stress granule assembly is evidently a universal process across cell types and in response to a wide variety of stresses, it has recently become apparent that the composition of stress granules is context-dependent with respect to cell type and the initiating stress (35). Our study builds on this perspective by demonstrating context-dependent mechanisms of stress granule elimination that depend on both the type and duration of stress. This context is particularly important when considering how we investigate the relationship of stress granules to disease: We must be attentive to disease-relevant contexts, including the use of specific cell types or the administration of different types of stress.
Indeed, deconvolving the complexity of the existing literature on mechanisms of stress granule elimination requires careful consideration of the type and duration of the initiating stress. As demonstrated here, persistent stress granules, such as those that arise through chronic stress or disease mutations, are eliminated by autophagy-dependent degradation. In contrast, short-lived granules are rapidly disassembled, which permits recycling of constituents. Thus, we propose that prolonged stress or disease mutations result in persistence of stress granules that become gradually less dynamic and are eventually cleared through an autophagy-dependent degradative process, consistent with previous reports that clearance of such granules requires VCP and autophagy machinery such as ATG7 (6, 11, 43).
Autophagy-dependent clearance of stress granules results in degradation and permanent loss of constituent proteins and RNAs. In contrast, if cellular stress is removed relatively quickly, while stress granules remain dynamic, they are disassembled by decondensation – a reversal of LLPS – that occurs when sum of protein-protein, protein-RNA, and RNA-RNA interactions falls below the percolation threshold. This decrease in the sum total of interactions within the stress granule network could occur through several mechanisms, including a decrease in the concentration of individual constituents or altered post-translational modifications of constituent proteins that weakens the interaction network. In contrast to stress granule clearance, disassembly is a nondegradative process wherein the individual constituents disassociate from one another and are recycled, including liberation of the mRNAs to rejoin the translating pool (13).
The importance of context also helps to resolve earlier apparent inconsistencies regarding the role of ubiquitination in stress granule dynamics, because ubiquitination had previously been reported to be dispensable for the formation and disassembly of arsenite-induced stress granules (15). A striking example of the importance of context can also be seen in the use of VCP adaptors in the process of stress granule clearance. Elimination of arsenite-induced stress granules requires the adaptor ZFAND1 but not FAF2, whereas elimination of heat shock-induced stress granules requires FAF2 but not ZFAND1.
The cytosolic surface of the ER is a nexus for coordinating responses to heat stress, including the integrated stress response (44), the UPR (45), ERAD (46) and, now, disassembly of stress granules. The basis for disassembly of heat shock-induced stress granules at the ER surface appears to involve the ER-associated protein FAF2, which is essential for VCP to engage ubiquitinated G3BP1. Interestingly, FAF2 is not only an important factor in stress granule disassembly, but also in ERAD (33) and ER-associated metabolism of lipid droplets (37, 47). The role of ER localization in dismantling stress granules in response to other stresses remains unclear. A recent report describes “fission” of arsenite-induced stress granules occurring at contact sites with ER, interpreted as ER membrane-mediated mechanical cleavage of stress granules, which may be somehow related to disassembly (48).
Much remains to be learned about the distinct mechanisms of stress granule disassembly, and in particular how these mechanisms may be impacted by disease-causing mutations. Indeed, although disease mutations in VCP impair autophagy-dependent stress granule clearance (11), here we found that these mutations also impair autophagy-independent disassembly. Similar TAR DNA-binding protein 43 (TDP-43) laden, spontaneously arising, poorly dynamic stress granules are found in cells expressing disease mutant forms of RNA-binding proteins, such as FUS, hnRNPA1, hnRNPA2, and TIA-1 (43). Thus, mutations in VCP may impair its ability to disassemble stress granules, representing a mechanistic intersection for multiple, distinct disease-causing mutations.
