Abstract
We cloned and characterized a phosphatidylcholine-hydrolyzing phospholipase C (PC-PLC) gene from Burkholderia pseudomallei. DNA sequence analysis of the gene indicated an open reading frame coding for 700 amino acids with a 34-amino-acid signal peptide. When cleaved, this yields a secreted 73-kDa mature protein. The deduced amino acid sequence exhibited 48% similarity to that of a nonhemolytic PLC from Pseudomonas aeruginosa. The expressed PC-PLC was heat stable, nonhemolytic for sheep erythrocytes, and active between pH 2 and 8. Western blot analysis with sera from melioidosis patients indicated that they produced immunoglobulin M antibodies against this PC-PLC protein.
Melioidosis is a potentially fatal disease that is endemic in northern Australia and in Southeast Asia (particularly in northeastern Thailand) (5, 6). The average incidence of human melioidosis in Ubon Ratchatani, northeastern Thailand during the period 1987 through 1991 was 4.4 cases per 100,000 inhabitants (22). The causative agent is a gram-negative bacillus, Burkholderia pseudomallei (formerly known as Pseudomonas pseudomallei) (25). The most common clinical manifestation of this disease is pneumonia. In severe cases, patients die within 48 h of the onset of symptoms (1).
Phospholipases of the C type (PLC) are enzymes which cleave the phosphodiester bond of phospholipids to yield diacylglycerol and a water-soluble phosphate ester. B. pseudomallei, unlike most gram-negative bacteria, produces a PLC which results in a zone of opalescence around colonies grown on egg yolk emulsion-supplemented agar (2, 9, 23). Previous studies have implicated PLCs as virulence factors involved in infections by pathogenic bacteria such as Listeria monocytogenes (16, 21), Clostridium perfringens (19), and Pseudomonas aeruginosa (14, 15). In L. monocytogenes pathogenesis, two different PLC enzymes play a role in escape of the pathogen from the phagosome membrane and invasion of adjacent cells (16, 21).
This study characterized the gene encoding a nonhemolytic PLC from a strain of B. pseudomallei. The biological properties of the PLC and its similarity to a PLC from P. aeruginosa were examined.
Cloning and expression of the B. pseudomallei PLC gene.
B. pseudomallei SptI was isolated from the sputum of a patient by the Division of Bacteriology, Department of Microbiology, Faculty of Medicine Siriraj Hospital, Bangkok, Thailand. DNA was subsequently isolated by the method of Ausubel et al. (3). The isolated genomic DNA was digested with restriction enzymes (EcoRI, PstI, SacI, SacII, and XhoI), and the digests were hybridized with a PLC-specific probe. The PLC-specific probe (600 bp) was generated by using genomic DNA of P. aeruginosa as the template with PCR primers 5′ CGACATTCCCTACTAC 3′ (PLC-L) and 5′ CGCCGGCGGTGCTGAC 3′ (PLC-R), designed from the P. aeruginosa hemolytic PLC gene (GenBank accession no. M13047), nucleotide positions 455 to 470 and 1164 to 1179, respectively. The PCR was carried out on a thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) for 35 cycles of melting (94°C, 1 min), annealing (50°C, 1 min), and extension (72°C, 2 min). The PCR product was labeled with fluorescein-dUTP according to protocol of the Fluorescein Gene Images labeling system (Amersham International plc, Little Chalfont, Buckinghamshire, England). Hybridization was performed at 62°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (20) in hybridization buffer for 16 h. The stringent wash performed with 0.5× SSC containing 0.1% sodium dodecyl sulfate at 62°C, and the hybridized probe was detected by using the Fluorescein Gene Images detection system (Amersham). A 4.4-kb EcoRI DNA fragment of B. pseudomallei that hybridized with the probe was inserted into the EcoRI site of the pKSII(−) vector (Stratagene, Heidelberg, Germany) and introduced into Escherichia coli DH5α. The recombinant plasmid obtained was designated pSN-1, and the cloned fragment was designated SN-1. A restriction map of SN-1 is shown in Fig. 1. The assay used for detection of phosphatidylcholine-hydrolyzing PLC (PC-PLC) activity has been described previously (4, 10) and is based on enzymatic hydrolysis of p-nitrophenylphosphorylcholine (NPPC; Sigma Chemical Company) to liberate phosphatidylcholine and the yellow chromogenic compound p-nitrophenol. Only samples with PC-PLC activity turned bright yellow after incubation with the NPPC substrate (0.25 M Tris-HCl [pH 7.2], 6% glycerol, 1.0 μM ZnCl2, and 0.01 M NPPC). NPPC hydrolysis was detected in culture supernatants and cell lysates of E. coli carrying pSN-1 (data not shown), indicating that pSN-1 carried the PC-PLC gene from B. pseudomallei. PC-PLC activity was first detected at the beginning of exponential growth (2 h), and it increased continuously thereafter until the stationary phase (16 h) (data not shown). In every assay, positive and negative controls (E. coli harboring plasmids pDR540 and pKSII(−), respectively) were included. Plasmid pDR540 (14) contained the gene encoding the hemolytic PLC from P. aeruginosa and was kindly provided by M. L. Vasil (University of Colorado Health Sciences Center, Denver).
