Abstract
Stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most widespread and destructive fungal diseases of wheat worldwide. The cultivation and growth of resistant wheat varieties are the most economical, effective, and environmental friendly methods to control stripe rust. Therefore, it is necessary to use new resistance genes to breed resistant wheat varieties. A single dominant gene temporarily designated as YrM8664-3, from a wheat-Leymus mollis introgression line M8664-3 highly resistant to Chinese predominant Pst races, is a potentially valuable source of stripe rust resistance for breeding. Herein, based on previous YrM8664-3 chromosome location results (bin 4AL13-0.59-0.66 close to 4AL12-0.43-0.59) and expression change information of candidate genes and bioinformatics analysis, several candidate genes with significantly different expression changes were then selected and verified by virus-induced gene silencing (VIGS). Two of the candidate genes temporarily designated as TaFBN [containing plastid lipid-associated proteins (PAP)_fibrillin domain in its protein] and Ta_Pes_BRCT [containing Pescadillo and breast cancer tumour suppressor protein C-terminus (BRCT) domain in its protein], produced the most significant resistance changes in the wheat-Pst interaction system after silencing. These two genes were further verified by Agrobacterium-mediated wheat genetic transformation technology. According to the identification of disease resistance, the resistance function of the candidate gene TaFBN was further verified. Then, the expression of TaFBN under hormone treatment indicated that TaFBN may be related to the salicylic acid (SA) and abscisic acid (ABA) signaling pathways. Combined with the expression of TaFBN in response to environmental stress stimulation, it can be reasonably speculated that TaFBN plays an important role in the resistance of wheat to Pst and is involved in abiotic stress pathways.
Keywords: functional verification, stripe rust, resistance, wheat-Leymus mollis, YrM8664-3
Introduction
Wheat (Triticum aestivum L.) is one of the most important food crops in the world. Wheat stripe rust, caused by Pst, is one of the most important diseases that affects wheat production. It has the characteristics of rapid outbreaks that cause regional epidemics and subsequent extensive and severe harm to wheat crops, and it has been found in almost all major wheat-producing areas in the world. When wheat is susceptible to Pst infection, the yield loss is ~10–20%, but it can exceed 50% or even result in no harvest in severe cases (Wan et al., 2007). Since the 1950s, several nationwide stripe rust epidemics have been recorded in China, among which, a yield loss of 6, 3.2, 2.65, and 1.3 billion kg of wheat occurred in 1950, 1964, 1990, and 2002, respectively (Wan et al., 2004). In 2017, wheat stripe rust was once again epidemic throughout China, affecting an area of 5.56 million hm2, which was the largest annual occurrence since 2002 (Huang et al., 2018). The most effective, economical, and environmental friendly method to control stripe rust is to breed and grow disease-resistant wheat varieties.
Disease resistance in wheat varieties, like all plants, often depends on pattern-triggered immunity (PTI) brought about by microbial patterns via pattern-recognition receptors (PRRs) localized on cell surfaces, and effector-triggered immunity (ETI) activated by pathogen effector proteins via predominantly intracellular localized receptors called nucleotide-binding, leucine-rich repeat receptors (NLRs) (Jones and Dangl, 2006; Cui et al., 2015; Yu et al., 2017; Yuan et al., 2021). The disease resistance reaction produced by plants is a complex and orderly process, and is also regulated by multiple genes, especially resistance (R) genes (Jia et al., 2000; Dodds et al., 2006; Luo et al., 2011).
By analyzing protein sequences encoded by cloned R genes, it was found that R genes targeted at different sources and pathogens possessed similar characteristic domains, such as nucleotide binding site (NBS), leucine-rich repeat (LRR), toll-interleukin-1 receptor (TIR), coiled-coil (CC), protein kinase (PK), and the transmembrane domain (TM) (Jones et al., 2016; Monteiro and Nishimura, 2018; Ma et al., 2020). In addition, plant disease resistance is closely related to hormone signal transduction and environmental stress stimulation (Denance et al., 2013; Derksen et al., 2013). At present, salicylic acid (SA), jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), and other signaling molecules involved in plant disease resistance have been extensively studied (Schenk et al., 2000; Denance et al., 2013; Derksen et al., 2013). Therefore, studies on plant disease resistance genes will assist in expanding our understanding of the mechanism of resistance at a deeper level, and will provide some reference for disease control.
Histopathological studies on the interaction between wheat and Pst are the basis for revealing the detailed process of Pst infection and host resistance. Kang et al. (2002) found that the growth of a pathogenic fungus was inhibited in resistant varieties compared with that in susceptible varieties. Reactive oxygen species (ROS) bursting is one of the fastest and most effective disease resistance reactions in the interaction between plants and their pathogens. By dyeing leaf tissues with diaminobenzidine (DAB), Wang et al. (2007) found that ROS were produced in guard cells of both compatible and incompatible combinations after the interaction. In terms of biochemistry, the resistance of wheat varieties to Pst is mainly reflected in protective enzymes, such as superoxide dismutase (SOD) and phenylalanine ammonia-lyase (PAL), as well as the increase in activities of defensive enzymes, such as antimicrobial hydrolase and chitinase, and the increase in resistant proteins and lignin in host cell walls (Asthir et al., 2011; Zheng et al., 2020).
Virus-induced gene silencing is an RNA interference-based technology that transiently knocks down a target gene expression using modified plant viral genomes. When a targeted gene is inserted into a viral genome and a plant is inoculated with viruses, plant cells recognize the threat of the invading viruses and use protective defense mechanisms to destroy any foreign genes carried by viruses and viral vectors. Loss of function phenotype or decreased expression activity of the target gene occurs, and then the function of the target gene can be identified according to phenotypic changes (Scofield et al., 2005; Feng et al., 2015).
The identification of wheat disease-resistance genes and studies on disease-resistance mechanisms are the basis of wheat disease-resistance breeding and disease control. M8664-3 is the hybrid offspring of common wheat cultivar 7182 and wheat-related species Leymus mollis (Trin) Hara. Our previous studies have shown that the dominant gene YrM8664-3 in M8664-3 confers all-stage resistance to Chinese prevalent Pst race CYR33. YrM8664-3 was located in bin 4AL13-0.59-0.66 close to 4AL12-0.43-0.59 on chromosome 4AL and flanked by single-nucleotide polymorphism markers AX111655681 and AX109496237 with genetic distances of 5.3 and 2.3 centimorgans, respectively (Chao et al., 2018). However, because of the alien chromosome fragment that may exist in the YrM8664-3 region, it is difficult to further finely map and clone the gene. The sequencing of the entire genome of wheat variety Chinese Spring has been completed, making it possible to identify candidate genes and perform functional verification analysis of YrM8664-3 in the reference genome region corresponding to the located chromosome interval.
To identify the candidate genes involved in stripe rust resistance in M8664-3, in this study, the resistance of M8664-3 to CYR33 was investigated and then analyzed. Combined with histological and histochemical techniques, the invasion and infection processes of CYR33 on M8664-3 were studied in detail, and the resistance process of M8664-3 to CYR33 was comprehensively analyzed. Based on the physical location of YrM8664-3, the genes related to disease resistance were selected from the annotated database of the Chinese Spring genome according to the structure domains related to disease resistance and function prediction information. Then, a functional validation analysis was carried out to identify the disease-resistant genes. The finding in this study could provide a basis for wheat stripe rust resistance breeding.
Materials and Methods
Plants and Pathogens
The wheat-Leymus mollis introgression line M8664-3 used in this study was provided by Professor Jie Fu, College of Agronomy, Northwest A&F University, Yangling, China.