Stress granule disassembly is mediated by extraction of ubiquitinated G3BP, a finding that underscores the central role for G3BP in maintaining the stress granule interaction network. Moreover, because cells produce distinct patterns of ubiquitination in response to different stressors (14), our study illustrates how even a single protein within a stress-specific ubiquitinome can be pursued experimentally to demonstrate meaningful consequences for cellular function.
Materials and Methods
Plasmid, siRNA and transfection
Synthetic G3BP1 fragments with K36/50/59/64/76/123R (NTF2L 6KR), K353/357/376/393R (RRM 4KR), and K36/50/59/64/76/123/353/357/376/393R (NTF2L/RRM 10KR) mutations were inserted into HindIII and BamHI sites of pEGFP-C3 (Clontech) by Bio Basic Inc. pEGFP-C3 G3BP1 K50R, K36/50R, K50/59R, K36/50/59R, K50/59/64R, K36/50/59/64R, K50/76R, K50/123R, and K50/76/123R mutants were generated by site-directed mutagenesis using a Q5 Site-Directed Mutagenesis kit (New England Biolabs E0054S). pRK5-HA-ubiquitin-WT, pRK5HA-ubiquitin-K48R, pRK5-HA-ubiquitin-K63R, pRK5-HA-ubiquitin-K48, and pRK5-HA-ubiquitin-K63 (Addgene 17608, 17604, 17606, 17605, and 17606, respectively) were kindly provided by T. Dawson. VCP WT, VCP R155H, and VCP A232E were inserted into BamHI and HindIII sites of pmCherry-N1 (Clontech) (49). pCLi40w-MND-G3BP1-GFP and pCLiw40-MND-G3BP1-PAGFP plasmids, used to generate G3BP-GFP stable U2Os cells via lentiviral transduction, were constructed by releasing dsRedEX2-EIF1α-GFP from pCLEG-MND-dsRedEX2 (Vector Development and Production, SJCRH) and inserting G3BP1-GFP or G3BP1-PAGFP into the EcoRI and BsrGI positions. G3BP1-GFP and G3BP1-PAGFP inserts were released from peGFP-N1-G3BP and pePAGFP-N1-G3BP by EcoRI and dBsrGI digestion. G3BP1 was inserted in peGFP-N1 or pePAGFP-N1 (Clontech) with primers containing EcoRI and BamHI sites in the 5′ and 3′ sites of G3BP. Similarly, the pCLi40w-MND-TIAL1-PAGFP construct was generated using EcoRI and KpnI. pRK5-FLAG-FAF2 (Addgene 53777) was kindly provided by Y. Ye. pRK5-FLAG-FAF2 1–356, pRK5-FLAG-FAF2 1–264, pRK5-FLAG-FAF2 1–138, pRK5-FLAG-FAF2 49–445, and pRK5-FLAG-FAF2 90–264 were inserted into SalI and NotI sites of pRK5-FLAG-FAF2. For knockdown experiments, the following siRNA constructs were purchased from Horizon Discovery: pooled nontargeting siRNA (D-001810–10), VCP (L-008728–00), FAF2 (L-010649–02), UFD1L (L-017918–00), NPL4 (L-020796–01), DERL1 (L-010733–02), PLAP (L-016215–00), UBXN4 (L-014184–01), and ZFAND1 (L-009638–02). Transporter 5 (Polysciences 26008) and FuGENE 6 (Promega E2691) were used for transient transfections of cDNA into HEK293T and U2OS cells, respectively, according to the manufacturer’s instructions. Lipofectamine RNAiMax (Thermo Fisher Scientific 13778150) was used for transfection of siRNA according to the manufacturer’s instructions.
Cell culture
HEK293T (CRL-3216) and U2OS (HTB-96) cells were purchased from ATCC, cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum (HyClone; SH30396.03), 1X GlutaMAX (Thermo Fisher Scientific; 35050061), penicillin (50 U/ml)-streptomycin (50 μg/ml) (Gibco; 15140–122) and maintained at 37°C in a humidified incubator with 5% CO2. U2OS G3BP1/2 KO cells have been previously described in (50). U2OS cells stably expressing GFP-G3BP1 have been previously described in (51).