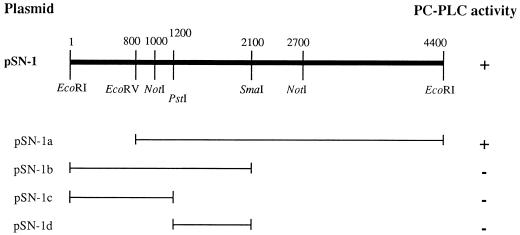
FIG. 1.
Restriction map of the plasmid clone pSN-1 and its derivatives. The presence (+) or absence (−) of PC-PLC activity is also indicated. Plasmids pSN-1a, -1b, -1c, and -1d were derived from pSN-1 by restriction enzyme digestion at the sites indicated in the map (in nucleotides) and cloned into pKSII(−).
The cloning vector pSN-1 contained an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. When induced by IPTG, the culture supernatants from E. coli harboring pSN-1 showed PC-PLC activity. Activity was also present without IPTG induction (data not shown), suggesting that the SN-1 insert carried a promoter that was recognized by the E. coli RNA polymerase. When pSN-1 plasmid subclones were constructed and assayed for PC-PLC activity, only pSN-1a with insert SN-1a exhibited PC-PLC activity (Fig. 1). Thus, it appeared that the PC-PLC gene was located between EcoRV and EcoRI restriction sites.
DNA sequence analysis.
The nucleotide sequence of the 3.6-kb EcoRV-EcoRI fragment SN-1a containing the PC-PLC gene was determined by using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Inc., Foster City, Calif.). Analysis of the data revealed an open reading frame of 2,100 bp which encoded a protein of 700 amino acids. A potential ribosome binding site, AGGAAG, was identified 7 bp upstream of an ATG codon. The gene for PC-PLC on fragment SN-1a had a G+C content of 68%, which closely resembled that estimated for chromosomal DNA from B. pseudomallei (69%) (18). A putative signal peptide sequence (34 amino acids in length) with a polar C-terminal region that ended with the sequence Ala-Leu-Ala, 9 amino acids after the hydrophobic core, was identified. Signal peptidase cleavage was expected to occur at this position. Comparison of the putative amino acid sequence encoded by the PC-PLC open reading frame with amino acid sequence data deposited in the GenBank database revealed 48 and 44% similarity to the nonhemolytic and the hemolytic PLCs from P. aeruginosa, respectively.
Further comparison of the P. aeruginosa (13) and B. pseudomallei nonhemolytic PLCs revealed several similar properties. The entire proposed signal peptide from B. pseudomallei comprised 34 amino acids, close to the 35-amino-acid signal peptide in P. aeruginosa (13) and longer than that usual procaryotic signal sequences of 20 or 23 residues (8, 24). The B. pseudomallei sequence also contains the amino acid phenylalanine, as does the P. aeruginosa sequence but usually not other procaryotic signal sequences (13). In contrast, the predicted pI values of B. pseudomallei and P. aeruginosa nonhemolytic PLCs were quite different. That of P. aeruginosa was 8.8 (basic protein) (13), whereas that of B. pseudomallei was 6.7 (acidic protein). The difference could be explained by the smaller number of lysine and arginine residues in the B. pseudomallei PC-PLC (data not shown).
Biological properties of the PC-PLC protein.
PLCs can be classified as hemolytic or nonhemolytic depending on their ability to lyse sheep erythrocytes. The hemolytic activity was tested as previously described (12). Culture supernatants and cell lysates of E. coli harboring pSN-1a did not lyse sheep erythrocytes but did hydrolyze NPPC to liberate a yellow chromogen (data not shown), indicating that pSN-1a encoded a nonhemolytic PC-PLC. The result also suggested that this enzyme cannot hydrolyze sphingomyelin because the major phospholipid components of the outer leaflet of the erythrocyte membrane are phosphatidylcholine and sphingomyelin.
The stability of the expressed PC-PLC was studied by incubating the culture supernatant of clone pSN-1a at different temperatures (4, 37, 65, and 80°C) for different periods of time (1, 3, 5, and 24 h). It was heat stable at 65°C, but heating to 80°C for 15 min caused a 100% loss of activity. With respect to the effect of pH, the PC-PLC enzyme functioned from pH 2 to 8 but activity was lost above pH 8. NPPC hydrolysis under acidic conditions was not due to nonspecific hydrolysis, since the negative control (NPPC mixed with culture broth) did not give a positive reaction. However, we cannot rule out the possibility that the lack of activity above pH 8 was due to instability of the NPPC substrate under alkaline conditions.
