Abstract
Gene-expression profiling can be used to classify human tumors into molecular subtypes or risk groups, representing potential future clinical tools for treatment prediction and prognostication. However, it is less well-known how prognostic gene signatures derived in one malignancy perform in a pan-cancer context. In this study, a gene-rule-based single sample predictor (SSP) called classifier for lung adenocarcinoma molecular subtypes (CLAMS) associated with proliferation was tested in almost 15 000 samples from 32 cancer types to classify samples into better or worse prognosis. Of the 14 malignancies that presented both CLAMS classes in sufficient numbers, survival outcomes were significantly different for breast, brain, kidney and liver cancer. Patients with samples classified as better prognosis by CLAMS were generally of lower tumor grade and disease stage, and had improved prognosis according to other type-specific classifications (e.g. PAM50 for breast cancer). In all, 99.1% of non-lung cancer cases classified as better outcome by CLAMS were comprised within the range of proliferation scores of lung adenocarcinoma cases with a predicted better prognosis by CLAMS. This finding demonstrates the potential of tuning SSPs to identify specific levels of for instance tumor proliferation or other transcriptional programs through predictor training. Together, pan-cancer studies such as this may take us one step closer to understanding how gene-expression-based SSPs act, which gene-expression programs might be important in different malignancies, and how to derive tools useful for prognostication that are efficient across organs.
Keywords: pan-cancer, lung adenocarcinoma, molecular subtypes, gene expression, single sample predictor, prognosis
Introduction
Cancer is the second leading cause of death globally and estimated to account for 9.6 million deaths in 2018 (www.who.int). Cancer outcome is consistently improved through better diagnostics, clinical management and novel therapeutics. To further enhance cancer survival, new molecular-based tools are needed beyond existing clinical markers to refine prognostication and treatment prediction for individual patients.
Gene-expression profiling is a molecular technique that allows stratification of cancer patients for subtyping and risk assessment prediction based on the expression levels of specific gene sets, commonly referred to as gene signatures. In breast cancer, gene-expression-based molecular subtypes and treatment prediction signatures have now reached clinical use based on analyses of large clinical trials [1–3]. While gene-expression profiling has been successful in some diseases, clinical usefulness in other malignancies has been limited. An example of the latter is lung cancer, which is the most common form of cancer today with over 2 million new cases estimated for 2018 [4]. Molecular subtypes, prognostic signatures and also treatment predictive signatures based on gene expression have been proposed in lung cancer [5–10], but to our knowledge none are yet in clinical use. Notably, detailed analyses of lung adenocarcinoma gene signatures have demonstrated an overlap of signatures with respect to their classifications (e.g. overlap in risk predictions), as well as the existence of a common component related to tumor cell proliferation in many of the reported prognostic signatures [7, 11].
Several reasons may account for the lack of clinical translation of a gene signature. These include issues with performance reproducibility and applicability to fixated tissue that often is subjected to RNA degradation, but also issues related to the predictor development and implementation process itself. Concerning the latter, most gene signatures have been developed in a way that often requires gene-centering of input data to assure comparable relative gene-expression levels across samples. To circumvent this need, predictors based on gene rules (comparing the expression of gene pairs) have been proposed (see e.g. [12] for discussion and references), in which samples are classified truly independently of each other, often without any need of data processing, thereby making them true single sample predictors (SSPs).
To try to overcome the problems with applying gene expression signatures in a clinical context in lung cancer, we recently reported a classifier for lung adenocarcinoma molecular subtypes (CLAMS) [13]. CLAMS is a true SSP that can identify lung adenocarcinoma gene-expression subtypes as proposed by The Cancer Genome Atlas (TCGA) consortium [6] either in a three-group (Terminal Respiratory Unit (TRU), Proximal-Proliferative (PP) and Proximal-Inflammatory (PI)) or two-group (TRU, nonTRU) context. CLAMS was successfully trained and evaluated using a variety of different technical gene-expression platforms including Affymetrix, Illumina and Agilent microarrays, as well as RNA sequencing. Independent validation of CLAMS prediction in lung adenocarcinoma showed that predicted classes were associated with specific clinicopathological and molecular features, as well as with patient outcome with TRU-classified tumors showing superior survival compared to nonTRU cases [13]. For the two-class (TRU/nonTRU) classification approach, CLAMS uses the expression of 36 genes that are highly associated with cell proliferation arranged into 18 pair rules. Since increased cell proliferation is a hallmark of cancer and prognostically important also for other malignancies (e.g. for breast cancer [14]), we hypothesized that CLAMS would be able to effectively stratify patients with solid cancer types other than lung adenocarcinoma into groups with noticeably different disease progression. Interestingly, the application and performance of rule-based SSPs like CLAMS in a massive pan-cancer context has not been reported to date. We believe the application of rule-based SSPs outside the malignancy they were developed in may broaden our understanding of how these classifiers work, their limitations and strengths.
To resolve the knowledge gap of how rule-based SSP models work pan-cancer, we collected publicly available gene-expression data from nearly 15 000 patients representing 32 solid cancer types. For each malignancy, we classified tumors according to the reported two-class CLAMS SSP [13] into better (TRU used as acronym) or worse (nonTRU used as acronym) prognosis subgroups and compared the overall survival of patients in these groups. In this context, we used CLAMS as a case example of applying a rule-based SSP pan-cancer. Apart from lung cancer, we found statistically significant differences between CLAMS prognosis groups in breast, brain, kidney and liver cancer. In comparison with clinical markers and disease specific molecular classifications, we observed both concordance and discordance between classification methods. In addition, based on pan-cancer data, we could showcase the potential of using the existing wealth of public genomic data to facilitate future development of SSPs in malignancies with currently less data available. Taken together, pan-cancer studies such as ours may take us one step closer to understanding how gene-expression-based SSPs act, which gene-expression programs might be important in different malignancies, and how to derive tools useful for prognostication that are efficient across organs.