Generation of G3BP1 add-back cell lines
U2OS G3BP1/2 dKO cells were transfected with pEGFP-C3 G3BP1 WT, NTF2L 6KR, RRM 4KR, or NTF2L/RRM 10KR constructs using FUGENE 6 (Promega); 48 hours after transfection, G418 sulfate (500 μg/ml; Thermo Fisher Scientific 10131035) was added to culture media without penicillin and streptomycin for selection. After pharmacological selection, GFP-positive cells were purified using cell sorting to produce stable cell lines.
Heat shock and drug treatments
For heat shock, cells were transferred to a 43°C humidified incubator with 5% CO2. Chemicals dissolved in DMSO were prepared and added to cells as follows: sodium arsenite (0.5 mM, Millipore Sigma 1062771000), TAK-243 (1 μM, ChemieTek CT-M7243), CB-5083 (1 μM, Cayman Chemical 19311), bafilomycin A1 (10 nM, Cayman Chemical 11038), cycloheximide (100 μg/ml, Sigma-Aldrich C7698), and bortezomib (1 μM, Millipore Sigma 5043140001). D-Sorbitol (0.4 M, Millipore Sigma S1876) was dissolved in culture media and prewarmed before treatment. Eeyarestatin I was used at a final concentration of 56 μM (30 min pretreatment).
Immunoblotting
Cells were washed twice with PBS and lysed with RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; Thermo Scientific 89901) supplemented with 1 mM EDTA (Invitrogen 15575020) and proteinase inhibitor cocktail (Roche 1183617001). Lysates were centrifuged for 15 min at 4°C at 20,000g. 4× NuPAGE LDS sample buffer (Thermo Fisher Scientific NP0007) was added to the supernatant and samples were boiled at 90°C for 5 min. Samples were separated in 4 to 12% NuPAGE Bis-Tris gels (Thermo Fisher Scientific NP0336BOX or NP0321BOX) and transferred to PVDF membranes (Thermo Fisher Scientific IB24001) using an iBlot 2 transfer device (Thermo Fisher Scientific). Membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences 927–50000) and then incubated with primary antibodies at 4°C overnight: G3BP1 (Proteintech 13057–2-AP), ubiquitin (Santa Cruz Biotechnology sc-8017), HA tag (Invitrogen 715500), GFP (Invitrogen A11122), FLAG (Proteintech 20543–1-AP), VCP (Santa Cruz Biotechnology sc-20799 or Thermo Fisher Scientific MA3–004), FAF2 (Proteintech 16251–1-AP), UFD1L (Proteintech 10615–1-AP), NPL4 (Proteintech 11638–1-AP), DERL1 (Thermo Scientific PA553444), PLAP (Santa Cruz Biotechnology sc-390454), UBXN4 (Thermo Fisher Scientific PA5577611), ZFAND1 (Sigma-Aldrich HPA023383), β-actin (Santa Cruz Biotechnology sc-47778 or Sigma-Aldrich A5316), and GAPDH (Santa Cruz Biotechnology sc-32233 or Sigma-Aldrich G9545). Membranes were washed three times with TBS-T (0.05% Tween) and further incubated with IRDye 680RD/800CW-labeled secondary antibodies (LI-COR Biosciences 926–68073 or 926–32212) at a dilution of 1:10,000. Membranes were visualized with an Odyssey Fc imaging system (LI-COR Biosciences) and quantified using ImageJ software (NIH).
