Abstract
Objective
Real-time reverse transcription-polymerase chain reaction is the gold standard for the diagnosis of COVID-19, but it is necessary to utilize other tests to determine the burden of the disease and the spread of the outbreak such as IgG-, IgM-, and IgA-based antibody detection using enzyme-linked immunosorbent assay (ELISA).
Materials and Methods
We developed an indirect ELISA assay to quantitatively measure the amount of COVID-19 IgG, IgM, and IgA antibodies present in patient serum, dried blood, and plasma.
Results
The population cutoff values for positivity were determined by receiver operating characteristic curves to be 1.23 U/mL, 23.09 U/mL, and 6.36 U/mL for IgG, IgM, and IgA, respectively. After albumin subtraction, the specificity remained >98% and the sensitivity was 95.72%, 83.47%, and 82.60%, respectively, for IgG, IgM, and IgA antibodies to the combined spike subunit 1 receptor binding domain and N proteins in serum. Plasma and dried blood spot specimens were also validated on this assay.
Conclusion
This assay may be used for determining the seroprevalence of SARS-CoV-2 in a population exposed to the virus or in vaccinated individuals.
Keywords: COVID-19, SARS-CoV-2, iELISA, S1 RBD, nucleocapsid protein, serum/plasma, dried blood
In December 2019, the novel coronavirus SARS-CoV-2 emerged in Wuhan, China, and has led to a global pandemic, with the COVID-19 disease affecting >61 million individuals worldwide and accounting for nearly 300,000 deaths in the United States to date.1,2 The transmissibility and disease severity are much higher compared to the original SARS virus, and effective control over the disease is still lacking. The fight against SARS-CoV-2 has been focused on the prevention of infection, detection of cases of infection, and diagnosis and monitoring of the disease. Prevention of infection is predominantly accomplished through vaccination, social distancing, hand hygiene, and masking. Unfortunately, a recommended, global therapy is still not currently available, although the US Food and Drug Administration had approved one treatment (remdesivir) as of November 2020 and has issued emergency use authorizations for several other treatments.3 Molecular tests have been the gold standard for the detection and diagnosis for cases of infection; however, large-scale, continued implementation has been hindered because of cost, feasibility, speed, and reagent availability. Analytical issues with reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays have been well documented and include high false-positive and false-negative rates because of contamination, specimen integrity, or cross-reactivity issues.4-6 To develop diagnostics, therapeutics, and vaccines against SARS-CoV-2, a better understanding of the immunogenicity and pathobiology of SARS-CoV-2 infections is needed. Immunological assays have become a good alternative in this regard.
An indirect enzyme-linked immunosorbent assay (iELISA) is a common biochemical technique that is most suitable for determining total antibody concentrations in a specimen. This method is commonly utilized to diagnose infection and to quantify antibodies against an invading antigen rather than to detect a virus itself. Infection with the SARS-CoV-2 virus elicits the development of IgM- and IgG-specific antibodies, which are the most available antibodies for assessing response, whereas less is known about the response of IgA in the blood. Previous studies have shown variable isotype responses to SARS-CoV-2, with 1 study noting that 92.7% of patients tested positive for anti-SARS-CoV-2 nuclear capsid IgA, whereas only 85.4% had IgM and only 77.9% tested positive for IgG.7 Current testing for the SARS-CoV-2 virus is limited, and compared to RT-qPCR, ELISA is a less complex procedure that uses more affordable and available equipment. Similarly, antigens and antibodies are considerably more stable than RNA, which reduces the potential of false-negative results. The ability to collect specimens from many places in the body (and not being restricted to nasal swabs) improves testing accuracy as well. Although iELISA is not ideal for early diagnosis, it has been used to (1) diagnose patients who are more than 1 week post-symptom onset, (2) determine potential immunity and risk of infection, (3) advance contact tracing, and (4) understand the extent of COVID-19 spread and immunity in communities through epidemiological studies that are particularly important for fighting COVID-19 while minimizing economic impact.8,9 Similarly, with the many different antibody serology tests commercially available, a number of studies assessing their performance have been conducted suggesting that the antibody isotype and timing should be carefully considered to optimize the diagnostic accuracy and usefulness of the assay.9-17
The SARS-CoV-2 virus contains approximately 27 proteins, including 4 structural proteins: the spike proteins (S protein), membrane proteins, envelope proteins, and nucleocapsid proteins (N protein). The S protein, through the receptor-binding domain (RBD) of the S1 subunit, is understood to be the viral faction that binds with the host cell receptors and facilitates viral entry into the cell.18 The N protein is the most abundant viral protein and was characterized first after emergence of the virus.9 Thus, the S1 RBD and N proteins are of the most interest as candidates for diagnosis and antibody determination. Here, we report the development of the recombinant SARS-CoV-2 antigen that is used in production and iELISAs to the N protein, the S1 RBD protein, and the combination of both the N and S1 RBD proteins. The determination of the sensitivity and specificity, dynamics, and magnitude of IgG, IgM, and IgA antibody responses in patients with COVID-19 are presented and the antibody responses in a variety of biological specimens including serum, plasma, and dried blood spot are discussed.
Materials and Methods
Cloning and DNA Preparation
We cloned cDNA fragments encoding the S1 RBD protein (GenBank accession number QHD43416, Arg319-Phe541) and the full-length N protein (GenBank accession number QHD43423, Met1-Ala419) of SARS-CoV-2 (Wuhan-Hu-1 strain) into a mammalian cell expression vector, pExpR10, with an N-terminal signaling peptide to direct the secretion of the recombinant proteins into the cell culture medium and a C-terminal His-tag for protein affinity purification. The recombinant expression constructs were confirmed by DNA sequencing. A large amount of endotoxin-free cell transfection-grade plasmid DNA was extracted using the Thermo Invitrogen GigaPrep plasmid preparation kits according to the manufacturer’s protocol (Thermo Fisher Scientific, New York, NY).