Chinese predominate Pst race CYR33 was used for seedling tests. After the identification on differential hosts of Chinese Pst, CYR33 was increased in susceptible variety Mingxian169.
Seedling Tests, RNA Extraction
Seedling tests were conducted under controlled greenhouse conditions according to Wan et al. (2007) and Bansal et al. (2017). M8664-3 and Mingxian 169 were planted in 7 7 7 cm pots with 15–20 seeds per pots. When the first leaves fully expanded, fresh CYR33 urediniospores were inoculated onto wheat leaves by the smear method, and sterile water was used as MOCK-inoculation control (Roelfs et al., 1992). Approximately 14–16 days post inoculation (dpi), when obvious uredinia were observed on the leaves of Mingxian169, the types of infection types were recorded according to a 0–9 scale. Also, leaf samples were collected at 0, 12, 24, 48, 72, and 96 h post inoculation (hpi). RNA extraction was performed using a Magen plant total RNA extraction kit (Magen Biotech, Guangzhou, China). The concentration and purity of each RNA sample were evaluated using a micro-ultraviolet spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific, Wilmington, DE, United States), and the integrity was determined by 1% agarose gel electrophoresis. The first strand of DNA was synthesized with HiScript II Q-RT SuperMix for qPCR (+ gDNA wiper) (Vazyme Biotech, Nanjing, China).
Candidate Gene Selection and Quantitative Real-Time PCR (Qrt-PCR) Analysis
The sequence of the linked markers of YrM8664-3 was blasted against the genome sequence of Chinese Spring IWGSC RefSeq v1.0 Genome (IWGSC et al., 2018) (https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies), and the gene was located in the range of 41.6 × 107-63.9 × 107 bp (base pair) on wheat chromosome 4AL (Chao et al., 2018). Referring to the IWGSC RefSeq v1.0 annotation (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/), genes near the linked markers containing conserved domains of disease resistance, such as NBS, LRR, and PK, or genes hit by the linked markers were selected.
To measure the transcriptional expression level of selected genes by qRT-PCR, specific primers (Supplementary Table 1) were designed using the Primer Premier 5.0 software (Li et al., 2011). Using TaEF-1α (GenBank accession number Q03033) as an internal reference gene, the relative transcription expression level of target genes was determined. All the qRT-PCR reactions were performed in a 20-μl reaction mixture containing 10 μl Cham QTM SYBR® qPCR Master Mix (Vazyme Biotech, Nanjing, China), 0.2 μl each of the forward and reverse gene-specific primers (10 μM), and 2 μl of diluted cDNA (1:10). A Bio-Rad iQ5 Real Time PCR (Bio-Rad, Hercules, CA, United States) system was used to generate cycle threshold (CT) values for the quantification of relative gene expression using the comparative 2−ΔΔCt method (Livak and Schmittgen, 2001). All the samples were analyzed in three biological replications, and all the PCR analyses were replicated three times.
Sequence Amplification, Identification, and Bioinformatics Analysis
According to the primers (Supplementary Table 2) at the positive and negative ends of the open reading frame (ORF) region, the cDNA of M8664-3 was used as the template for amplification with gene-specific primers as follows: pre-denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final incubation at 72°C for 2 min. The PCR-amplified products were examined by 1.2% agarose gel electrophoresis. The specific target bands were recovered and sequenced by Tsingke (Xi'an, China).
The online BLAST1 program from the National Center for Biotechnology Information (NCBI) was used to analyze the cDNA sequence. The amino acid sequence was analyzed with ProtParam2 and TMPred3 to detect the primary structure and position information of the polypeptide transmembrane region, respectively. TargetP-2.04 and Plant-mPLoc5 in Cell-PLoc 2.0 were used to predict the subcellular location of the possible protein structure of the amino acid sequence. The conserved domain was identified by Pfam6 and simple modular architecture research tool (SMART7). Multiple sequence alignments were performed using DNAMAN8.0 (Lynnon BioSoft, San Ramon, CA, United States). To reveal phylogenetic relationships and potential functional characteristics of targeted proteins from different species, corresponding proteins were collected from different species according to the domain, namely Arabidopsis thaliana, Oryza sativa, Zea mays, Brachypodium virgatum, and Hordeum vulgare. The phylogenetic relationship was inferred with the neighbor-joining (NJ) method, and a midpoint rooted base tree was drawn in MEGA 7.0 with 1,000 bootstrap iterations (Kumar et al., 2016).
Virus-Induced Gene Silencing in Wheat
Virus-induced gene silencing, mediated by barley stripe mosaic virus (BSMV), was performed to reveal the function of nine candidate genes during the interaction between wheat M8664-3 and CYR33 (Fitzmaurice et al., 2002; Scofield et al., 2005). The fragment of target gene with PacI and NotI was derived from its coding sequence and amplified by RT-PCR (Supplementary Table 1) to construct the BSMV:γ plasmid for gene silencing. After amplification of the target gene fragment and vector linearization, the target gene and linearized vector were recombined, connected, linearized with a restriction enzyme, and transcribed in vitro (RiboMAX TM Large-Scale RNA Production System-T7 and Ribo m7G Cap Analog; Promega, Madison, WI, United States) to form a recombinant virus containing the target gene (Petty and Jackson, 1990). BSMV:γ was used as a blank control and γ-PDS as a positive control by friction inoculation, and the second leaf of wheat seedlings was infected with BSMV, with three biological replicates for each gene.
After 24-h incubation in the dark in an artificial climate incubator, the seedlings were placed in a growth chamber at 25°C with 60–80% humidity. When the photo-bleaching phenotype was observed on the BSMV: γ-PDS plants at ~10 dpi, the fourth leaves of γ-gene silencing plants were inoculated with CYR33. Leaf samples were collected at 0, 24, 48, 96, and 120 hpi for qRT-PCR analysis and histological observation (Li et al., 2011). The infection phenotype of Pst was observed at ~14 dpi.
Histological Observation and Statistical Analysis
Inoculated and control leaves of both Pst seedling test and VIGS-induced gene silencing were sampled at different time points for histological observation. For each time point, ~15 blades were sampled (Ayliffe et al., 2011). The H2O2 burst for ROS was observed by DAB (MP Biomedicals, Solon, OH, United States) staining (Xiao et al., 2003; Zou et al., 2018). WGA-Alexa 488 (Invitrogen, Carlsbad, CA, United States) was used to fluorescently stain the Pst infestation structure in wheat leaf tissue. The infestation site was determined by the production of germ tubes by Pst and the formation of substomatal vesicle in the stomata. For each treatment, 50 infection sites were randomly examined using an Olympus BX-53 microscope (Olympus Corporation, Tokyo, Japan), and the area of ROS, hypha branches, hypha length, and necrotic areas were observed and measured.
The standard error of deviation was calculated using Microsoft Excel. The statistical significance was evaluated by Student's t test (P < 0.05) using the SPSS software (SPSS, Inc., Chicago, IL, United States).