Immunoprecipitation
Cells were washed twice with PBS and lysed with IP lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol; Thermo Scientific 87787) supplemented with 20 mM N-ethylmaleimide (NEM) (Sigma-Aldrich E3876), 50 μM PR-619 (Sigma-Aldrich, 662141) and proteinase inhibitor cocktail (Roche 1183617001). Lysates were centrifuged at 4°C for 15 min at 20,000g. The supernatants were incubated with normal mouse IgG (Santa Cruz Biotechnology sc-2025), GFP antibody (Santa Cruz Biotechnology sc-9996), VCP antibody (Santa Cruz Biotechnology sc-57492), FLAG antibody (Santa Cruz Biotechnology sc-166355), or G3BP1 antibody (BD 611127) conjugated with protein A/G magnetic beads (Thermo Fisher Scientific 88803) at 4°C overnight. Beads were washed three times with wash buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.05% Tween-20, 20 mM NEM and 50 μM PR-619) and treated with 0.1 M glycine pH 2.7 (Teknova G4527) at room temperature for 10 min to elute proteins. The resulting samples were analyzed by immunoblotting.
TUBE pulldown
Lysates were prepared using the same procedures as when performing immunoprecipitation. Lysates were incubated with Halo-beads or Halo-4xUBAUBQLN1-beads at 4°C overnight. Beads were washed three times with wash buffer, mixed with 4X NuPAGE LDS sample buffer, and boiled at 90°C for 5 min. The resulting samples were analyzed by immunoblotting.
Deubiquitination assay
Lysates were prepared using the same procedures as when performing immunoprecipitation, except that NEM and PR-619 were excluded from the tubes where the deubiquitination reaction was performed. Lysates were incubated with or without 2.72 μg of USP2 (LifeSensors DB501) overnight and further incubated with normal mouse IgG or G3BP1 antibody conjugated with protein A/G magnetic beads for 2 hours at 4°C. Beads were washed three times with wash buffer and treated with 0.1 M glycine pH 2.7 at room temperature for 10 min to elute proteins. The resulting samples were analyzed by immunoblotting.
Di-GLY TMT ubiquitinome analysis
Proteomic profiling of the ubiquitinome was performed with a previously reported protocol (52) with modification. Briefly, for each sample, HEK293T cells from five 15-cm dish were harvested, washed with ice-cold PBS, and collected by centrifugation. Cells were lysed in a buffer (50 mM HEPES, pH 8.5, 8 M urea, and 0.5% sodium deoxycholate) with deubiquitinating enzyme (DUB) inhibitors (50 μM PR-619 and 10 mM iodoacetamide) using probe sonication (15 s ×3 cycles). Iodoacetamide-induced pseudo-di-GLY peptides were not detected at room temperature (53) and were not recognized by di-GLY antibodies during the enrichment (54). To further minimize the impact of DUB activities, the cell lysis protocol was modified by immediately digesting the lysates with LysC (Wako) at an enzyme/substrate ratio of 1:100 (w/w) for 2 hours at room temperature, to achieve rapid DUB deactivation through proteolysis. Following this, the samples were quenched with DTT (30 mM for 30 min) and subjected to trypsin digestion and desalting. The desalted peptides (~4 mg) were resuspended in 400 μl of ice-cold IAP buffer (50 mM MOPS, pH 7.2, 10 mM sodium phosphate, and 50 mM NaCl) and centrifuged at 21,000g for 10 min at 4°C to remove any insoluble material. The peptides were then incubated with di-GLY antibody beads (Cell Signaling Technology) at an antibody-to-peptide ratio of 1:20 (w/w, optimized through a pilot experiment) for 2 hours at 4°C with gentle end-over-end rotation. The antibody beads were then collected by a brief centrifugation and washed three times with 1 ml of ice-cold IAP buffer and twice with 1 ml of ice-cold PBS, while the supernatants were carefully removed and saved. The di-GLY peptides were eluted at room temperature twice with 50 μl of 0.15% TFA, dried, and resuspended in 50 mM HEPES (pH 8.5) for 11-plex TMT labeling. The TMT-labeled peptides were combined equally among replicates, desalted, and analyzed by LC/LC-MS/MS using a similar protocol as described above except with the following differences: (i) a 60-min gradient used in the acidic pH LC-MS/MS with Orbitrap QE HF mass spectrometer (Thermo Fisher Scientific), (ii) database search using the COMET algorithm [v2018.013 (55)] with 0.02-Da mass tolerance for MS/MS ions, full trypticity with maximal five missed cleavages, maximal five dynamic modifications per peptide, and di-GLY modification on Lys (+114.04293 Da) as a dynamic modification.
