Abstract
Cardiometabolic diseases are frequently polygenic in architecture, comprising a large number of risk alleles with small effects spread across the genome1–3. Polygenic scores (PGS) aggregate these into a metric representing an individual’s genetic predisposition to disease. PGS have shown promise for early risk prediction4–7 and there is an open question as to whether PGS can also be used to understand disease biology8. Here, we demonstrate that cardiometabolic disease PGS can be used to elucidate the proteins underlying disease pathogenesis. In 3,087 healthy individuals, we found that PGS for coronary artery disease, type 2 diabetes, chronic kidney disease and ischaemic stroke are associated with the levels of 49 plasma proteins. Associations were polygenic in architecture, largely independent of cis and trans protein quantitative trait loci and present for proteins without quantitative trait loci. Over a follow-up of 7.7 years, 28 of these proteins associated with future myocardial infarction or type 2 diabetes events, 16 of which were mediators between polygenic risk and incident disease. Twelve of these were druggable targets with therapeutic potential. Our results demonstrate the potential for PGS to uncover causal disease biology and targets with therapeutic potential, including those that may be missed by approaches utilizing information at a single locus.
Subject terms: Vascular diseases, Type 2 diabetes, Metabolism, Population genetics, Functional genomics
Ritchie et al. use polygenic scores to identify plasma proteins with causal roles in cardiometabolic disease.
Main
Human genetic studies have identified numerous proteins involved in coronary artery disease (CAD), type 2 diabetes (T2D) and other cardiometabolic diseases through a combination of genome-wide association studies (GWAS), fine-mapping, colocalization and Mendelian randomization by overlaying information at strong cardiometabolic disease loci9–12. However, cardiometabolic diseases are polygenic in architecture since they depend on many thousands of variants across the genome, nearly all exerting small lifelong effects13–17. These variants are spread across many different pathways and likely exert their effects through multiple levels of regulation, including gene expression, proteins and their interactions, cell morphology and higher-order physiological processes18–20. PGS aggregate these small effects into a single number for each individual that captures a fraction of their disease susceptibility. The use of PGS for risk stratification has shown potential clinical utility for disease prevention21, yet the specific molecular consequences that precede disease risk for these polygenic effects are unknown. For example, proteins that are pathway-level hubs through which polygenic effects converge could be particularly promising targets for pharmaceutical intervention22,23.
In this study, we demonstrated how PGS can be used to identify proteins with causal roles in disease aetiology. The INTERVAL cohort consists of approximately 50,000 adult blood donors in England24,25, of which 3,087 participants have linked electronic hospital records, imputed genome-wide genotypes and quantitative levels of 3,438 plasma proteins26 (Supplementary Data 1 and 2). A schematic of the study is given in Extended Data Fig. 1. The characteristics of the participants are given in Extended Data Fig. 2; participants with a history of any cardiometabolic disease were excluded (Supplementary Table 1), reducing the potential for reverse causality in downstream analysis.
Extended Data Fig. 1. Study schematic.
Overview of the study design.
Extended Data Fig. 2. Cohort characteristics.
IQR: interquartile range. Body mass index (BMI) was computed from self-reported height and weight.
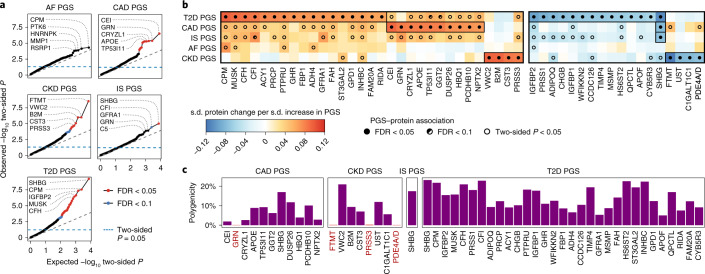
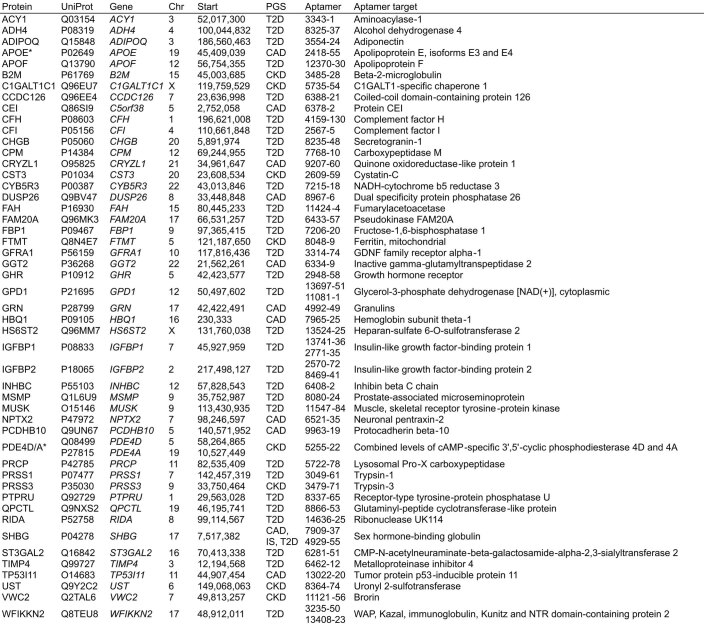
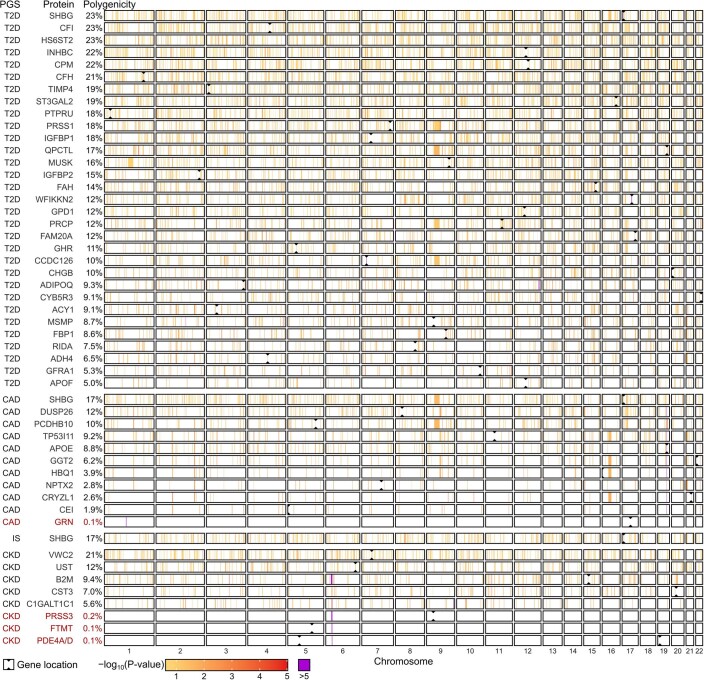
To quantify each participant’s relative polygenic risk of atrial fibrillation (AF), CAD, chronic kidney disease (CKD), ischaemic stroke (IS) and T2D, we applied externally derived genome-wide PGS consisting of 1.8–3.2 million variants. Using PGS, we identified 49 proteins whose levels differed with respect to polygenic risk at a false discovery rate (FDR) of 5% (Fig. 1a,b, Extended Data Figs. 3 and 4 and Supplementary Tables 2 and 3): 31 proteins for the T2D PGS; 11 proteins for the CAD PGS; 8 proteins for the CKD PGS; and 1 protein for the IS PGS. PGS–protein associations included proteins previously associated with cardiometabolic disease, such as cystatin-C (CST3) and beta2-macroglobulin (B2M), which are biomarkers for CKD27, and fructose-1,6-bisphosphatase 1 (FBP1), which plays a key role in glucose regulation and is a target of T2D drugs28. Associated proteins belonged to multiple non-overlapping pathways (Supplementary Information) and many are relatively understudied in the context of their respective diseases (Extended Data Fig. 5) thereby warranting future study.
Fig. 1. Proteins associated with polygenic risk for cardiometabolic disease.
a, Quantile–quantile plots of two-sided P values from linear regression testing associations between PGS and protein levels in n = 3,087 INTERVAL participants across all 3,438 tested proteins. Each plot compares the distribution of observed two-sided P values (y axes) to the distribution of expected two-sided P values under the null hypothesis for 3,438 tests (x axes) on a −log10 scale. Associations were fitted using linear regression adjusting for age, sex, ten genotype principal components, sample measurement batch and time between blood draw and sample processing. Full summary statistics including exact P values are provided in Supplementary Data 3a. The top five proteins by P value are labelled. b, Heatmaps showing the 49 proteins whose levels were significantly associated with at least one PGS after Benjamini–Hochberg FDR multiple-testing correction (FDR < 0.05) of the two-sided P values (statistical tests are as described in a). Each heatmap cell shows the s.d. change in protein levels per s.d. increase in PGS. Point estimates for the 49 FDR-significant proteins are detailed in Extended Data Fig. 3. Details about each protein are provided in Extended Data Fig. 4. c, Barplots showing the proportion of the genome (%) required to explain each PGS–protein association in n = 3,087 INTERVAL participants (polygenicity). Proteins are ordered from left to right by strength of PGS–protein association. Highlighted in red are PGS–protein associations that were explained by singular variants regulating protein levels, pQTLs, rather than polygenic. Percentages are detailed in Extended Data Fig. 3.
Extended Data Fig. 3. Summary statistics for PGS to protein to disease associations.