Gene Expression
Large-scale, transient cell transfection of human embryonic kidney (HEK) 293 suspension cells was performed using the VWR DNA Transfection Reagent following the manufacturer’s protocol (VWR, Philadelphia, PA). The cell culture was collected 72 hours post transfection and was centrifuged at 3000g for 20 minutes at 4ºC; the culture supernatants were further clarified by passing through a 0.45 μm membrane filter.
Protein Purification
His-tagged recombinant S1 RBD and N proteins were purified using immobilized metal affinity chromatography. Briefly, the clarified culture supernatant was diluted with an equal amount of 2× binding buffer (40 mM sodium phosphate buffer, pH 7.4) and loaded onto a 5 mL-Ni sepharose high performance nickel-charged column (GE Healthcare, Chicago, IL) with the flow rate at 5 mL/min, pre-equilibrated with 1× binding buffer (20 mM sodium phosphate buffer, pH 7.4). After specimen loading, the column was sequentially washed with 1× binding buffer (10 column volumes) and then 1× washing buffer (20 mM sodium phosphate buffer, pH 7.4, containing 40 mM imidazole; 10 column volumes). The Ni-column-bound proteins were eluted with a 1× elution buffer (20 mM sodium phosphate buffer, pH 7.4, containing 750 mM imidazole). All fractions were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining. Appropriate fractions were then pooled and concentrated using a Millipore Amicon Ultra-4 Centrifugal Filter Unit (MilliporeSigma, St Louis, MO). The final purified protein was thoroughly desalted by dialysis against 1× phosphate buffered saline (PBS; pH 7.4) at 4ºC overnight with 2 buffer changes. The protein concentration was determined with a Thermo Pierce bicinchoninic acid assay kit using bovine serum albumin as the protein standard (Thermo Fisher Scientific). The final protein products were aliquoted and stored at –80ºC until use.
Western Blot Analysis
Purified protein was mixed with a 5× SDS protein loading buffer and boiled for 10 minutes. For each individual specimen, 0.5 μg of purified S1 RBD or N protein was loaded per lane on a 10% SDS-PAGE. Proteins were electrotransferred to polyvinylidene fluoride (PVDF) membranes at a constant 100 V for 60 minutes. The PVDF membranes were blocked with 5% nonfat milk for 2 hours at room temperature and incubated with primary antibody (catalog numbers 130–10808, 130–10807, RayBiotech, Peachtree Corners, GA) at a 1:1000 dilution overnight at 4˚C, according to the manufacturer’s protocol. The membranes were washed with tris-buffered saline with Tween-20 (TBST) 5 times for 5 minutes per wash and incubated with horseradish peroxidase (HRP)–conjugated goat anti-mouse IgG antibody (catalog number 115–035–003, Jackson ImmunoResearch, West Grove, PA) at a 1:5000 dilution for 1 hour at room temperature. The membranes were washed with TBST again and visualized using an enhanced chemiluminescence reagent (CHMI-0300–2C, Surmodics, Eden Prairie, MN) and scanned using UVP Bioimaging systems (Wazobia Enterprise, Houston, TX).
Indirect ELISA
Indirect ELISA for the determination of IgG, IgM, and IgA antibodies to the S1 RBD and N protein in serum, plasma, and dried blood was developed. A 96-well microplate (Greiner Bio-One, Monroe, NC) was coated with 1.5 μg/mL of recombinant S1 RBD protein and/or 1 μg/mL N protein in 100 μL of 0.1 M Na2HPO4 buffer (pH 9.0) and incubated overnight at 4°C. Plates were washed 5 times with PBS and 1% Tween-20 (PBST) and then blocked by adding 120 μL of blocking buffer per well (Rockland, PA). The plates were incubated for 1.5 hours at room temperature. The blocking buffer was then discarded and 100 μL of the specimen was added into the wells and incubated for 1 hour at room temperature. Serum and plasma specimens were diluted at 1:1500 for IgG and 1:500 for IgM and IgA. Dried blood specimens were eluted at a 1:10 ratio in elution buffer and diluted with specimen diluent at a 1:10 ratio for IgG and a 1:100 ratio for IgM and IgA analysis. The wells were washed 5 times with PBST, and 100 μL of antihuman biotinylated IgG, IgM, or IgA was added (Jackson ImmunoResearch; Southern Biotech, Birmingham, AL) to each well and incubated for 30 minutes. After washing 5 times with PBST, 100 μL of HRP-streptavidin solution was added to each well and incubated for another 30 minutes. The wells were washed another 5 times, and 100 μL of TMB substrate (Surmodics, Eden Prairie, MN) was added to each well and incubated for 15 minutes. The enzymatic reaction was stopped by adding 50 uL of 0.2 M sulfuric acid to each well. The absorbance of each specimen was measured at 450 nm using an ELISA plate reader (BioTek, Winooski, VT).
Patient Specimens
Serum specimens from patients with COVID-19 were commercially sourced: Bioresource Technology (57 specimens; Weston, FL), Cantor Bioconnect (54 specimens; Santee, CA), Texas Direct Diagnostics (79 specimens; Irving, TX), and PanoHealth (41 specimens; Peachtree Corners, GA). Thirty-one and 27 specimens collected by PanoHealth had matched plasma and dried blood specimens, respectively. Another set of 20 specimens was collected in the Shunde Hospital at Guangzhou University of Chinese Medicine (Guangzhou, China). Patient COVID-19 status was determined with RT-PCR using nasopharyngeal or oropharyngeal swab specimens to detect SARS-CoV-2 nucleic acid, and specimens were at least 10 days post-symptom onset. Pre-COVID-19 serum specimens collected prior to 2019 and negative for COVID-19 were obtained from BioIVT (15 specimens; Westbury, NY) and RayBiotech Life (249 specimens). The serum specimens were separated after centrifugation at 1000 rpm for 10 minutes, and COVID-19-positive specimens were then inactivated with 0.5% Triton X-100 for 1.5 hours. All specimens were stored at –80°C until further use. Overall, 515 serum specimens (251 COVID-19-positive, 264 COVID-19-negative) were included for assay validation. Assay cross-reactivity was determined with 72 COVID-19 negative serum specimens from patients who were RT-PCR-positive for adenovirus, antinuclear antibodies of autoimmune disease, cytomegalovirus, hepatitis B virus, hepatitis C virus, human parainfluenza viruses, influenza B, MP virus, or respiratory syncytial virus. These pathogens were chosen based on the US Food and Drug Administration recommendations for validation studies to be conducted for SARS-CoV-2 serology tests related to the tested specimen types. A summary of the specimens used in this study are noted in Table 1. This study was approved by the institutional review board (number 8291-BZhang), and the Institutional Human Ethics Committee of the Shunde Hospital of Guangzhou University of Chinese Medicine approved this study (approval number KY-2020001).