Immunofluorescence
Cells were grown in 8-well chamber slides (Millipore PEZGS0816). Cells were fixed with 4% paraformaldehyde (Alfa Aesar J61899) in PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and blocked with 3% BSA for 1 hour. Samples were further incubated with primary antibodies as the following targets in blocking buffer at 4°C overnight: Lys48 linkage–specific ubiquitin (Millipore Sigma 05–1307), Lys63 linkage–specific ubiquitin (Millipore Sigma 05–1308), G3BP1 (BD 611127), G3BP1 (Proteintech 13057–2-AP), eIF3η (Santa Cruz Biotechnology sc-16377), VCP (BD Biosciences 612183), FAF2 (Proteintech 16251–1-AP), and calnexin (Thermo Fisher Scientific PA5–19169). Samples were washed three times with PBS and incubated with host-specific Alexa Fluor 488/555/647 secondary antibodies (Thermo Fisher Scientific) for 1 hour at room temperature. For microscopic imaging, slides were mounted with ProLong Gold Antifade reagent with DAPI (Thermo Fisher Scientific P36931). Images were captured using a Leica TCS SP8 STED 3× confocal microscope with a 63× oil objective. To stain ER-resident calnexin, cells were grown in fibronectin-coated coverslips (neuVitro GG18FIBRONECTIN) and permeabilized with 0.2% Triton X-100 in PBS for 1 min. Slides were mounted with ProLong Glass Antifade reagent (Thermo Fisher Scientific P36984).
Disassembly of stress granules, intracellular phase diagram, and FRAP using time-lapse live-cell microscopy
A Yokogawa CSU W1 spinning disk attached to a Nikon Ti2 eclipse with a Photometrics Prime 95B camera using Nikon Elements software was used in time-lapse live-cell imaging and FRAP. The light path was split between the port for the spinning disk/acquisition laser and the FRAP lasers, enabling FRAP to occur simultaneously while imaging. Imaging was taken using a 60× Plan Apo 1.4NA oil objective and Perfect Focus 2.0 (Nikon) engaged for the duration of the capture. During imaging, cells were maintained at 37°C and supplied with 5% CO2 using a Bold Line Cage Incubator (Okolabs) and an objective heater (Bioptechs).
To monitor the disassembly of heat shock–induced stress granules, multipoint images over 5 xy fields for each condition per one replicate were taken with the 555-nm laser. At 2 min into imaging, the objective temperatures were raised to 43°C for 60 min. After heat shock, the temperature was lowered back to 37°C to alleviate the stress, and cells were imaged after 2 hours had passed. Images were taken at each xy position every 30 s. Cells containing two or more stress granules larger than 0.54 μm were counted as stress granule–positive cells.
For intracellular phase diagrams, multipoint images over 25 xy fields for each condition per one replicate were taken with the 555-nm laser. At 2 min into imaging, the objective temperatures were raised to 43°C for 60 min. Images were taken at each xy position every 1 min. Phase diagrams were constructed by measuring GFP fluorescence intensity in each cell and assessing the presence of stress granules using Fiji software.
For fluorescence recovery after photobleaching, time lapses were acquired every 100 ms over the course of 45 s for stress granules with photobleaching with the 488-nm FRAP laser occurring 2 s into capture. Data were taken from at least n =10 different cells or lysate granules for each condition. In Nikon Elements, ROIs were generated in the photobleached region, a nonphotobleached cell, and the background for each time lapse, and the mean intensity of each was extracted. For photobleached regions, a 2.5-μm-diameter circle was used. Data were repeated in triplicate for each condition, with each replicate having at least n = 10 cells. The values retrieved from the ROIs were exported into Igor Pro 7.0 (WaveMetrics) and fit curves were generated after photobleach and background values were corrected.