Material and Methods
Gene-expression cohorts
Data were gathered from several public repositories to a grand total of 40 data sets representing 32 different cancer types (Supplementary Table S1). The majority of data sets (32 data sets representing 32 different cancer types) came from TCGA, which was restricted to include gene-expression data from only solid tumor types generated from HTSeq RNA sequencing (RNAseq V2). Replicates per patient and samples flagged by the Pan-Cancer Atlas (PanCanAtlas, gdc.cancer.gov/about-data/publications/pancanatlas) initiative were removed. Only primary tumor samples were kept, and sets had to contain at least 30 samples to be used. In total, 8729 patients with expression data remained based on these requirements and their files were downloaded from the National Cancer Institute’s Genomic Data Commons Data Portal (portal.gdc.cancer.gov). The information for each gene consisted of HTSeq-acquired Fragments Per Kilobase per Million reads mapped (FPKM), and no further processing of the public data was made. Clinical outcome data for the samples included in this study were obtained from the PanCanAtlas. Acronyms for cancer types follow TCGA nomenclature for TCGA data sets (see Figure 1).
Figure 2 .

Overall survival curves and hazard ratio of CLAMS groups. Kaplan–Meier survival curves showed significant differences between better (TRU, green) and worse (nonTRU, blue) prognosis groups within tumors in the breast (invasive carcinoma), brain (lower grade glioma), kidney (renal papillary cell carcinoma), liver (hepatocellular carcinoma) and lung (adenocarcinoma). Data sets are from TCGA unless stated otherwise. HR: hazard ratios for overall survival and 95% confidence interval between CLAMS groups (TRU as reference) from univariate Cox regression analysis.
Two other large publicly available breast cancer cohorts were added. The first one, the Gene-expression based Outcome for Breast cancer Online (GOBO), corresponds to gene expression quantification data from 1881 samples available online (co.bmc.lu.se/gobo). GOBO was created by pooling together 11 data sets that used Affymetrix U133A arrays as reported by [15]. For these samples, we processed corresponding Affymetrix CEL files as outlined in [16] to obtain raw, unnormalized gene-expression intensities used in subsequent analyses. The second data set, the Sweden Cancerome Analysis Network-Breast (SCAN-B), comprises RNA sequencing-based gene expression quantification data (FPKM) and clinicopathological data from 3520 samples of patients diagnosed between 2010 and 2015 that had primary surgery and no neoadjuvant treatment [17]. No additional preprocessing beyond the publicly available FPKM data was made. SCAN-B is representative of a population-based breast cancer cohort in Sweden and has available clinical treatment data obtained from the national quality registry for breast cancer [17], allowing sub-analyses to be performed in specific therapy groups. The addition of GOBO and SCAN-B breast cancer cohorts gave more depth to the study as they differ in, e.g. median overall survival time of patients from diagnosis to either death or censoring to the TCGA data set (27.5 months in TCGA, 97.3 in GOBO and 54.4 in SCAN-B).
Six other smaller published data sets with information regarding treatment response were obtained from original publications and are referred to by the first author of the respective publications. Three of these data sets were from breast cancer patients treated with neoadjuvant chemotherapy, adding 133 [18], 178 [19] and 97 [20] samples to our study. For these data sets, we used either the publicly available data from the publication directly (as for the Hess et al. data set [18]) or the generated raw, unnormalized data for samples analyzed by Affymetrix arrays as outlined above and in [16]. The remaining three treatment response data sets were from malignant melanoma patients treated with anti-PD-1 immune checkpoint inhibitors contributing with 51 [21], 86 [22] and 121 [23] samples. For these data sets, the gene-expression data deposited with the publication was used without further processing as raw sequence data were not available. Only samples with both gene-expression profiles and response information were used. When samples were collected at multiple time points for the same patient (as in the Riaz et al. data set [21]), those obtained before treatment start were kept.
Prognosis prediction
CLAMS [13] was installed as a package in RStudio running R v3.6.3 [24]. This SSP classifies lung adenocarcinoma samples into known expression subtypes in a two-class (TRU/nonTRU) or three-class (TRU/PP/PI) approach. In this study, the two-class CLAMS SSP was used to classify all samples from the above data sets into better prognosis (referred to as TRU in the original report for CLAMS [13]) or worse prognosis (originally referred to as nonTRU by CLAMS) groups. All 36 genes used by CLAMS for classification were present in all included data sets. These 36 genes are arranged into 18 pair rules, and whether these rules are fulfilled for a sample (i.e. whether the expression of gene A is lower than gene B) determines the likelihood of the sample belonging to one or the other prognosis category (Supplementary Table S2). In the current study, we used class labels as outputted by the two-class CLAMS (TRU/nonTRU) algorithm for consistency with the available software. It should be noted that TRU-classified non-lung adenocarcinoma cases are for instance not expected to share TRU lung adenocarcinoma specific features such as frequent EGFR mutations and increased surfactant expression, but they would be expected to share a generally lower expression of proliferation associated genes. Chi-square test or Fisher’s exact test was performed to see if CLAMS predictions were linked to patient/tumor characteristics within the different cancer types.
Survival analysis
Across all data sets, excluding treatment prediction cohorts, overall survival represented the common clinical endpoint suitable for survival analysis. Survival curves were estimated with the Kaplan–Meier method based on overall survival data censored at 5 years as the relevant event with the R package survminer [25]. Only data sets containing at least eight TRU samples from repositories that had survival information (TCGA, GOBO, SCAN-B) were included in this step. Log-rank test was used to compare the two groups (predicted better/worse prognosis) and a P-value threshold of 0.05 was set to be considered statistically significant. Correction for multiple testing was performed using the Benjamini–Hochberg method [26]. The univariate Cox proportional hazards regression model was used to estimate hazard ratios of overall survival of CLAMS groups together with a 95% confidence interval. Multivariate Cox regression was performed using gender, age, tumor stage, histological grade, tumor size and lymph node status as covariates whenever relevant and available.
In silico tumor proliferation scoring
All cancer samples were classified as high or low proliferative according to expression levels of genes commonly associated with cell proliferation based on a previous gene network analysis in lung cancer [27]. For values to be comparable across data sets, the analysis was restricted to the 9641 genes present in all cohorts. In cases where multiple transcripts were annotated with the same gene symbol, the highest expression value was kept. For each sample, genes were ranked from lower to higher expression levels. The ranks of 74 genes present in all data sets and previously associated with tumor proliferation in lung cancer [27] were extracted and summed to a rank score. A low rank score based on these 74 genes thus indicates a relative low (in silico) level of proliferation for a specific sample. Proliferation rank scores were also used to divide samples of each cancer type into low (values lower than the 33rd percentile), intermediate (between 33rd and 67th percentiles) and high proliferation groups (higher than the 67th percentile). Kaplan–Meier survival curves and univariate Cox proportional hazards regression model as explained above were performed on these groups to test whether proliferation was an important prognostic component in other malignancies.