PLC gene homology.
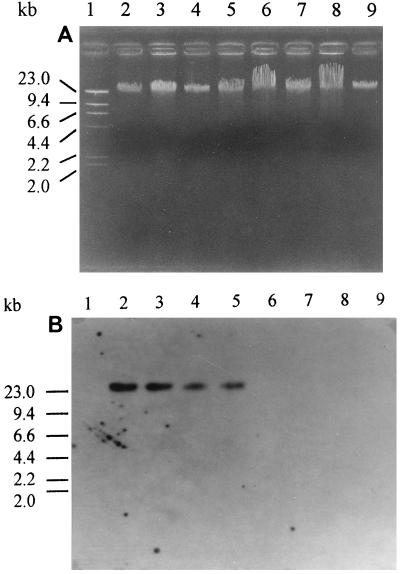
The relatedness of the PC-PLC gene from B. pseudomallei to those of other gram-positive and gram-negative bacteria was analyzed by Southern hybridization under medium-stringency conditions. A labeled 4.4-kb EcoRI fragment derived from the pSN-1 clone was used as the DNA probe. The probe hybridized with genomic DNAs from B. mallei, B. cepacia, and P. aeruginosa but not with those from L. monocytogenes, C. perfringens, or Bacillus cereus (Fig. 2). These data suggested that there is significant DNA similarity between the B. pseudomallei PC-PLC gene and genes of gram-negative bacteria but not genes of gram-positive bacteria.
FIG. 2.
(A) Agarose gel electrophoresis of genomic DNA from various PLC-producing bacteria. (B) Southern blot of DNA from panel A, probed with the labeled 4.4-kb DNA insert from pSN-1. Lanes: 2, B. pseudomallei; 3, B. mallei; 4, B. cepacia; 5, P. aeruginosa; 6, L. monocytogenes; 7, C. perfringens; 8, Bacillus cereus. A standard lambda/HindIII DNA marker and Salmonella paratyphi A genomic DNA (negative control) were included in lanes 1 and 9, respectively. The positions of molecular size markers are shown on the left of both panels.
Serum reactivity to PC-PLC protein.
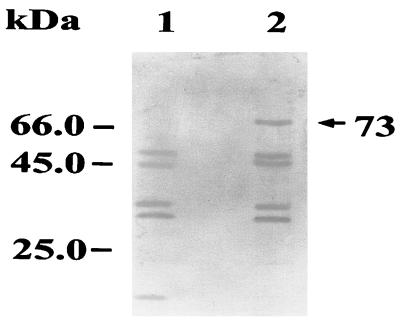
Cell lysates of E. coli harboring the plasmid pSN-1a or the vector control were electrophoresed, electroblotted (11) onto a nitrocellulose membrane (Micron Separation Inc., Wesborough, Mass.), and incubated with pooled sera from 10 melioidosis patients (a generous gift from S. Sirisinha, Chulabhorn Research Institute, Bangkok, Thailand), diluted 1:200. All of the 10 serum samples were obtained from septicemic patients in the area in which B. pseudomallei is endemic (Khon Kaen province, northeastern Thailand). Antibodies that bound to the blotted proteins were detected by using 1:500 dilutions of alkaline phosphatase-conjugated rabbit anti-human immunoglobulin M (IgM; Sigma). Positive reactions were visualized by the development of a red-purple color after addition of the substrate (7). It was found that a 73-kDa band was recognized in E. coli harboring pSN-1a but not in E. coli harboring the pKSII(−) control (Fig. 3). This result indicated that the molecular mass of the PC-PLC expressed from pSN-1a was 73 kDa and that it could induce IgM antibody production in melioidosis patients.
FIG. 3.
Western blot showing expression of recombinant PC-PLC in E. coli. Lanes: 1, E. coli carrying pKSII(−) without an insert; 2, E. coli containing the PC-PLC gene from B. pseudomallei. The blot was reacted with pooled sera from 10 melioidosis patients and then reacted with alkaline phosphatase-conjugated rabbit anti-human IgM and a chromogenic substrate. The numbers on the left correspond to the positions of molecular size markers. The arrow indicates the expressed 73-kDa PC-PLC protein.
Several features of melioidosis suggest that B. pseudomallei is a facultative intracellular pathogen. After the initial phase of infection, B. pseudomallei may persist in a dormant stage in macrophages for years (17). The mechanism by which it survives within human phagocytes is not known. In L. monocytogenes, phosphatidylinositol-hydrolyzing PLC and PC-PLC have been shown to be virulence factors involved in intracellular survival and cell-to-cell spread (16, 21). B. pseudomallei PC-PLC might play a similar role. The demonstration that PC-PLC of B. pseudomallei is expressed in melioidosis patients and is active even under acidic conditions provides sufficient grounds for further studies on its possible role as a virulence factor.