Photoactivation of G3BP1
For live imaging of photoactivatable GFP, U2OS cells expressing G3BP-GFP or G3BP1-PAGFP were plated on 40-mm #1.5 thick coverslips (Bioptechs) or chambered coverglass (Millipore). Cells were observed with a Marianas confocal microscope (Leica) with a 63× objective. For heat shock live imaging experiments 48 hours after transfection, the coverslip was transferred to a FCS2 chamber assembled according to the manufacturer’s instructions (Bioptechs). Media was perfused through the chamber, and then the chamber was placed into a Marianas spinning disk confocal system with a stage-top incubator and 63× objective with an objective heater (Bioptechs), both preheated to 37°C. The Microaqueduct Slide heater (FCS2 system) and the heated objective with 37°C immersion oil (Zeiss) were used to control the temperature. Movies were collected on a Marianas confocal microscope with 63× objective, with 40-s intervals for the live imaging experiments, 200-ms intervals for the FLAP experiments of the time period. Average and standard errors were calculated from three independent experiments measuring at least 35 cells.
ATP measurement
Cellular contents of ATP were measured using CellTiter-Glo 2.0 assay kit (Promega G9242) according to the manufacturer’s instructions; 200 mM 2-deoxy-D-gluose (Millipore Sigma D6134) was used to inhibit the glycolysis pathway.
Statistical analysis
Statistical analysis was performed in GraphPad Prism. Comparisons between two means were performed by two-tailed t test. Comparisons among multiple means over 2 were performed by one-way analysis of variance (ANOVA) with Tukey’s test. Mantel-Cox tests were used to compare the dissociation curves showing cells with stress granules in live-cell imaging.
Supplementary Material
Acknowledgements:
We thank N. Nedelsky for editorial assistance; the Center for Advanced Genome Engineering at St. Jude Children’s Research Hospital for assistance with CRISPR-Cas9 modified cell lines; and J. Messing and J. Temirov (both at St. Jude) for assistance with microscopy.
Funding: Supported by NIH grant R35NS097074, HHMI, and ALS Association grant 18-IIA-419 (J.P.T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests: J.P.T. is a consultant for 5AM and Third Rock Ventures.
Data and materials availability:
All data are available in the main text or the supplementary materials.
References and Notes:
- 1.Buchan JR, mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11, 1019–1030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomes E, Shorter J, The molecular language of membraneless organelles. J. Biol. Chem 294, 7115–7127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang P et al. , G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345 e328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders DW et al. , Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324 e328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guillen-Boixet J et al. , RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361 e317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu C, Pappu RV, Taylor JP, Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science 370, 56–60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedersha N et al. , G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol 212, 845–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P et al. , Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tauber D, Tauber G, Parker R, Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem. Sci 45, 764–778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchan JR, Kolaitis RM, Taylor JP, Parker R, Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B et al. , ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol. Cell 74, 742–757 e748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson P, Kedersha N, RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol 10, 430–436 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Maxwell BA et al. , Ubiquitination is essential for recovery of cellular activities following heat shock Science, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markmiller S et al. , Active Protein Neddylation or Ubiquitylation Is Dispensable for Stress Granule Dynamics. Cell Rep. 27, 1356–1363 e1353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turakhiya A et al. , ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol. Cell 70, 906–919 e907 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Swatek KN, Komander D, Ubiquitin modifications. Cell Res. 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtake F, Saeki Y, Ishido S, Kanno J, Tanaka K, The K48-K63 Branched Ubiquitin Chain Regulates NF-kappaB Signaling. Mol. Cell 64, 251–266 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Hornbeck PV et al. , PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amino acid abbreviations: Ala A; Asp D; Glu E; Gly G; His H; Lys K; Leu L; Asn N; Pro P; Gln Q; Ser S; Val V; Tyr Y.