Table 1.
Sample Information
| Positive Specimens | COVID-19, PCR Confirmed | 251 |
|---|---|---|
| Prepandemic specimens (collected before 2019) | Adenovirus infected | 7 |
| Antinuclear antibodies infected | 5 | |
| Cytomegalovirus infected | 5 | |
| Hepatitis B virus infected | 13 | |
| Hepatitis C virus infected | 15 | |
| Human parainfluenza viruses infected | 6 | |
| Influenza B infected | 5 | |
| MP virus infected | 6 | |
| Respiratory syncytial virus infected | 10 | |
| Healthy control sample | 179 | |
| Total | 515 |
PCR, polymerase chain reaction.
Statistical Analysis
The concentrations of antibodies against the SARS-CoV-2 S1 RBD and N proteins were summarized as the mean ± standard deviation by COVID-19 diagnosis and the interval between the specimen collection after the onset of COVID-19 symptoms. We conducted receiver operating characteristic analysis19 on the antibody concentration measured using iELISA to assess its diagnostic performance, with SARS-CoV-2 RT-PCR results as the reference standard. The cutoff with the best performance was selected based on the sensitivity and specificity; ie, the true positive rate among PCR-confirmed positive specimens and the true negative rate among pre-COVID-19 negative control specimens. The correlation of iELISA results among different specimen types (serum, plasma, and dried blood specimens) from the same individual were evaluated with Pearson’s correlation coefficient. The antibody concentrations in patients who were PCR-confirmed positive were plotted against time after symptom onset, with a smoothed line predicted by local weighted regression (loess)20 to reflect the trend of time-course shifting. All statistical analyses were implemented using R (version 3.6.3).21
Results
Production of Recombinant SARS-Cov-2 S1 RBD and N Proteins in Mammalian Cells
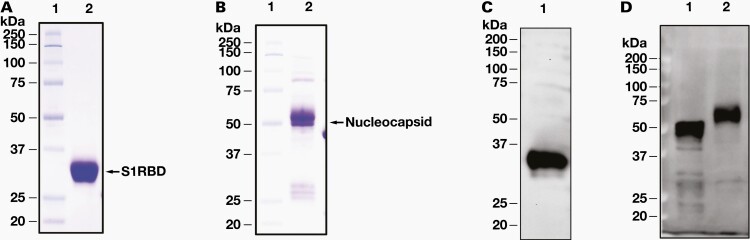
At 3 days post-transfection, recombinant S1 RBD and N proteins were successfully secreted into the serum-free HEK293 cell culture medium. The expressed and secreted recombinant proteins were further affinity-purified with the C-terminal-fused His-tag. The final desalted S1 RBD protein (RayBiotech; catalog number 230–30162) migrated as an approximately 30 kDa protein band on SDS-PAGE under dithiothreitol, beta-mercaptoethanol reducing conditions (Figure 1A), which was larger than the expected size of 25 kDa. The purified N protein (RayBiotech; catalog number 230–30164; Figure 1B) had 1 major band at approximately 55 kDa, which was larger than the expected size of 47 kDa. To investigate these differences, the N and S1 RBD proteins were deglycosylated using an enzyme mixture. After cleaving the glycans, major bands at the expected size were present, indicating that the untreated recombinant proteins were glycosylated (data not shown). The purified recombinant proteins were confirmed by Western blot using a mouse monoclonal antibody, anti-SARS-CoV-2 S1RBD (130–10808) and anti-N protein (130–10817) (Figure 1C and 1D).
Figure 1.
12% SDS-PAGE analysis of purified recombinant S1 RBD and N proteins. Purified S1 RBD (A) and N protein (B) were run at 10% SDS-PAGE and probed with mouse monoclonal anti-SARS-CoV-2 S1 RBD antibody (130–10808) (C) and anti-SARS-CoV-2 N protein antibody (130–10817) (D). Lane 1 is purified Escherichia coli-derived N protein and Lane 2 is HEK293-purified N protein. HEK, human embryonic kidney; N protein, nucleocapsid protein; RBD, receptor binding domain; S1, spike subunit 1 protein; SDS-PAGE, sodium docetyl sulfate-polyacrylamide gel electrophoresis.
Production and Comparison of S1 RBD and N Protein iELISAs
The iELISAs measuring the IgG, IgM, and IgA antibodies to the S1 RBD and N proteins of SARS-CoV-2 were developed for the serology testing of the patient specimens. Pre-COVID-19 (collected before 2019) negative control specimens and PCR-confirmed positive specimens were incubated on the S1 RBD and N protein coated plates. The anti-S1 RBD protein antibodies or anti-N protein antibodies present in the patient specimens were bound to the plate and unbound antibodies were removed. The specific, bound antibodies were detected with anti-human IgG, anti-human IgM, or anti-human IgA. To quantitatively measure the relative titer of patient antibodies, we selected a pool of 10 PCR-confirmed positive specimens with high titers (optical density [OD] > 1.0) of the IgG, IgM, and IgA antibodies and defined a certain dilution as an arbitrary unit. Then, we used a serial dilution of this positive control to determine the quantitative units of antibodies present in each unknown specimen tested.