Wheat Transformation and Functional Verification
Agrobacterium tumefaciens strain EHA105 harboring binary vector, pCAMBIA3301, was used to optimize the transformation system, with ubiquitin promoter and targeted gene replacing cauliflower mosaic virus 35S promoter and GUS gene encoding β-glucuronidase. The ORF of TaFBN and Ta_Pes_BRCT was amplified separately and inserted into the frame of an expression cassette within the T-DNA region of the pCAMBIA3301 vector digested with BamHI and SpeI. The construct was verified by DNA sequencing and introduced into Agrobacterium EHA105. Then, the Agrobacterium-mediated transformation method was applied to genetically transform the target gene (Li et al., 2019). The seeds of 14-day-pollinated immature wheat variety Fielder (scutellum size 1 mm) were treated with 75% alcohol for 30 s and 0.1% HgCl for 10 min, and the immature embryos were then removed with a dissecting needle on an aseptic work table. The immature embryos were infected with the obtained Agrobacterium, and then placed on the screening medium and co-cultured in darkness at 23°C for 3 days. Under a microscope, hypocotyls were excised from the contact between the hypocotyl and scutellum of the seeds, and the obtained scutellums were cultured on the screening medium for 14 days. After cutting the callus, a second selection was performed, and the tissue was cultured for 14 days. The healthy callus was transferred to a regeneration medium, and regenerated plantlets were obtained after 7 days. The 786-bp genomic fragment of TaFBN and the 1,791-bp genomic fragment of Ta_Pes_BRCT were introduced into wheat cultivar Fielder, and the T0 generation plants were screened with gene-specific primers by PCR amplification under the follow conditions: 1 min pre-denaturation at 94°C, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, elongation at 72°C for 45 s, and a final extension step of 10 min at 72°C. Then, the T2 generation plants derived from the positive T0 plant progeny were inoculated with CYR33 for functional analysis of the target genes.
Abiotic and Hormone Stress Treatments
In order to evaluate the expression level of TaFBN in wheat M8664-3 under hormone perception and environmental stress conditions, seedlings of M8664-3 were divided into eight groups. The first four groups were sprayed with methyl jasmonate (MeJA), ethylene (ET), salicylic acid (SA), or abscisic acid (ABA), at 100 mM each as a hormone treatment and cultivated at 16°C. The last four groups were treated with low temperature (4°C), high temperature (37°C), salt (200 mM NaCl), and drought (15% PEG6000) to experience environmental stress, and sterile water was sprayed at 16°C as a blank control. Leaf samples were collected for the expression level analysis of TaFBN 0, 1, 3, 6, 12, and 24 h after the different stimulus treatments.
Results
ROS Is Involved in Wheat Cultivar M8664-3 Resistance Against CYR33
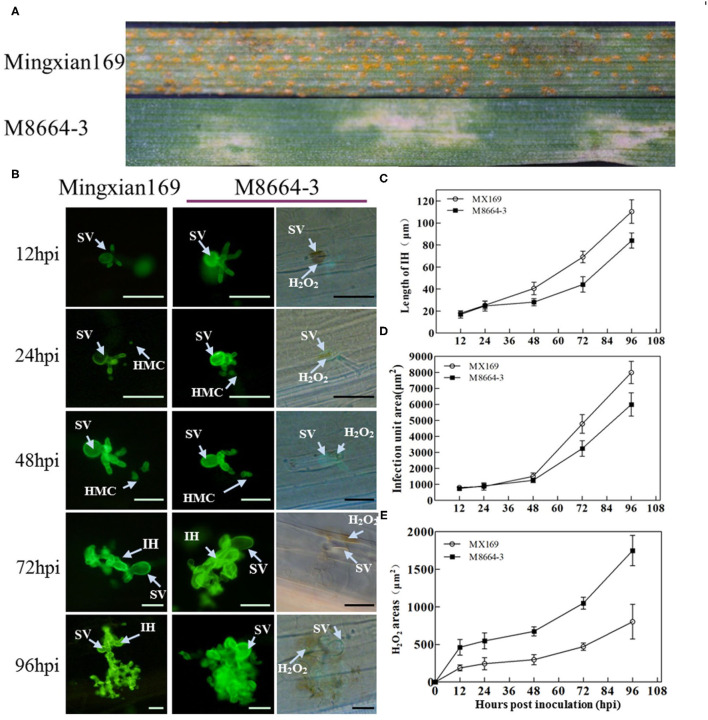
The infection type (IT) of wheat M8664-3 to CYR33 was 2 with large areas of necrosis on the leaves, whereas the IT of susceptible wheat Mingxian169 was 9 with fully expanded uredinium covered on the leaves at 15 dpi (Figure 1A).
Figure 1.
(A) Symptoms of wheat-Leymusmollis introgression line M8664-3 and susceptible control cultivar Mingxian169 after inoculation with Puccinia striiformis f. sp. tritici (Pst) race CYR33. (B) Wheat germ agglutinin (WGA) was used to stain the leaves to visualize pathogens. SV, substomatal vesicle; IH, infection hyphae; HMC, haustorial mother cell; ROS, reactive oxygen species. (C) Hyphal lengths were measured at different hpi. (D) Infection unit area at different hpi. (E) H2O2 area at different hpi. Error bars represent the standard deviations of three independent samples. Bar, 50 μm.
Histological observation showed that significant DAB staining appeared at the leaf infection sites at 12 and 96 hpi (Figure 1B). At 12 hpi, strong DAB staining appeared in the guard cells directly contacted by the substomatal vesicle, and there was a significant difference in the H2O2 staining area of the infestation sites between M8664-3 and Mingxian 169. Between 24 and 48 hpi, the H2O2 staining area of the M8664-3 and Mingxian169 infection sites continuously increased. Between 72 and 96 hpi, the staining area of the M8664-3 infection site expanded to include guard cells and surrounding areas. In the mesophyll cells of Mingxian169, the DAB staining expansion area was far smaller than that of M8664-3 (Figure 1).
Expression Analysis of Selected Candidate Genes
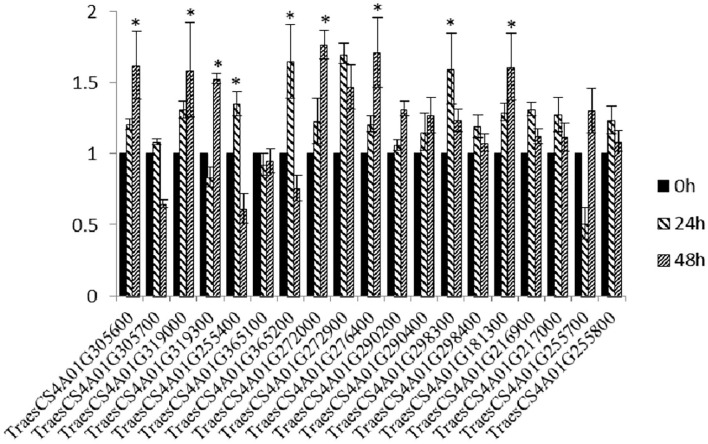
Through annotation analysis, 19 genes near the linked markers and containing disease resistance-related domains, or hit by the linked markers of YrM8664-3 were selected (Supplementary Figure 1; Supplementary Table 2). Among these, nine genes were significantly upregulated after inoculation with CYR33, as determined by qRT-PCR (Figure 2), namely, TraesCS4A01G305600 (Leucine-rich repeat receptor-like protein kinase family protein), TraesCS4A01G319000 (PGR5-like protein 1A, chloroplastic), TraesCS4A01G319300 (disease-resistance protein (NBS-LRR class) family), TraesCS4A01G365200 (Sn1-specific diacylglycerol lipase alpha), TraesCS4A01G272000 (plastid-lipid associated protein PAP/fibrillin family protein, which encodes TaFBN), TraesCS4A01G272900 (beta-glucosidase, which encodes Ta_Pes_BRCT), TraesCS4A01G276400 (Pescadillo homolog), TraesCS4A01G298300 (nonspecific phospholipase C), and TraesCS4A01G181300 (AP2-like ethylene-responsive transcription factor) (Table 1).
Figure 2.
Relative expression levels of 19 candidate genes at 24 and 48 h in M8664-3 after inoculation with Pst race CYR33. The data were normalized to the wheat TaEF-1α gene. Wheat leaves treated with distilled water were included as a control. Error bars represent the standard deviations of three independent samples. The significance of differences is indicated by asterisks and tested using Student's t-test (P < 0.05).