- 21.Jain S et al. , ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freibaum BD, Messing J, Yang P, Kim HJ, Taylor JP, High fidelity reconstitution of stress granules and nucleoli in mammalian cellular lysate. bioRxiv, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matus S, Bosco DA, Hetz C, Autophagy meets fused in sarcoma-positive stress granules. Neurobiol. Aging 35, 2832–2835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krisenko MO et al. , Syk Is Recruited to Stress Granules and Promotes Their Clearance through Autophagy. J. Biol. Chem 290, 27803–27815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrone L et al. , Isogenic FUS-eGFP iPSC Reporter Lines Enable Quantification of FUS Stress Granule Pathology that Is Rescued by Drugs Inducing Autophagy. Stem Cell Reports 10, 375–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto A et al. , Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct 23, 33–42 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Patterson GH, Lippincott-Schwartz J, A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Meyer H, Bug M, Bremer S, Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Stolz A, Hilt W, Buchberger A, Wolf DH, Cdc48: a power machine in protein degradation. Trends Biochem. Sci 36, 515–523 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Yeung HO et al. , Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans 36, 62–67 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Ye Y, Tang WK, Zhang T, Xia D, A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front Mol Biosci 4, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Y, Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J. Struct. Biol 156, 29–40 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Meyer H, Weihl CC, The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci 127, 3877–3883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burana D et al. , The Ankrd13 Family of Ubiquitin-interacting Motif-bearing Proteins Regulates Valosin-containing Protein/p97 Protein-mediated Lysosomal Trafficking of Caveolin 1. J. Biol. Chem 291, 6218–6231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markmiller S et al. , Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youn JY et al. , High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 69, 517–532 e511 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Olzmann JA, Richter CM, Kopito RR, Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. U. S. A 110, 1345–1350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloppsteck P, Ewens CA, Forster A, Zhang X, Freemont PS, Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim. Biophys. Acta 1823, 125–129 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Lee JN et al. , Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proc. Natl. Acad. Sci. U. S. A 107, 21424–21429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL, SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. U. S. A 105, 12325–12330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Y et al. , Pathogenic mutation of UBQLN2 impairs its interaction with UBXD8 and disrupts endoplasmic reticulum-associated protein degradation. J. Neurochem 129, 99–106 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Zhou HL, Geng C, Luo G, Lou H, The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP. Genes Dev. 27, 1046–1058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nedelsky NB, Taylor JP, Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol 15, 272–286 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Costa-Mattioli M, Walter P, The integrated stress response: From mechanism to disease. Science 368, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams CJ, Kopp MC, Larburu N, Nowak PR, Ali MMU, Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front Mol Biosci 6, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopata A, Kniss A, Lohr F, Rogov VV, Dotsch V, Ubiquitination in the ERAD Process. Int. J. Mol. Sci 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zehmer JK et al. , Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J. Cell Sci 122, 3694–3702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JE, Cathey PI, Wu H, Parker R, Voeltz GK, Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tresse E et al. , VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6, 217–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang K et al. , Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell 173, 958–971 e917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figley MD, Bieri G, Kolaitis RM, Taylor JP, Gitler AD, Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J. Neurosci 34, 8083–8097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose CM et al. , Highly Multiplexed Quantitative Mass Spectrometry Analysis of Ubiquitylomes. Cell Syst 3, 395–403 e394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu P et al. , Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bustos D, Bakalarski CE, Yang Y, Peng J, Kirkpatrick DS, Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol. Cell. Proteomics 11, 1529–1540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eng JK et al. , A deeper look into Comet--implementation and features. J. Am. Soc. Mass Spectrom 26, 1865–1874 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.