Nucleotide sequence accession number.
The complete nucleotide sequence of the PC-PLC gene of B. pseudomallei has been deposited in the GenBank database under accession no. AF107252.
Acknowledgments
This work was supported by grant 75-348-227 from the Siriraj-China Medical Board.
Computer analysis of the DNA sequences was accomplished with the kind support of M. Juricek (Institute of Science and Technology for Research and Development, Mahidol University, Bangkok, Thailand). We thank T. W. Flegel for critical reading of the manuscript.
REFERENCES
- 1.Ashdown L R, Johnson R W, Koehler J M, Cooney C A. Enzyme-linked immunosorbent assay for the diagnosis of clinical and subclinical melioidosis. J Infect Dis. 1989;160:253–260. doi: 10.1093/infdis/160.2.253. [DOI] [PubMed] [Google Scholar]
- 2.Ashdown L R, Koehler J M. Production of hemolysin and other extracellular enzymes by clinical isolates of Pseudomonas pseudomallei. J Clin Microbiol. 1990;28:2331–2334. doi: 10.1128/jcm.28.10.2331-2334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1987. pp. 2.4.1–2.4.5. [Google Scholar]
- 4.Berka R M, Gray G L, Vasil M L. Studies of phospholipase C (heat-labile hemolysin) in Pseudomonas aeruginosa. Infect Immun. 1981;34:1071–1074. doi: 10.1128/iai.34.3.1071-1074.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaowagul W, White N J, Dance D A B, Wattanagoon Y, Naigowit P, Davis T M E, Looareesuwan S, Pitawatchara N. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 6.Dance D A B. Melioidosis: the tip of the iceberg? Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekpo P, Sarasombath S, Banchuin N, Pongsunk S, Korbsrisate S, Sirisinha S. Monoclonal antibodies against protein antigens of salmonellae causing paratyphoid fever and their diagnostic application. Asian Pac J Allergy Immunol. 1995;13:63–70. [PubMed] [Google Scholar]
- 8.Emr S D, Silhavy J. Importance of secondary structure in the signal sequence for protein secretion. Proc Natl Acad Sci USA. 1983;80:4599–4603. doi: 10.1073/pnas.80.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esselmann M T, Liu P V. Lecithinase production by gram-negative bacteria. J Bacteriol. 1961;81:939–945. doi: 10.1128/jb.81.6.939-945.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurioka S, Matsuda M. Phospholipase C assay using p-nitrophenylphosphorylcholine together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal Biochem. 1976;75:281–289. doi: 10.1016/0003-2697(76)90078-6. [DOI] [PubMed] [Google Scholar]
- 11.Laemli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–688. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Nakazawa T, Yamada Y, Ishibashi M. Characterization of hemolysin in extracellular products of Pseudomonas cepacia. J Clin Microbiol. 1987;25:195–198. doi: 10.1128/jcm.25.2.195-198.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostroff R M, Vasil A I, Vasil M L. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J Bacteriol. 1990;172:5915–5923. doi: 10.1128/jb.172.10.5915-5923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostroff R M, Wretlind B, Vasil M L. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect Immun. 1989;57:1369–1373. doi: 10.1128/iai.57.5.1369-1373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plotkowski M C, Meirelles M N. Concomitant endosome-phagosome fusion and lysis of endosomal membranes account for Pseudomonas aeruginosa survival in human endothelial cells. J Submicrosc Cytol Pathol. 1997;29:229–237. [PubMed] [Google Scholar]
- 16.Portnoy D A, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruksachartvuthi S, Aswapokee N, Thankerngpol K. Survival of Pseudomonas pseudomallei in human phagocytes. J Med Microbiol. 1990;31:109–114. doi: 10.1099/00222615-31-2-109. [DOI] [PubMed] [Google Scholar]
- 18.Redfearn M S, Palleroni N J, Stanier R Y. A comparative study of Pseudomonas pseudomallei and Bacillus mallei. J Gen Microbiol. 1966;43:293–313. doi: 10.1099/00221287-43-2-293. [DOI] [PubMed] [Google Scholar]
- 19.Saint-Joanis B, Garnier T, Cole S. Gene cloning shows the alpha-toxin of Clostridium perfringens to contain both sphingomyelinase and lecithinase activities. Mol Gen Genet. 1989;219:453–460. doi: 10.1007/BF00259619. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suputtamongkol Y, Hall A J, Dance D A B, Chaowagul W, Rajchanuvong A, Smith M D, White N J. The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int J Epidemiol. 1994;23:1082–1090. doi: 10.1093/ije/23.5.1082. [DOI] [PubMed] [Google Scholar]
- 23.Titball R W. Bacterial phospholipases C. Microbiol Rev. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 25.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]