Abstract
Eukaryotic translation initiation factor 5 (eIF5) interacts with the 40S initiation complex (40S–eIF3–AUG–Met-tRNAf–eIF2–GTP) to promote the hydrolysis of ribosome-bound GTP. eIF5 also forms a complex with eIF2 by interacting with the β subunit of eIF2. In this work, we have used a mutational approach to investigate the importance of eIF5-eIF2β interaction in eIF5 function. Binding analyses with recombinant rat eIF5 deletion mutants identified the C terminus of eIF5 as the eIF2β-binding region. Alanine substitution mutagenesis at sites within this region defined several conserved glutamic acid residues in a bipartite motif as critical for eIF5 function. The E346A,E347A and E384A,E385A double-point mutations each caused a severe defect in the binding of eIF5 to eIF2β but not to eIF3-Nip1p, while a eIF5 hexamutant (E345A,E346A,E347A,E384A,E385A,E386A) showed negligible binding to eIF2β. These mutants were also severely defective in eIF5-dependent GTP hydrolysis, in 80S initiation complex formation, and in the ability to stimulate translation of mRNAs in an eIF5-dependent yeast cell-free translation system. Furthermore, unlike wild-type rat eIF5, which can functionally substitute for yeast eIF5 in complementing in vivo a genetic disruption of the chromosomal copy of the TIF5 gene, the eIF5 double-point mutants allowed only slow growth of this ΔTIF5 yeast strain, while the eIF5 hexamutant was unable to support cell growth and viability of this strain. These findings suggest that eIF5-eIF2β interaction plays an essential role in eIF5 function in eukaryotic cells.
Eukaryotic initiation factor 5 (eIF5), a monomeric protein of 49 kDa in mammals (9, 10, 21) and 46 kDa in the yeast Saccharomyces cerevisiae (5, 6), plays an essential role in the initiation of protein synthesis. Following scanning of mRNA by the 40S preinitiation complex (40S–eIF3–Met-tRNAf–eIF2–GTP) and positioning of the initiator Met-tRNAf at the AUG codon of the mRNA to form the 40S initiation complex (eIF3–40S–AUG–Met-tRNAf–eIF2–GTP), the initiation factor eIF5 interacts with the 40S initiation complex to effect the hydrolysis of ribosome-bound GTP. Hydrolysis of GTP causes the release of eIF2-GDP, Pi, and eIF3 from the 40S initiation complex, which is essential for the subsequent joining of the 60S ribosomal subunit to the 40S complex to form a functional 80S initiation complex (80S–mRNA–Met-tRNAf) that is active in peptidyl transfer (for reviews, see references 16, 18, and 19). eIF5-dependent GTP hydrolysis has also been shown to play an important role in the selection of the AUG start codon (15).
An interesting feature of the derived amino acid sequence of mammalian (rat and human) and yeast eIF5 proteins (26) is the presence of sequence motifs at the N-terminal region of eIF5 that have weak homology to characteristic domains present in proteins belonging to the GTPase superfamily (3). However, unlike these proteins, eIF5 neither binds nor hydrolyzes free GTP or GTP bound to the (Met-tRNAf–eIF2–GTP) ternary complex in the absence of 40S ribosomal subunits (4, 7). eIF5 promotes GTP hydrolysis only when the nucleotide is bound to eIF2 in the 40S initiation complex (Met-tRNAf–eIF2–GTP–eIF3–40S–AUG) (4, 7). These results suggest that eIF5 must interact with one or more components of the 40S initiation complex to cause hydrolysis of GTP. In agreement with this hypothesis, we observed that mammalian eIF5 forms a complex with mammalian eIF2 (7), a component of the 40S initiation complex, and that eIF5-eIF2 complex formation occurs through the β subunit of eIF2 (11). Complex formation between eIF5 and Nip1p subunit of eIF3 has also been reported (1, 2).
In the case of eIF5 and eIF2β interaction, deletion studies have shown that the N-terminal region of eIF2β binds eIF5 and that the conserved stretch of lysine residues in this region plays an important role in this interaction (11). Similar interaction between yeast eIF5 and yeast eIF2β was also reported from this laboratory (11) and later by others (1), indicating that the interaction domains of eIF5 and the β subunit of eIF2 are conserved through evolution. In later studies, Asano et al. (1) observed that a bipartite motif at the C-terminal region of yeast eIF5 containing conserved aromatic and acidic residues is required for binding to both eIF2β and the Nip1p subunit of eIF3. However, the important question remained as to whether the interaction of eIF5 with eIF2β is required for eIF5-dependent GTP hydrolysis and is thus a key molecular interaction in the translation initiation pathway.
In the work presented here, we demonstrate that mammalian eIF5 interacts with mammalian eIF2β through a conserved C-terminal region. We have carried out a systematic mutational analysis of conserved residues in the C-terminal eIF2β-binding region of rat eIF5 to generate mutants which are defective in binding to eIF2β but are active in binding to Nip1p. We show that these mutants are also defective in eIF5-dependent GTP hydrolysis and consequently in 80S initiation complex formation as well as in in vitro protein synthesis. Furthermore, whereas mammalian eIF5 can functionally substitute for the homologous yeast protein in vivo in yeast cells (17), the mutant eIF5 proteins that are defective in binding to eIF2β are unable to complement a genetic disruption in the chromosomal copy of the TIF5 gene in vivo. Taken together, our results suggest that the interaction between eIF5 and the β subunit of eIF2 is required for eIF5-dependent hydrolysis of GTP during translation initiation and consequently is essential for overall protein synthesis.
MATERIALS AND METHODS
tRNA, ribosomes, purified proteins, and antibodies.
The preparation of 35S-labeled rabbit liver initiator Met-tRNAf (30,000 to 50,000 cpm/pmol) and 40S and 60S ribosomal subunits from Artemia salina eggs were described previously (4, 9). Purified eIF2 from rabbit reticulocyte lysates and recombinant rat eIF5 were isolated as described elsewhere (4, 7). Immunoglobulin G antibody specific for recombinant rat eIF5 was isolated from rabbit antisera raised against the purified protein as described elsewhere (12). Immunoblot analysis was carried out as described previously (8, 12). Mouse anti-glutathione S-transferase (GST) antibodies were a kind gift from Charles Weaver of our institution. Rabbit anti-yeast Nip1p antibodies were a kind gift of David Goldfarb, University of Rochester, Rochester, N.Y. The mixture of protease inhibitors added to buffer solutions used during purification of recombinant proteins from bacterial cell extracts consisted of leupeptin (0.5 μg/ml), pepstatin A (0.7 μg/ml), aprotinin (2 μg/ml), and freshly prepared phenylmethylsulfonyl fluoride (PMSF; 1 mM).
Construction of plasmids and yeast strains.