Beta: standard deviation change in protein levels per standard deviation increase in PGS (from Fig. 1b) in linear regression adjusting for age, sex, 10 genotype PCs, sample measurement batch, and time between blood draw and sample processing. FDR: Benjamini-Hochberg false discovery rate corrected P-value. FDR correction was applied separately for each PGS to all 3,438 P-values from linear regression of each of the 3,438 measured proteins on the respective PGS. Polygenicity: proportion of the genome (%) required to explain the PGS to protein association (from Fig. 1c). HR: hazard ratio for 7.7 year risk of hospitalisation with the respective disease conferred per standard deviation increase in protein levels (from Fig. 2b) in cox proportional hazard models using follow-up as time scale and adjusting for age, sex, sample measurement batch, and time between blood draw and sample processing. Associations highlighted in red indicate significant associations after Bonferroni correction for the 42 tests (P < 0.0012). Associations dulled in grey indicate P > 0.05. % PGS Mediated: Percentage of total association between the respective PGS and 7.7 year risk of hospitalisation with the respective disease mediated by the respective protein (from Fig. 2d). Highlighted in red indicates mediation was significant after Bonferroni correction for the 42 tests (P < 0.0012). Entries dulled in grey indicate P > 0.05. Linear regression, polygenicity, cox proportional hazard models, and mediation analysis were all performed in the same n = 3,087 independent INTERVAL participants. In each instance, 95% CI corresponds to the 95% confidence interval of the respective point estimate. All P-values are two-sided. 95% confidence intervals and P-values could not be formulated for the polygenicity tests. For proteins measured by more than one SomaLogic aptamer (GPD1, IGFBP1, IGFBP2, SHBG, and WFIKKN2) effect sizes were averaged and two-sided P-values were obtained from averaged Z-scores, and aptamer-specific summary statistics are detailed in Supplementary Table 3.
Extended Data Fig. 4. Information about each PGS associated protein.
Aptamer: Sequence ID for the SomaLogic aptamer(s) targeting the protein. A * next to the protein name indicates the aptamer(s) binds to specific isoforms of the listed protein or binds to multiple proteins; see Aptamer target column. Extended details on aptamer sensitivity and specificity can be found in Supplementary Table 2.
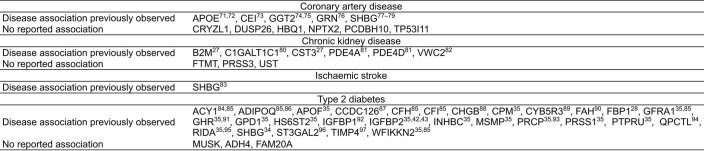
Extended Data Fig. 5. Previous evidence for PGS-associated proteins in disease.
Citations provided where association with the respective disease has been previously observed71–97.
PGS–protein associations were robust to technical, physiological and environmental confounding. We observed directional consistency and strong correlation of effect sizes when utilizing an orthogonal proteomics technology in independent samples (Extended Data Fig. 6a–c and Supplementary Information). Protein levels and PGS–protein associations were also temporally stable over 2 years of follow-up (Extended Data Fig. 6c,d and Supplementary Information). PGS–protein associations were also robust to circadian and seasonal effects, inclusion of participants with any prevalent cardiometabolic disease and body mass index (BMI), with the exception of six T2D PGS–protein associations that were partially mediated by BMI (Extended Data Fig. 6f,g).
Extended Data Fig. 6. Robustness of PGS to protein associations.
a-c) Robustness and longitudinal stability of PGS to protein associations to proteomics technology. d-e) Robustness and longitudinal stability of protein levels to proteomics technology. f) Robustness of PGS-protein associations to environmental and physiological confounding. g) Mediation of PGS-protein associations through body mass index (BMI) for six proteins associated with T2D PGS. a) Compares PGS-protein associations from Fig. 1b in n = 3,087 INTERVAL participants in which protein levels were measured with the SomaLogic platform (x-axis) to PGS-protein associations tested in an independent set of n = 418 INTERVAL participants in which protein levels were measured with the Olink Explore platform (y-axis). In total 1,463 proteins were quantified by the Olink Explore platform, including 907 quantified by the SomaLogic platform, and among these 16 of the 49 PGS-associated proteins from Fig. 1b. b) Compares PGS-protein associations from Fig. 1b (x-axis) to PGS-protein associations tested in an independent set of n = 3,848 INTERVAL participants in which protein levels were measured with the Olink T96 platform (y-axis). In total 265 proteins were quantified by the Olink T96 platform, including 224 quantified by the SomaLogic platform, and among these 4 of the 49 PGS-associated proteins from Fig. 1b. c) Compares PGS-protein associations tested in n = 646 INTERVAL participants in which protein levels were measured with both the SomaLogic platform (x-axis) and, after two-years of follow-up, the Olink T96 platform (y-axis). a-c) Data shown correspond to the beta estimates from linear regression (points) and their 95% confidence interval (bars), indicating standard deviation change in protein levels per standard deviation increase in the respective PGS (denoted by colour). Solid points indicate two-sided P-value < 0.05 for the test on the y-axis. Linear regression on both axes were adjusted for age (at protein measurement), sex, 10 genotype PCs, and platform-specific technical covariates. Full summary statistics including exact P-values are detailed in Supplementary Data 3,b for linear regression tests on y-axes, and in Supplementary Data 3,a for linear regression tests on x-axes. d) Compares protein levels quantified by the SomaLogic platform (x-axes) to protein levels quantified by the Olink T96 platform (y-axes) after two years of follow-up in n = 646 INTERVAL participants. e) Compares protein levels quantified by the Olink T96 platform (x-axes) to protein levels quantified by the Olink Explore platform (y-axes) in n = 418 INTERVAL participants. f) Compares PGS-protein associations from Fig. 1b in n = 3,087 INTERVAL participants (x-axes) to PGS-protein associations (1) additionally adjusted for circadian effects (time of day of blood draw), (2) additionally adjusted for seasonal effects (date of blood draw), (3) when including 87 additional participants with prevalent cardiometabolic disease (n = 3,174 on y-axis), and (4) when adjusting for BMI (n = 3,072 participants with non-missing BMI on y-axis). All associations were testing using linear regression adjusting for age, sex, 10 genotype PCs, sample measurement batch, and time between blood draw and sample measurement in addition to the covariates noted above. Data shown correspond to the beta estimates from linear regression (points) and their 95% confidence interval (bars), indicating standard deviation change in protein levels per standard deviation increase in the respective PGS (denoted by colour). Full summary statistics including exact P-values in these sensitivity analyses are detailed in Supplementary Data 3,c. g) For the six proteins whose association with T2D PGS was attenuated by adjustment for BMI (P > 0.05; Extended Data Fig. 6f) gives, from mediation analysis, the estimated effect of T2D PGS on the protein levels through BMI (standard deviation change in protein levels through BMI per standard deviation increase in T2D PGS), percentage of T2D PGS to protein levels mediated by BMI, and the estimated effect of T2D PGS on protein levels independent of BMI in n = 3,072 INTERVAL participants. All P-values are two-sided.
Most PGS–protein associations were not explained by protein quantitative trait loci (pQTLs) (Supplementary Table 4); instead, they were highly polygenic (Fig. 1c). Each protein required a median 12% of the genome to explain its association with a PGS. Only 4 associations could be explained by pQTLs and contributing loci were spread across the genome for the remaining 46 associations (Extended Data Fig. 7).
Extended Data Fig. 7. Polygenicity of PGS to protein associations.
Linkage disequilibrium (LD) blocks contributing to each PGS to protein association in polygenicity tests. Briefly, each PGS was partitioned into 1,703 approximately independent LD blocks54 then tested for association with each protein in linear regression adjusting for age, sex, 10 genotype PCs, sample measurement batch, and time between blood draw and sample processing in 3,087 INTERVAL participants. Full summary statistics including exact two-sided P-values for these tests are detailed in Supplementary Data 3,e. Next, to obtain the set of LD blocks contributing to each PGS to protein association, LD blocks were sequentially removed from the PGS in ascending order by association P-value (two-sided) until the association between resulting PGS and protein levels were attenuated (two-sided P > 0.05). Full summary statistics including exact two-sided P-values for these tests are detailed in Supplementary Data 3,f. The polygenicity of PGS to protein association (% of genome) shown on the left (and in Fig. 1c) was subsequently computed based on the sum of lengths of all contributing LD blocks (in base pairs) as a proportion of the genome. Here, associations (−log10 two-sided P-values) between protein levels and LD blocks contributing to the PGS to protein association are shown. Regions in white contain LD blocks that did not contribute to the PGS to protein association. PGS to protein associations listed in red are those explained by pQTLs (cis and/or trans) rather than polygenic.
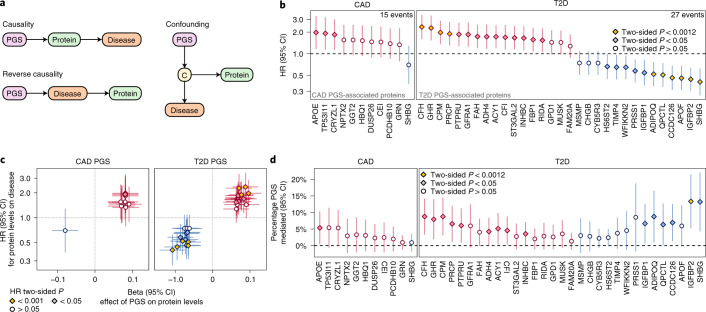
Three possible scenarios could explain a PGS–protein association29: (1) the protein plays a causal role in disease; (2) protein levels are changing in response to disease processes but are not themselves causal (reverse causality); and (3) protein levels are correlated with some other causal factor (confounding) (Fig. 2a). Utilizing a median of 7.7 years of follow-up in nationwide electronic hospital records, we examined whether levels of PGS-associated proteins were associated with risk of onset of the respective cardiometabolic disease, then performed mediation analysis30–32 to identify the proteins that mediate the PGS–disease associations and thereby play causal roles in disease pathogenesis.
