Abstract
Online, open access databases for biological knowledge serve as central repositories for research communities to store, find and analyze integrated, multi-disciplinary datasets. With increasing volumes, complexity and the need to integrate genomic, transcriptomic, metabolomic, proteomic, phenomic and environmental data, community databases face tremendous challenges in ongoing maintenance, expansion and upgrades. A common infrastructure framework using community standards shared by many databases can reduce development burden, provide interoperability, ensure use of common standards and support long-term sustainability. Tripal is a mature, open source platform built to meet this need. With ongoing improvement since its first release in 2009, Tripal provides full functionality for searching, browsing, loading and curating numerous types of data and is a primary technology powering at least 31 publicly available databases spanning plants, animals and human data, primarily storing genomics, genetics and breeding data. Tripal software development is managed by a shared, inclusive governance structure including both project management and advisory teams. Here, we report on the most important and innovative aspects of Tripal after 11 years development, including integration of diverse types of biological data, successful collaborative projects across member databases, and support for implementing FAIR principles.
Keywords: community databases, Tripal, open source software, community governance, genomics, genetics, breeding, FAIR
Introduction
Biological knowledge data portals (commonly referred to as ‘databases’ or ‘community databases’) have been integral resources since the first online research databases were established in the 1990s. These portals often include genetic, genomic, trait and/or breeding data from model organisms, and their continuing and growing importance has been well-described in recent reviews by Oliver et al. (2016) [1] and Leonelli and Ankeny (2012) [2]. Early trials for agricultural community databases resulted in a handful of successful projects, including MaizeGDB [3], CottonDB (CottonGen) [4] and Dendrome (TreeGenes) [5]. Early examples in the biomedical and model organism community included FlyBase [6], Wormbase [7] and The Arabidopsis Information Resource [8]. As the need for data integration and community support became evident, online resources for single organisms, taxonomic clades or specific types of data were developed. Collectively, they enable researchers to access high-quality, curated public data from thousands of research projects.
The majority of biological data portals enable data search and exploration through a variety of visualization and analytic tools. Related data are linked or integrated, providing a more complete view of variants, genes, genomes and metabolic networks. For example, many databases offer gene pages that display DNA and RNA sequences, protein products, isoforms, function, orthologs, publications, germplasm, expression data and relevant metabolic networks. With similar use cases, early databases recognized the need for shared software tools and standards, leading to the development and rapid adoption of the Generic Model Organism Database (GMOD; [9]).
As genomic and phenomic data grow in size and complexity, and as new data types and research methods emerge, individual communities want their own curated repositories. The requirements of a modern biological data portal far exceeds what any single investigator-led team can accomplish in isolation, even with the support and funding of a larger research community. If individual groups are successful in generating a new database, they must consider how to fund a team of developers to maintain both the framework and data curation activities. Even well-established databases face tremendous challenges when upgrading to new technology platforms, implementing new storage designs (schema modifications) for new data types, and maintaining hardware and software backends.
Development of the open source Tripal platform began in 2008 under the auspices of the GMOD project (http://gmod.org/) to address these challenges. Tripal is a generic software platform that provides the means for linking the physical storage of biological information such as genetic, genomic and breeding data to an easy-to-use User Interface (UI) for searching, browsing, loading and curating data [10–12]. To enable data curation and site interface management by non-technical staff, it is built on the popular, open source content management system (CMS) Drupal (https://www.drupal.org/). The platform is customizable in appearance and functionality, as well as extendable for each site’s unique needs. Tripal supports existing biological standards, including controlled vocabularies, common biological file formats, metadata standards and the standardized object relational database schema, Chado [13]. Since its inception, Tripal has maintained an active developer community which is recognized as the source of its long-term success. The following will examine the current scope and usage of Tripal, as well as the future development needed to sustain and expand its utility.
Tripal overview
Tripal was publicly released in 2009 [10] and by 2011 had been adopted by at least seven public data portals (Figure 1). The current release (version 3.3) has at least 31 public sites (Table 1). Anonymous installation tracking by Drupal reports 130 public instances worldwide [14, 15].