For expression of eIF5 as a GST fusion protein, the open reading frame (ORF) of rat eIF5 cDNA (10) was synthesized by one-stage PCR using pET-5a-eIF5 (7) as the template and appropriate oligonucleotide primers corresponding to the N-terminal and C-terminal ends of the eIF5 ORF. Both primers had BamHI overhangs. The N-terminal primer introduced an in-frame methionine codon following the BamHI site, while the C-terminal primer had a translation stop site preceding the BamHI overhang. It should be noted here that a similar strategy of introducing start and stop codons was used in the cloning of all PCR fragments described below. The PCR product was digested with BamHI and cloned into the same site of the vector pGEX-KG (Pharmacia Biotech Inc.) in order to express eIF5 as a GST fusion protein. Deletion mutants of eIF5 were generated by one-stage PCR amplification of eIF5 ORF sequences using pGEX-KG-eIF5 as the template and appropriate oligonucleotide primers containing BamHI/EcoRI overhangs. A BamHI/EcoRI restriction fragment of each PCR-amplified deletion mutant was inserted at the same restriction sites of the vector pGEX-KG. The resulting constructs expressed deleted eIF5 mutants as GST fusion proteins. For expression of histidine-tagged yeast Nip1p protein (13), a pYES2/GS plasmid containing the Nip1p ORF was bought from Invitrogen, and the ORF of the Nip1p cDNA was PCR amplified using appropriate oligonucleotide primers containing NheI-BamHI overhangs. The PCR product was digested with restriction enzymes NheI and BamHI and cloned into the same sites in the vector pRSET-C. The construction of yeast centromeric plasmids pRS316-TIF5, pTM100-EIF5, and pUB-TIF5R and the haploid yeast strains TMY101 and TMY201R has been described by Maiti and Maitra (17). The construction of plasmid pGEX-2T-eIF2β and the preparation of GST-eIF2β fusion protein have been described elsewhere (11).
The yeast strains used in this study (17) are W303α (MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100), TMY101 (MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif5::TRP1[pRS316-TIF5]), and TMY201R (MATα leu2-3,112 his 3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif5::TRP1[pUB-TIF5R]). The media for yeast cell growth were prepared as reported previously (17).
Site-directed mutagenesis of rat eIF5 coding sequence and expression of mutant rat eIF5 proteins in yeast.
Point mutations within the coding sequence of eIF5 present in the yeast centromeric plasmid pTM100-EIF5 or the bacterial expression plasmid pGEX-KG-eIF5 were constructed by one-stage PCR using a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. We designed appropriate 26- to 30-mer mutagenic oligonucleotide primers to create the desired mutations by maintaining the reading frame of eIF5. All mutated ORFs were sequenced to confirm the desired mutations and ensure error-free DNA synthesis in other regions of the ORF. To detect expression of wild-type or mutant rat eIF5 proteins in yeast cells, the haploid yeast strain TMY101, containing the chromosomal copy of the TIF5 gene disrupted with the TRP1 marker gene and harboring the URA3 plasmid pRS316-TIF5, was transformed with the pTM100-EIF5 series of plasmids containing either a wild-type or mutant rat eIF5 ORF, respectively, under the transcriptional control of GAL1 promoter. Trp+ Ura+ Leu+ transformants were selected on SGal plates lacking tryptophan, leucine, and uracil (SGal-Trp-Leu-Ura) plates. In each case, one selected transformant was then grown in 3 ml of SGal-Trp-Leu-Ura medium to an A600 of about 0.8. The cells were then harvested, and lysates prepared from these cells were subjected to immunoblot analysis by an adaptation of the procedure of Sachs and Deardorff (23) using rabbit anti-mammalian eIF5 (12) antibodies as probes.
Expression and purification of recombinant wild-type and mutant rat eIF5 proteins.
Escherichia coli XL1-Blue cells transformed with recombinant pGEX-KG plasmids containing either the wild-type or mutant eIF5 coding sequences were grown in 2 liters of 2YT medium to an A600 of 0.9 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested 2.5 h postinduction, washed with ice-cold 0.9% NaCl, quick-frozen in a dry ice-ethanol bath, and stored at −70°C until use. The cell yield in each case was about 10 g (wet weight).
For isolation of recombinant eIF5, the frozen cells were suspended in 30 ml of sonication buffer containing 20 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 100 mM KCl, 1 mM EDTA, 10 mM 2-mercaptoethanol, and 0.5 mM PMSF, treated with lysozyme (final concentration of 100 μg/ml), incubated for 20 min at 0 to 4°C, and then disrupted by sonication. After the cell debris was removed by centrifugation at 15,000 × g for 20 min, the supernatant was treated with a mixture of protease inhibitors and then incubated with 30 μg of pancreatic DNase I for 30 min at 0°C. After centrifugation at 15,000 × g for 30 min, the clear supernatant was mixed with 2 ml of a suspension of glutathione (GSH)-Sepharose beads (Pharmacia) previously equilibrated in 20 mM Tris-HCl (pH 7.5)–0.1 mM EDTA–1 mM dithiothreitol–10% glycerol (buffer B) containing 100 mM KCl and 0.5 mM PMSF. The mixture was gently incubated in a rotator at 4°C for 1 h, and the liquid containing unabsorbed protein was then removed from the beads by pouring the suspension into a 5-ml column. The column containing GST-eIF5-bound beads was then washed sequentially with (i) buffer B plus 280 mM KCl (until the A280 was below 0.1) and (ii) 10 ml of 10 mM potassium phosphate (pH 7.0)–150 mM NaCl (phosphate-buffered saline [PBS]). The beads were then suspended in 3 ml of PBS and incubated with 200 U of thrombin (Pharmacia) overnight at 0°C, resulting in the release of eIF5 from GST-eIF5 fusion protein bound to the beads into the supernatant. The released eIF5 was isolated free of beads by pouring the entire mixture onto a small column and collecting the flowthrough liquid. The beads in the column were washed with 2 ml of PBS, and the resulting flowthrough fraction was mixed with the initial flowthrough fraction. The pooled fractions were dialyzed against 600 ml of buffer B containing 80 mM KCl for about 2 h and then applied to a 1-ml-bed-volume FPLC-MonoQ column (Pharmacia BioTech) equilibrated in buffer B–100 mM KCl. After the column was washed with this buffer, bound proteins were eluted with a linear gradient (0.5 ml/min) of 15 ml (total volume) from buffer B–100 mM KCl to buffer B–500 mM KCl. Fractions of 0.5 ml were collected and assayed for eIF5 by Western blotting using polyclonal anti-eIF5 antibodies. Fractions containing eIF5 (eluting at about 360 mM KCl) were pooled and dialyzed against 600 ml of 20 mM Tris-HCl (pH 7.5)–100 mM KCl–0.1 mM EDTA–1 mM dithiothreitol–60% glycerol for about 6 h and then stored at −20°C. Under these conditions, eIF5 activity was stable for at least 6 months.
Preparation of His6-tagged yeast Nip1p.
The pRSET-C-yNip1 plasmid containing the yeast Nip1p coding sequence (13) fused to His6-tag at its N terminus was transformed into E. coli BL21(DE3) cells. Expression of His6-Nip1p was induced by the addition of 0.5 mM IPTG to an exponentially growing 250-ml bacterial culture. Cells were harvested 2 h postinduction, suspended in 3 ml of a mixture containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 5 mM 2-mercaptoethanol, 5 mM potassium imidazole, and a mixture of protease inhibitors (buffer C), and then disrupted by sonication. Following addition of Triton X-100 to 1% (final concentration), the cell lysate was clarified by centrifugation at 15,000 × g for 20 min. The supernatant was incubated with Ni-nitrilotriacetic acid (NTA) agarose beads, preequilibrated in buffer C containing 25 mM potassium imidazole (pH 6.0) for 1 h at 4°C. The beads containing His6-Nip1p fusion protein were washed three times with 1 ml of buffer C–25 mM potassium imidazole. The amount of protein present in the washed beads was quantitated by the Bio-Rad method. The beads were stored in small aliquots at −70°C until use.
GST-eIF2β fusion protein binding assay.