Treatment prediction
Six previously published breast cancer and malignant melanoma data sets (see above) had their samples classified by CLAMS into better (TRU) or worse (nonTRU) prognosis groups. Fisher’s exact test was used to analyze whether association between CLAMS-predicted subtypes and treatment response existed. Response categories included in the analysis were: for breast cancer, (i) pathologic complete response, (ii) near pathologic complete response and (iii) residual disease; for melanoma, (i) complete response, (ii) partial response, (iii) stable disease and (iv) progressive disease following RECIST v1.1 [28].
Data availability
All gene-expression data used in the current study and any sample information are available through original studies. CLAMS classification of all samples is available as Supplementary Table S3. R code used in the analyses conducted in this study can be found at https://github.com/StaafLab/CLAMS-pan-cancer.
Results
Application of CLAMS in a pan-cancer context
The CLAMS SSP was used to classify 40 data sets with a total of 14 796 samples from patients with 32 different cancer types into better (referred to as TRU by CLAMS) or worse (nonTRU) prognosis groups based on gene expression profiles. Of the 40 data sets, six were included for treatment response assessment and consequently analyzed separately, whereas the remaining data sets were included in the patient outcome analysis (Figure 1A). Analyzed data sets included both primary (neoadjuvant biopsies or surgical resections) and metastatic disease (biopsies) (Supplementary Table S1). The proportion of samples classified as TRU varied greatly between malignancies, ranging from 61.4% in the thyroid carcinoma set to no sample at all for several cancer types (Figure 1B). Of data sets included for prognostic evaluations (34 data sets representing 32 cancer types), all samples were classified exclusively as worse prognosis (nonTRU) for 11 TCGA data sets (representing 11 cancer types), while nine other TCGA data sets had eight or less samples classified into the better prognosis group. As survival analysis was a focus of this study, these 20 TCGA data sets (and therefore 20 cancer types) were not analyzed further. This left 14 data sets representing 12 different cancer types with enough samples to be used in prognostic evaluations. A full summary of classifications on data set level and individual sample level is available in Supplementary Tables S1 and S3.
Figure 1 .

Study design and pan-cancer classification of samples with CLAMS. (A) Flowchart of study highlighting the three main analyses performed and which of the 40 data sets comprising a total of 14 796 samples were used in each. (B) Proportion by data set of samples classified by CLAMS as better (TRU, green) or worse (nonTRU, blue) prognosis based on gene-expression data. Data sets are from TCGA unless stated otherwise in square brackets. Data sets marked with circles were used for subsequent overall survival analysis given that over eight samples classified as TRU existed; data sets marked with triangles were only used for prediction of treatment response and analyzed separately. Values on the right correspond to the number of TRU samples over the total number of samples (n).
Association between CLAMS and patient outcome in malignancies other than lung cancer
Of the remaining 14 data sets (12 different cancer types, n = 10 087 tumors) with patient survival data, differences in overall survival between the two predicted prognosis groups were statistically significant after multiple testing correction using the Benjamini–Hochberg method for four other malignances besides lung adenocarcinoma: invasive breast cancer (breast), lower grade glioma (brain), renal papillary cell carcinoma (kidney) and hepatocellular carcinoma (liver) (Figure 2, Supplementary Figure S1). Consistent with lung adenocarcinoma, a classification as poor prognosis by CLAMS was associated with a significant increase in the risk of death for all four malignancies (Figure 2).
CLAMS classification of breast cancer
CLAMS was used to classify 6473 breast invasive carcinoma (BRCA) samples available in three different large data sets (TCGA, GOBO and SCAN-B). Of these, 763 (11.8%) were classified as better prognosis (TRU) and 5710 (88.2%) as worse prognosis (nonTRU). Even though the predictor divided samples into two groups in all data sets, the TCGA cohort presented a lower proportion of TRU-classified cases (3.6% compared to 10.6% for GOBO and 14.9% for SCAN-B; Table 1). PAM50 subtypes and clinicopathological relevant groups were available for most samples and differed considerably between data sets (Table 1). Most samples marked as better prognosis by CLAMS belonged to groups expected to have better prognosis by those classifications as well [17, 29–31]: 541 (70.9%) TRU-classified samples were of the PAM50 subtype Luminal A, and 481 (63%) were positive for either estrogen (ER) or progesterone receptor (PR) or both, and negative for human epidermal growth factor receptor 2 (HER2). An additional 212 samples were regarded as PAM50 normal-like. The normal-like group has the lowest prevalence of all PAM50 subtypes and its clinical relevance is less clear, being therefore excluded from many studies [31–33]. When excluding normal-like tumors from our analysis, 98.2% of TRU-classified BRCA samples had breast cancer specific classifications of better prognosis (such as Luminal A, ER-positive, PR-positive and HER2-negative). In opposite, of the 2787 Luminal A tumors present in the three cohorts, only 19.4% were classified as TRU by CLAMS, indicating that CLAMS identifies a subset of lower proliferative Luminal A tumors (Wilcoxon rank sum test: P < 0.001; Supplementary Figure S2A).
Table 1.