Fig. 2. PGS-associated proteins influence 7.7-year risk of CAD and T2D.
a, Possible models of causality for PGS–protein–disease associations. C, causal disease factor upstream of the protein that induces a correlation between protein levels and disease. b, Association of PGS-associated proteins with 7.7-year risk of hospitalization with CAD or T2D in n = 3,087 INTERVAL participants in Cox proportional hazards models adjusting for age, sex, sample measurement batch and time between blood draw and sample processing. The data shown correspond to the HR for CAD or T2D conferred per s.d. increase in protein levels (points) and its 95% CI (vertical bars). P < 0.0012 indicates that the association was significant after Bonferroni correction for the 42 tests. c, Comparison of associations between protein levels and the CAD PGS or T2D PGS from Fig. 1b (x axes) to HRs for protein levels for 7.7-year risk of hospitalization with CAD or T2D from Fig. 2b (y axes). Beta estimates (points; x axes) correspond to s.d. change in protein levels per s.d. increase in CAD PGS or T2D PGS in the linear regression described in Fig. 1b. HRs (y axes) are as described in b. Linear regression and Cox proportional hazards models were fitted in the same n = 3,087 samples. The point shape and colour correspond to the P value in b. d, Percentage of PGS–disease association mediated by each protein in causal mediation analysis in n = 3,087 INTERVAL participants adjusting for age, sex, ten genotype principal components, sample measurement batch and time between blood draws. The data shown are the percentage of association between the PGS and hospitalization with CAD or T2D after 7.7 years of follow-up in Cox proportional hazard models mediated by each respective protein (points) and the 95% CI of this percentage (vertical bars). Proteins are ordered from left to right by their HR in Fig. 1b and coloured red where protein levels increased with PGS or blue where protein levels decreased with PGS in Fig. 1b. P < 0.0012 indicates that mediation was significant after Bonferroni correction for the 42 tests. Full summary statistics including exact two-sided P values for b–d are detailed in Extended Data Fig. 3.
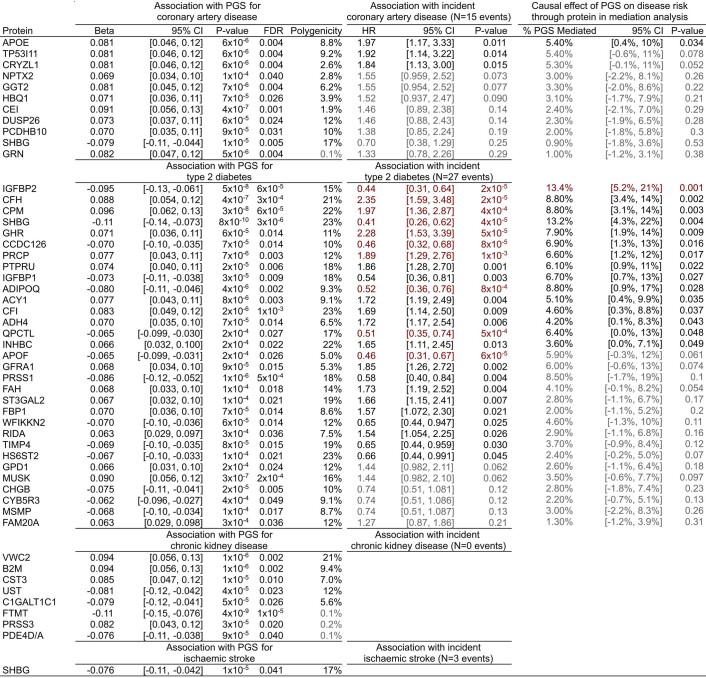
During follow-up of the participants with PGS and plasma proteomics, there were 27 incident T2D events and 15 incident CAD events, enabling us to evaluate the CAD and T2D PGS and their corresponding 42 associated proteins. Ten of 31 (32%) T2D PGS-associated proteins were significantly associated (P < 0.0012, Bonferroni correction for the 42 tested proteins) and a further 15 proteins were nominally significantly associated (P < 0.05) with increased risk of T2D (Fig. 2b and Extended Data Fig. 3). For the CAD PGS, no proteins were Bonferroni significant and 3 of 11 (27%) proteins were nominally significant. Notably, there was clear directional consistency between the effects of PGS on protein levels and hazard ratios (HRs) for protein levels on incident disease risk (Fig. 2c). Using mediation analysis, we found one protein, insulin-like growth factor-binding protein 2 (IGFBP2), that was a significant mediator (P < 0.0012) of polygenic T2D risk (Fig. 2d and Extended Data Fig. 3), indicating a causal role in disease pathogenesis. A further 1 and 14 proteins were nominally significant mediators of polygenic CAD and T2D risk, respectively. Protein–disease associations and causal mediation for IGFBP1 and IGFBP2 on polygenic T2D risk were replicated in independent samples where four PGS-associated proteins (IGFBP2, IGFBP1, progranulin (GRN) and tissue inhibitor of metalloproteinases 4 (TIMP4)) were measured with orthogonal proteomics technology (Supplementary Information and Supplementary Table 5).
Since polygenic disease risk is estimated from population-level data, it is unlikely that any single protein explains polygenic risk. In this study, we found that IGFBP2 explained 13.4% of the association between T2D PGS and incident T2D (Extended Data Fig. 3). Across all nominally significant mediators, proteins explained a median of 6.6% of PGS–disease associations (Extended Data Fig. 3), with the 1 CAD PGS mediator (apolipoprotein E (APOE)) explaining 5.4% of CAD polygenic risk–incident CAD association and the 15 T2D PGS mediators explaining 27% of the T2D polygenic risk–incident T2D association.
A complementary approach for causal inference, Mendelian randomization33, also supported causal effects on T2D for one protein, sex hormone-binding globulin (SHBG) (Extended Data Fig. 9 and Supplementary Tables 6 and 7), which is consistent with our mediation analysis and previous Mendelian randomization analysis of SHBG on T2D34. Notably, only 11 (22%) of the proteins associated with PGS could be tested with Mendelian randomization due to a lack of cis-pQTLs as genetic instruments, highlighting the complementarity of our PGS–protein association and mediation approach for identifying causal proteins.
Extended Data Fig. 9. Mendelian randomisation analysis.
a) Causal effects of protein levels on disease risk estimated through two-sample Mendelian randomisation analysis of pQTL summary statistics and disease GWAS summary statistics. OR: consensus estimate of the odds ratio conferred per standard deviation increase in protein levels across five Mendelian randomisation methods. * Estimated causal effect is directionally consistent with PGS-protein associations in Fig. 1b. 95% CI: 95% confidence interval. P-value: Two-sided P-value obtained by averaging Z-scores across five Mendelian randomisation methods. Entries are greyed out where P > 0.05, and red where P < 0.0038 (Bonferroni correction for 13 tests). Pleiotropy P-value: two-sided P-value for the intercept term in Egger regression, indicating where P < 0.05 confounding of the causal estimate by horizontal pleiotropy. Full summary statistics including exact P-values are detailed in Supplementary Table 6. b) Dose response curves showing the estimated causal effect of changes in protein levels on disease risk for each protein and disease. Points on each plot show the cis-pQTLs used as genetic instruments for each test. On the x-axes, points show the standard deviation change in protein levels per copy of the minor allele in the pQTL summary statistics, and horizontal bars show + /- the standard error. On the y-axes, points show odds ratio conferred per copy of the minor allele in the GWAS summary statistics, and vertical bars indicate show + /- the standard error. Effect sizes, standard errors, and exact two-sided P-values from pQTL and GWAS summary statistics are detailed in Supplementary Table 7. The slope of the orange dashed line corresponds to the estimated causal effect (consensus Odds Ratio from a). The yellow ribbon shows the 95% confidence interval for the estimated causal effect (slope), accounting also for the 95% confidence interval for the intercept term in Egger regression.
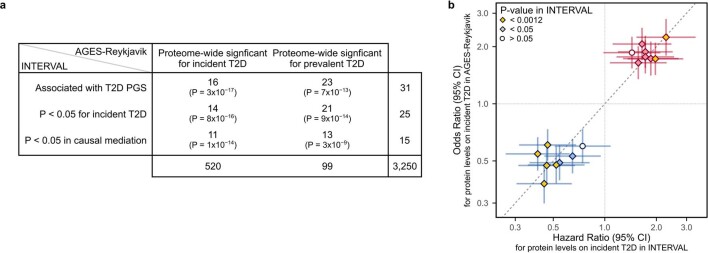
Our findings are also consistent with a previous observational study of plasma proteins and T2D risk in the Age, Gene/Environment Susceptibility-Reykjavik Study (AGES-Reykjavik), a cohort of 5,438 older Icelanders with 654 prevalent T2D cases and 112 incident T2D cases in 2,940 participants with 5 years of follow-up and free of T2D at baseline35. Of the 31 proteins associated with the polygenic risk of T2D in our healthy, pre-symptomatic cohort, 23 were associated with prevalent T2D and 16 were associated with incident T2D in the AGES-Reykjavik Study (Extended Data Fig. 10a and Supplementary Table 8). Notably, HRs for incident T2D in our INTERVAL analyses were directionally consistent and of similar magnitude to the odds ratios for incident T2D in the AGES-Reykjavik Study (Extended Data Fig. 10b), with a significant overlap between the significant proteins from our causal mediation analysis in INTERVAL and those previously associated with incident T2D (Extended Data Fig. 10a).