A typical binding reaction mixture (200 μl) contained 20 mM potassium phosphate (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, 0.5 mM PMSF, 10 μl of a 30% suspension of GSH beads containing bound GST-eIF2β (12 μg of total protein), and about 9 μg of either purified wild-type or mutant eIF5 protein. Reaction mixtures were gently mixed in a rotator at 4°C for 1 h and then centrifuged. The supernatant containing unbound proteins was discarded, and the beads were then washed two times with 1 ml of reaction buffer containing 1% Triton X-100 for 20 min. The washed beads were resuspended in 20 μl of 1% sodium dodecyl sulfate (SDS)–gel loading buffer and heated in a boiling water bath for 3 min, and the released proteins were resolved on 0.1% SDS–15% polyacrylamide gels. The separated polypeptides were then transferred onto a polyvinylidene difluoride membrane, which was analyzed by Western blotting using anti-eIF5 polyclonal antibodies.
Assay of binding of mammalian eIF5 to Ni-NTA agarose beads containing bound yeast Nip1p.
A typical binding reaction mixture contained 200 μl of buffer B, 100 mM KCl, 1% Triton-X, 0.5 mM PMSF, and 10 μl of a suspension of Ni-NTA agarose beads containing 5 μg of yeast Nip1p and 5 μg of recombinant rat eIF5 (wild type or mutant). Each reaction mixture was gently mixed in a rotator at 4°C for 1 h and then centrifuged. The supernatant was discarded; the beads were washed two times with buffer B containing 100 mM KCl and 1% Triton X-100 (20 min each time), suspended in 20 μl of 1% SDS–gel loading buffer, and incubated for 3 min in a boiling water bath; the released polypeptides were separated on 0.1% SDS–15% polyacrylamide gels followed by Western blotting using anti-eIF5 antibodies. In control reactions, Ni-NTA agarose beads containing bound yeast Nip1p (5 μg) were incubated with 5 μg of recombinant human eIF1A (8). Following incubation, these beads were treated similarly and the washed beads containing bound proteins were analyzed in Western blots using anti-eIF1A antibodies (8).
Cell-free translation.
The haploid yeast strain TMY201R, which has the disrupted chromosomal copy of the TIF5 gene, but harboring plasmid pUB-TIF5R, which carries out conditional expression of a functional but rapidly degradable form of yeast eIF5 as a ubiquitin-conjugated eIF5 fusion protein, was used as the source of cell-free protein-synthesizing extract (17). An exponentially growing culture of this strain in YPGal medium supplemented with adenine sulfate (0.4 mg/ml) was harvested, and cells were suspended in 1 liter of YPD medium containing adenine sulfate (0.4 mg/ml) such that the initial A600 was about 0.03 and grown at 30°C for about 22 h (about two generations in YPD medium), at which time cell growth was nearly completely arrested. Cells were then harvested, cell translation extracts were prepared, and mRNA-dependent cell-free translation was performed as described elsewhere (17).
Other methods.
The 40S initiation complex containing bound [γ-32P]GTP was prepared and isolated free of unreacted reaction components by sucrose density centrifugation as described elsewhere (4, 7). eIF5-mediated [γ-32P]GTP hydrolysis reactions and 80S initiation complex formation were measured as described previously (7, 21). 35S-labeled eIF2β was expressed in vitro from plasmid pET-5a-eIF2β (11) in S-30 bacterial TNT extracts (Promega) using the manufacturer's protocol. Yeast transformations were performed as described by Rose et al. (22). Methods for plasmid and genomic DNA preparations, restriction enzyme digestion, DNA ligation, cloning, and bacterial transformations were according to standard protocols (24).
RESULTS
Interaction of rat eIF5 deletion mutants with the β subunit of eIF2.
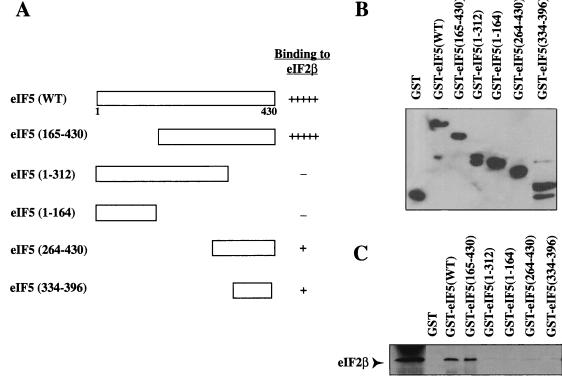
Mammalian eIF5 forms a specific complex with eIF2 (7), and this complex formation occurs by interaction of eIF5 with the β subunit of eIF2 (11). To map the region of rat eIF5 involved in binding eIF2β, combinations of PCR-amplified C-terminal and N-terminal deletions of rat eIF5 coding sequences were individually cloned into the pGEX-KG expression vector (Fig. 1A) for expression of both the wild-type and deletion fragments of eIF5 as GST fusion proteins in E. coli XL1-Blue cells. Lysates prepared from IPTG-induced bacteria were then immobilized on GSH-Sepharose beads, and the presence of GST-eIF5 fusion proteins on these beads was detected by Western blot analysis using anti-GST monoclonal antibody (Fig. 1B). The GSH beads containing bound GST-eIF5 proteins were then tested for the ability to bind 35S-labeled eIF2β expressed in vitro in bacterial extracts (Materials and Methods) as shown in Fig. 1C. We observed that when amino acids 1 to 164 were deleted from eIF5, GST-eIF5(165-430) still bound eIF2β nearly as efficiently as wild-type eIF5, whereas GST-eIF5(1-164) did not bind eIF2β. When 118 amino acids were deleted from the C terminus of eIF5, the resulting GST-eIF5(1-312) also did not bind eIF2β. GST immobilized on GSH-Sepharose beads did not bind eIF2β, as expected (Fig. 1C). These results suggested that a region at the C terminus of GST-eIF5(165-430) contains the eIF2β binding site. Deletion of additional amino acids from both N and C termini of eIF5(165-430) caused the resultant mutants GST-eIF5(264-430) and GST-eIF5(334-396) to bind eIF2β with greatly reduced efficiency (Fig. 1C). The very weak binding exhibited by both GST-eIF5(264-430) and GST-eIF5(334-396) compared to GST-eIF5(165-430) suggests that although a 63-amino-acid stretch between amino acids 334 and 396 of eIF5 can bind eIF2β, albeit with low efficiency, the region encompassing amino acids 165 to 263 may be necessary for optimal binding of eIF2β. It should be noted that the fragment eIF5(165-263), by itself, did not bind eIF2β (data not shown). The reduced efficiency of binding exhibited by GST-eIF5(264-430) and GST-eIF5(334-396) compared to GST-eIF5(165-430) may also be due to improper folding of the shorter fragments which partially buries the eIF2β binding site in these deletion mutants, making this site less accessible to eIF2β. Using the S. cerevisiae system, Asano et al. (1) have also shown that yeast eIF2β expressed in TNT lysates binds to the C-terminal region of yeast eIF5.
FIG. 1.
Deletion analysis of eIF5 to determine the eIF2β-binding domain. (A) Schematic representation of deletion mutants of eIF5 expressed as GST fusion proteins in E. coli XL1-Blue cells. The efficiency with which each deletion mutant of eIF5 binds 35S-labeled eIF2β, determined by autoradiography, is shown by + and −. WT, wild type. (B) Expression of GST fusion proteins of eIF5 deletion mutants immobilized on GSH beads was measured by Western blot analysis of 3 μg of protein bound to these beads using anti-GST antibodies. It is not immediately apparent why in case of GST-eIF5(1-312) a polypeptide of the predicted size was not observed. (C) The same eIF5 deletion mutants immobilized on GSH beads (6 μg of bound protein) were incubated at 4°C with 35S-labeled eIF2β synthesized in vitro in bacterial S-30 extracts (Promega) at 4°C for 1 h. The proteins bound to the washed beads were subjected to SDS–15% polyacrylamide gel electrophoresis followed by autoradiography of the dried gel. In the first lane, 35S-labeled eIF2β alone was electrophoresed.