CLAMS classification of breast cancer data sets
| TCGA (n = 1072) | GOBO (n = 1881) | SCAN-B (n = 3520) | |||||
|---|---|---|---|---|---|---|---|
| CLAMS | TRU | NonTRU | TRU | NonTRU | TRU | NonTRU | |
| Overall proportion | 3.6% | 96.4% | 10.6% | 89.4% | 14.9% | 85.1% | |
| CLAMS stratified by PAM50 | Prognosis | ||||||
| Luminal A | Better prognosis | 79.5% | 47.7% | 44.0% | 22.4% | 80.5% | 45.9% |
| Luminal B | Intermediate prognosis | 0% | 19.0% | 0.5% | 28.0% | 0% | 26.0% |
| Normal-like | Intermediate prognosis | 17.9% | 2.9% | 53.0% | 11.8% | 18.9% | 3.7% |
| HER2-enriched | Worse prognosis | 0% | 7.6% | 0.5% | 14.2% | 0% | 10.6% |
| Basal | Worse prognosis | 0% | 17.1% | 0.5% | 18.0% | 0.2% | 11.5% |
| Unclassified/other | 2.6% | 5.7% | 1.5% | 5.6% | 0.4% | 2.1% | |
| CLAMS stratified by clinically relevant groups | Prognosis | ||||||
| PR/ER+ HER2- | Better prognosis | 48.7% | 45.0% | 0% | 0% | 88.2% | 69.1% |
| HER2+ PR/ER- | Intermediate prognosis | 0% | 2.9% | 0% | 1.3% | 1.0% | 4.3% |
| HER2+ PR/ER+ | Intermediate prognosis | 0% | 7.2% | 0% | 5.4% | 3.4% | 10.3% |
| Triple negative | Worse prognosis | 2.6% | 11.5% | 0% | 0% | 2.3% | 10.9% |
| Unclassified/other | 48.7% | 33.4% | 100% | 93.3% | 5.2% | 5.3% | |
Concerning overall survival, CLAMS classification showed significant difference in only two of the three breast cancer cohorts, GOBO and SCAN-B (P < 0.001; Figure 2). The TCGA breast cancer cohort did not reach statistical significance (P = 0.30; Supplementary Figure S1). The same pattern was observed using univariate Cox regression: while being classified as nonTRU significantly increased the risk of dying for patients in the first two cohorts (Figure 2), there was no such increase in the TCGA data set (hazard ratio (HR) = 2.35, 95% confidence interval (CI) = 0.58–9.53, P = 0.23, reference = TRU). When age, tumor size, grade and lymph node status were included as covariates in a multivariate Cox analysis, patients classified as nonTRU by CLAMS still showed an increase in risk of death for GOBO (HR = 3.73, 95% CI = 1.50–9.27, P = 0.005) and SCAN-B (HR = 1.73, 95% CI = 1.06–2.83, P = 0.027). However, when only SCAN-B patients with ER/PR-positive and HER2-negative tumors going through adjuvant endocrine treatment (n = 1579) were considered, CLAMS classification was not statistically significant in the multivariate analysis (P = 0.29).
CLAMS classification of brain tumors
Lower grade gliomas (LGG) comprise grade II and grade III tumors as defined by the World Health Organization for central nervous system malignancies [34]. Of the 508 LGG cases, 90 (17.7%) were classified as TRU and 418 (82.3%) as nonTRU by CLAMS. Samples of this cancer type classified by CLAMS as better prognosis (TRU) were significantly more represented in grade II tumors (n = 63, Chi-square test: P < 0.001), which show better prognosis than grade III tumors [35]. The TCGA cohort also contains histological information, even though it might not be up to date with what is currently used in the field (e.g. using oligoastrocytoma, which is discouraged as diagnose [34]). No histological type was particularly overrepresented among cases classified as better prognosis by CLAMS (Chi-square: P = 0.66) since TRU-classified samples were distributed between astrocytomas (n = 31), oligoastrocytomas (n = 22) and oligodendrogliomas (n = 37).
LGG can also be divided based on mutations in the IDH gene coupled to the codeletion of chromosome arms 1p and 19q (1p/19q), which is linked to disease progression [35]. Three subtypes are accepted: (i) presence of mutation and codeletion (better prognosis), (ii) presence of mutation and absence of deletion (intermediate prognosis) and (iii) absence of mutation (IDH wildtype) (worse prognosis). Subtypes were available for 277 LGG samples, 50 (18.1%) of which were classified as better prognosis (TRU) by CLAMS. No particular trends were observed since TRU cases corresponded to 15–20% of samples for each of the three subtypes (n = 14, n = 25, n = 11, respectively; Chi-square: P = 0.92). This indicates that CLAMS prediction transcends proposed molecular risk groups in LGG, i.e. low-proliferate cases are likely present in each LGG risk group. For individual molecular LGG subtypes, CLAMS classification was statistically significant only within the worse prognosis IDH wildtype subtype (P = 0.0057; Supplementary Figure S3), which has been reported to be heterogeneous in terms of survival [36]. In a multivariate Cox regression, the difference in survival between CLAMS classes was not significant when gender, age and tumor grade were included as covariates (P = 0.11). It needs to be acknowledged that more samples are needed for better stratified survival analysis, as groups of TRU classified samples divided by molecular subtypes contained less than 25 patients each and small numbers of events.
CLAMS classification of kidney cancer
Renal papillary cell carcinoma (KIP) is the second most common form of kidney cancer. Of the 277 KIP cases included in the study, 105 (37.9%) were classified as better prognosis (TRU) and 172 (62.1%) as worse prognosis (nonTRU) by CLAMS (Figure 1). Regarding tumor staging, TRU-classified samples from this malignancy were more represented in less aggressive stage I tumors (n = 80, Chi-square: P < 0.001). KIP has recently been divided into four genomic subtypes associated with better to worse patient survival respectively: (i) type-1-enriched group a (P-e.1a), (ii) type-1-enriched group b (P-e.1b), (iii) type-2-enriched (P-e.2) and (iv) ‘CpG island methylator phenotype’ (P.CIMP-e) [37]. Among TRU-classified cases, 87 (82.9%) belonged to type-1-enriched groups and none was assigned to P.CIMP-e, thus showing congruence between prognosis groups of both classifications (Chi-square: P < 0.001). Similar to LGG, worse prognosis (nonTRU) samples showed increased risk of death when analyzed with univariate Cox proportional hazards (Figure 2), but this increase was not statistically significant in a multivariate analysis including gender, age and tumor stage as covariates (P = 0.46).