Figure 1 .
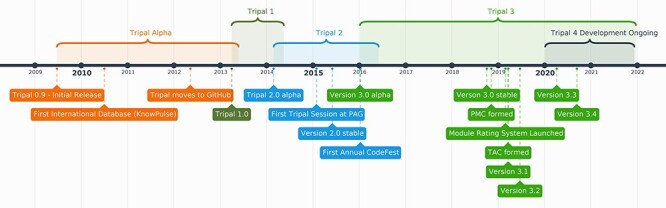
Timeline of events in Tripal’s software and community development. The progression of Tripal software versions can be seen along the top and the community milestones are shown along the bottom. Tripal version 1.0, the first version to provide generic support for all of Chado, was released in 2013. Version 2, which represented an upgrade from Drupal 6 to Drupal 7, was released in 2015, and version 3 with a redesigned data storage based on controlled vocabularies was released in October of 2018. The current version of Tripal, version 3.3, was released April 2020 and Tripal version 4 (an upgrade to support Drupal 8 and 9) is under development and scheduled for alpha release in 2021. Abbreviations: PAG (Plant and Animal Genome Conference).
Table 1.
Data portals partially or wholly utilizing the Tripal software platform. This table highlights the diversity in data types, described organisms and location of the developer community found among existing databases utilizing Tripal as of October, 2020
| Data Portal | Kingdom | Survey Response (Y/N) | Reported Data Types | Primary Location |
|---|---|---|---|---|
| Banana Genome Hub (http://banana-genome.cirad.fr/) [19] | Plantae | Y | Gene families, Genomes, Genotypes, Ontologies, Phylogenetics, Synteny, Transcriptomes | Montpellier, France |
| Cacao Genome Database (http://www.cacaogenomedb.org/) | Plantae | N | Genetic maps, Genomes, Genotypes, Ontologies, Pathways, Transcriptomes | Ames, IA, USA |
| CGD (http://www.citrusgenomedb.org/) | Plantae | Y | Contacts, Expression, Genetic maps, Genomes, Genotypes, Germplasm, Pedigrees, Phenotypes, Synteny, Transcriptomes, QTLs | Pullman, WA, USA |
| CorkOakDB (http://corkoakdb.org/) | Plantae | N | Expression, Genomes, Publications, Transcriptomes | Oeiras, Portugal |
| CottonGen (http://www.cottongen.org/) [4] | Plantae | Y | Contacts, Genetic maps, Genomes, Genotypes, Germplasm, Images, Ontologies, Pedigrees, Phenotypes, Publications, Synteny, Transcriptomes, QTLs | Pullman, WA, USA |
| Cucurbit Genomics (http://cucurbitgenomics.org/) [20] | Plantae | N | Genomes, Genotypes, Ontologies, Pathways, Synteny, Transcriptomes | Ithaca, NY, USA |
| GeneNet Engine (http://gene-networks.org/) [21] | Plantae | N | Gene co-expression networks, Ontologies | Pullman, WA, USA |
| GDR (https://www.rosaceae.org/) [22] | Plantae | Y | Contacts, Environmental data, Expression, Gene families, Genetic maps, Genomes, Genotypes, Germplasm, Haplotypes, Images, Ontologies, Pedigrees, Phenotypes, Publications, Synteny, Transcriptomes, QTLs | Pullman, WA, USA |
| GDV (http://ww.vaccinium.org/) | Plantae | Y | Contacts, Expression, Genetic maps, Genomes, Genotypes, Germplasm, Pedigrees, Phenotypes, Publications, Transcriptomes, QTLs | Pullman, WA, USA |
| GrainGenomes (development version) | Plantae | Y | Genetic maps, Genomes, Genotypes, Germplasm, Images, Pedigrees, Phenotypes, Publications, QTLs | Ithaca, NY, USA |
| Grass Genome Hub (https://grass-genome-hub.southgreen.fr/) | Plantae | Y | Gene families, Genomes, Synteny, Transcriptomes, QTLs | Montpellier, France |
| Hardwood Genomics Project (http://www.hardwoodgenomics.org/) | Plantae | Y | Expression, Gene families, Genomes, Ontologies, Transcriptomes | Knoxville, TN, USA |
| I5K Workspace (https://i5k.nal.usda.gov/) [23] | Animalia | Y | Genomes | Beltsville, MD, USA |