Strategy for mutational analysis of conserved residues in the eIF2β-binding region of eIF5.
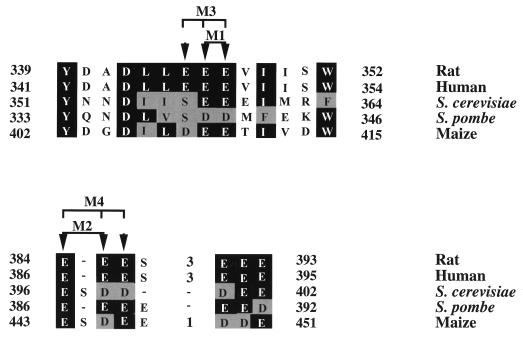
The aim of this work was to create mutations in mammalian eIF5 that would affect the binding of eIF5 with eIF2β and to test whether eIF5 bearing these mutations also affect eIF5-dependent hydrolysis of GTP bound to the 40S initiation complex and consequently overall protein synthesis in vitro. Our mutagenesis strategy was guided by our previous observation (11) that the mammalian eIF5-binding region of eIF2β lies at the N-terminal region of eIF2β that contains stretches of conserved lysine residues which have been shown to be necessary for eIF5-eIF2β interaction (1, 11). Such stretches of conserved lysine residues are not present in the Nip1p subunit of eIF3 (1). Based on these observations, we reasoned that the eIF2β-binding region of eIF5 may consist of conserved acidic amino acid residues. Comparison of the amino acid sequence of the C-terminal region of rat eIF5 (which contains the mammalian eIF2β-binding region of rat eIF5) with those from other species revealed the presence of an acidic amino acid-rich bipartite motif consisting of a number of conserved glutamic acid and aspartic acid residues, most notably in the region lying between amino acids 339 and 352 and between amino acids 384 and 393 in rat eIF5 (Fig. 2). We introduced alanine substitution mutations at the positions of these conserved acidic amino acid residues. Since mammalian eIF5 has been shown (17) to functionally substitute for yeast eIF5 in sustaining yeast cell growth and viability, we initially examined whether these mutations affect the ability of mammalian eIF5 to substitute for yeast eIF5 in a ΔTIF5 haploid yeast strain. The rationale behind this strategy was that if eIF5-eIF2β interaction were required for the essential eIF5 function in yeast cells, eIF5 mutants defective in its interaction with eIF2β would be lethal in yeast cells. We therefore introduced single- and double-point mutations in both halves of the bipartite motif in the eIF2β-binding region of rat eIF5 whereby conserved acidic amino acid residues were mutated to alanine. The ability of these mutant eIF5 proteins to support growth of ΔTIF5 yeast cells was then tested.
FIG. 2.
Conservation of amino acid residues at the C-terminal bipartite motif of rat eIF5 from different species and locations of alanine substitution mutations within this conserved region. The amino acid sequences of the C-terminal bipartite motif of rat eIF5 (amino acids 339 to 352 and amino acids 384 to 393) were aligned with the corresponding regions in human, S. cerevisiae, Schizosaccharomyces pombe, and maize eIF5 for maximum homology using the program DNASTAR. The sequences of rat, human, and S. cerevisiae eIF5 are from reference 26; the sequences of S. pombe and maize eIF5 were obtained from SWISSPROT (accession no. Q09689, and P55876, respectively). The highly conserved amino acid residues between eIF5 of all species are highlighted with dark shading, and the moderately conserved residues are highlighted with light shading. Gaps are represented by broken lines. Residues in rat eIF5 that were targeted for mutagenesis in this study to generate eIF5 mutants M1 to M4 are indicated by arrowheads.
Effect of alanine substitution mutations in rat eIF5 on its ability to sustain yeast cell growth and viability.
To test the function of the eIF5-Ala mutants in yeast cells, the eIF5-Ala mutants were generated using a PCR-based site-directed mutagenesis protocol with the yeast expression plasmid pTM100-EIF5 (17) as the template as described in Materials and Methods. In the LEU2-based CEN plasmid pTM100-EIF5, the wild-type rat eIF5 ORF (gene designation EIF5) is under the transcriptional control of galactose-inducible GAL1 promoter. We used the plasmid shuffling technique to determine if the mutant rat eIF5 proteins could functionally substitute for the corresponding yeast protein in vivo. For this purpose, we used the haploid yeast strain TMY101, which carries an inactive TIF5 allele disrupted with the TRP1 marker gene and is kept viable by maintenance of a centromeric URA3 plasmid, pRS316-TIF5, that contains the yeast wild-type TIF5 gene under the transcriptional control of its natural promoter. The host strain TMY101 was transformed individually with both the wild-type and mutant recombinant eIF5 expression plasmids and the parental vector, pTM100. Trp+ Ura+ Leu+ transformants were selected on SGal plates and replica plated onto another similar plate which also contained uracil and 5-fluoroorotic acid (5-FOA) to select against retention of the URA3-based plasmid pRS316-TIF5, expressing wild-type yeast eIF5.
Figure 3A shows that cells transformed with plasmid pTM100-EIF5 (which expresses wild-type rat eIF5 from the GAL1 promoter) grew on 5-FOA plates, in agreement with the results reported previously from this laboratory (17). Under the same conditions, cells transformed with the vector plasmid pRS315 failed to grow as expected. Cells transformed with pTM100-EIF5(mutant) each carrying a single point mutation at Glu-346, Glu-347, Glu-384, or Glu-385 residue in eIF5 also grew on 5-FOA plates as efficiently as cells that were transformed with pTM100-EIF5 (wild type) (Fig. 3A). Alanine substitution mutation at Asp-342 of eIF5 also did not affect cell growth on 5-FOA plates (data not shown). These results were not surprising if we reason that the eIF5-binding region of eIF2β consists of a stretch of lysine residues that presumably make direct physical interaction with a stretch(es) of conserved acidic amino acid residues at the C-terminal end of eIF5. Presumably, mutation of a single acidic amino acid residue, e.g., a glutamic acid residue in the bipartite motif, still allowed eIF2β to make effective physical interaction with other glutamic acid residues present in this motif. Based on this reasoning, we introduced double-point mutations in both halves of the bipartite motif whereby two consecutive glutamic acid residues, Glu-346,Glu-347 and Glu-384,Glu-385, were mutated to alanine. The resultant mutant eIF5 proteins were designated M1 and M2, respectively. TMY101 cells were transformed with pTM100-EIF5(M1) and pTM100-EIF5(M2) separately, and the Trp+ Ura+ Leu+ transformants selected on SGal plates (Fig. 3B) were replica plated onto similar plates containing 5-FOA. We observed that cells transformed with pTM100-EIF5(M1) and pTM100-EIF5(M2) produced microcolonies (Fig. 3B), indicating a very slow growth phenotype. These two strains were recovered from 5-FOA plates, and their growth rates were determined in SGal-Trp-Leu liquid medium. We observed that while yeast cells expressing wild-type mammalian eIF5 grew with a doubling time of 5 h, yeast cells expressing mutants M1 and M2 grew with doubling times of 13.4 and 7 h, respectively. However, when all six glutamic acid residues (Glu-345, Glu-346, Glu-347, Glu-384, Glu-385, and Glu-386) in both halves of the bipartite motif were mutated to alanine, the resultant mutant eIF5, designated M5, failed to sustain yeast cell growth on 5-FOA plates (Fig. 3B). These results indicate that eIF5 mutants M1 (E346A,E347A) and M2 (E384A,E385A) were partially defective, while mutant M5 (E345A,E346A,E347A,E384A,E385A,E386A) was severely defective in functionally substituting for yeast eIF5 in maintaining yeast cell growth and viability. This functional defect of the rat eIF5 proteins M1, M2, and M5 was not due to lack of expression of mutant eIF5 in yeast cells. When cell extracts prepared from Trp+ Ura+ Leu+ transformants harboring both pRS316-TIF5 and pTM100-EIF5 (wild-type as well as mutant) plasmids were analyzed by Western blotting using rabbit anti-rat eIF5 antibodies, mutant eIF5 proteins M1, M2, and M5 were found to be expressed at levels comparable to wild-type rat eIF5 (compare lanes d to f with lane b in Fig. 3C). Extracts prepared from TMY101 yeast cells expressing only yeast eIF5 from pRS316-TIF5 and not harboring any recombinant vector showed no polypeptide band immunoreactive with anti-rat eIF5 antibodies, as expected (Fig. 3C, lane c).