Extended Data Fig. 10. Overlap of results with proteome-wide T2D associations in AGES-Reykjavik.
a) Contingency table tabulating the overlap in results from our study detailed in Extended Data Fig. 3 (rows) with proteome-wide significant associations with incident and prevalent T2D in AGES-Reykjavik in Gudmundsdottir et al. 202035 (columns). One-sided P-values from Fisher’s exact tests are given in each cell testing whether the overlap is greater than expected by chance. Row totals and column totals indicate the number of proteins in each row and column group, and the total overlap in proteins present in both studies (3,250) is given in the bottom right. b) For the 16 of 31 proteins nominally associated with T2D PGS in INTERVAL (Fig. 2b) and proteome-wide significant for incident T2D in AGES-Reykjavik, compares hazard ratios (points; x-axis) for incident T2D in INTERVAL (N = 27 cases over 7.7 years of follow-up in 3,087 participants) to odds ratios (points; y-axis) for incident T2D in AGES-Reykjavik (N = 112 cases after 5 years of follow-up in 2,940 participants). Cox proportional hazards models in INTERVAL were fit with follow-up as time scale, adjusting for age, sex, 10 genotype PCs, sample measurement batch, and time between blood draw and sample processing. Logistic regression in AGES-Reykjavik were fit adjusting for age and sex35. Horizontal and vertical bars correspond to the 95% confidence intervals of the hazard ratios and odds ratios respectively. Two-sided P < 0.0012 indicates association with incident T2D in INTERVAL from Fig. 2b was significant after Bonferroni correction for the 42 tested protein to disease associations. Summary statistics including exact two-sided P-values from both analyses are given in Supplementary Table 8.
Finally, we examined the druggability of proteins mediating polygenic disease risk using the druggable genome22. We found that 12 of the 16 proteins mediating the polygenic disease risk were also druggable targets (Table 1). Nine of these were targets of, or interacted with, 76 compounds in the DrugBank database36 (Supplementary Table 9). These results suggest therapeutic potential for these proteins as modulators of risk for T2D or CAD and indicate high priority targets for further investigation.
Table 1.
Druggable proteins that were nominally significant mediators of polygenic risk
| Protein | PGS/ disease | Evidence tiera | Small-molecule targetb | Biological targetc | ADMEd | DrugBank compoundse | Summary of therapeutic uses for licensed drugs |
|---|---|---|---|---|---|---|---|
| ADH4 | T2D | Tier 1 | Y | N | Y | 3 | Female reproductive disorders, infection control |
| GHR | T2D | Tier 1 | N | Y | N | 3 | Acromegaly, dwarfism, idiopathic short stature, human immunodeficiency virus weight loss |
| PRCP | T2D | Tier 1 | Y | N | N | 0 | |
| SHBG | T2D | Tier 1 | Y | Y | N | 68 | Fertility and reproductive treatments, cancers, mental health, developmental disorders, hypertension, high cholesterol |
| CPM | T2D | Tier 2 | Y | Y | N | 0 | |
| IGFBP1 | T2D | Tier 2 | Y | Y | N | 1 | Growth failure due to insulin-like growth factor 1 deficiency |
| IGFBP2 | T2D | Tier 2 | Y | Y | N | 1 | Growth failure due to insulin-like growth factor 1 deficiency |
| ADIPOQ | T2D | Tier 3A | N | Y | N | 0 | |
| APOE | CAD | Tier 3A | N | Y | N | 5 | Zinc deficiency |
| CFH | T2D | Tier 3A | N | Y | N | 5 | Zinc deficiency, malnutrition, ear and respiratory infections |
| CFI | T2D | Tier 3A | N | Y | N | 3 | Zinc deficiency, malnutrition, ear and respiratory infections |
| INHBC | T2D | Tier 3A | N | Y | N | 0 |
List of PGS-associated proteins with nominal evidence (P < 0.05) of causal disease effects in mediation analysis (Fig. 2d) that are part of the druggable genome. Full details of each drug and interaction are provided in Supplementary Table 9. Y, yes; N, no.
aEvidence of druggability in Finan et al.22. Tier 1: targets of approved small molecules, biotherapeutic drugs and clinical-phase drug candidates. Tier 2: targets with known bioactive drug-like small-molecule binding partners as well as those with ≥50% identity (over ≥75% of the sequence) with approved drug targets. Tier 3A: secreted or extracellular proteins, proteins with more distant similarity to approved drug targets and members of key druggable gene families not already included in tier 1 or 2, with genes that were in proximity (±50 kb) to a GWAS SNP and had an extracellular location.
bThe protein is targeted, or predicted to be targeted, by a small molecule.
cThe protein is targeted, or predicted to be targeted, by a biotherapeutic (monoclonal antibody/enzyme or other protein).
dThe protein is involved in absorption, distribution, metabolism or excretion (ADME) of a compound. The information in these preceding columns was obtained from Table S1 in Finan et al.22.
eThe number of drugs or compounds in DrugBank database v.5.17 (https://go.drugbank.com/releases/5-1-7) that interact with the protein.
Polygenic disease scores are explicitly constructed to maximize risk prediction, typically without consideration of the underlying biology. However, PGS also hold additional promise for identifying molecular pathways in the development and progression of disease8,29. In this study, we identified 49 plasma proteins significantly associated with PGS for cardiometabolic disease in a healthy pre-disease cohort. Twenty-eight of these proteins were associated with increased risk of future disease and 16 were nominally significant mediators of T2D or CAD, including 12 druggable targets, suggesting that their modulation may potentially attenuate disease risk.
The vast majority of PGS–protein associations were highly polygenic, including for several well-known cardiometabolic disease proteins. This polygenicity was driven by aggregate modest polygenic effects on protein levels from across the genome, which were independent of cis- and trans-pQTLs and also present for proteins without pQTLs or for which sample sizes have not yet been sufficient for pQTL detection. This highlights the complementarity of PGS to established approaches that utilize information at a single locus, such as Mendelian randomization, colocalization and fine-mapping. However, it is important to recognize that mediation analysis provides weaker evidence of causality than these established single-locus approaches since it is more difficult to rule out confounding (either from measured or unmeasured factors)37, especially since PGS by design capture horizontal pleiotropy.
Our findings identify new potential targets of cardiometabolic disease that both are supported by human genetic evidence of causality and may be amenable to pharmacological manipulation. Our strongest results were for IGFBP2, a druggable target22, as a replicable mediator of polygenic risk and incident T2D. IGFBP2 is involved in the regulation of glucose uptake into adipocytes and is associated with increased insulin sensitivity and decreased adipogenesis38–41. Increased plasma IGFBP2 levels have been associated with lower T2D risk35,42,43; our findings are directionally consistent and indicate that this association is likely causal. Ten additional druggable proteins were found to be new mediators between polygenic risk and incident T2D.
Twelve new protein associations were found for CAD, CKD or T2D. Among these, the strongest evidence was for alcohol dehydrogenase 4 (ADH4), which is involved in a number of metabolic pathways44 and was found to be both a mediator of polygenic risk and incident T2D and a druggable target. Furthermore, several new associations concerned proteins with sparse literature on their function; for example, crystallin zeta like 1 (CRYZL1) was associated with polygenic risk and incident CAD; however, little is known about CRYZL1 beyond its gene identification45.
Proteomic data are becoming increasingly available in cohorts of large sample sizes, such as the planned proteomic profiling of UK Biobank participants. Proteomic platforms are also increasing their coverage of the human proteome. Therefore, we anticipate that our PGS mediation analysis approach will enable the identification of further causal proteins for cardiometabolic and other polygenic diseases in future studies.
Overall, this study demonstrates that PGS can be utilized to elucidate new disease biology with therapeutic potential and provides a useful study design for future studies into the molecular drivers of polygenic disease.
Methods
INTERVAL cohort
INTERVAL is a cohort of approximately 50,000 participants nested within a randomized trial studying the safety of varying the frequency of blood donation24,25. Participants were blood donors aged 18 years and older (median 44 years of age; 49% women) recruited between June 2012 and June 2014 from 25 centres across England. The collection of their blood samples for research purposes was done using standard protocols24: blood samples for research purposes were collected in 6-ml EDTA tubes using standard venepuncture protocols. The tubes were inverted three times and transferred at ambient temperature to the UK Biocentre for processing. Plasma was extracted into two 0.8-ml plasma aliquots by centrifugation and subsequently stored at −80 °C before use. Participants gave written informed consent and this study was approved by the National Research Ethics Service (no. 11/EE/0538).
Electronic health records were obtained for all INTERVAL participants from the January 2021 release of the National Health Service (NHS) Hospital Episode Statistics database (https://digital.nhs.uk/data-and-information/data-tools-and-services/data-services/hospital-episode-statistics) for all events up to 8 February 2020, before the onset of the COVID-19 pandemic in England. The median and maximum follow-up times were 6.9 years and 7.7 years, respectively. The earliest available hospital record for any INTERVAL participant was from 25 March 1999, with a maximum retrospective follow-up of 13.6 years. These records came in the form of International Statistical Classification of Diseases and Related Health Problems, 10th revision (ICD-10) codes46 and were subsequently made available to analysts after summarization into 301 end points using the CALIBER rule-based phenotyping algorithms47 (https://www.caliberresearch.org/portal). ICD-10 codes contributed to each event regardless of whether they coded for primary or non-primary diagnoses in the hospital records.