CLAMS classification of liver cancer
Of the 368 TCGA liver hepatocellular carcinoma (LIHC) cases, 38 (10.3%) were classified as better prognosis (TRU) and 330 (89.7%) as worse prognosis (nonTRU) by CLAMS. TRU-classified LIHC cases were significantly more represented in grade I and grade II tumors (n = 14 and n = 20, respectively; Fisher’s exact test: P < 0.001), but not in any particular tumor stage (Fisher’s: P = 0.51). In the past two decades, several studies have identified molecular subtypes and prognostic genomic signatures in LIHC, some of which can be associated across classifications (reviewed in [38]). A recent multi-platform analysis divided a subset of TCGA samples from this malignancy into three clusters, with one of them (iCluster1) being associated with overexpression of proliferation marker genes, showing significantly worse prognosis than the other two [39]. Between patients with both cluster and CLAMS classification (n = 183), there were 22 TRU-classified samples present mainly in iCluster1 (n = 10) and iCluster2 (n = 11), showing no congruence between the two classification methods. The CLAMS groups had, however, significant survival differences within both of those clusters (P = 0.017 and P = 0.046, respectively; Supplementary Figure S3). In addition, an increase in the risk of death for patients classified as worse prognosis (nonTRU) was also seen in a multivariate Cox regression including all LIHC samples, and gender, age, tumor stage, and grade as covariates (HR = 2.96, 95% CI = 1.07–8.25, P = 0.037, reference = TRU), an association which remained significant within iCluster1 as well (HR = 12.84, 95% CI = 1.19–138.3, P = 0.035).
Another recent study divided LIHC into four subtypes based on integrated methylation and gene expression data: HS1 (better prognosis), HS2, HS3 and HS4 (worse prognosis, associated with iCluster1) [40]. Between patients with both HS1-HS4 and CLAMS classification available (n = 366), there were 38 TRU-classified samples mainly in the intermediate (HS3, n = 25) and better prognosis groups (HS1, n = 9), again showing no congruence between CLAMS and molecular LIHC subtypes. However, within the HS3 group, CLAMS classes showed a significant difference in survival outcome (P = 0.026, Supplementary Figure S3) associated with an increase in the risk of dying for patients classified as nonTRU (HR = 3.55, 95% CI = 1.08–11.65, P = 0.037, reference = TRU), though CLAMS classification was not significant in a multivariate Cox regression including gender, age, tumor stage and grade as covariates (P = 0.09). Here again it needs to be acknowledged that more samples are needed for better stratified survival analysis of liver cancer patients.
Link between better prognosis predicted by CLAMS and lower in silico-determined tumor proliferation
The gene-pair rules used by CLAMS to classify samples into TRU/nonTRU subtypes are comprised mainly of genes associated with tumor proliferation [13]. To test if samples classified as better prognosis (TRU) indeed showed signs of lower proliferation than those classified as worse prognosis (nonTRU), we compared the rank-based expression of a set of highly correlated genes previously associated with tumor proliferation in lung cancer through gene network analysis (e.g. AURKA, CCNB1, TOP2A) [27] across all 40 data sets included in the study. This analysis revealed intrinsically different in silico tumor proliferation levels across malignancies (Figure 3). While some cancer types appear to be low proliferative (e.g. THCA, thyroid carcinoma), others are restricted to higher proliferation values (e.g. UCS, uterine carcinosarcoma). Some cancer types present, however, intermediate proliferation and a wider range of values, as exemplified by lung adenocarcinoma (LUAD) and breast invasive carcinoma (BRCA) in our data.
Figure 3 .
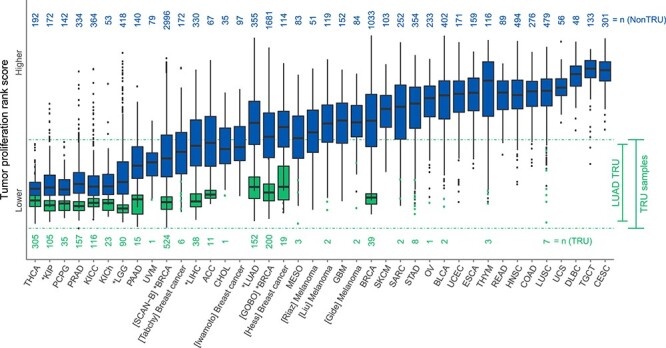
Tumor proliferation rank scores of better and worse prognosis groups by CLAMS. Cancer samples allocated into better (TRU, green) or worse (nonTRU, blue) prognosis groups by CLAMS were classified as more or less proliferative according to expression levels of genes linked to proliferation in lung cancer [27]. Numbers above and below boxplots refer to sample sizes (n). Sample sizes smaller than 10 are represented by dots. Cancer types that had significantly different survival outcomes between CLAMS groups are marked with an asterisk. Dotted lines correspond to the upper and lower proliferation limits of TRU samples. For acronyms, see Figure 1.
Irrespective of cancer type, patients with better prognosis as predicted by CLAMS had lower in silico tumor proliferation gene rank scores than those predicted with a worse prognosis (Wilcoxon rank sum test: P < 0.001 for each cancer type separately, Figure 3). The same pattern in proliferation scores between CLAMS groups could be seen when malignancies were divided into the molecular subtypes mentioned previously, i.e. TRU cases had lower gene rank scores than nonTRU. In fact, an association between in silico tumor proliferation gene rank scores and prognosis was observed within the different molecular subtypes in multiple cancer types: in BRCA, LGG and KIP (but not LIHC), molecular subtypes linked to better prognosis tended to show higher numbers of samples with low proliferative scores (Supplementary Figure S2B). In addition, almost all (99.1%) non-LUAD samples classified as TRU by CLAMS had in silico proliferation gene rank scores within the ranges observed for LUAD TRU samples (Figure 3).
Pan-cancer association of in silico tumor proliferation gene rank scores with patient outcome irrespective of CLAMS
Tumor proliferation is known to be a prognostic gene-expression component for malignancies such as LUAD [11] and BRCA [14], and is an important component of gene signatures like the PAM50 subtypes in breast cancer [31]. Our pan-cancer analysis with CLAMS revealed other cancer types where this seems to also be the case. To test whether in silico proliferation scores could be considered prognostic in cancer types with proliferation spans where CLAMS would not apply, samples for each malignancy were divided into three equally sized proliferation groups based on the previously computed rank scores: low, intermediate and high (Figure 4). As expected, the five cancer types that had significantly different survival outcomes between CLAMS prognosis groups (BRCA [seen in all cohorts], LGG, KIP, LIHC, LUAD) also had significant differences between survival of proliferation groups (Supplementary Figure S4). In addition to these, five other malignancies showed an increase in the risk of dying associated with the high-proliferation group (low as reference) and have therefore a potential prognostic component in tumor cell proliferation (Figure 4): adrenocortical carcinoma (ACC), kidney renal clear cell carcinoma (KICC), mesothelioma (MESO), pancreatic adenocarcinoma (PAAD) and sarcoma (SARC).