FIG. 3.
Effects of mutations in rat eIF5 on growth of haploid yeast transformants expressing rat eIF5. (A) Haploid yeast strain TMY101 (see Materials and Methods) was transformed separately with different recombinant eIF5 expression plasmids, pTM100-EIF5 expressing either wild-type eIF5 (17) or alanine-mutant eIF5 protein, each containing a single point mutation at E346A, E347A, E384A, or E385A from a GAL1 promoter, and also the vector plasmid pRS315 as indicated. Transformants were initially selected on SGal-Trp-Leu-Ura plates and then replica plated onto both SGal-Trp-Leu-Ura (left) and SGal containing 5-FOA and uracil (SGal-Trp-Leu+Ura + 5-FOA; right). Cells were allowed to grow on these plates for 5 days. (B) Haploid yeast strain TMY101 was transformed separately with mutant eIF5-expressing plasmids pTM100-EIF5(M1), pTM100-EIF5(M2), and pTM100-EIF5(M5) as indicated. Transformants selected on SGal-Trp-Leu-Ura plates (left) were replica plated on SGal-Trp-Leu+Ura + 5-FOA (right). Cells were allowed to grow on the 5-FOA plates for 5 days. (C) Immunoblot analysis of eIF5 in lysates of yeast cells expressing both yeast eIF5 and wild-type (WT) or mutant mammalian eIF5 proteins M1, M2, and M5 from the recombinant plasmids. Yeast cells harboring both the URA3 plasmid pRS316-TIF5 (which expresses yeast eIF5 from its own natural promoter) and the different recombinant LEU expression plasmids expressing either the wild-type or mutant mammalian eIF5 were grown to mid-logarithmic phase in synthetic medium containing 2% galactose as the sole source of carbon. Cell lysates were prepared as described in Materials and Methods and analyzed by Western blotting using rabbit polyclonal anti-rat eIF5 antibodies. Lane a, purified recombinant rat eIF5 as a marker; lanes b to f, extracts from tif5::TRP1 yeast cells harboring yeast eIF5 expression plasmid pRS316-TIF5 and LEU2-based rat eIF5 expression plasmids as follows: lane b, pTM100-EIF5; lane c, pRS315 (vector control); lane d, pTM100-EIF5(M1); lane e, pTM100-EIF5(M2); lane f, pTM100-EIF5(M5).
Analysis of eIF5 mutants for the ability to bind the β subunit of eIF2.
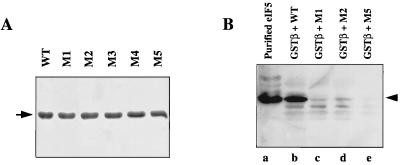
The eIF5 mutants M1, M2, and M5, which were defective in eIF5 function in yeast cells, were tested for the ability to participate in various in vitro partial reactions attributed to eIF5. We first examined the ability of the eIF5 mutants to bind the β subunit of eIF2. For this purpose, both wild-type and mutant eIF5 fusion proteins expressed in bacteria were purified to apparent electrophoretic homogeneity (Fig. 4A) as described in Materials and Methods. (eIF5 mutants M3 and M4 correspond to alanine substitution mutations at Glu-345,Glu-346,Glu-347 and Glu-384,Glu-385,Glu-387, respectively.) The ability of each purified protein to bind to GST-eIF2β immobilized on GSH-Sepharose beads was examined. Figure 4B shows that wild-type eIF5 bound GST-eIF2β efficiently (lane b). In contrast, mutant eIF5 proteins M1 and M2 showed very poor binding (<5%) to eIF2β (lanes c and d), while mutant eIF5 M5 showed virtually no binding to eIF2β (lane e). As expected, wild-type eIF5 did not bind to GST alone immobilized on GSH-Sepharose beads (data not shown). These results show that the mutant eIF5 proteins were severely defective in their interaction with eIF2β.
FIG. 4.
Interaction between eIF5 mutants and the β subunit of eIF2. (A) Recombinant wild-type (WT) and mutant eIF5 proteins M1 to M5 were purified from IPTG-induced XL1-Blue cell lysates as described in Materials and Methods. Purified recombinant eIF5 proteins (3 μg of each) were subjected to SDS-polyacrylamide gel electrophoresis (15% gel) and visualized by Coomassie blue staining. The arrow indicates the position of purified eIF5. (B) Purified recombinant wild-type (lane b) and mutant eIF5 proteins M1 (lane c), M2 (lane d), and M5 (lane e) (9 μg of each) were separately incubated with 12 μg of GST-eIF2β fusion protein immobilized on GSH beads. Following incubation at 4°C with gentle shaking, reaction mixtures were centrifuged, and the beads were washed, suspended in 1× Laemmli buffer, and subjected to Western blot analysis using polyclonal anti-eIF5 antibodies. In lane a, purified rat eIF5 was electrophoresed as a marker and probed with anti-eIF5 antibodies.
eIF5 mutants defective in binding eIF2β are also defective in eIF5-dependent GTP hydrolysis and 80S initiation complex formation.
The purified eIF5 mutant proteins were also tested for their ability to promote hydrolysis of GTP bound to the 40S initiation complex (Fig. 5A). In agreement with the results previously published from this laboratory (7), wild-type recombinant rat eIF5 mediated rapid hydrolysis of GTP bound to the 40S initiation complex. In contrast, under similar experimental conditions, mutant eIF5 proteins M1 and M2 showed four- to fivefold reductions in activity in promoting GTP hydrolysis (Fig. 5A). When three glutamic acid residues—Glu-345,Glu-346,Glu-347 and Glu-384,Glu-385,Glu-386—in each half of the bipartite motif were mutated to alanine, the resulting eIF5 mutant proteins M3 (E345A,E346A,E347A) and M4 (E384A,E385A,E386A) were even less active in the GTP hydrolysis reaction (Fig. 5A). When eIF5 mutant M5 (E345A,E346A,E347A,E384A,E385A,E386A), in which six glutamic acid residues in the bipartite motif were mutated to alanine, was tested in the GTPase reaction, the release of 32Pi from the 40S initiation complex was even lower (Fig. 5A).
FIG. 5.