| Kiwifruit Genome Database (http://kiwifruitgenome.org/) [24] | Plantae | N | Genomes, Ontologies, Synteny, Transcriptomes | Hefei, China |
| KnowPulse (http://knowpulse.usask.ca) [25] | Plantae | Y | Genetic maps, Genomes, Genotypes, Germplasm, Ontologies, Pedigrees, Phenotypes, Publications, QTLs | Saskatoon, SK, Canada |
| Legume Information System (http://legumeinfo.org/) [26] | Plantae | Y | Expression, Gene families, Genetic maps, Genomes, Genotypes, Germplasm, Ontologies, Pan-genomes, Phenotypes, Phylogenetics, Publications, Synteny, Transcriptomes, QTLs | Ames, IA, USA |
| LiceBase (https://licebase.org/) | Animalia | N | Contacts, Genomes, Genotypes, Images, Phenotypes, Publications, Stocks | Bergen, Norway |
| Mimubase (http://mimubase.org/) | Plantae | Y | Contacts, Genomes, Genotypes, Ontologies, Publications | Storrs, CT, USA |
| Musa Germplasm Information System (https://www.crop-diversity.org/mgis/) [27] | Plantae | Y | Genotypes, Germplasm, Phenotypes, Stocks | Montpellier, France |
| NanDeSyn Database (http://nandesyn.single-cell.cn/) [28] | Plantae | N | Expression, Gene families, Genomes, Genotypes, Ontologies, Phenotypes, Publications, Synteny | Beijing, China |
| PeanutBase (http://peanutbase.org/) [29] | Plantae | Y | Expression, Gene families, Genetic maps, Genomes, Genotypes, Germplasm, Images, Ontologies, Pan-genomes, Phenotypes, Phylogenetics, Publications, Stocks, Synteny, Transcriptomes, QTLs | Ames, IA, USA |
| Planarian Educational Resource (https://cuttingclass.stowers.org) [30] | Animalia | Y | Expression, Genes, Ontologies, Transcriptomes | Kansas City, MO |
| Planosphere (https://planosphere.stowers.org/) [31] | Animalia | Y | Expression, Genomes, Images, Ontologies, Transcriptomes | Kansas City, MO, USA |
| PCD (https://www.pulsedb.org/) | Plantae | Y | Contacts, Genetic maps, Genomes, Genotypes, Germplasm, Pedigrees, Phenotypes, Publications, Synteny, QTLs | Pullman, WA, USA |
| Rice Genome Hub (https://rice-genome-hub.southgreen.fr/) | Plantae | Y | Genomes, Ontologies, Publications, Synteny, Transcriptomes | Montpellier, France |
| RNAStructurome (https://structurome.bb.iastate.edu/) [32] | Human | N | RNA structures | Ames, IA, USA |
| SeriolaDB (https://www.serioladb.org/) | Animalia | N | Genomes, Ontologies, QTLs | Ames, IA, USA |
| SIMRBase (https://simrbase.stowers.org) [33] | Animalia | Y | Genomes, Transcriptomes | Kansas City, MO, USA |
| SpinachBase (http://spinachbase.org/) [34] | Plantae | N | Genomes, Ontologies, Pathways, Transcriptomes | Ithaca, NY, USA |
| TreeGenes (https://treegenesdb.org/) [5] | Plantae | Y | Contacts, Environmental data, Gene families, Genetic maps, Genomes, Genotypes, Ontologies, Phenotypes, ublications, Stocks, Transcriptomes, QTLs | Storrs, CT, USA |
| US-SPD (https://swinepathogendb.org/) | Animalia | Y | Genomes | Ames, IA, USA |
| Zeamap (http://www.zeamap.com/) [35] | Plantae | N | Expression, Genetic maps, Genomes, Genotypes, Phenotypes, Synteny, Transcriptomes, QTLs | Wuhan, China |
Tripal supports the Chado data schema [13] to house genetic, genomic and breeding data in a Relational Database Management System [16]. Chado is a part of the GMOD and provides a generic and flexible relational model for complex biological data. It was developed first by FlyBase [17] and is used wholly or in part by many data portals today. However, as of Tripal version 3, users can opt to use a different database backend [12].