Figure 4 .
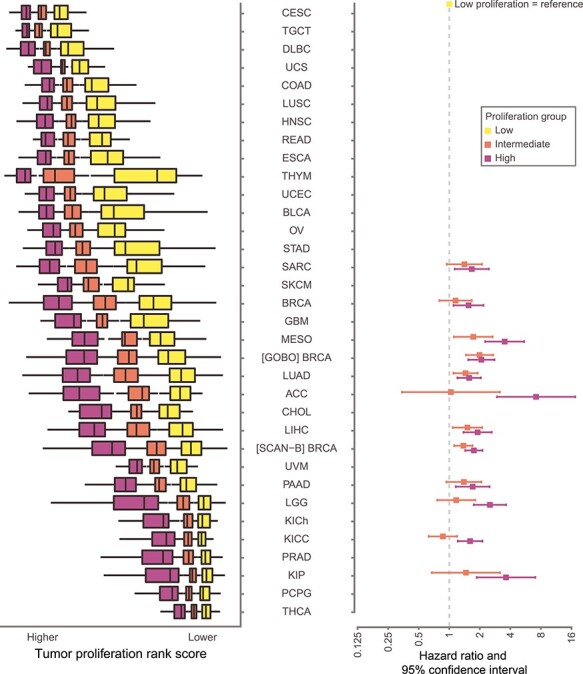
Forest plot of hazard ratios of overall survival between proliferation groups. Each malignancy was divided in three equally sized proliferation groups represented by boxplots (outliers are not shown). Only significant hazard ratios for overall survival and 95% confidence intervals between low (reference, yellow) and high (pink) proliferation groups for each cancer type from univariate Cox regression analysis are shown, along with the values for the corresponding intermediate proliferation group (low as reference). Log values above 1onemean an increased risk of dying for that group. Note that mostly less proliferative cancer types in a pan-cancer context show proliferation as a potential prognostic predictor. For acronyms, see Figure 1. For values, see Supplementary Figure S4.
CLAMS and response to treatment
Certain patient and tumor characteristics can be considered predictive of patient response to specific treatments. To investigate the predictive performance of CLAMS classification, we applied it to samples from six data sets containing information regarding treatment response (e.g. pathological complete response to therapy by RECIST [28]). The chosen malignancies were breast tumors (n = 408) and malignant melanomas (n = 258), accounting for two different treatment approaches: neoadjuvant chemotherapy and immune checkpoint inhibitor treatment of metastatic disease, respectively. Most samples were classified as worse prognosis (nonTRU) by CLAMS in the breast cancer and melanoma cohorts (93.9% and 98.4%, respectively). Moreover, there was no correlation between the few samples considered to have better prognosis according to CLAMS and response to treatment (Supplementary Table S4).
Discussion
With an increasing number of targeted therapies available, characterization of patients’ tumors becomes increasingly important to direct treatment efforts. Cancer is a heterogeneous disease, but several key components have been identified as common across multiple cancer types. Cell proliferation, one of the hallmarks of cancer, is the main gene-expression component of CLAMS [13] and a prognostically important trait in various malignancies [11, 14, 41, 42]. Since lung adenocarcinoma (LUAD) presents one of the widest distributions of proliferation (rank) scores in a pan-cancer context (Figure 3), it stands to reason that CLAMS, a gene expression-based SSP developed specifically for this malignancy, could also show prognostic power in other malignancies. Molecular classifiers that can be applied pan-cancer are still lacking, and it is unclear how rule-based classifiers (like CLAMS) work outside the malignancy they were derived in. Rule-based SSPs are conceptually more suitable for clinical implementation due to platform and cohort normalization independence. Taken together, this was the rationale behind testing our SSP in almost 15 000 samples from 32 different cancer types, representing to the best of your knowledge the first application of a rule-based SSP in a pan-cancer context.
As the bulk of the analyzed cohorts originated from the TCGA consortium, it needs to be acknowledged that these may not be optimal for prognostic evaluations as shown previously [43], representing a potential source of bias in this study. The potential shortcomings of the TCGA cohorts became evident when comparing the three larger breast cancer cohorts: TCGA, GOBO and SCAN-B. Within these, CLAMS classification was relevant for survival outcomes in the latter two, but no significant differences could be seen in TCGA samples. While the SCAN-B cohort has been demonstrated to closely mimic a breast cancer population from southern Sweden [17], the TCGA breast cancer cohort is skewed towards tumors with more aggressive clinical features and molecular subtypes. In addition, incomplete treatment information and shorter follow-up times also represent challenges in outcome analyses.
CLAMS classification of tumors from different solid malignancies (predominantly early stage resected tumors) identified four other malignancies in which CLAMS classes appeared to have prognostic association: invasive breast cancer, lower grade glioma, renal papillary cell carcinoma and hepatocellular carcinoma. For these four cancer types, we observed a general agreement in risk prediction between CLAMS groups and existing clinical markers such as tumor grade and stage: samples classified as TRU by CLAMS (better prognosis) were often of lower tumor grade/stage, which is consistent with the fact that lower grade/stage tumors are commonly smaller and less proliferative than higher stage tumors. A perhaps more interesting observation concerns proposed molecular subtypes in the four malignancies. Here, we observed an agreement between CLAMS classification and proposed low-risk subtypes in both breast and kidney cancer, while in lower grade glioma and liver cancer better prognosis (TRU) cases according to CLAMS were observed across different molecular subtypes. This illustrates different usage potentials dependent on cancer type and is also likely a reflection of how strongly the different molecular subtypes are associated with cell proliferation. As an example, TRU-classified breast cancer samples represented a low proliferation subset of the Luminal A subtype, which has also been shown to be typically low proliferative when compared to other PAM50 subtypes [17] (Supplementary Figure S2).