Analysis of eIF5 mutants for the ability to mediate hydrolysis of GTP bound to the 40S initiation complex and to form the 80S initiation complex. (A) eIF5-mediated GTP hydrolysis. Reaction mixtures (50 μl) contained 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM KCl, and 1 mM dithiothreitol (buffer R), isolated 40S initiation complex (Met-tRNAf–eIF2–[γ-32P]GTP–40S–AUG) containing 2.5 pmol of bound [γ-32P]GTP (38,500 cpm/pmol) isolated as described previously (4), and 20 ng of purified recombinant wild-type (WT) or mutant eIF5 proteins M1 (E346A,E347A), M2 (E384A,E385A), M3 (E345A,E346A,E347A), M4 (E384A,E385A,E386A), and M5 (E345A,E346A,E347A,E384A,E385A,E386A). Following incubation at 25°C, aliquots (8 μl) were removed at each indicated time point and the amount of 32Pi released by the hydrolysis of [γ-32P]GTP was measured by the ammonium phosphomolybdate method as described elsewhere (4). A reaction lacking eIF5 was also included, and the amount of 32Pi released in this control reaction mixture (<0.1 pmol) was subtracted from the results shown. The results shown represent the total amount of 32Pi formed per 50-μl reaction mixture. (B) 80S initiation complex formation. Reaction mixtures containing buffer R, 3 pmol of preformed [35S]Met-tRNAf–eIF2–GTP ternary complex (25,000 cpm/pmol), 0.5 A260 unit of 40S ribosomal subunits, and 0.1 A260 unit of the AUG codon were incubated for 5 min at 37°C to form the 40S initiation complex ([35S]Met-tRNAf–eIF2–GTP–40S–AUG). The chilled reaction mixtures were supplemented with 0.8 A260 unit of 60S ribosomal subunits and purified wild-type or mutant eIF5 at the indicated amount. Following incubation at 37°C for 5 min, the chilled reaction mixtures were sedimented through a 5-ml linear 7.5 to 30% (wt/vol) sucrose gradient in buffer R for 105 min at 48,000 rpm at 4°C in a Beckman 50.1 rotor. Fractions (0.25 ml) collected from the bottom of each tube were counted for 35S radioactivity to quantitate formation of the 80S initiation complex. A control reaction mixture lacking eIF5 formed <0.1 pmol of 80S initiation complex. This value was subtracted from the results shown.
Since hydrolysis of GTP is a stringent prerequisite for the joining of 60S ribosomal subunits to the 40S initiation complex (16, 18, 19), we also tested these mutant eIF5 proteins for the ability to mediate the conversion of the 40S initiation complex to the 80S initiation complex. Figure 5B shows that purified wild-type eIF5 mediated nearly quantitative conversion of the 40S initiation complex (Met-tRNAf–eIF2–GTP–40S–AUG) to the 80S initiation complex (80S–AUG–Met-tRNAf). In contrast, under the same experimental conditions, mutant proteins M1 and M2 were both less effective in forming the 80S initiation complex. Mutant eIF5 proteins M3 and M4 were even less active (data not shown). In eIF5 mutant M5, in which six glutamic acid residues in the bipartite motif of the eIF2β-binding region of eIF5 were mutated to alanine, the amount of 80S initiation complex formed was reduced to <10%. These results, which are in agreement with those obtained in the GTP hydrolysis reaction, indicate that the interaction of eIF5 with eIF2β is required for eIF5 to function in the GTP hydrolysis reaction.
eIF5 mutants defective in binding to eIF2β are also defective in in vitro translation.
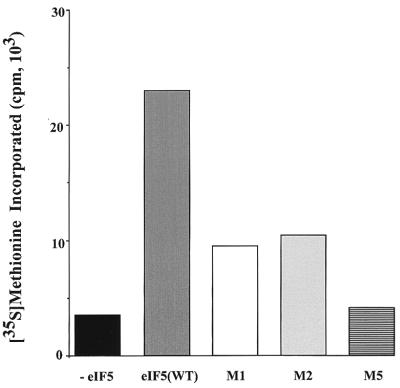
We previously described a yeast cell-free translation system which was dependent on exogenously added purified yeast or mammalian eIF5 for translation of mRNAs in vitro (17). We used such an eIF5-depleted yeast cell-free translation system (17) to examine the effect of addition of purified mutant eIF5 proteins for the ability to restore translation of yeast mRNAs in vitro. Figure 6 shows that in the absence of exogenously added eIF5, these extracts showed poor activity in translation of yeast mRNA. Translation could be restored in these lysates by the addition of purified wild-type rat eIF5 (Fig. 6). In the absence of mRNA, addition of eIF5 had virtually no effect (data not shown). In contrast to wild-type rat eIF5, mutant eIF5 proteins M1 and M2 were about threefold less active in restoring translation (Fig. 6). The eIF5 mutant M5, in which six glutamic acid residues were mutated to alanine, was virtually inactive in restoring translation in these extracts (Fig. 6). These results suggest that the binding of eIF5 to eIF2β plays an essential role in the function of eIF5 in translation of mRNAs.
FIG. 6.
Effect of eIF5-Ala mutations on in vitro translation of total yeast RNA. eIF5-depleted cell translation extracts were prepared from TMY201R cells (17), incubated, and analyzed for [35S]methionine incorporation into proteins as described in Materials and Methods. Each reaction mixture (50 μl) contained 15 μCi of [35S]methionine (11 Ci/mmol), 25 μg of total yeast RNA, and where indicated either 100 ng of purified recombinant rat wild-type eIF5 [eIF5(WT)] or 100 ng each of purified mutant eIF5 M1, M2, or M5 protein. Following incubation at 25°C for 40 min, aliquots (10 μl) were withdrawn and analyzed for [35S]methionine incorporation into proteins. A control reaction mixture lacking total yeast RNA and exogenously added eIF5 was also incubated and analyzed. The amount of [35S]methionine incorporated into proteins in this control reaction mixture was subtracted, and the results are shown.
eIF5 mutants defective in binding eIF2β are not defective in binding the Nip1p (p93) subunit of eIF3.
In addition to interacting with eIF2β, eIF5 has been shown to specifically interact with the multisubunit initiation factor eIF3 in both yeast (1, 20) and mammalian (2) cells. Asano et al. (1) demonstrated that in the yeast S. cerevisiae system, the C-terminal 165 amino acids of eIF5 were sufficient for its interaction with both eIF2β and eIF3-p93 (Nip1p). It was therefore of interest to determine whether mutations in the bipartite motif in the C terminus of mammalian eIF5, which disrupts the eIF5-eIF2β interaction, also affect eIF5 and eIF3-Nip1p interaction. However, the binding region of mammalian eIF5 to mammalian eIF3-Nip1p has not yet been identified. For this reason, we investigated the interaction between mammalian eIF5 with bacterially expressed recombinant yeast Nip1p based on the assumption that if interaction between eIF5 and Nip1p were physiologically important, the interaction domains could be evolutionarily conserved between yeast and mammals.
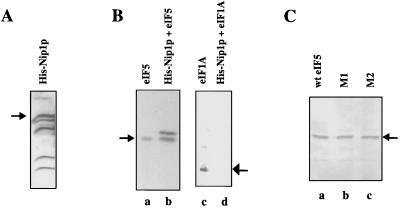
We expressed His-tagged Nip1p in bacterial cells and immobilized the expressed protein on Ni-NTA agarose beads as described in Materials and Methods. Western blot analysis using rabbit anti-yeast Nip1p antibodies as a probe showed that the beads contained bound intact Nip1p as well as several lower-molecular-weight immunoreactive polypeptides (Fig. 7A). These lower-molecular-weight polypeptides arose presumably due to proteolysis of Nip1p in bacterial cell extracts. When the beads containing bound Nip1p were incubated with wild-type recombinant rat eIF5 and the washed beads were subjected to Western blot analysis using anti-rat eIF5 antibodies, we observed that wild-type rat eIF5 was retained on these beads (Fig. 7B, lane b). As a control, the 17-kDa initiation factor eIF1A, which is known not to interact with Nip1p, was not retained, as expected (Fig. 7B, lane d). These results show that rat eIF5, like its yeast counterpart, interacts with yeast Nip1p, indicating that the interaction between eIF5 and eIF3-Nip1p is evolutionarily conserved. When rat eIF5 mutants M1 and M2, which are defective in binding to eIF2β, were tested for binding to yeast Nip1p under the same experimental conditions, they also bound yeast Nip1p to the same extent as wild-type rat eIF5 (Fig. 7C). Likewise, in GST pull-down assays, both GST-eIF5M1 and GST-eIF5M2 retained Nip1p to a similar extent as GST-eIF5(wild-type) (data not shown). These results suggest that the regions of eIF5 responsible for binding eIF2β are distinct from those involved in binding eIF3-Nip1p, indicating that the interactions of eIF5 with eIF2β and eIF3-Nip1p may have distinct functions in the initiation of translation.