Genotyping, quality control and imputation of INTERVAL participants were performed as described previously48: participants were genotyped using the Affymetrix UK Biobank Axiom array in ten batches. Samples were removed if they had sex mismatch, had extreme heterozygosity, were of non-European ancestry or were duplicate samples. Related samples were removed by excluding one sample from each pair of close relatives (first- or second-degree; identity by descent ). Genotyped variants were removed if they were monomorphic, bi-allelic and had Hardy–Weinberg equilibrium P < 5 × 10−6 or call rate <99%. SHAPEIT3 was used to phase variants; imputation to the UK10K/1000 Genomes panel was performed using the Sanger Imputation Server (https://imputation.sanger.ac.uk).
Protein levels in INTERVAL were quantified using SOMAscan assays, processed and quality-controlled as described previously26: relative concentrations of 4,034 SOMAscan aptamers were measured in 3,562 INTERVAL participants in two batches by SomaLogic using v.3 of the SOMAscan platform. Aptamer concentrations (relative fluorescence units) were natural log-transformed and then adjusted within each batch for participant age, sex, the first three genetic principal components and time between blood draw and sample processing (<1 or >1 day); the residuals were then inverse rank normal-transformed. In this study, we further adjusted the normalized protein levels used in previous studies for batch number and filtered to 3,793 high-quality aptamers targeting 3,438 proteins after obtaining the latest information about aptamer sensitivity and specificity from SomaLogic. Aptamers were excluded if, in v.4 of the SOMAscan platform, they (1) targeted non-human proteins, (2) measured the fusion construct rather than the target protein or (3) measured a contaminant. A curated information sheet for all 4,034 aptamers is provided in Supplementary Data 1. The distributions of aptamer levels and associations with covariates before and after quality control are given in Supplementary Data 2.
In total, 3,087 INTERVAL participants, without prevalent cardiometabolic disease (see below) and with matched genotype, proteomic and electronic health record data available for the primary analyses, passed quality control.
Prevalent disease exclusion
The NHS Blood and Transplant blood donation eligibility criteria (https://www.blood.co.uk/who-can-give-blood/) meant that there were built-in exclusions for the INTERVAL cohort for people with a history of major diseases, recent illness or infection. Specifically, for cardiometabolic diseases, the blood donation eligibility criteria excluded individuals who had been diagnosed with AF, had a history of any stroke or a history of major heart disease, including heart failure, coronary thrombosis, myocardial infarction, cardiomyopathy, ischaemic heart disease and arrhythmia, or surgery for non-congenital heart conditions. Use of aspirin or other blood thinners to control elevated blood pressure (hypertension) also made people ineligible to donate blood and participate in the INTERVAL cohort. Individuals with T2D were ineligible unless their T2D was well controlled by diet alone, did not require regular insulin treatment and the individual had not required insulin treatment for at least 4 weeks before attempting blood donation. Extended details on the blood donation criteria eligibility for specific diseases, medications and lifestyle factors can be found at https://my.blood.co.uk/knowledgebase.
In addition to intrinsic exclusion due to the blood donation eligibility criteria, participants were excluded from the analyses if they had any events relating to cardiometabolic disease before baseline assessment. Among the 301 CALIBER end points, we classified 48 as cardiometabolic disease or having potential to introduce reverse causality by modifying risk for incident AF, CAD, CKD, IS or T2D (Supplementary Table 1). In total, 87 participants (2.7%) were excluded, predominantly due to prevalent hypertension (n = 57 events; 66% of excluded participants) and prevalent diabetes (n = 11 events; 13% of excluded participants), with all others accounting for less than 5% of excluded participants (Supplementary Table 1).
PGS
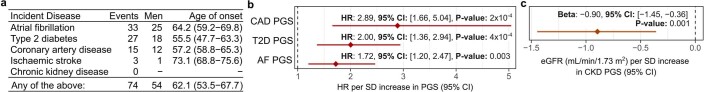
PGS were derived in a consistent manner by linkage disequilibrium (LD) thinning, at an r2 threshold of 0.9, the latest GWAS summary statistics for each respective disease (Supplementary Information). The GWAS summary statistics used to derive the AF, CKD and T2D PGS were those published by Nielsen et al.13 (GCST006414), Wuttke et al.14 (GCST008065) and Mahajan et al.15 (GCST007517), respectively. The PGS for CAD and IS used in this study were our previously published CAD49 and stroke50 meta-PGS. The CAD PGS was derived from the meta-analysis of three PGS for CAD, including a PGS derived as described above from the GWAS summary statistics published by Nikpay et al.51. The IS PGS was derived from the meta-analysis of PGS for IS and its risk factors, including a PGS derived as described above from the GWAS summary statistics for IS published by Malik et al.16. Each PGS comprised 1.75–3.23 million single-nucleotide polymorphisms (SNPs) genome-wide and is available to download through the Polygenic Score Catalog52 (https://www.pgscatalog.org/) with accession nos PGS000727 (AF), PGS000018 (CAD), PGS000728 (CKD), PGS000039 (IS) and PGS000729 (T2D). All PGS were derived from the GWAS summary statistics including only individuals with European ancestry. See Supplementary Information and Extended Data Fig. 8 for details on PGS validation.
Extended Data Fig. 8. Incident disease and PGS validity.
a) Incident disease events over the 7.7 year of follow-up in the n = 3,087 INTERVAL participants. Endpoint: incident disease definition available in INTERVAL for the relevant PGS, as defined by CALIBER phenotyping algorithms. Age of onset: median age of first hospitalisation with the respective endpoint. Numbers in brackets gives the interquartile range. b) Hazard ratio (HR) (points) and 95% confidence interval (95% CI) (horizontal bar) for 7.7 year risk of hospitalisation with the respective endpoint per standard deviation increase in the respective PGS in cox proportional hazards models using follow-up as time scale and adjusting for age, sex, 10 genotype PCs, sample measurement batch, and time between blood draw and sample processing in n = 3,087 INTERVAL participants. P-values are two-sided. c) Association between CKD PGS with estimated glomerular filtration rate (eGFR), a marker of renal function used in chronic kidney disease diagnosis: decreased eGFR is indicative of reduced renal function98. EGFR was computed from serum creatinine in n = 3,307 participants using the CKD-EPI equation99. Association was fit with linear regression adjusting for age and sex, and 10 genotype PCs. The point corresponds to the change in eGFR per standard deviation increase in CKD PGS, and the horizontal bar corresponds to the 95% CI. P-values are two-sided.
The levels of each PGS (sum of dosages × weights) were computed in INTERVAL from probabilistic dosage data using PLINK v.2 (ref. 53) after mapping PGS variants to those available in the INTERVAL genotype data (Supplementary Information). The levels of each PGS were adjusted for the first ten principal components of the imputed genotype data and standardized to have mean of 0 and s.d. of 1 before downstream statistical analyses.
PGS–protein associations
Each of the five PGS was tested for association with each of the 3,793 aptamers using linear regression (Fig. 1a,b and Extended Data Fig. 3). PGS and proteins were adjusted for covariates and normalized before model fitting (see above). Linear regression coefficients were averaged where multiple high-quality aptamers targeted the same protein (Supplementary Information). FDR correction was subsequently applied across the 3,438 P values (1 per protein) for each PGS separately. Details on aptamer specificity and sensitivity are given in Supplementary Table 2 for the 54 aptamers targeting the 49 PGS-associated proteins; aptamer-specific estimates of PGS on protein levels are detailed in Supplementary Table 3 for the 5 PGS-associated proteins targeted by more than one aptamer (WFIKKN2, GPD1, IGFBP1, IGFBP2 and SHBG).
Polygenicity of PGS–protein associations
To quantify the polygenicity of PGS–protein associations (Fig. 1c and Extended Data Fig. 7), we performed a multistep experiment to determine the proportion of the genome required to explain that association. First, we split the given PGS into separate scores for each of the 1,703 approximately independent LD blocks estimated in Europeans from the 1000 Genomes reference panel by Berisa and Pickrell54 (https://bitbucket.org/nygcresearch/ldetect-data/src/master/EUR/fourier_ls-all.bed). Next, we tested each of these 1,703 scores for association with the given protein (Supplementary Data 3e). Then, we retested the PGS to protein association, progressively removing independent LD blocks, at each step removing the LD block whose score had the strongest association with the protein. From this, we quantified polygenicity (Fig. 1c) based on the LD blocks needed to be removed from the given PGS to attenuate the PGS–protein association (so that association P > 0.05; Supplementary Data 3f) as the sum of removed LD block sizes/sum of all LD block sizes (that is, the proportion of the genome removed). Extended Data Fig. 7 shows the independent LD blocks contributing to the polygenicity of each PGS–protein association.
Independent contributions of PGS and pQTLs to protein levels
Multivariable linear regression models were fitted for each protein on PGS levels and pQTL dosages to estimate their independent contributions to protein levels (Supplementary Table 4). The pQTLs used for each protein were (1) conditionally independent pQTLs mapped in INTERVAL and published by Sun et al.26, which included both cis-pQTLs (within 1 Mb of the encoding gene) and trans-pQTLs passing the trans significance threshold of P < 1.5 × 10−11; (2) trans-pQTLs with P < 1.5 × 10−11 (lead variant only) for proteins not published in Sun et al.26 (B2M, DUSP26 and FTMT); and (3) hierarchically significant55,56 cis-pQTLs (lead variant only) mapped in this study (Supplementary Data 4 and Supplementary Information) for proteins without cis-pQTLs passing the trans-pQTL significance threshold above (ACY1, ADIPOQ, APOE, CST3, GPD1, PTPRU, SHBG and UST).