Table 1.
Nineteen candidate genes and markers linked to resistance gene YrM86664-3 on chromosome 4AL.
| Position (bp) | No. of selected genes/Markers of YrM8664-3 | Gene accession | Annotation |
|---|---|---|---|
| 415683496–415683891 | BE446584 | ||
| 456290064–456292784 | C15 | TraesCS4A01G181300 | AP2-like ethylene-responsive transcription factor |
| 456292321–456292391 | AX111655681 | ||
| 515779130–515782750 | C16 | TraesCS4A01G216900 | ABC transporter G family member |
| 516343042–516343722 | C17 | TraesCS4A01G217000 | Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein |
| 567479603–567482101 | C5 | TraesCS4A01G255400 | Serine/threonine-protein kinase |
| 567493169–567495275 | C18 | TraesCS4A01G255700 | Ethylene-dependent gravitropism-deficient and yellow-green-like 2 |
| 567646583–567646825 | AX109496237 | ||
| 567650074–567650374 | AX109001562 | ||
| 567650160–567654340 | C19 | TraesCS4A01G255800 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| 583754800–583757140 | C8 | TraesCS4A01G272000 | Plastid-lipid associated protein PAP / fibrillin family protein |
| 583756221–583756371 | BE403251 | ||
| 583949402–583953409 | C9 | TraesCS4A01G272900 | Beta-glucosidase |
| 583949552–583949670 | BE403721 | ||
| 584817688–584823187 | C10 | TraesCS4A01G276400 | Pescadillo homolog |
| 584818692–584818992 | AX86179210 | ||
| 594162606–594167272 | C11 | TraesCS4A01G290200 | Ankyrin repeat protein-like |
| 594165370–594165403 | BE406959 | ||
| 594212051–594212608 | BE591356 | ||
| 594213511–594217122 | C12 | TraesCS4A01G290400 | Phosphomethylpyrimidine synthase |
| 597023208–597024827 | C13 | TraesCS4A01G298300 | Non specific phospholipase C |
| 597024543–597025097 | BE637642 | ||
| 597052406–597054154 | C14 | TraesCS4A01G298400 | Cation calcium exchanger |
| 600913413–600914954 | C1 | TraesCS4A01G305600 | Leucine-rich repeat receptor-like protein kinase family protein |
| 600917368–600917668 | AX109895154 | ||
| 600918067–600918777 | C2 | TraesCS4A01G305700 | Leucine-rich repeat receptor-like protein kinase family protein |
| 607633308–607635652 | C3 | TraesCS4A01G319000 | PGR5-like protein 1A, chloroplastic |
| 607888036–607888131 | BV211529 | ||
| 608267651–608271046 | C4 | TraesCS4A01G319300 | Disease resistance protein (NBS-LRR class) family |
| 638240325–638241099 | C6 | TraesCS4A01G365100 | SHAGGY-like kinase |
| 638661695–638661817 | Xgpw2331 | ||
| 638791957–638796803 | C7 | TraesCS4A01G365200 | Sn1-specific diacylglycerol lipase alpha |
Silencing of TaFBN and Ta_Pes_BRCT Weakens Wheat Resistance Against CYR33
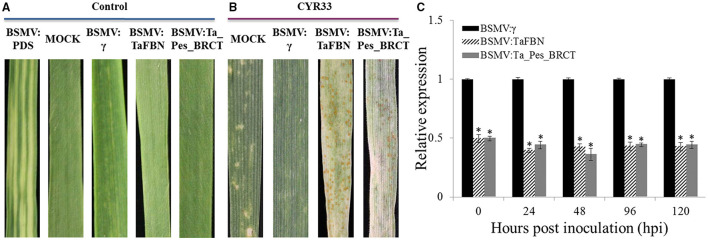
Unique fragments were designed to knock down these nine candidate genes using primers specified in Supplementary Table 1. All of the BSMV-inoculated plants displayed chlorotic mosaic symptoms at 10 dpi, but there were no obvious defects in further leaf growth, while the leaves inoculated with BSMV: TaPDS exhibited photobleaching (Figure 3A), indicating that the BSMV induced gene silencing system functions well. After knocking down these genes with the VIGS method, M8664-3 became susceptible in the TaFBN- and Ta_Pes_BRCT-silenced system (Figure 3B), which indicated that these two genes may be involved in the resistance of M8664-3 to Pst. To determine the efficiency of VIGS, qRT-PCR was performed to examine the relative transcript levels of TaFBN and Ta_Pes_BRCT in the fourth leaves of infected plants. Compared with control inoculations, transcript levels of TaFBN-knockdown plants were reduced by 49, 39, 42, 43, and 42% at 0, 24, 48, 96, and 120 hpi, and Ta_Pes_BRCT knockdown plants also showed a stable efficiency by reducing to 49, 44, 36, 44, and 44% at 0, 24, 48, 96, and 120 hpi with CYR33, respectively (Figure 3C).
Figure 3.
Transient silencing of TaFBN and Ta_Pes_BRCT with the BSMV-VIGS method. (A) Wheat leaves treated with 1× Fes buffer (MOCK) show no phenotypic changes. Mild chlorotic mosaic symptoms were detected on the plants inoculated with BSMV:γ, BSMV:PDS, BSMV:TaFBN, or BSMV:Ta_Pes_BRCT. (B) Phenotypes of the fourth leaves infected with uredospores of Pst race CYR33 at 10 days post inoculation. (C) Silencing efficiency assessment of TaFBN in the TaFBN-knockdown plants and Ta_Pes_BRCT in the Ta_Pes_BRCT-knockdown plants. Error bars represent the standard deviations of three independent samples. The significance of differences is indicated by asterisks and tested using Student's t-test (P < 0.05).
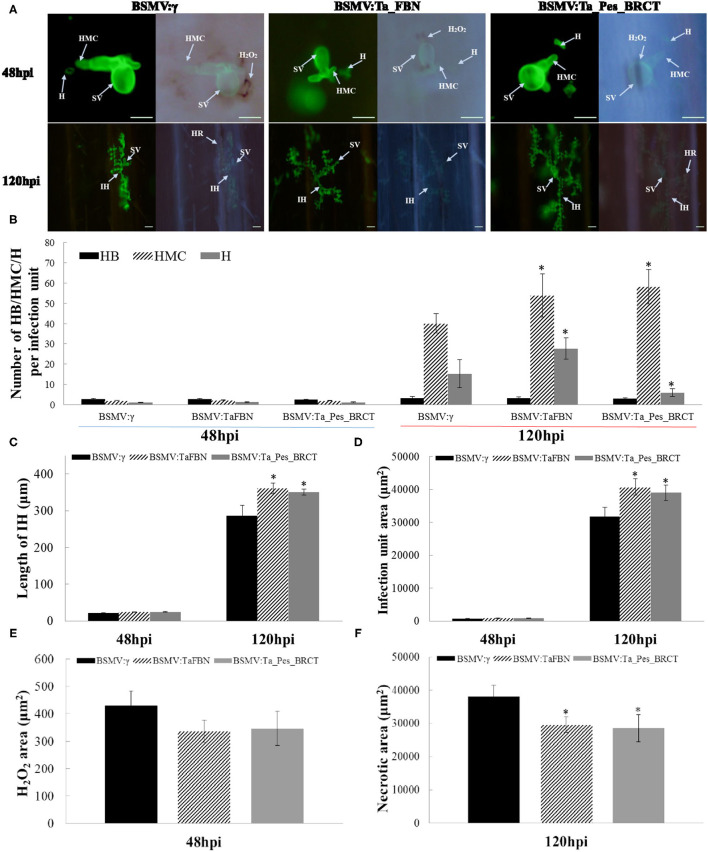
Histological observations showed that the number of haustorial mother cells, hypha length, and Pst growth area in TaFBN- and Ta_Pes_BRCT-silenced leaves all slightly increased as compared with that of the unsilenced treatment 48 hpi, but the DAB-reactive oxygen staining area did not significantly change, and no necrotic cells were found. At 120 hpi, the area of Pst growth, length of hyphae, and number of hyphae branches, haustorial mother cells, and haustorium in TaFBN- and Ta_Pes_BRCT-silenced leaves were all increased, and the difference was extremely significant. DAB-reactive oxygen staining decreased to 0, and the area of necrotic cells also significantly decreased compared with the unsilenced treatment at 120 hpi (Figure 4). In summary, the silencing of TaFBN and Ta_Pes_BRCT reduced disease resistance and made plants more susceptible to Pst.