FIG. 7.
Expression of His-Nip1p in bacteria and interaction between His-Nip1p and eIF5. (A) Ni-NTA agarose beads containing bound His-Nip1p were analyzed by Western blot using rabbit anti-yeast Nip1p polyclonal antibodies. The position of intact His-Nip1p is indicated by an arrow. (B) Ni-NTA agarose beads containing 5 μg of His-Nip1p were incubated with 5 μg of either purified recombinant rat eIF5 (lane b) or purified rat eIF1A as a negative control (lane d) for 1 h at 4°C. Proteins bound to the washed beads were analyzed by Western blot using rabbit anti-rat eIF5 (left) and anti-rat eIF1A (right) antibodies. Purified rat eIF5 (lane a) and purified rat eIF1A (lane c) were also run as markers on the same Western blots. (C) Ni-NTA agarose beads containing 5 μg of His-Nip1p were incubated with 5 μg of either purified wild-type (wt) rat eIF5 (lane a) or the mutant proteins M1 (lane b) and M2 (lane c) for 1 h at 4°C. After incubation, the beads were washed and proteins bound to the washed beads were analyzed by Western blot using rabbit anti-rat eIF5 antibodies. The arrow indicates the position of purified rat eIF5. It should be noted that two immunoreactive polypeptides were observed in panels B (lane b) and C. The slower-migrating polypeptide arises from His-Nip1p preparation (data not shown). The intensity of this band varies from preparation to preparation.
DISCUSSION
It is now well established that hydrolysis of GTP during translation initiation occurs only when eIF5 interacts with GTP bound to eIF2 as a Met-tRNAf–eIF2–GTP ternary complex in the 40S initiation complex (Met-tRNAf–eIF2–GTP–eIF3–40S–AUG) and that eIF5 by itself does not hydrolyze either free GTP or GTP bound to eIF2 as a Met-tRNAf–eIF2–GTP ternary complex (4, 7, 21). These results suggest that eIF5 interacts with one or more components of the 40S initiation complex to effect the hydrolysis of bound GTP. Subsequent studies, showing that eIF5 forms a specific complex with eIF2 (7) by interacting with the β subunit of eIF2 (1, 11), led us to hypothesize that protein-protein interaction between eIF5 and the 40S subunit-bound eIF2 may be critical for the hydrolysis of GTP bound to the 40S initiation complex (11). To demonstrate such a correlation, we carried out mutational analysis of eIF5 to identify the amino acid residues in the protein critical for its interaction with eIF2β. The eIF5 mutants defective in such interactions were then analyzed for the ability to promote the hydrolysis of GTP during translation initiation.
In the work presented in this paper, we first carried out deletion analysis of eIF5 to demonstrate that the C-terminal region of rat eIF5 binds eIF2β (Fig. 1). In view of our previous observation (11) that a 22-amino-acid region at the N-terminal of mammalian eIF2β containing stretches of conserved lysine residues is involved in the binding of eIF2β to eIF5, we reasoned that a stretch of acidic amino acid residues in the C-terminal eIF2β-binding region of eIF5 may be involved in its interaction with mammalian eIF2. Alanine substitution mutagenesis within this region defined several glutamic acid residues, which are highly conserved between species, as important for binding to eIF2β. The E346A,E347A and E384A,E385A double-point mutations each caused a profound decrease in the specific binding of eIF5 to eIF2β, while eIF5 mutant M5, in which all six glutamic acid residues in the two halves of the bipartite motif were mutated to alanine, showed barely detectable binding to eIF2β. It is therefore likely that these conserved glutamic acid residues which constitute a bipartite motif in eIF5 make direct contacts with the conserved lysine residues in the polylysine stretches of eIF2β and is indeed a component of the eIF2β binding site of eIF5. Further characterization of these two eIF5 mutants showed that the purified expressed proteins containing each of the two double-point mutations were severely defective in eIF5-dependent hydrolysis of GTP bound to the 40S initiation complex and consequently defective also in 80S initiation complex formation. These mutants were also defective in stimulating translation of yeast mRNAs in an eIF5-dependent yeast cell-free translation system. The importance of eIF5-eIF2β interaction in eIF5 function was further confirmed by our demonstration that while wild-type rat eIF5 can substitute for yeast eIF5 function in ΔTIF5 haploid yeast cells, the mutant rat eIF5 proteins M1 and M2, when expressed in such ΔTIF5 yeast cells, showed severe growth defects. It appears that the mutant M1 is much more defective in growth (doubling time of 13.4 h) than the mutant M2 (doubling time of 7 h), indicating that the first motif comprising glutamic acid residues 345 to 347 plays a more important role in eIF5-eIF2β interaction and consequently eIF5 function than the second motif comprising the glutamic acid residues 384 to 386. It is interesting to note here that the first motif is more conserved than the second one (Fig. 2). Furthermore, mutant eIF5 M5 containing alanine substitution mutations in all six glutamic acid residues in the bipartite motif that showed barely detectable binding to eIF2β was unable to maintain cell growth and viability of such yeast cells. These findings suggest that interaction of eIF5 with eIF2β is required for eIF5 function in vivo and in vitro.
Asano et al. (1) have previously demonstrated that the yeast eIF2β-binding region of yeast eIF5 contains a bipartite motif at the C terminus of eIF5. Our observation that eIF2β-binding region of rat eIF5 also contains a bipartite motif is in agreement with their work. We show that this bipartite motif consists of two regions, one surrounding glutamic acid residue 345 to 347 and the other surrounding glutamic acid residues 384 to 386. While mutagenesis of the glutamic acid residues in any one region caused profound decrease both in the binding of eIF5 to eIF2β as well as in eIF5-mediated GTP hydrolysis from the 40S initiation complex, neither of these two reactions was completely abolished under these conditions. However, when the glutamic acid residues in both regions of the bipartite motif were mutated, the resulting mutant eIF5 was virtually inactive in eIF5-dependent GTP hydrolysis and in 80S initiation complex formation. These observations suggest that in the native eIF5 molecule, these two regions of the bipartite motif must come together in interacting with the polylysine-rich region of the eIF5-binding site of eIF2β. Presumably, mutagenesis of any one region of the bipartite motif still allows eIF5 to bind weakly to the polylysine-rich eIF5-binding region of eIF2β via the glutamic acid residues of the other region and allow slow GTP hydrolysis. Additionally, it should be noted that the assays used to study binding of eIF5 to eIF2β measure stoichiometric interaction between the two initiation factors and are at best semiquantitative. In contrast, eIF5-dependent hydrolysis of GTP bound to the 40S initiation complex measures the rate of GTP hydrolysis in which eIF5 is known to act catalytically (12). This may explain our observation that eIF5 mutants M1 and M2, which bind very weakly to eIF2β, still exhibit slow GTP hydrolysis activity that is about 20% of the activity of wild-type eIF5.