The IgG antibodies to the N protein were determined in 246 pre-COVID-19 negative control serum specimens and 81 PCR-confirmed positive serum specimens, and the IgM and IgA antibodies were determined in 56 pre-COVID-19 negative control serum specimens and 10 PCR-confirmed positive serum specimens. The IgG antibodies to the S1 RBD protein were determined in 264 pre-COVID-19 negative control serum specimens and 114 PCR-confirmed positive serum specimens, and the IgM and IgA antibodies were determined in 264 pre-COVID-19 negative control serum specimens and 115 PCR-confirmed positive serum specimens. The positivity cutoff values for the serum IgG antibodies to the S1 RBD and N proteins were determined to be 23.84 U/mL and 22.81 U/mL, respectively. The positivity cutoff values for the serum IgM antibodies to the S1 RBD and N proteins were 507.06 U/mL and 668.83 U/mL, respectively. Finally, the positivity cutoff values for the serum IgA antibodies to the S1 RBD and N proteins were 329.93 U/mL and 33.72 U/mL, respectively. The specificity and sensitivity for each protein and each antibody are shown in Table 2. The specificity for the IgG, IgM, and IgA antibodies to both proteins was >98%, whereas the sensitivity ranged, with IgG antibodies being the highest at 76% for both proteins. The sensitivity of IgM to the S1 RBD was only 66% and the sensitivity to the N protein was only 10%. The sensitivity of IgA was 80% to the N protein but was only 26% to the S1 RBD protein.
Table 2.
Comparison Sensitivity and Specificity of S1 RBD and N Protein iELISAs for IgG, IgM, and IgA Antibodies
| Target | Specificity | False Positives | Sensitivity | False Negatives | |
|---|---|---|---|---|---|
| S1 RBD | IgG | 98.48% | 4 | 76.31% | 27 |
| N | IgG | 98.37% | 4 | 76.54% | 19 |
| S1 RBD | IgM | 98.10% | 5 | 66.09% | 39 |
| N | IgM | 98.21% | 1 | 10.00% | 9 |
| S1 RBD | IgA | 98.10% | 5 | 26.08% | 85 |
| N | IgA | 98.21% | 1 | 80.00% | 2 |
RBD, receptor binding domain; S1, spike subunit 1 protein.
Interestingly, most of the pre-COVID-19 negative control specimens and PCR-confirmed positive specimens were accurately identified for IgG antibodies to both the N and S1 RBD proteins; however, a subset of specimens was inaccurately identified as either a false negative or a false positive. All the false-positive specimens for each IgG, IgM, and IgA antibody were different between the S1 RBD and N protein assays. Of the larger number of false-negative specimens, only 1 IgG and IgM specimen and 2 IgA specimens were the same for both the S1 RBD and N protein assays. These data strongly suggest that a subset of individual specimens will react differently to the S1 RBD or N proteins. Given these data, we decided to combine both the S1 RBD and N proteins together on a plate to increase the sensitivity and minimize the rate of false negatives for each specimen.
Comparison of the Combined S1 RBD and N Protein iELISA
To test whether the specificity and sensitivity could be enhanced by using a combination of the S1 RBD and N proteins, the 2 proteins were first mixed, and then the mixture of proteins was coated into 96-well plates. The IgG antibodies to the combined S1 RBD and N proteins were measured in 260 pre-COVID-19 negative control serum specimens and 117 PCR-confirmed positive serum specimens; the IgM and IgA antibodies to the combined S1 RBD and N proteins were measured in 264 pre-COVID-19 negative control serum specimens and 115 PCR-confirmed positive serum specimens. The positivity cutoff values for the serum IgG, IgM, and IgA antibodies to the combined S1 RBD and N proteins were 6.13 U/mL, 146.97 U/mL, and 68.32 U/mL, respectively. The data showed that the specificity remained >98% for all antibodies and the sensitivity increased for IgG from 76% up to 94.87%. The sensitivity for IgM was 62.60%, which was decreased slightly from 66.09% compared to the S1 RBD protein–alone assay but significantly increased from 10% compared to the N protein–alone assay. The sensitivity for IgA was 51.30%, which was significantly increased from 26.08% compared to the S1 RBD protein–alone assay and was decreased from 80% compared to the N protein–alone assay (Table 3). Although we found that this approach minimized the number of false negatives/positives, we noticed a significant level of background in the pre-COVID-19 negative control specimens that was decreasing the sensitivity and specificity of the assay. To overcome this issue, we included an albumin-coated plate. The same 264 pre-COVID-19 negative control serum specimens and 115 PCR-confirmed positive serum specimens were incubated on both the combined S1 RBD and N protein coated plates and an albumin-coated plate, and the assay was run simultaneously. The resulting ODs for each specimen on the albumin plate were subtracted from the ODs of the respective specimens on the S1 RBD and N protein-coated plates before we degermined the quantitative units of antibodies present. After albumin subtraction, the specificity remained >98% and the sensitivity was significantly increased to 95.72%, 83.47%, and 82.60%, respectively for the IgG, IgM, and IgA antibodies (Table 4). A population cutoff for positivity was determined for each antibody to be 1.23 U/mL, 23.09 U/mL, and 6.36 U/mL for IgG, IgM, and IgA, respectively. Thus, an unknown serum specimen above the respective U/mL cutoff was considered positive.
Table 3.
Sensitivity and Specificity of the Combined S1 RBD and N Protein iELISA for IgG, IgM, and IgA Antibodies
| Target | Specificity | False Positives | Sensitivity | False Negatives |
|---|---|---|---|---|
| IgG | 98.07% | 5 | 94.87% | 6 |
| IgM | 98.11% | 5 | 62.60% | 43 |
| IgA | 98.11% | 5 | 51.30% | 56 |
Table 4.
Sensitivity and Specificity of the Combined S1 RBD and N Protein and Albumin Subtracted iELISA for IgG, IgM, and IgA Antibodies
| IgG | IgM | IgA | |
|---|---|---|---|
| AUC | 0.9839 | 0.9606 | 0.9441 |
| Specificity | 98.07% | 98.48% | 98.10% |
| Sensitivity | 95.72% | 83.47% | 82.60% |
| Accuracy | 96.34% | 93.93% | 93.40% |
| Population positivity cutoff | 1.23 U/mL | 23.09 U/mL | 6.36 U/mL |
| Kappa | 0.95 | 0.9 | 0.89 |
AUC, area under the curve.