Figure 4.
Histological observations of Pst race CYR33 infection in TaFBN-knockdown and Ta_Pes_BRCT-knockdown wheat plants. (A) WGA was used to stain the leaves to visualize pathogens. (B) Number of HB/HMC/H; (C) length of IH; (D) infection unit area; (E) H2O2 area; and (F) necrotic area staining by 3,3′-diaminobenzidine (DAB) were measured with DP-BSW software in TaFBN-knockdown plants at 48 and 120 hpi after inoculation. SV, substomatal vesicle; IH, initial hyphae; HMC, haustorial mother cell; SH, secondary hyphae. Error bars represent the standard deviations of three independent samples. The significance of differences is indicated by asterisks and tested using Student's t-test (P < 0.05). Bar, 50 μm.
TaFBN or Ta_Pes_BRCT Encodes Proteins Related to Defense Response
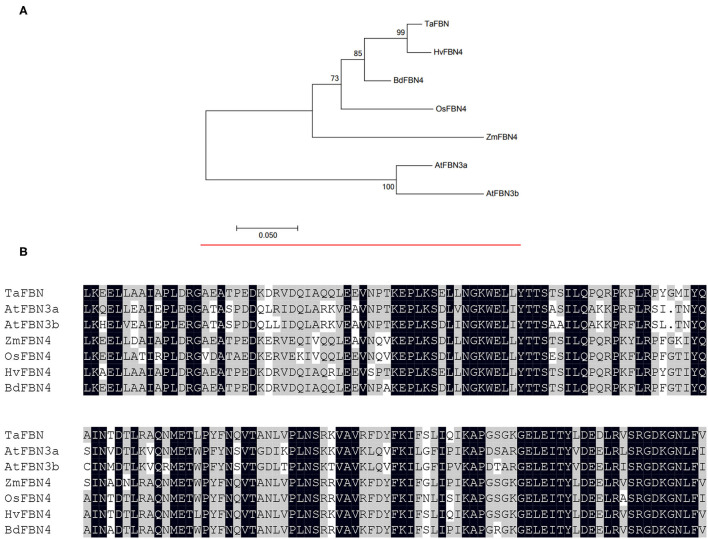
TaFBN encodes a protein composed of 261 amino acids, with a molecular weight of 28.59 kDa, an isoelectric point (PI) of 9.34, and an average hydrophobicity of −0.330, which suggested that it may be a hydrophilic protein. There was a PAP_fibrillin domain at positions 90–251 of the amino acid; therefore, it was temporarily named TaFBN. The phylogenetic analysis of TaFBN with H. vulgare (HvFBN4, KAE8820120), Brachypodium distachyon (BdFBN4, XP_003560711), O. sativa (OsFBN4, XP_015632312), A. thaliana (AtFBN3a, NM_113511; AtFBN3b, BT020596), and Z. mays (ZmFBN4, ACG27798) resulted in the clustering of TaFBN with HvFBN4, BdFBN4, OsFBN4, and ZmFBN4, all of which are members of FBN4 proteins in monocotyledons (Figure 5A). A nucleic acid sequence analysis revealed that TaFBN shared 96.55% identity with HvFBN4 from H. vulgare. Multiple amino acid sequence alignments of TaFBN with HvFBN4, BdFBN4, OsFBN4, and ZmFBN4 showed that TaFBN4 is predicted to encode proteins with the unique conserved domains of PAP complex FBN4 (Figure 5B). Therefore, it was determined that TaFBN and FBN4 were clustered together, and that the functional annotations of the latter on the UniProt website were related to resistance to bacterial diseases and ozone.
Figure 5.
(A) Phylogenetic analysis of fibrillin (FBN) proteins. A neighbor-joining tree of FBN in Triticuma estivum (TaFBN, KAF7095220), Hordeum vulgare (HvFBN4, KAE8820120), Brachypodium distachyon (BdFBN4, XP_003560711), Oryza sativa (OsFBN4, XP_015632312), Arabidopsis thaliana (AtFBN3a/AT3G26070, NM_113511; AtFBN3b/At3g26080, BT020596), and Zea mays (ZmFBN4, ACG27798). (B) Multiple amino acid sequence alignments of TaFBN with OsFBN4, ZmFBN4, AtFBN3a, AtFBN3b, and HvFBN4. Amino acid identity (black boxes) and similarity (gray boxes) are shown within the protein kinase domain.
Ta_Pes_BRCT encodes a protein composed of 596 amino acids, with a molecular weight of 68.32 kDa, PI of 7.68, and an average hydrophobicity of −0.615, which indicated that it may be a hydrophilic protein. The amino acid coded by Ta_Pes_BRCT has a Pescadillo (PES) domain at positions 9–277, and the 339–417 amino acids contain a BRCT domain. The predicted function may be related to ribosomes; therefore, it was temporarily named Ta_Pes_BRCT. The phylogenetic analysis and multiple amino acid sequence alignment of Pes_BRCT proteins indicated that Ta_Pes_BRCT is predicted to encode proteins with conserved domains of the Pes_BRCT complex (Supplementary Figure 2). The functional annotations of BRCT on the UniProt website were related to DNA repair under stress. As predicted by the TMpred program, TaFBN and Ta_Pes_BRCT have no transmembrane domain. As predicted by TargetP-2.0 and Plant-mPLoc, the location of the protein encoded by Ta_FBN was predicted in Chloroplast; the location of the protein encoded by Ta_Pes_BRCT was predicted in Nucleus.
Functional Verification of Transgenic Plants
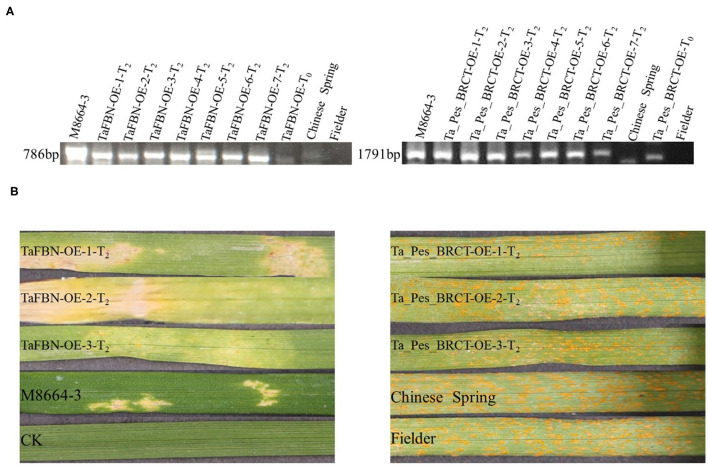
In order to further analyze the function of TaFBN and Ta_Pes_BRCT in stripe rust resistance, the two genes were introduced into the susceptible bread wheat variety Fielder by an Agrobacterium-mediated transformation method. The individuals of T0 generation were identified by PCR analysis (Figure 6A), and seven independent T2 lines obtained from the positive T0 progeny were further used to conduct disease resistance tests and PCR detection procedures. Three replicates were tested for each line. The T2 generation of TaFBN transgenic plants was resistant to CYR33, while all of the Ta_Pes_BRCT transgenic plants were susceptible to CYR33 (Figure 6B), which indicated that TaFBN confers more important resistance to CYR33.