An important property of eIF5-dependent GTP hydrolysis reaction is that in addition to eIF2 and eIF5, 40S ribosomal subunits also play a key role in GTP hydrolysis during translation initiation. It is likely that when the ternary complex is transferred to the 40S ribosomal subunits, eIF2 acquires a conformation such that its interaction with eIF5 via the β subunit of eIF2 activates the latent GTPase activity of eIF2. Alternatively, 40S ribosomes may play a more direct role in GTP hydrolysis and could be a coeffector. Kozak has postulated (16) that the 40S ribosomal subunit may have a “GTPase-activating center,” analogous to the presence of a similar domain in the 50S ribosomal subunit of prokaryotes that mediates GTP hydrolysis by the prokaryotic initiation factor IF2 and elongation factors EFTu and EFG. In prokaryotes, both IF2 and EFTu have been shown to have a weak GTPase activity (16, 18) that is markedly stimulated by 50S ribosomal subunits. It remains unknown whether eIF2 possesses an intrinsic GTPase activity. In analogy with proteins of the GTPase superfamily, the γ subunit of eIF2, which contains the consensus GTP-binding domains (14) and is presumably involved in binding of GTP by eIF2, may also possess latent GTPase activity, although this has not been demonstrated experimentally. We postulate that the interaction of eIF5 with the β subunit of eIF2 bound as the Met-tRNAf–eIF2–GTP ternary complex on the 40S ribosomal subunit induces a conformational change in eIF2 resulting in the activation of the latent GTPase activity of the γ subunit of eIF2. In this respect, eIF5 acts as a GTPase-activating protein (GAP). It should, however, be noted that typical GAPs e.g., Rho GAPs and Ras GAPs, that have been characterized extensively contain sequence motifs that are necessary for their GTPase-stimulating activity, in addition to motifs that are necessary for interacting with their G proteins (25). However, eIF5 has no apparent homology with any member of the GAP family. It remains to be determined whether eIF5 possesses any additional sequence motifs that are required for its GTPase-activating function.
Finally, Asano et al. (1) have shown that the C-terminal one-third of yeast eIF5 contains an acidic and aromatic amino acid-rich bipartite motif that is necessary for the binding of both eIF2β and the Nip1p subunit of yeast eIF3. Mutations (one mutant containing 7 mutations and the other containing 12 mutations of conserved amino acid residues) in this motif which disrupt eIF5-eIF2β interaction also disrupt eIF5-Nip1p interaction (1). Data presented in this paper suggest, however, that the amino acid residues critical for eIF5-eIF2β interaction may be distinct from those that are critical for eIF5-Nip1p interaction although the binding domains of eIF2β and eIF3-Nip1p may be in the same region of eIF5 and may in fact overlap. It is quite likely that the interactions between eIF5-eIF2β and eIF5-Nip1p are not mutually exclusive and each interaction may play a distinct role in eIF5 function. Clearly additional structure-function studies will be necessary to establish the nature of the eIF5-Nip1p interaction and its role in eIF5 function.
ACKNOWLEDGMENTS
We are grateful to Jerard Hurwitz and Stewart Shuman, Sloan-Kettering Cancer Research Institute, New York, N.Y., for critically reading the manuscript.
This work was supported by grant GM15399 from the National Institutes of Health and by Cancer Core Support Grant P30CA13330 from the National Cancer Institute.
REFERENCES
- 1.Asano K, Krishnamoorthy T, Phan L, Pavitt G D, Hinnebusch A G. Conserved bipartite motifs in yeast eIF5 and eIF2Bɛ, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay A, Maitra U. Cloning and characterization of the p42 subunit of mammalian translation initiation factor 3 (eIF3): demonstration that eIF3 interacts with eIF5 in mammalian cells. Nucleic Acids Res. 1999;27:1331–1337. doi: 10.1093/nar/27.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti A, Maitra U. Function of eukaryotic initiation factor 5 in the formation of an 80S ribosomal polypeptide chain initiation complex. J Biol Chem. 1991;266:14039–14045. [PubMed] [Google Scholar]
- 5.Chakravarti D, Maiti T, Maitra U. Isolation and immunochemical characterization of eukaryotic translation initiation factor 5 from Saccharomyces cerevisiae. J Biol Chem. 1993;268:5754–5762. [PubMed] [Google Scholar]
- 6.Chakravarti D, Maitra U. Eukaryotic initiation factor 5 from Saccharomyces cerevisiae. J Biol Chem. 1993;268:10524–10533. [PubMed] [Google Scholar]
- 7.Chaudhuri J, Das K, Maitra U. Purification and characterization of bacterially expressed mammalian translation initiation factor 5 (eIF5): demonstration that eIF5 forms a specific complex with eIF2. Biochemistry. 1994;33:4794–4799. doi: 10.1021/bi00182a007. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri J, Si K, Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J Biol Chem. 1997;272:7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 9.Chevesich J, Chaudhuri J, Maitra U. Characterization of mammalian initiation factor 5 (eIF5) J Biol Chem. 1993;268:20659–20667. [PubMed] [Google Scholar]
- 10.Das K, Chevesich J, Maitra U. Molecular cloning and expression of cDNA for mammalian translation initiation factor 5. Proc Natl Acad Sci USA. 1993;90:3058–3062. doi: 10.1073/pnas.90.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Maiti T, Das K, Maitra U. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the β subunit of eIF2. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S, Chevesich J, Maitra U. Further characterization of eukaryotic initiation factor 5 from rabbit reticulocytes. Immunochemical characterization and phosphorylation by casein kinase II. J Biol Chem. 1989;264:5134–5140. [PubMed] [Google Scholar]
- 13.Gu G, Moerschell R P, Sherman F, Goldfarb D S. N1P1, a gene required for nuclear transport in yeast. Proc Natl Acad Sci USA. 1992;89:10355–10359. doi: 10.1073/pnas.89.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannig E M, Cigan A M, Freeman B A, Kinzy T G. GCD11, a negative regulator of GCN4 expression, encodes the γ subunit of eIF2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H-K, Yoon H, Hannig E M, Donahue T F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 17.Maiti T, Maitra U. Characterization of translation initiation factor 5 (eIF5) from Saccharomyces cerevisiae. Functional homology with mammalian eIF5 and the effect of depletion of eIF5 on protein synthesis in vivo and in vitro. J Biol Chem. 1997;272:18333–18340. doi: 10.1074/jbc.272.29.18333. [DOI] [PubMed] [Google Scholar]
- 18.Maitra U, Stringer E A, Chaudhuri A. Initiation factors in protein biosynthesis. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- 19.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 31–69. [Google Scholar]
- 20.Phan L, Zhang X, Asano K, Anderson J, Vornlocher H P, Greenberg J R, Qin J, Hinnebusch A G. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raychaudhuri P, Chaudhuri A, Maitra U. Eukaryotic initiation factor 5 from calf liver is a single polypeptide chain protein of Mr = 62,000. J Biol Chem. 1985;260:2132–2139. [PubMed] [Google Scholar]
- 22.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sachs A B, Deardorff J A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Scheffzek K, Ahmadian M R, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 26.Si K, Das K, Maitra U. Characterization of multiple mRNAs that encode mammalian translation initiation factor 5 (eIF5) J Biol Chem. 1996;271:16934–16938. doi: 10.1074/jbc.271.28.16934. [DOI] [PubMed] [Google Scholar]