Incident disease associations
PGS and protein levels were tested for association with incident disease using Cox proportional hazards models adjusting for age and sex (Fig. 2b and Extended Data Fig. 8) using the survival package (version 3.2-7) in R. The timescale used was time from baseline to first event of the relevant disease or to the latest available date in the hospital records (8 February 2020). PGS and proteins were adjusted for covariates and normalized before model fitting (see above). Cox model coefficients were averaged where multiple high-quality aptamers targeted the same protein (Supplementary Information).
Incident disease events for AF, CAD, CKD, IS and T2D were defined as the first hospital episode for the closest matching CALIBER phenotype47. Incident AF events were defined as any hospital episode with the ICD-10 code I48. Incident IS events were defined as any hospital episode with the ICD-10 code I63 or I69.3. Incident CAD events were defined as any hospital episode with ICD-10 code I21–I23, I24.1 or I25.2 (CALIBER end point myocardial infarction). The closest matching CALIBER phenotype for T2D was for diabetes more broadly, including ICD-10 codes for any hospital episode for T1D or T2D or complications thereof: E10–E14, G59.0, G63.2, H28.0, H36.0, M14.2, N08.3 or O24.0–O24.3. However, we note that individuals with T1D are not eligible to donate blood and adult onset of T1D is relatively rare compared to T2D57. The closest matching CALIBER phenotype for CKD was for end-stage renal disease more broadly, defined as any hospital episode with the ICD-10 codes N16.5, N18.5, T82.4, T86.1, Y60.2, Y61.2, Y84.1, Z49.1, Z49.2, Z94.0 and Z99.2.
Mediation analysis
Mediation analysis was used to identify causal proteins by identifying the PGS-associated proteins that partially mediate the association of PGS on disease (Fig. 2d). This approach uses the counterfactual framework to infer causal effects30–32 and can be adapted to this setting because the arrow of causality between PGS and any associated phenotype can only flow in one direction since the PGS is fixed at conception (that is, the underlying alleles in each person cannot be modified later in life by protein levels or the development of cardiometabolic disease). In this study, we used the natural effects model developed by Vansteelandt et al.58, which is available in the medflex R package (version 0.6-7 used in this study)59, to estimate natural indirect effects (effects of PGS on disease through protein levels) on the log odds scale by imputing unobserved counterfactuals. Standard errors were computed using the robust sandwich estimator60, from which 95% confidence intervals (CIs) and P values were calculated. The percentage of PGS–disease associations mediated by each protein and 95% CIs were subsequently computed as the natural indirect effect and its 95% CI was divided by the total effect estimated by each mediation test. Multiple mediation analysis61 was performed using the R package mma (version 10.3.2)62 to quantify the proportion of PGS–disease association mediated by the 15 causal T2D proteins.
Mendelian randomization
Two-sample Mendelian randomization33 was also performed as an orthogonal approach to identify proteins that may play a causal role in disease (Extended Data Fig. 9 and Supplementary Tables 6 and 7). PGS-associated proteins were tested provided they had three or more independent, as determined by LD (r2 < 0.1), hierarchically significant cis-pQTLs after mapping cis-pQTLs to the GWAS summary statistics (Supplementary Information) using five different Mendelian randomization methods63–66, each of which makes use of information across three or more instruments to estimate causal effects, with each method differentially robust to different sources of bias, to obtain a consensus (median) estimate of causal effects of protein levels on disease risk (Supplementary Information). Hierarchically significant cis-pQTLs and tagging variants (LD r2 > 0.1) were excluded where they encoded changes to protein structure67 (for example, missense mutations) and therefore potentially reflected differences in aptamer binding affinity rather than regulation of protein levels (Supplementary Information). Aptamers were also excluded if they had similar affinity for/comparable binding to multiple proteins or differential binding to specific isoforms (Supplementary Table 3 and Supplementary Information).
In total, 11 of the 49 PGS-associated proteins could be tested. GWAS summary statistics were obtained from Nelson et al.17 for CAD (GCST004787), Wuttke et al.14 for CKD (GCST008065), Malik et al.16 for IS (GCST006906) and Mahajan et al.15 for T2D (GCST007518). In all cases, we used the GWAS summary statistics for the samples of recent European ancestry. For T2D, we used the BMI-adjusted GWAS summary statistics to avoid false positive causal estimates arising where pQTLs influence T2D risk through BMI rather than through the tested protein (horizontal pleiotropy). We considered there to be a significant causal effect where P < 0.05 along with no significant evidence that causal effects were due to associations of the pQTLs with some other causal risk factor (horizontal pleiotropy; Egger intercept66 P > 0.05). Analysis was performed using the R package MendelianRandomization (version 0.5.0)68. Colocalization analysis69 was also performed where the cis-pQTL instruments had P < 1 × 10−6 in the respective GWAS (Supplementary Table 7 and Supplementary Information).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Notes and Methods, references and legends for Supplementary Tables and Data.
Supplementary Tables 1–9.
Supplementary Data 1–4.
Acknowledgements
Participants in the INTERVAL randomized controlled trial were recruited with the active collaboration of NHS Blood and Transplant (www.nhsbt.nhs.uk), which has supported fieldwork and other elements of the trial. DNA extraction and genotyping were co-funded by the National Institute for Health Research (NIHR), the NIHR BioResource (http://bioresource.nihr.ac.uk) and the NIHR Cambridge Biomedical Research Centre (BRC) (no. BRC-1215-20014). Olink Proteomics assays were funded by Biogen. SomaLogic assays were funded by Merck and the NIHR Cambridge BRC (no. BRC-1215-20014). The academic coordinating centre for INTERVAL was supported by core funding from the NIHR Blood and Transplant Research Unit in Donor Health and Genomics (no. NIHR BTRU-2014-10024), UK Medical Research Council (MRC) (no. MR/L003120/1), British Heart Foundation (nos SP/09/002, RG/13/13/30194 and RG/18/13/33946) and the NIHR Cambridge BRC (no. BRC-1215-20014). A complete list of the investigators and contributors to the INTERVAL trial is provided in ref. 25. The academic coordinating centre thanks blood donor centre staff and blood donors for participating in the INTERVAL trial. This work was supported by Health Data Research UK, which is funded by the UK MRC, Engineering and Physical Sciences Research Council (EPSRC), Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome. This study was also supported by the Victorian Government’s Operational Infrastructure Support programme. This work was performed using resources provided by the Cambridge Service for Data Driven Discovery operated by the University of Cambridge Research Computing Service (https://www.hpc.cam.ac.uk/high-performance-computing), provided by Dell EMC and Intel using tier-2 funding from the EPSRC (capital grant no. EP/P020259/1), and DiRAC funding from the Science and Technology Facilities Council (www.dirac.ac.uk). This work uses data provided by patients and collected by the NHS and Public Health England as part of their care and support. Data on hospital episode statistics, mortality and cancer registration were obtained from NHS Digital (data sharing agreement reference no. DARS-NIC-156334-711SX). S.C.R. and J.M. were funded by the NIHR Cambridge BRC (no. BRC-1215-20014). S.A.L. is supported by a Canadian Institutes of Health Research postdoctoral fellowship (no. MFE-171279). G.A. was supported by a National Health and Medical Research Council of Australia Early Career Fellowship (no. 1090462). S.B. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (no. 204623/Z/16/Z). A.V.K. was supported by grants from the National Human Genome Research Institute (award nos 1K08HG010155 and 5UM1HG008895), an institutional grant from the Broad Institute of MIT and Harvard (variant2function) and a Hassenfeld Scholar Award from Massachusetts General Hospital. J.D. holds a British Heart Foundation Professorship and an NIHR Senior Investigator Award. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The views expressed in this manuscript are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Extended data
Author contributions
S.C.R., S.Z., M. Chaffin, B.G.D., A.C.C., N.S., S.K., A.V.K., A.S.B. and M.I. conceptualized the study. S.C.R., M.A., Y.L. and A.S.B. curated the data. S.C.R. carried out the formal analysis. J.D. and M.I. acquired the funding. S.C.R., M.A., Y.L., B.G.D., A.C.C. and M. Chaffin carried out the investigation. S.C.R., S.A.L., S.M.T., S.L., P.S., J.M., G.A. and M.I. devised the methodology. A.V.K., S.K., A.S.B. and M.I. administered the project. W.H.O., D.J.R., N.A.W., B.G.D., A.C.C., E.D.A., M. Chapman, J.D., A.S.B. and M.I. curated the resources. S.C.R. managed the software. G.A., B.G.D., A.C.C., E.D.A., S.K., A.S.B. and M.I. supervised the study. S.C.R., S.A.L., S.M.T., S.L., P.S., J.M., G.A., S.B. and A.V.K. validated the data. S.C.R. visualized the data. S.C.R. and M.I. wrote the original manuscript draft. S.C.R., S.A.L., S.M.T., S.L., P.S., J.M., G.A., B.G.D., A.C.C., N.S., S.B., A.V.K., J.D., A.S.B. and M.I. reviewed and edited the manuscript draft.