Figure 6.
Stable transformation of the susceptible bread wheat cultivar Fielder with TaFBN confers resistance to Pst race CYR33. (A) Identification by PCR of individuals transformed with TaFBN and Ta_Pes_BRCT. Wild-type Fielder was used as a negative control; M8664-3 containing the full sequence of TaFBN and Ta_Pes_BRCT was used as a positive control. (B) Fielder T2 plants transformed with TaFBN were resistant to CYR33 but those transformed with Ta_Pes_BRCT were susceptible to CYR33. Wild-type Fielder, Chinese Spring, and M8664-3 were used as susceptible and resistant controls.
TaFBN Responds to Abiotic Stress and Hormone Treatments
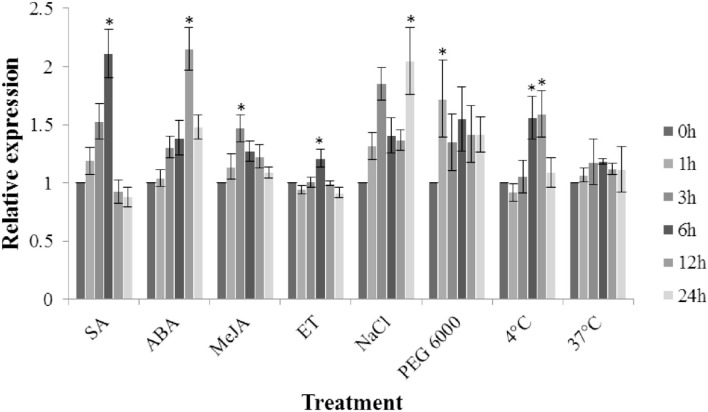
Under different hormone treatments (MeJA, ET, SA, and ABA), the transcription level of TaFBN in the leaves of M8664-3 wheat seedlings was determined (Figure 7). The expression level of TaFBN significantly increased under SA and ABA treatments, and peaked (more than twice) at 6 and 12 hpi, respectively. The expression of TaFBN also slightly increased after MeJA and ET treatment. Under abiotic stress, the expression of TaFBN significantly increased after NaCl, PEG6000, and 4°C treatment, but its expression in 37°C-high temperature treatment did not significantly increase. In summary, TaFBN might be induced by SA, ABA, high salt, drought, and low temperature to increase its expression.
Figure 7.
Expression analysis of TaFBN under different hormones and abiotic stresses. SA, salicylic acid; ABA, abscisic acid; MeJA, methyl jasmonate; ET, ethylene. Error bars represent the variations among three independent replicates. The data were normalized to the TaEF-1α gene. Wheat leaves treated with distilled water were included as a control. Error bars represent the standard deviations of three independent samples. The significance of differences is indicated by asterisks and tested using Student's t-test (P < 0.05).
Discussion
In our previous study, YrM8644-3 was located in bin 4AL13-0.59-0.66 near 4AL12-0.43-0.59 on wheat chromosome 4A. However, it was challenging to clone this gene because of YrM8664-3 derived from the wheat-L. mollis introgression line M8664-3 and the complexity of the wheat hexaploid genome. Fortunately, the continuous improvement in the whole genome sequencing of wheat “Chinese Spring” provided great convenience for gene cloning and functional analysis (Avni et al., 2017; IWGSC et al., 2018).
In this study, based on the chromosome location of YrM8664-3 in our previous study, the sequences of the linked markers of YrM8664-3 were assigned against the Chinese Spring IWGSC RefSeq V1.0 Reference Genome (IWGSC et al., 2018), and then qRT-PCR was performed to analyze the expression level of candidate genes under Pst infection. Among the 19 selected genes that are near the linked markers and containing resistance domains or just hit by linked markers of YrM8664-3, the expression level of nine genes were significantly enhanced after inoculation with CYR33. The VIGS system was used to characterize gene function, and only two candidate genes, TaFBN and Ta_Pes_BRCT, silencing plants were found to be significantly weakened in disease resistance to CYR33. VIGS is a technique for rapid gene function analysis based on the principle of specific degradation of endogenous mRNA sequences caused by post-transcriptional gene silencing (PTGS). BSMV-VIGS is widely used in the rapid analysis of gene function of monocotyledonous plants, particularly barley and wheat. Feng et al. (2015) selected six unigenes from transcriptome analysis and achieved transient silencing of the six unigenes individually through VIGS using the BSMV vector. The results showed that the six unigenes inhibited the vernalization of wheat, and that during silencing or down-regulation, the genes promoted flower development in wheat. Liu et al. (2020) used transient expression and BSMV-mediated TabHLH49 gene silencing, and discovered that TabHLH49 positively regulated WZY2 dehydrogenase expression and increased the resistance of wheat to drought.
The most direct and effective verification method for gene function is transgenic technology. By transferring the HvBADH1 gene from H. vulgare into T. aestivum via traditional Agrobacterium tumefaciens-mediated transformation, Li et al. (2019) found that the overall salt tolerance of target plants was significantly improved, and that the damaging effect of high salt was significantly reduced after overexpression of the HvBADH1 gene. In cereals, Agrobacterium-mediated transgenic sites are generally considered to be cleaner, with fewer copies and rearrangements than biologically generated transgenic sites (Wu et al., 2006). Horvath et al. (2003) subjected the stem-rust-susceptible barley cv. Golden Promise to Agrobacterium-mediated transformation with the Rpg1 gene, and characterized their seedling infection response to pathotype Pgt-MCC of the stem rust fungus. This demonstrated that susceptible barley can become resistant by transformation with a cloned resistant gene. In this study, the TaFBN and Ta_Pes_BRCT genes were transformed into the susceptible variety Fielder by Agrobacterium-mediated transformation. The TaFBN transgenic plants exhibited obvious resistance after inoculation with CYR33, which indicates that TaFBN may be involved in stripe rust resistance in M8664-3.
TaFBN has a conserved fibrillin (FBN) domain that was named fibrils, because those related proteins were first detected in fibrils in the chromoplasts of Rosa rugosa and Capsicum annuum fruit (Newman et al., 1989; Deruere et al., 1994; Kim et al., 2015). FBN proteins participate in a variety of important biological functions, in addition to photosynthesis and structural roles, which also respond to numerous abiotic and biotic stresses, especially oxidative stress (Youssef et al., 2010; Kim et al., 2015). Leitner-Dagan et al. (2006) showed that the Chrc (FBN1) in cucumber leaves was induced by Sphaerotheca fuliginea and LeChrc (FBN1) in tomato plants infected with Botrytis cinerea. LeChrc expression is a necessary condition for resistance to B. cinerea. Using the tomato plant system, transgenic plants with LeChrc inhibition were more susceptible to infection both in vitro with isolated leaves and in growth chambers with intact leaves and stems (Cooper et al., 2003; Leitner-Dagan et al., 2006). Similarly, studies (Singh et al., 2010; Jiang et al., 2020) have shown that both Arabidopsis and apple FBN4 gene T-DNA insertion mutants are more sensitive to bacterial infections Pseudomonas syringae pathovar tomato and Erwinia amylovora, respectively. In Arabidopsis, pathogen-associated molecular pattern (PAMP) induces the phosphorylation of FBN4, and it is speculated that FBN4 may be involved in plant disease resistance response.