However, as illustrated in Figure 3, all TRU-classified samples show similar in silico proliferation scores irrespective of malignancy type. This is an important observation for several reasons. Firstly, it demonstrates that true SSPs like CLAMS can be applied pan-cancer and also across many different technical platforms as CLAMS-like predictors are not dependent on different pre-processing steps, e.g. normalization, scaling or gene centering. Secondly, the observation that 99.1% of TRU classified non-LUAD cancers were comprised within the in silico proliferation range of TRU samples from LUAD is a powerful indicator of the potential of tuning SSPs to identify specific levels of, e.g. tumor proliferation. Since CLAMS was trained on gene-expression profiles from LUAD samples based on an existing molecular subtype classification [44], it becomes intrinsically adjusted to the biological proliferation expressed by those specific samples (which is in turn captured by the gene-expression measurements), and it is bound by the original TRU/nonTRU classification. However, based on Figure 3, it appears that tuning the SSP could change the position and width of the in silico proliferation scores associated with the TRU class by re-training within LUAD or even in a pan-cancer setting. Furthermore, we believe the constraints set by the original CLAMS training are also the reason why malignancies with high in silico proliferation scores such as uterine carcinosarcoma do not present samples classified as better prognosis by CLAMS, they simply do not reach proliferation values as low as those of TRU lung adenocarcinomas. In addition, it cannot be ruled out that cancer types may use different proliferation pathways that could lead to skewed proliferation values for genes included in a gene signature. Together, these observations are thus illustrating the limitations/applicability of CLAMS specifically in a pan-cancer context. Furthermore, these observations are also likely generalizable to general pan-cancer usefulness of rule-based SSPs when applied to malignancies other than the ones they were derived in. The observed in silico proliferation band of TRU samples in Figure 3 is also a likely explanation to why CLAMS has no treatment predictive capabilities: more aggressive diseases subjected to, e.g. neoadjuvant therapy often has higher intrinsic cell proliferation. These cases are not the ones that CLAMS identify as better prognosis given the current tuning of the predictor.
It is important to acknowledge that while many tumor types did not present any samples classified as TRU in our analysis, this does not imply that characteristics such as tumor proliferation do not carry prognostic information for these malignancies. If CLAMS would have been tuned differently, prognostic association could have been found in other cancer types, but with a tradeoff of potentially losing significance on the ones shown in this study due to dilution of low-proliferative samples with more high-proliferative ones. To test this concept, we divided each malignancy/data set into three proliferation subsets based on the in silico scores shown in Figure 3 and analyzed the survival of each group. Reassuringly, the malignancies significantly stratified by CLAMS showed differences also between proliferation groups, along with five additional cancer types. Moreover, these malignancies were mainly low proliferative in silico when considered pan-cancer, suggesting that proliferation itself may not be the main prognostic gene expression component above some threshold. Consistently, other transcriptional programs representing tumorigenic processes and/or the tumor microenvironment, such as expression of genes associated with immune response, have been shown to be prognostic and/or predictive in several malignancies (e.g. malignant melanoma) [14, 45–48].
Taken together, the results presented in this study lead us to postulate that SSPs similar to CLAMS could likely be trained and tuned using a pan-cancer data set to find relevant survival groups in specific cancer types and patient subgroups thereof. This novel pan-cancer training approach for true SSP methods would be especially interesting for malignancies that have a low number of profiled samples available, which limits the possibility for appropriate usage of large training and validation cohorts in predictor development. In addition, whether working with in silico tumor proliferation values or any other trait, it would be recommended to include samples from multiple batches, platforms and cohorts in the training step, as done for CLAMS [13] according to previous suggestions [12].
In summary, we tested a true single sample prognosis predictor developed for lung adenocarcinoma in multiple malignancies through a pan-cancer approach. We demonstrate that a lung cancer gene signature has prognostic capability in other cancer types, a finding that likely extends to other SSPs that involve a prognostic component related to cell proliferation developed in other malignancies. Extending the results beyond a single gene-expression signature, we showcase the potential of using the existing wealth of public genomic data to facilitate future development of SSPs in malignancies with currently less data available. Pan-cancer studies such as ours take us one step closer to understanding how gene-expression-based SSPs act, which gene-expression programs might be important in different malignancies, and how to derive tools useful for prognostication that are efficient across organs.
Key Points
Single sample predictors (SSPs) developed for predicting prognosis in one malignancy can be useful in other malignancies given that they capture biological components important for prognosis in both.
Classifier for lung adenocarcinoma molecular subtypes, an SSP developed to predict prognosis in lung adenocarcinoma, identifies tumors of generally lower proliferation when considered pan-cancer, which is verified for instance for breast cancer.
SSPs can potentially be trained on data from multiple cancer types to separate specific groups in a target malignancy that has less samples available.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Shamik Mitra and Martin Lauss at the Division of Oncology and Pathology for providing malignant melanoma data sets.
Deborah F. Nacer is a PhD candidate at Lund University, Sweden. Her research is focused on bioinformatics aspects of lung and breast cancer multi-omics.
Helena Liljedahl, PhD, is an assistant researcher at Lund University, Sweden. Her research is focused on developing algorithms for prediction of molecular subtypes in lung cancer.
Anna Karlsson, PhD, is an assistant researcher at Lund University, Sweden. Her research is focused on genomic characterization of lung cancer.
David Lindgren, PhD, is assistant researcher at Lund University, Sweden. His research is focused on genomic characterization of kidney cancer.
Johan Staaf, PhD, is an associate professor of experimental oncology at Lund University, Sweden. His research is focused on translational breast and lung cancer genomics.
Contributor Information
Deborah F Nacer, Lund University, Sweden.
Helena Liljedahl, Lund University, Sweden.
Anna Karlsson, Lund University, Sweden.
David Lindgren, Lund University, Sweden.
Johan Staaf, Lund University, Sweden.