Data availability
All data used in this study are publicly available or deposited in a public repository. The INTERVAL cohort data are available via the European Genome-phenome Archive with study accession no. EGAS00001002555. Dataset access is subject to approval by an independent data access committee since the data contain potentially identifying and sensitive patient information. Response times from the data access committee are typically within 1 week. All other data used in this study are publicly available without restriction. The PGS used in this study are available to download through the Polygenic Score Catalog (https://www.pgscatalog.org/) with accession nos PGS000727 (AF), PGS000018 (CAD), PGS000728 (CKD), PGS000039 (IS) and PGS000729 (T2D). The GWAS summary statistics used to generate new PGS for CKD, T2D and AF in this study are available to download through the GWAS Catalog (https://www.ebi.ac.uk/gwas/) with study accession nos GCST008065 (for the CKD GWAS published by Wuttke et al.14), GCST007517 (for the T2D GWAS published by Mahajan et al.15) and GCST006414 (for the AF GWAS published by Nielsen et al.13). The additional GWAS summary statistics used for Mendelian randomization analysis are also available through the GWAS Catalog with study accession nos GCST004787 (for the CAD GWAS published by Nelson et al.17), GCST006906 (for the IS GWAS published by Malik et al.16) and GCST007518 (for the T2D GWAS adjusted for BMI published by Mahajan et al.15). Full pQTL summary statistics published by Sun et al.26 for all SomaLogic SOMAscan aptamers are available to download from https://www.phpc.cam.ac.uk/ceu/proteins/. The DrugBank database is publicly available to download at https://www.drugbank.ca/releases/latest. Summary statistics for all statistical tests are available in Supplementary Data 3; the additional cis-pQTLs mapped in this study are provided in Supplementary Data 4.
Code availability
The code used to generate the results of this study, along with a detailed list of software and versions, is available on GitHub (https://github.com/sritchie73/cardiometabolic_prs_plasma_proteome/), which is permanently archived by Zenodo70 at 10.5281/zenodo.4762747.
Competing interests
Several authors are now employed by or run pharmaceutical companies. All significant contributions to this study were made before these roles and the named companies had no role in the study. M.A. is an employee of AstraZeneca. P.S. is an employee of Roche. J.M. is an employee of Genomics PLC. G.A. is an employee of CSL Limited. S.K. is the chief executive officer of Verve Therapeutics. The other authors declare no competing interests.
Footnotes
Peer review information Nature Metabolism thanks Matthew Nelson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Isabella Samuelson.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Scott C. Ritchie, Email: sr827@medschl.cam.ac.uk
Michael Inouye, Email: mi336@medschl.cam.ac.uk.
Extended data
is available for this paper at 10.1038/s42255-021-00478-5.
Supplementary information
The online version contains supplementary material available at 10.1038/s42255-021-00478-5.
References
- 1.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh P-R, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 2019;28:R133–R142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- 5.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy MI, Mahajan A. The value of genetic risk scores in precision medicine for diabetes. Expert Rev. Precis. Med. Drug Dev. 2018;3:279–281. doi: 10.1080/23808993.2018.1510732. [DOI] [Google Scholar]
- 8.International Common Disease Alliance Recommendations and White Paper v.1.0 (ICDA Organizing Committee and Working Groups, 2020); https://drive.google.com/file/d/16SVJ5lbneN9hB9E03PZMhpescAN527HO/view
- 9.Erdmann J, Kessler T, Munoz Venegas L, Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc. Res. 2018;114:1241–1257. doi: 10.1093/cvr/cvy084. [DOI] [PubMed] [Google Scholar]
- 10.Visscher PM, et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018;19:491–504. doi: 10.1038/s41576-018-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020;52:1122–1131. doi: 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen JB, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuttke M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2019;51:957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan A, et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik R, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CP, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 18.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinnott-Armstrong N, Naqvi S, Rivas M, Pritchard JK. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. eLife. 2021;10:e58615. doi: 10.7554/eLife.58615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Li YI, Pritchard JK. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177:1022–1034.e6. doi: 10.1016/j.cell.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, et al. Polygenic risk scores in cardiovascular risk prediction: a cohort study and modelling analyses. PLoS Med. 2021;18:e1003498. doi: 10.1371/journal.pmed.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finan C, et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017;9:eaag1166. doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghoussaini M, et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49:D1311–D1320. doi: 10.1093/nar/gkaa840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore C, et al. The INTERVAL trial to determine whether intervals between blood donations can be safely and acceptably decreased to optimise blood supply: study protocol for a randomised controlled trial. Trials. 2014;15:363. doi: 10.1186/1745-6215-15-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Angelantonio E, et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. Lancet. 2017;390:2360–2371. doi: 10.1016/S0140-6736(17)31928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovanović D, Krstivojević P, Obradović I, Durdević V, Dukanović L. Serum cystatin C and beta2-microglobulin as markers of glomerular filtration rate. Ren. Fail. 2003;25:123–133. doi: 10.1081/JDI-120017475. [DOI] [PubMed] [Google Scholar]
- 28.van Poelje PD, Dang Q, Erion MD. Fructose-1,6-bisphosphatase as a therapeutic target for type 2 diabetes. Drug Discov. Today Ther. Strateg. 2007;4:103–109. doi: 10.1016/j.ddstr.2007.10.003. [DOI] [Google Scholar]
- 29.Holmes MV, Davey Smith G. Can Mendelian randomization shift into reverse gear? Clin. Chem. 2019;65:363–366. doi: 10.1373/clinchem.2018.296806. [DOI] [PubMed] [Google Scholar]
- 30.Imai K, Keele L, Tingley D. A general approach to causal mediation analysis. Psychol. Methods. 2010;15:309–334. doi: 10.1037/a0020761. [DOI] [PubMed] [Google Scholar]
- 31.Imai K, Keele L, Tingley D, Yamamoto T. Unpacking the black box of causality: learning about causal mechanisms from experimental and observational studies. Am. Polit. Sci. Rev. 2011;105:765–789. doi: 10.1017/S0003055411000414. [DOI] [Google Scholar]
- 32.Hernán MA. A definition of causal effect for epidemiological research. J. Epidemiol. Community Health. 2004;58:265–271. doi: 10.1136/jech.2002.006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding EL, et al. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N. Engl. J. Med. 2009;361:1152–1163. doi: 10.1056/NEJMoa0804381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudmundsdottir V, et al. Circulating protein signatures and causal candidates for type 2 diabetes. Diabetes. 2020;69:1843–1853. doi: 10.2337/db19-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanderWeele TJ. Mediation analysis: a practitioner’s guide. Annu. Rev. Public Health. 2016;37:17–32. doi: 10.1146/annurev-publhealth-032315-021402. [DOI] [PubMed] [Google Scholar]
- 38.Russo VC, Azar WJ, Yau SW, Sabin MA, Werther GA. IGFBP-2: the dark horse in metabolism and cancer. Cytokine Growth Factor Rev. 2015;26:329–346. doi: 10.1016/j.cytogfr.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Assefa B, et al. Insulin-like growth factor (IGF) binding protein-2, independently of IGF-1, induces GLUT-4 translocation and glucose uptake in 3T3-L1 adipocytes. Oxid. Med. Cell. Longev. 2017;2017:3035184. doi: 10.1155/2017/3035184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheatcroft SB, et al. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes. 2007;56:285–294. doi: 10.2337/db06-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedbacker K, et al. Antidiabetic effects of IGFBP2, a leptin-regulated gene. Cell Metab. 2010;11:11–22. doi: 10.1016/j.cmet.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Rajpathak SN, et al. Insulin-like growth factor axis and risk of type 2 diabetes in women. Diabetes. 2012;61:2248–2254. doi: 10.2337/db11-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wittenbecher C, et al. Insulin-like growth factor binding protein 2 (IGFBP-2) and the risk of developing type 2 diabetes. Diabetes. 2019;68:188–197. doi: 10.2337/db18-0620. [DOI] [PubMed] [Google Scholar]
- 44.Yin S-J, Chou C-F, Lai C-L, Lee S-L, Han C-L. Human class IV alcohol dehydrogenase: kinetic mechanism, functional roles and medical relevance. Chem. Biol. Interact. 2003;143–144:219–227. doi: 10.1016/S0009-2797(02)00167-9. [DOI] [PubMed] [Google Scholar]
- 45.Kim MY, et al. Identification of a zeta-crystallin (quinone reductase)-like 1 gene (CRYZL1) mapped to human chromosome 21q22.1. Genomics. 1999;57:156–159. doi: 10.1006/geno.1998.5714. [DOI] [PubMed] [Google Scholar]
- 46.International Statistical Classification of Diseases and Related Health Problems: Instruction Manual (World Health Organization, 2004).