During plant growth and development stages, when plants are under abiotic stresses (such as drought, cold, heat, bright light, and wound management) or hormone induction (with gibberellin, jasmonic acid, and abscisic acid), the expression of fibrillins is varied and complex (Pruvot et al., 1996; Kuntz et al., 1998; Langenkamper et al., 2001; Leitner-Dagan et al., 2006; Simkin et al., 2008). When red pepper fruits were treated with gibberellic acid, FBN1 mRNA and protein levels decreased (Deruere et al., 1994). Conversely, the FBN1 and FBN2 proteins are involved in the jasmonate biosynthesis pathway in Arabidopsis in response to light and cold stress (Youssef et al., 2010). In addition, when tomato flacca mutant plants were subjected to drought stress, ABA biosynthesis was defective, and FBN protein accumulation decreased, because ABA treatment can induce FBN protein levels (Gillet et al., 1998). Herein, we observed an induction of TaFBN upon SA and ABA treatment, suggesting that TaFBN may be an effector associated with the SA and ABA signaling pathways. Taken together with our observation of enhancement of TaFBN expression in response to environmental stress stimuli (high salt, drought, cold, heat), it is reasonable to hypothesize that TaFBN functions at the nexus of biotic and abiotic stress pathways.
BRCT motifs were originally identified in the breast cancer tumor suppressor protein BRCA1 by Koonin et al. (1996), and now have been identified in numerous proteins involved in DNA repair and cell cycle checkpoints (Mathilde et al., 2003). Roy et al. (2015) indicated the importance of BRCT in regulating the stability of proteins under genotoxic stress in plants. Pst-infected wheat may produce oxygen-free radicals, such as O2− and H2O2, that damage cells, which subsequently produce metabolic byproducts that cause DNA base damage. Ta_Pes_BRCT is a BRCT-domain-containing protein that may be involved in DNA repair and that could be induced to up-regulate by Pst. However, after transgenic verification, it was found that Ta_Pes_BRCT is a related gene in the process of wheat resistance to Pst that plays less important role than TaFBN.
In summary, this study selected candidate genes of YrM8664-3 by bioinformatics analysis, and verified the resistant function of the candidate genes TaFBN and Ta_Pes_BRCT by qRT-PCR, BSMV-VIGS, and genetic transformation. Finally, it was validated that TaFBN may be involved in YrM8664-3 stripe rust resistance as an important gene via the SA and ABA signaling pathways.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
BW and QL designed the experiments. PJ, KC, JL, and ZW performed the experiments and analyzed the data. PJ, JL, KC, ZW, PC, QL, and BW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key R&D Program of China (Grant Nos: 2018YFD0200403 and 2018YFD0200501), the Open Project Program of State Key Laboratory of Crop Stress Biology for Arid Areas, NWAFU, Yangling, Shaanxi, 712100, China (CSBAA2019007), the Technical Guidance Project of Shaanxi Province (Grant No: 2017CGZH-HJ-01), the National Science Foundation of China (Grant No: 31501620), and the China Ministry of Education 111 Project (Grant No: B07049).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the reviewers for helpful comments and valuable suggestions during the revision of the early version of the manuscript.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.754823/full#supplementary-material
References
- Asthir B., Koundal A., Bains N. S. (2011). Kinetic properties of cell wall bound superoxide dismutase in leaves of wheat (Triticum aestivum L.) following stripe rust (Puccinia striiformis) infection. Indian J. Biochem. Biophys. 48, 341–345. 10.1016/j.plrev.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Avni R., Nave M., Barad O., Baruch K., Twardziok S. O., Gundlach H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. 10.1126/science.aan0032 [DOI] [PubMed] [Google Scholar]
- Ayliffe M., Devilla R., Mago R., White R., Talbot M., Pryor A., et al. (2011). Nonhost resistance of rice to rust pathogens. Mol. Plant Microbe Interact. 24, 1143–1155. 10.1094/MPMI-04-11-0100 [DOI] [PubMed] [Google Scholar]
- Bansal M., Kaur S., Dhaliwal H. S., Bains N. S., Bariana H. S., Chhuneja P., et al. (2017). Mapping of Aegilops umbellulata-derived leaf rust and stripe rust resistance loci in wheat. Plant Pathol. 66, 38–44. 10.1111/ppa.12549 [DOI] [Google Scholar]
- Chao K. X., Yang J. Y., Liu H., Jing J. X., Li Q., Wang B. T., et al. (2018). Genetic and physical mapping of a putative Leymus mollis-derived stripe rust resistance gene on wheat chromosome 4A. Plant Dis. 102, 1001–1007. 10.1094/PDIS-05-17-0671-RE [DOI] [PubMed] [Google Scholar]
- Cooper B., Clarke J. D., Budworth P., Kreps J., Hutchison D., Park S., et al. (2003). A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. USA. 100, 4945–4950. 10.1073/pnas.0737574100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H. T., Tsuda K., Parker J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. 10.1146/annurev-arplant-050213-0400 [DOI] [PubMed] [Google Scholar]
- Denance N., Sanchez-Vallet A., Goffner D., Molina A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. 10.3389/fpls.2013.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207, 79–87. 10.1016/j.plantsci.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Deruere J., Romer S., d'Harlingue A., Backhaus R. A., Kuntz M., Camara B. (1994). Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6, 119–133. 10.1105/tpc.6.1.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N., Lawrence G. J., Catanzariti A. M., Teh T., Wang C. I., Ayliffe M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA. 103, 8888–8893. 10.1073/pnas.0602577103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. L., Wang K. T., Ma C., Zhao Y. Y., Yin J. (2015). Virus-induced gene silencing-based functional verification of six genes associated with vernalization in wheat. Biochem. Bioph. Res. Commun. 458, 928–933. 10.1016/j.bbrc.2015.02.064 [DOI] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Holzberg S., Lindbo J. A., Padgett H. S., Palmer K. E., Wolfe G. M., et al. (2002). Epigenetic modification of plants with systemic RNA viruses. OMICS 6, 137–151. 10.1089/153623102760092742 [DOI] [PubMed] [Google Scholar]
- Gillet B., Beyly A., Peltier G., Rey P. (1998). Molecular characterization of CDSP 34, a chloroplastic protein induced by water deficit in Solanum tuberosum L. plants, and regulation of CDSP 34 expression by ABA and high illumination. Plant J. 16, 257–262. 10.1046/j.1365-313x.1998.00292.x [DOI] [PubMed] [Google Scholar]
- Horvath H., Rostoks N., Brueggeman R., Steffenson B., von Wettstein D., Kleinhofs A. (2003). Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc. Natl. Acad. Sci. USA. 100, 364–369. 10.1073/pnas.0136911100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Y., Steffenson B. J., Sela H., Stinebaugh K. (2018). Resistance of Aegilops longissima to the rusts of wheat. Plant Dis. 102, 1124–1135. 10.1094/PDIS-06-17-0880-RE [DOI] [PubMed] [Google Scholar]