Detection of IgG, IgM, and IgA Antibodies in Different Specimen Matrices
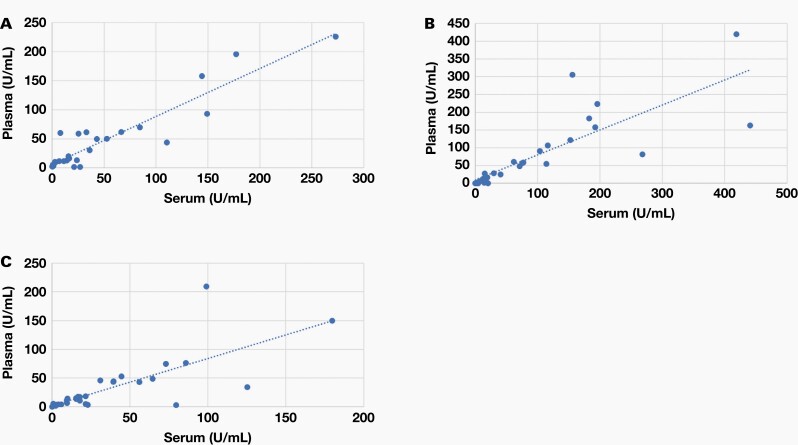
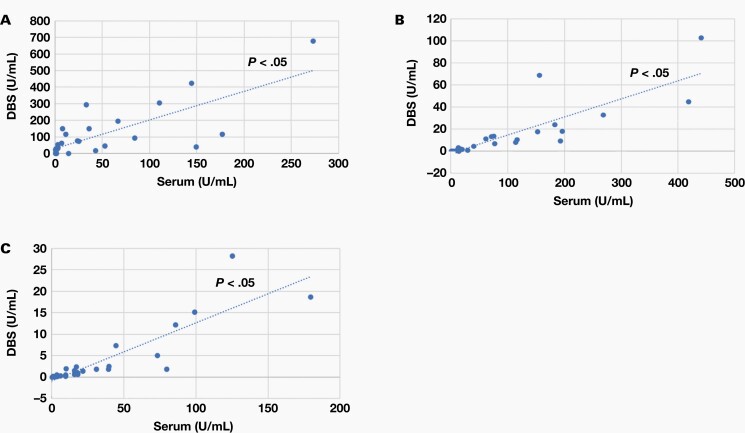
Analysis of COVID-19 in several different specimen types provide a few advantages. Dried blood specimens in particular have a minimally invasive specimen collection, minimal specimen processing requirements, low cost, long-term stability in shipping or storage, and increased feasibility for remote sampling, which makes it an ideal specimen type for large-scale serological profiling of patients with COVID-19. To test whether this quantitative approach can be used to effectively measure antibodies present in specimen types other than serum, we also tested plasma and dried blood specimens. The amount of IgG, IgM, and IgA antibodies present in the combined S1 RBD and N proteins using albumin-subtracted ELISA in 31 matched, PCR-confirmed positive serum and plasma specimens and 27 matched, PCR-confirmed positive serum and dried blood specimens was moderately correlated between all 3 specimen types (Figure 2 and Figure 3). These data suggest that this approach is well suited and can be easily adapted to different specimen types.
Figure 2.
Correlations between serum and plasma. We analyzed 31 positive specimens using a scatterplot; each blue dot represents a specimen. The x axis represents U/mL for serum and the y axis represents U/mL for plasma. Correlations for IgG (A), IgM (B), and IgA (C) between serum and plasma, respectively.
Figure 3.
Correlations between serum and DBS. We analyzed 27 positive specimens using a scatterplot; each blue dot represents a sample. The x axis represents U/mL for serum and the y axis represents U/mL for DBS. Correlations for IgG (A), IgM (B), and IgA (C) between serum and DBS, respectively. DBS, dried blood specimen.
Cross-Reactivity and Double-Blind Validation of the Combined S1 RBD and N Proteins Using Albumin-Subtracted Assay
In addition to validating this assay’s use in multiple specimen types, we wanted to validate the assay for lack of cross-reactivity to other viruses and autoimmune diseases. We examined the IgG, IgM, and IgA antibody levels to the SARS-CoV-2 S1 RBD and N proteins using albumin-subtracted iELISA in serum specimens from 72 patients with confirmed virus infection, including those with adenovirus, antinuclear antibodies of autoimmune disease, cytomegalovirus, hepatitis B virus, hepatitis C virus, human parainfluenza viruses, influenza B, MP virus, and respiratory syncytial virus. As shown in Table 5, among the 72 specimens, none cross-reacted for IgG, and only 2 specimens, a hepatitis C virus–infected specimen and a respiratory syncytial virus–infected specimen, cross-reacted for IgM. Only 1 specimen, a respiratory syncytial virus–infected specimen, cross-reacted for IgA. These data show low cross-reactivity of this assay with several common viruses.
Table 5.
Cross-Reactivity of the Combined S1 RBD and N Protein and Albumin Subtracted iELISA for IgG, IgM, and IgA Antibodies with Several Common Viruses
| Virus | Numbers of Specimens | Cross-Reactivity result (positive/negative) | ||
|---|---|---|---|---|
| IgG | IgM | IgA | ||
| Adenovirus | 7 | 0/7 | 0/7 | 0/7 |
| Antinuclear antibodies | 5 | 0/5 | 0/5 | 0/5 |
| Cytomegalovirus | 5 | 0/5 | 0/5 | 0/5 |
| Hepatitis B virus | 13 | 0/13 | 0/13 | 0/13 |
| Hepatitis C virus | 15 | 0/15 | 1/14 | 0/15 |
| Human parainfluenza viruses | 6 | 0/6 | 0/6 | 0/6 |
| Influenza B | 5 | 0/5 | 0/5 | 0/5 |
| MP virus | 6 | 0/6 | 0/6 | 0/6 |
| Respiratory syncytial virus | 10 | 0/10 | 1/9 | 1/9 |
We also performed a double-blinded study with a unique specimen set of 30 serum specimens, containing 10 pre-COVID-19 negative control specimens and 20 PCR-confirmed positive specimens each. The established cutoffs of 1.23 U/mL, 23.09 U/mL, and 6.36 U/mL for IgG, IgM, and IgA, respectively, were used for assessment of positivity. The specificities of both double-blinded studies were >90% for all antibodies and the sensitivities were 90.00%, 75.00%, and 70.00% for IgG, IgM, and IgA, respectively. These data were similar, despite the small sample size, to the original specimen set used to establish the cutoffs (Table 6). Taken together, these data show the robustness of the assay in accurately determining the SARS-CoV-2 antibodies present in a patient specimen.