- 47.Kuan V, et al. A chronological map of 308 physical and mental health conditions from 4 million individuals in the English National Health Service. Lancet Digit. Health. 2019;1:e63–e77. doi: 10.1016/S2589-7500(19)30012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inouye M, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abraham G, et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat. Commun. 2019;10:5819. doi: 10.1038/s41467-019-13848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikpay M, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambert SA, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nature. 2021;53:420–425. doi: 10.1038/s41588-021-00783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berisa T, Pickrell JK. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson CB, Bogomolov M, Benjamini Y, Sabatti C. TreeQTL: hierarchical error control for eQTL findings. Bioinformatics. 2016;32:2556–2558. doi: 10.1093/bioinformatics/btw198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang QQ, Ritchie SC, Brozynska M, Inouye M. Power, false discovery rate and winner’s curse in eQTL studies. Nucleic Acids Res. 2018;46:e133. doi: 10.1093/nar/gky780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vansteelandt S, Bekaert M, Lange T. Imputation strategies for the estimation of natural direct and indirect effects. Epidemiol. Methods. 2012;1:130–158. doi: 10.1515/2161-962X.1014. [DOI] [PubMed] [Google Scholar]
- 59.Steen J, Loeys T, Moerkerke B, Vansteelandt S. medflex: an R package for flexible mediation analysis using natural effect models. J. Stat. Softw. 2017;76:1–46. doi: 10.18637/jss.v076.i11. [DOI] [Google Scholar]
- 60.Liang K-Y, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. doi: 10.1093/biomet/73.1.13. [DOI] [Google Scholar]
- 61.Yu Q, Fan Y, Wu X. General multiple mediation analysis with an application to explore racial disparities in breast cancer survival. J. Biom. Biostat. 2014;5:1–9. [Google Scholar]
- 62.Yu Q, Li B. mma: an R package for mediation analysis with multiple mediators. J. Open Res. Softw. 2017;5:11. doi: 10.5334/jors.160. [DOI] [Google Scholar]
- 63.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie, S. sritchie73/cardiometabolic_prs_plasma_proteome: Nature revisions round 3. Zenodo10.5281/zenodo.4762747 (2021).
- 71.Rasmussen KL. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: a review. Atherosclerosis. 2016;255:145–155. doi: 10.1016/j.atherosclerosis.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 72.Sofat R, et al. Circulating apolipoprotein E concentration and cardiovascular disease risk: meta-analysis of results from three studies. PLoS Med. 2016;13:e1002146. doi: 10.1371/journal.pmed.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikpay M, Soubeyrand S, Tahmasbi R, McPherson R. Multiomics screening identifies molecular biomarkers causally associated with the risk of coronary artery disease. Circ. Genom. Precis. Med. 2020;13:e002876. doi: 10.1161/CIRCGEN.119.002876. [DOI] [PubMed] [Google Scholar]
- 74.Ruttmann E, et al. γ-Glutamyltransferase as a risk factor for cardiovascular disease mortality: an epidemiological investigation in a cohort of 163 944 Austrian adults. Circulation. 2005;112:2130–2137. doi: 10.1161/CIRCULATIONAHA.105.552547. [DOI] [PubMed] [Google Scholar]
- 75.Lee DS, et al. Gamma glutamyl transferase and metabolic syndrome, cardiovascular disease, and mortality risk: the Framingham Heart Study. Arterioscler. Thromb. Vasc. Biol. 2007;27:127–133. doi: 10.1161/01.ATV.0000251993.20372.40. [DOI] [PubMed] [Google Scholar]
- 76.Kojima Y, et al. Progranulin expression in advanced human atherosclerotic plaque. Atherosclerosis. 2009;206:102–108. doi: 10.1016/j.atherosclerosis.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 77.Pugeat M, et al. Interrelations between sex hormone-binding globulin (SHBG), plasma lipoproteins and cardiovascular risk. J. Steroid Biochem. Mol. Biol. 1995;53:567–572. doi: 10.1016/0960-0760(95)00102-6. [DOI] [PubMed] [Google Scholar]
- 78.Sutton-Tyrrell K, et al. Sex-hormone-binding globulin and the free androgen index are related to cardiovascular risk factors in multiethnic premenopausal and perimenopausal women enrolled in the Study of Women Across the Nation (SWAN) Circulation. 2005;111:1242–1249. doi: 10.1161/01.CIR.0000157697.54255.CE. [DOI] [PubMed] [Google Scholar]
- 79.Liu PY, Death AK, Handelsman DJ. Androgens and cardiovascular disease. Endocr. Rev. 2003;24:313–340. doi: 10.1210/er.2003-0005. [DOI] [PubMed] [Google Scholar]
- 80.Li G-S, et al. Do the mutations of C1GALT1C1 gene play important roles in the genetic susceptibility to Chinese IgA nephropathy? BMC Med. Genet. 2009;10:101. doi: 10.1186/1471-2350-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshida T, et al. Association of gene polymorphisms with chronic kidney disease in high- or low-risk subjects defined by conventional risk factors. Int. J. Mol. Med. 2009;23:785–792. doi: 10.3892/ijmm_00000193. [DOI] [PubMed] [Google Scholar]
- 82.Foster MC, Yang Q, Hwang S-J, Hoffmann U, Fox CS. Heritability and genome-wide association analysis of renal sinus fat accumulation in the Framingham Heart Study. BMC Med. Genet. 2011;12:148. doi: 10.1186/1471-2350-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madsen TE, et al. Circulating SHBG (sex hormone-binding globulin) and risk of ischemic stroke: findings from the WHI. Stroke. 2020;51:1257–1264. doi: 10.1161/STROKEAHA.120.028905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baumeier C, et al. Caloric restriction and intermittent fasting alter hepatic lipid droplet proteome and diacylglycerol species and prevent diabetes in NZO mice. Biochim. Biophys. Acta. 2015;1851:566–576. doi: 10.1016/j.bbalip.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 85.Ngo D, et al. Proteomic profiling reveals novel biomarkers and pathways in type 2 diabetes risk. JCI Insight. 2021;6:e144392. doi: 10.1172/jci.insight.144392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spranger J, et al. Adiponectin and protection against type 2 diabetes mellitus. Lancet. 2003;361:226–228. doi: 10.1016/S0140-6736(03)12255-6. [DOI] [PubMed] [Google Scholar]
- 87.Lau W, Andrew T, Maniatis N. High-resolution genetic maps identify multiple type 2 diabetes loci at regulatory hotspots in African Americans and Europeans. Am. J. Hum. Genet. 2017;100:803–816. doi: 10.1016/j.ajhg.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suckale J, Solimena M. The insulin secretory granule as a signaling hub. Trends Endocrinol. Metab. 2010;21:599–609. doi: 10.1016/j.tem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 89.Kim-Muller JY, et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat. Commun. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guevara-Aguirre J, et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rajwani A, et al. Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes. 2012;61:915–924. doi: 10.2337/db11-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu S, Lind L, Zhao L, Lindahl B, Venge P. Plasma prolylcarboxypeptidase (angiotensinase C) is increased in obesity and diabetes mellitus and related to cardiovascular dysfunction. Clin. Chem. 2012;58:1110–1115. doi: 10.1373/clinchem.2011.179291. [DOI] [PubMed] [Google Scholar]
- 94.Grarup N, Sandholt CH, Hansen T, Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57:1528–1541. doi: 10.1007/s00125-014-3270-4. [DOI] [PubMed] [Google Scholar]
- 95.Dwinovan J, Colella AD, Chegeni N, Chataway TK, Sokoya EM. Proteomic analysis reveals downregulation of housekeeping proteins in the diabetic vascular proteome. Acta Diabetol. 2017;54:171–190. doi: 10.1007/s00592-016-0929-y. [DOI] [PubMed] [Google Scholar]
- 96.Lopez PH, et al. Mice lacking sialyltransferase ST3Gal-II develop late-onset obesity and insulin resistance. Glycobiology. 2017;27:129–139. doi: 10.1093/glycob/cww098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato N. Insights into the genetic basis of type 2 diabetes. J. Diabetes Investig. 2013;4:233–244. doi: 10.1111/jdi.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Levey AS, Coresh J. Chronic kidney disease. Lancet. 2012;379:165–180. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 99.Levey AS, et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Notes and Methods, references and legends for Supplementary Tables and Data.
Supplementary Tables 1–9.
Supplementary Data 1–4.
Data Availability Statement
All data used in this study are publicly available or deposited in a public repository. The INTERVAL cohort data are available via the European Genome-phenome Archive with study accession no. EGAS00001002555. Dataset access is subject to approval by an independent data access committee since the data contain potentially identifying and sensitive patient information. Response times from the data access committee are typically within 1 week. All other data used in this study are publicly available without restriction. The PGS used in this study are available to download through the Polygenic Score Catalog (https://www.pgscatalog.org/) with accession nos PGS000727 (AF), PGS000018 (CAD), PGS000728 (CKD), PGS000039 (IS) and PGS000729 (T2D). The GWAS summary statistics used to generate new PGS for CKD, T2D and AF in this study are available to download through the GWAS Catalog (https://www.ebi.ac.uk/gwas/) with study accession nos GCST008065 (for the CKD GWAS published by Wuttke et al.14), GCST007517 (for the T2D GWAS published by Mahajan et al.15) and GCST006414 (for the AF GWAS published by Nielsen et al.13). The additional GWAS summary statistics used for Mendelian randomization analysis are also available through the GWAS Catalog with study accession nos GCST004787 (for the CAD GWAS published by Nelson et al.17), GCST006906 (for the IS GWAS published by Malik et al.16) and GCST007518 (for the T2D GWAS adjusted for BMI published by Mahajan et al.15). Full pQTL summary statistics published by Sun et al.26 for all SomaLogic SOMAscan aptamers are available to download from https://www.phpc.cam.ac.uk/ceu/proteins/. The DrugBank database is publicly available to download at https://www.drugbank.ca/releases/latest. Summary statistics for all statistical tests are available in Supplementary Data 3; the additional cis-pQTLs mapped in this study are provided in Supplementary Data 4.
The code used to generate the results of this study, along with a detailed list of software and versions, is available on GitHub (https://github.com/sritchie73/cardiometabolic_prs_plasma_proteome/), which is permanently archived by Zenodo70 at 10.5281/zenodo.4762747.