- IWGSC Appelss R., Eversole K., Stein N., Feuillet C., Keller B., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 10–1126. 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Jia Y., McAdams S. A., Bryan G. T., Hershey H. P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. 10.1093/emboj/19.15.4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Y., Hu H. C., Ma Y. H., an Zhou J. L. (2020). Genome-wide identification and characterization of the fibrillin gene family in Triticum aestivum. PeerJ 8:9225. 10.7717/peerj.9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Vance R. E., Dangl J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:6316. 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kang Z., Huang L. L., Buchenauer H. (2002). Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis. J. Plant Dis. Protect. 109, 25–37. [Google Scholar]
- Kim E. H., Lee Y., Kim H. U. (2015). Fibrillin 5 is essential for plastoquinone-9 biosynthesis by binding to solanesyl diphosphate synthases in Arabidopsis. Plant Cell 27, 2956–2971. 10.1105/tpc.15.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Altschul S. F., Bork P. (1996). Functional motifs. Nat. Genet 13, 266–268. 10.1038/ng0796-266 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz M., Chen H. C., Simkin A. J., Romer S., Shipton C. A., Drake R., et al. (1998). Upregulation of two ripening-related genes from a nonclimacteric plant (pepper) in a transgenic climacteric plant (tomato). Plant J. 13, 351–361. 10.1046/j.1365-313X.1998.00032.x [DOI] [Google Scholar]
- Langenkamper G., Manac'h N., Broin M., Cuine S., Becuwe N., Kuntz M., et al. (2001). Accumulation of plastid lipid-associated proteins (fibrillin/CDSP34) upon oxidative stress, ageing and biotic stress in Solanaceae and in response to drought in other species. J. Exp. Bot. 52, 1545–1554. 10.1093/jexbot/52.360.1545 [DOI] [PubMed] [Google Scholar]
- Leitner-Dagan Y., Ovadis M., Shklarman E., Elad Y., David D. R., Vainstein A. (2006). Expression and functional analyses of the plastid lipid-associated protein CHRC suggest its role in chromoplastogenesis and stress. Plant Physiol. 142, 233–244. 10.1104/pp.106.082404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. F., Cai J., Luo X., Chang T. L., Li J. X., Zhao Y. W., et al. (2019). Transformation of wheat Triticum aestivum with the HvBADH1 transgene from hulless barley improves salinity-stress tolerance. Acta Physiol. Plant 41, 1–14. 10.1007/s11738-019-2940-8 [DOI] [Google Scholar]
- Li X. Y., Wang X., Zhang S. P., Liu D. W., Duan Y. X., Dong W. (2011). Comparative profiling of the transcriptional response to soybean cyst nematode infection of soybean roots by deep sequencing. Chin. Sci. Bull. 56, 1904–1911. 10.1007/s11434-011-4510-3 [DOI] [Google Scholar]
- Liu H., Yang Y., Liu D. D., Wang X. Y., Zhang L. S. (2020). Transcription factor TabHLH49 positively regulates dehydrin WZY2 gene expression and enhances drought stress tolerance in wheat. BMC Plant Biol. 20, 1–10. 10.1186/s12870-020-02474-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luo S., Peng J., Li K., Wang M., Kuang H. (2011). Contrasting evolutionary patterns of the Rp1 resistance gene family in different species of Poaceae. Mol. Biol. Evol. 28, 313–325. 10.1093/molbev/msq216 [DOI] [PubMed] [Google Scholar]
- Ma S., Lapin D., Liu L., Sun Y., Song W., Zhang X., et al. (2020). Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370:6521. 10.1126/science.abe3069 [DOI] [PubMed] [Google Scholar]
- Mathilde G., Ghislaine G., Daniel V., Georges P. (2003). The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 35, 465–475. 10.1046/j.1365-313X.2003.01820.x [DOI] [PubMed] [Google Scholar]
- Monteiro F., Nishimura M. T. (2018). Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 56, 243–267. 10.1146/annurev-phyto-080417-045817 [DOI] [PubMed] [Google Scholar]
- Newman L. A., Hadjeb N., Price C. A. (1989). Synthesis of two chromoplast-specific proteins during fruit development in Capsicum annuum. Plant Physiol. 91, 455–458. 10.1104/pp.91.2.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty I. T., Jackson A. O. (1990). Mutational analysis of barley stripe mosaic virus RNA beta. Virology 179, 712–718. 10.1016/0042-6822(90)90138-h [DOI] [PubMed] [Google Scholar]
- Pruvot G., Cuine S., Peltier G., Rey P. (1996). Characterization of a novel drought-induced 34-kDa protein located in the thylakoids of Solanum tuberosum L. plants. Planta 198, 471–479. 10.1007/BF00620065 [DOI] [PubMed] [Google Scholar]
- Roelfs A. P., Huertaespino J., Marshall D. (1992). Barley stripe rust in Texas. Plant Dis. 76, 538–538. 10.1094/PD-76-0538C [DOI] [Google Scholar]
- Roy S., Banerjee V., Das K. P. (2015). Understanding the physical and molecular basis of stability of Arabidopsis DNA Pol lambda under UV-B and high NaCl stress. PLoS ONE 10:e0133843. 10.1371/journal.pone.0133843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P. M., Kazan K., Wilson I., Anderson J. P., Richmond T., Somerville S. C., et al. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA. 97, 11655–11660. 10.1073/pnas.97.21.11655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S. R., Huang L., Brandt A. S., Gill B. S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. 10.1104/pp.105.061861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A. J., Moreau H., Kuntz M., Pagny G., Lin C. W., Tanksley S., et al. (2008). An investigation of carotenoid biosynthesis in Coffea canephora and Coffea arabica. J. Plant Physiol. 165, 1087–1106. 10.1016/j.jplph.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Singh D. K., Maximova S. N., Jensen P. J., Lehman B. L., Ngugi H. K., McNellis T. W. (2010). FIBRILLIN4 is required for plastoglobule development and stress resistance in Apple and Arabidopsis. Plant Physiol. 154, 1281–1293. 10.1104/pp.110.164095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan A. M., Chen X. M., He Z. H. (2007). Wheat stripe rust in China. Aust. J. Agric. Res. 58, 607–619. 10.1071/AR06142 [DOI] [Google Scholar]
- Wan A. M., Zhao Z. H., Chen X. M., He Z. H., Jin S. L., Jia Q. Z., et al. (2004). Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 88, 896–904. 10.1094/PDIS.2004.88.8.896 [DOI] [PubMed] [Google Scholar]
- Wang C. F., Huang L. L., Buchenauer H., Han Q. M., Zhang H. C., Kang Z. S. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2− and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant 71, 230–239. 10.1016/j.pmpp.2008.02.006 [DOI] [Google Scholar]
- Wu H. X., Sparks C. A., Jones H. D. (2006). Characterisation of T-DNA loci and vector backbone sequences in transgenic wheat produced by Agrobacterium-mediated transformation. Mol. Breed. 18, 195–208. 10.1007/s11032-007-9090-1 [DOI] [Google Scholar]
- Xiao S., Brown S., Patrick E., Brearley C., Turner J. G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15, 33–45. 10.1105/tpc.006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef A., Laizet Y., Block M. A., Marechal E., Alcaraz J. P., Larson T. R., et al. (2010). Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 61, 436–445. 10.1111/j.1365-313X.2009.04067.x [DOI] [PubMed] [Google Scholar]
- Yu X., Feng B. M., He P., Shan L. B. (2017). From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. 10.1146/annurev-phyto-080516-035649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Jiang Z., Bi G., Nomura K., Liu M., Wang Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Chen L., Zhong S., Wei X., Zhao Q., Pan Q., et al. (2020). A Cu-only superoxide dismutase from stripe rust fungi functions as a virulence factor deployed for counter defense against host-derived oxidative stress. Environ. Microbiol. 22, 5309–5326. 10.1111/1462-2920.15236 [DOI] [PubMed] [Google Scholar]
- Zou S., Wang H., Li Y., Kong Z., Tang D. (2018). The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 218, 298–309. 10.1111/nph.14964 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.