Table 6.
Double-Blind Test Results of the Combined S1 RBD and N Protein and Albumin Subtracted iELISA for IgG, IgM, and IgA Antibodies
| Target | Specificity | False Positives | Sensitivity | False Negatives |
|---|---|---|---|---|
| IgG | 90.00% | 1 | 90.00% | 2 |
| IgM | 100.00% | 0 | 75.00% | 5 |
| IgA | 100.00% | 0 | 70.00% | 6 |
Time Course of Antibody Response During COVID-19
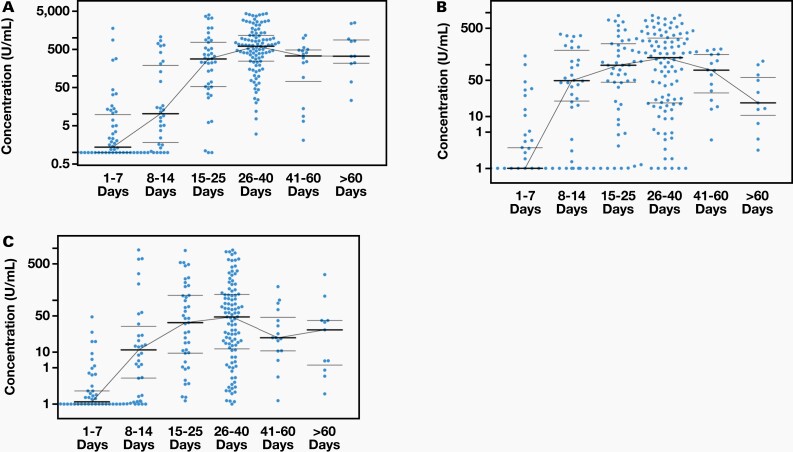
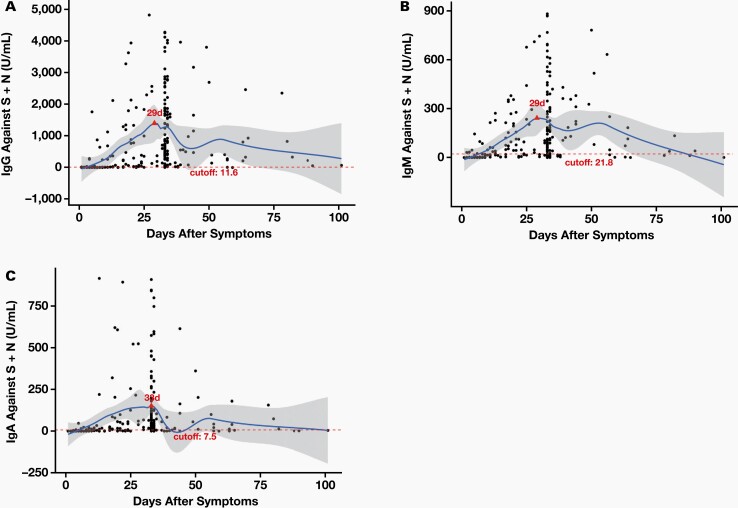
The quantitative nature of the developed assay allowed for the determination of the time course of the antibody response while a patient has COVID-19 disease. We assessed the IgG, IgM, and IgA antibody response in 251 PCR-confirmed positive serum specimens: 50 specimens were 1 to 7 days post-symptom onset, 30 specimens were 8 to 14 days post-symptom onset, 41 specimens were 15 to 25 days post-symptom onset, 103 specimens were 26 to 40 days post-symptom onset, 16 specimens were 41 to 60 days post-symptom onset, and 11 specimens were >60 days post-symptom onset. The levels of IgG, IgM, and IgA antibodies to the combined S1 RBD and N proteins using albumin-subtracted assay are shown in Figure 4 and Figure 5. These data show that most specimens have a similar response for IgG, IgM, and IgA antibodies, in terms of peak response and particularly after 10 days post-symptom onset.
Figure 4.
Time course for IgG (A), IgM (B), and IgA (C). We analyzed 251 positive specimens. Each blue dot represents a specimen; the thick black line represents the median for each group and the thin black line is for error bars. The x axes represent days after symptoms; there are 6 groups: 1–7 days, 8–14 days, 15–25 days, 26–40 days, 41–60 days, and >60 days. The y axes represent concentration (U/mL) for IgG, IgM, and IgA, respectively.
Figure 5.
Time course for IgG (A), IgM (B), and IgA (C). We analyzed 223 positive specimens. Each black dot represents a specimen. The x axes represent days after symptoms. The y axes represent concentration (U/mL) for IgG, IgM, and IgA, respectively. The red dotted line indicates the cutoff value, and the blue smooth line indicates the median.
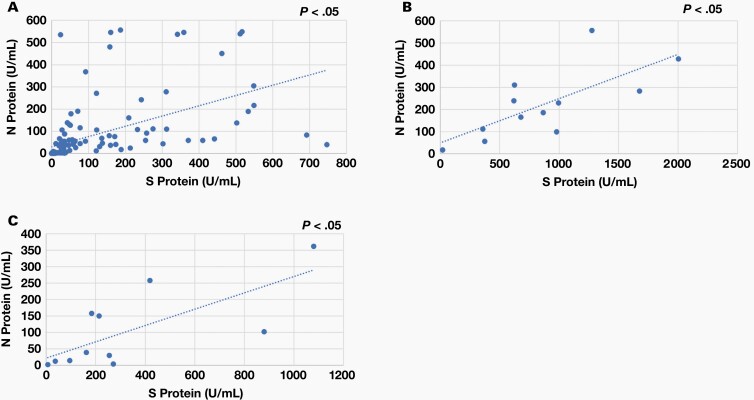
Correlation of Antibody Responses Against S1 RBD and N Proteins
To have a better understanding of the correlation of the immune response between the S1 RBD and N proteins, we analyzed 135 PCR-confirmed positive specimens for IgG and 12 PCR-confirmed positive specimens for IgM and IgA using a scatterplot. As shown in Figure 6, the anti-S1 RBD protein IgG, IgM, and IgA response correlations with the anti-N protein IgG, IgM, and IgA antibody response were r2 = .3045, .5264, and .5185, respectively (P <.05). Overall, the S1 RBD protein–specific IgM and IgA responses were well correlated with the N protein–specific IgM and IgA responses, whereas the S1 RBD protein–specific IgG response was weakly correlated with the N protein–specific IgG response.