Funding
Swedish Cancer Society; the Sjöberg Foundation; the Fru Berta Kamprad Foundation; BioCARE, a Strategic Research Program at Lund University; Gustav V:s Jubilee Foundation and The National Health Services (Region Skåne/ALF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cardoso F, van't Veer LJ, Bogaerts J, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med 2016;375:717–29. [DOI] [PubMed] [Google Scholar]
- 2. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26. [DOI] [PubMed] [Google Scholar]
- 3. Filipits M, Nielsen TO, Rudas M, et al. The PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancer. Clin Cancer Res 2014;20:1298–305. [DOI] [PubMed] [Google Scholar]
- 4. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research N . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research N . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ringner M, Staaf J. Consensus of gene expression phenotypes and prognostic risk predictors in primary lung adenocarcinoma. Oncotarget 2016;7:52957–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst 2010;102:464–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang H, Wang S, Xiao G, et al. Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies. Ann Oncol 2017;28:733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ringner M, Jonsson G, Staaf. Prognostic and chemotherapy predictive value of gene-expression phenotypes in primary lung adenocarcinoma. Clin Cancer Res 2016;22:218–29. [DOI] [PubMed] [Google Scholar]
- 12. Cirenajwis H, Lauss M, Planck M, et al. Performance of gene expression-based single sample predictors for assessment of clinicopathological subgroups and molecular subtypes in cancers: a case comparison study in non-small cell lung cancer. Brief Bioinform 2020;21:729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liljedahl H, Karlsson A, Oskarsdottir GN, et al. A gene expression-based single sample predictor of lung adenocarcinoma molecular subtype and prognosis. Int J Cancer 2021;148:238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle 2006;5:2198–202. [DOI] [PubMed] [Google Scholar]
- 15. Ringner M, Fredlund E, Hakkinen J, et al. GOBO: gene expression-based outcome for breast cancer online. PLoS One 2011;6:e17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Staaf J, Ringner M, Vallon-Christersson J, et al. Identification of subtypes in human epidermal growth factor receptor 2--positive breast cancer reveals a gene signature prognostic of outcome. J Clin Oncol 2010;28:1813–20. [DOI] [PubMed] [Google Scholar]
- 17. Vallon-Christersson J, Hakkinen J, Hegardt C, et al. Cross comparison and prognostic assessment of breast cancer multigene signatures in a large population-based contemporary clinical series. Sci Rep 2019;9:12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol 2006;24:4236–44. [DOI] [PubMed] [Google Scholar]
- 19. Tabchy A, Valero V, Vidaurre T, et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res 2010;16:5351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iwamoto T, Bianchini G, Booser D, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst 2011;103:264–72. [DOI] [PubMed] [Google Scholar]
- 21. Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with Nivolumab. Cell 2017;171:934–49e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gide TN, Quek C, Menzies AM, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 2019;35:238–55 e236. [DOI] [PubMed] [Google Scholar]
- 23. Liu D, Schilling B, Liu D, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25:1916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. R Core Team . 2020. R: A Language and Environment for Statistical Computing. Version 3.6.3. https://www.R-project.org/.
- 25. Kassambara A, Kosinski M, Biecek P. 2020. Survminer: Drawing Survival Curves Using 'ggplot2'. R Package Version 0.4.8. https://CRAN.R-project.org/package=survminer.
- 26. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol 1995;57:289–300. [Google Scholar]
- 27. Karlsson A, Jonsson M, Lauss M, et al. Genome-wide DNA methylation analysis of lung carcinoma reveals one neuroendocrine and four adenocarcinoma epitypes associated with patient outcome. Clin Cancer Res 2014;20:6127–40. [DOI] [PubMed] [Google Scholar]
- 28. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 29. Falato C, Tobin NP, Lorent J, et al. Intrinsic subtypes and genomic signatures of primary breast cancer and prognosis after systemic relapse. Mol Oncol 2016;10:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caan BJ, Sweeney C, Habel LA, et al. Intrinsic subtypes from the PAM50 gene expression assay in a population-based breast cancer survivor cohort: prognostication of short- and long-term outcomes. Cancer Epidemiol Biomarkers Prev 2014;23:725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weigelt B, Mackay A, A'Hern R, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol 2010;11:339–49. [DOI] [PubMed] [Google Scholar]
- 33. Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 2006;7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016;131:803–20. [DOI] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower grade gliomas. N Engl J Med 2015;372:2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu F, Li GZ, Liu HJ, et al. Molecular subtyping reveals immune alterations in IDH wild-type lower grade diffuse glioma. J Pathol 2020;251:272–83. [DOI] [PubMed] [Google Scholar]
- 37. Chen F, Zhang Y, Senbabaoglu Y, et al. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep 2016;14:2476–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee SH, Yim SY, Shim JJ, et al. Molecular subtypes and genomic signatures of hepatocellular carcinoma for prognostication and therapeutic decision-making. In: Hoshida Y (ed). Hepatocellular Carcinoma: Translational Precision Medicine Approaches. Cham (CH): Humana Press. 2019, 109–23. [PubMed] [Google Scholar]
- 39. Cancer Genome Atlas Research N . Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–41e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang X, Yang C, Wang J, et al. Integrative analysis of DNA methylation and gene expression reveals distinct hepatocellular carcinoma subtypes with therapeutic implications. Aging (Albany NY) 2020;12:4970–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226–39e1224. [DOI] [PubMed] [Google Scholar]
- 42. Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004;40:667–76. [DOI] [PubMed] [Google Scholar]
- 43. Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilkerson MD, Yin X, Walter V, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One 2012;7:e36530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Das S, Camphausen K, Shankavaram U. Cancer-specific immune prognostic signature in solid tumors and its relation to immune checkpoint therapies. Cancers (Basel) 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jonsson G, Busch C, Knappskog S, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res 2010;16:3356–67. [DOI] [PubMed] [Google Scholar]
- 47. Harbst K, Staaf J, Lauss M, et al. Molecular profiling reveals low- and high-grade forms of primary melanoma. Clin Cancer Res 2012;18:4026–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cabrita R, Lauss M, Sanna A, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All gene-expression data used in the current study and any sample information are available through original studies. CLAMS classification of all samples is available as Supplementary Table S3. R code used in the analyses conducted in this study can be found at https://github.com/StaafLab/CLAMS-pan-cancer.