Figure 6.
Correlation of antibody response between the S1 RBD and N proteins. (A) IgG: We analyzed 135 positive specimens. Each blue dot represents a specimen. The x axis represents the anti-S1 RBD protein IgG concentration (U/mL). The y axis represents the anti-N protein IgG concentration (U/mL). (B) IgM: We analyzed 12 positive specimens. Each blue dot represents a specimen. The x axis represents the anti-S1 RBD protein IgM concentration (U/mL). The y axis represents the anti-N protein IgM concentration (U/mL). (C) IgA: We analyzed 12 positive specimens. Each blue dot represents a specimen. The x axis represents the anti-S1 RBD protein IgA concentration (U/mL). The y axis represents the anti-N protein IgA concentration (U/mL). N protein, nucleocapsid protein; RBD, receptor binding domain; S1, spike subunit 1 protein.
Discussion
Here, we have established a comprehensive approach to measure antibodies against SARS-CoV-2. We have developed a quantitative immunoassay to detect IgG, IgM, and IgA antibodies to SARS-CoV-2 with high specificity and sensitivity and low cross-reactivity to several common viruses. This novel approach features a mixture of both the S1 RBD and N proteins and an albumin-coated plate that allows for the capture of a broad antibody response and at the same time a reduction of the nonspecific background. We were also able to confirm the suitability of this assay in several specimen types, including serum, plasma, and dried blood specimens, which broadens the scope of its potential use.
In this assay, after albumin subtraction, the specificity remained >98% and the sensitivity was significantly increased to 95.72%, 83.47%, and 82.60%, respectively, for IgG, IgM, and IgA antibodies to the combined S1 RBD and N proteins. Several immunoassays targeting IgG, IgM, or IgA antibodies have been developed with varying degrees of accuracy. Some of this variation results from the use of different antigens and protocols or from measuring different antibodies. Those different approaches may have different applications: eg, S1 RBD antibody detection can be used to indicate potential neutralization activity. In this study, we found differences in the response of some specimens to the S1 RBD protein vs the N protein and between the antibody isotypes. A previous study testing the performance of several chemiluminescence immunoassays showed that when using the S1 RBD protein, the responses of IgM and IgA were higher compared to the use of the N protein antigen.22 Liu et al23 showed varying sensitivities and specificities of IgG and IgM antibodies, with the S protein having higher sensitivity and an earlier antibody response compared to the N protein. Another study also confirmed differing antibody responses in the same specimen along with differing assay sensitivity and specificity.13 By combining different SARS-CoV-2 antigens, we were able to detect a broader antibody response. Based on these data, we expected that the combined detection of both the S1 RBD and the N proteins may improve the detection of the antibody response.
Cai et al10 showed that combining IgM and IgG isotypes improved the positivity of the assay. Several studies examined the accuracy of different lateral-flow immunoassays and ELISAs and found the highest accuracy in detection to be with combined IgG, IgM, and IgA analyses.11,12,14,15 A meta-analysis of the diagnostic performance of serology tests for COVID-19 showed that the combination of antigens—S, S1 RBD, and/or N proteins—also improved the accuracy of detection compared with any single antigen alone.24 Krishnamurthy et al25 also tested the IgG, IgM, and IgA antibody responses to the S1, S1 RBD, S2, and N antigens and found that the sensitivity varied among the tested antigens and antibodies and showed the highest sensitivity and specificity when combining all antigens and antibodies. Like the authors of these studies, we found that by combining the S1 RBD and N antigens and an albumin plate, we were able to improve the sensitivities of each antibody isotype in this assay.
The quantitative nature of the assay also allowed the determination of the time course of the antibody response while a patient has COVID-19 disease. Because most serological tests have tested specimens or groups of specimens collected at different time points, it is difficult to access the correct timeline for antibody responses against SARS-CoV-2 infection. We tested 251 PCR-confirmed positive serum specimens: 50 specimens were 1 to 7 days post-symptom onset, 30 specimens were 8 to 14 days post-symptom onset, 41 specimens were 15 to 25 days post-symptom onset, 103 specimens were 26 to 40 days post-symptom onset, 16 specimens were 41 to 60 days post-symptom onset, and 11 specimens were >60 days post-symptom onset; we found similar responses for IgG, IgM, and IgA antibodies in terms of peak response and particularly after 10 days post-symptom onset. Previous studies suggested that the sensitivity of antibody tests within the first week of disease is not ideal. However, sensitivity vastly increases in the second week.9,16,17 Similar to these and our results, Hu et al26 also observed a maximum antibody response on days 19 to 21, with the highest IgM response to be at days 16 to 18 and the highest IgG response to be at days 19 to 21 post-symptom onset. However, it is not yet clear how long the SARS-CoV-2–specific antibodies remain within a patient, or if the presence of those antibodies is an indicator of protection against COVID-19 disease. Because the likely path out of the current pandemic is through pharmaceutical means, it is essential to know the immune response for appropriate planning of vaccine strategies and protocols.
The antibody response at the site of infection or circulating systemically has not been well characterized. Thus, the ideal specimen type for use in serological analysis remains unknown. In this study, we confirmed the suitability of this assay in several specimen types, including serum, plasma, and dried blood specimens, which broadens the scope of its potential use. We found that the IgG, IgM, and IgA antibody responses were well correlated among tested specimen types. Krishnamurthy et al25 also compared clinically paired dried blood and serum specimens and found >90% correlation. Plasma has also been used successfully to measure antibody responses in patients with COVID-19.7,27,28
Our data indicate that the IgM and IgA responses to both the S1 RBD and N proteins have a significant and moderately strong correlation (r2 = .5264 and .5185; P <.05), whereas a weak correlation was seen in the IgG response to both the S1 RBD and N proteins (r2 = 0.3045, P <.05). This finding was consistent with previous studies.29,30 In contrast, Chen et al31 found a weak correlation of the IgM response between the S1 RBD and N proteins (r = .2928, corrected; P <.0001), whereas the IgG response between the S1 RBD and N proteins was significantly correlated (r = .5549, corrected; P <.001). Differences could be caused by specimen collection biases, the small number of specimens tested, or the different time frames of expression of each of the antibodies post-infection.
Conclusion
We have shown the quantitative detection of SARS-CoV-2 IgG, IgM, and IgA antibodies to the combined N and S1 RBD proteins with an albumin signal subtracted at a high sensitivity and specificity in serum, plasma, and dried blood specimens. We were also able to determine the temporal dynamics and magnitude of IgG, IgM, and IgA antibody responses in patients with COVID-19. Future studies that assess additional specimen types such as saliva or other oral specimens will be beneficial. Further analyses into the potential of these detected antibodies to be neutralizing can increase therapeutic and vaccine strategies in the future.
Acknowledgments
This research was funded by the RayBiotech Life innovative research fund, the Guangzhou Innovation Leadership Team (CXLJTD-201602), and The Science and Technology Innovation Project of Foshan Municipality, China (2020001000431).
Glossary
Abbreviations
- ELISA
enzyme-linked immunosorbent assay
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- iELISA
indirect enzyme-linked immunosorbent assay
- S protein
spike protein
- N protein
nucleocapsid protein
- RBD
receptor binding domain
- HEK
human embryonic kidney
- SDS-PAGE
sodium docetyl sulfate-polyacrylamide gel electrophoresis
- PBS
phosphate buffered saline
- PVDF
polyvinylidene fluoride
- TBST
tris-buffered saline with Tween-20
- HRP
horseradish peroxidase
- PBST
phosphate buffered saline with Tween-20
Disclosure
Some authors (Shuhong Luo, Chih Yun Cho, Kelly C. Whittaker, Xingqi Wang, Jie Feng, Meng Wang, Jianmin Fang, and Ruo-Pan Huang) are employees of and have a financial stake in RayBiotech.
REFERENCES
- 1.Worldometer. Reported cases and deaths by country or territory. https://www.worldometers.info/coronavirus/#countries. Accessed September 13, 2021.
- 2.Centers for Disease Control and Prevention. COVID-19: data and surveillance. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/index.html. Accessed September 13, 2021.
- 3.U.S, Food & Drug Administration. FDA combating COVID-19 with therapeutics. https://www.fda.gov/media/136832/download. Accessed September 13, 2021. [Google Scholar]
- 4. Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020;58(7):1070–1076. [DOI] [PubMed] [Google Scholar]
- 5. Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pascarella G, Strumia A, Piliego C, et al. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288(2):192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Padoan A, Bonfante F, Pagliari M, et al. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62:103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ejazi SA, Ghosh S, Ali N. Antibody detection assays for COVID-19 diagnosis: an early overview. Immunol Cell Biol. 2021;99(1):21–33. [DOI] [PubMed] [Google Scholar]
- 10. Cai XF, Chen J, Li Hu J, et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J Infect Dis. 2020;222(2):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espejo AP, Akgun Y, Al Mana AF, et al. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol. 2020;154(3):293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma H, Zeng W, He H, et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by chemiluminescence immunoanalysis. Preprint. Posted online April 30, 2020. medRxiv. 10.1101/2020.04.17.20064907. [DOI] [Google Scholar]
- 13. Qian C, Zhou M, Cheng F, et al. Development and multicenter performance evaluation of fully automated SARS-CoV-2 IgM and IgG immunoassays. Clin Chem Lab Med. 2020;58(9):1601–1607. [DOI] [PubMed] [Google Scholar]
- 14. Van Elslande J, Houben E, Depypere M, et al. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26(8):1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitman JD, Hiatt J, Mowery CT, et al. Test performance evaluation of SARS-CoV-2 serological assays. Preprint. Posted online May 17, 2020. medRxiv. doi: 10.1101/2020.04.25.20074856. [DOI] [Google Scholar]
- 16. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiao AT, Gao C, Zhang S. Profile of specific antibodies to SARS-CoV-2: the first report. J Infect. 2020;81(1):147–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. [DOI] [PubMed] [Google Scholar]
- 20. Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74(368):829–836. [Google Scholar]
- 21. R Core Team. 2017. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 22. Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virusdisease. 2020;31(1):13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H, Ai J, Loeffelholz MJ, Tang YW, Zhang W. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect. 2020;9(1):2200–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krishnamurthy HK, Jayaraman V, Krishna K, et al. Antibody profiling and prevalence in US patients during the SARS-CoV2 pandemic. PLoS One. 2020;15(11):e0242655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu Q, Cui X, Liu X, et al. The production and clinical implications of SARS-CoV-2 antibodies. Preprint. Posted online April 24, 2020. medRxiv. doi: 10.1101/2020.04.20.20065953. [Google Scholar]
- 27. Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130(10):5235–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ogata AF, Maley AM, Wu C, et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 patients with severe disease. Clin Chem. 2020;66(12):1562–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Algaissi A, Alfaleh MA, Hala S, et al. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci Rep. 2020;10(1):16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brochot E, Demey B, Touzé A, et al. Anti-spike, anti-nucleocapsid and neutralizing antibodies in SARS-CoV-2 inpatients and asymptomatic individuals. Front Microbiol. 2020;11:584251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Y, Tong X, Li Y, et al. A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog. 2020;16(9):e1008796. [DOI] [PMC free article] [PubMed] [Google Scholar]