The Drupal [18] CMS enables contributors without web development experience to build static information pages in Tripal through a simple interface. This allows users to tailor the look and feel of the website through a page templating system. Drupal provides a wealth of basic functionality such as user authentication, database connectivity and support for community built extension modules, including web-form security, news feeds and event calendars. Drupal has a mature open-source community that continues to maintain, develop and upgrade the platform, including documentation and best practices.
All web applications require active security management, and this is particularly critical for sites that have password-protected user accounts and private data. Fortunately, Drupal has a dedicated world-wide team of experts focused on security issues with fast turn around and a secure workflow for providing security releases. It is important for Tripal sites to monitor these security updates and keep their Drupal version current. In contrast, data web portals which do not use a well-established CMS need to handle security concerns on their own.
Tripal’s modular architecture consists of a set of Drupal modules that provide the building blocks of a biological data portal with built-in web-based content management and display customization. Any biological data store can be tied to Tripal, with support for Chado offered as a built-in option. The Tripal Core module provides functionality required by most biological data portals with a set of centralized services. Tripal’s functionality can be extended by building new modules or by extending existing ones. Developers are encouraged to share their modules through the Tripal website (https://tripal.readthedocs.io/en/latest/extensions.html). This architecture provides structure for the member databases and is a key innovation of the Tripal system (Figure 2). Tripal Core provides reliable, centralized services that benefit the majority of member databases, meeting the goals of reducing duplicative development, sharing best practices and exchanging data across community databases. In contrast, the extension modules can accommodate custom development for specific tools or data types. This allows individual databases to develop their own solutions without waiting on the full community for formal approval.
Figure 2 .
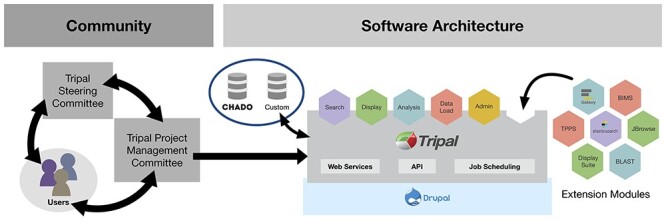
Tripal software architecture (middle panel) depends on Drupal, a popular CMS for building websites. Tripal Core communicates with the standard Drupal database as well as the Chado database or other user-installed database. Extension modules can build and extend from the Core module to provide support for new functions, such as importing/displaying new data types, providing analysis or search services to users and/or interacting with outside software packages such as JBrowse. Tripal Core is directly governed (left panel) by a PMC, which receives input and recommendations from the TAC. The Tripal community works together to develop Tripal and contribute extension modules (right panel), which are given the badges of bronze, silver or gold indicating compliance with a list of best practices and standards.
Tripal is open source, well-documented and community driven. The primary information portal is http://tripal.info, the Tripal source code and issue queue are available on GitHub (https://github.com/orgs/tripal) and documentation is available via Read the Docs [16] (https://tripal.readthedocs.io).
Unlike many open source science software projects, Tripal is not coordinated by a single group. Since the overall goal is to support diverse research groups and database developers with both shared and unique requirements, an open community with strong communication and shared governance is essential. From its inception, the project has maintained an active user and developer community that contributes to Core development, extension module development and community steering. This communication includes monthly meetings, emails, GitHub issues and dedicated Slack channels. Annual codefests have been offered since 2017.
To unify the growing international community, a governance structure was established in 2018, consisting of the Tripal Project Management Committee (PMC) and the Tripal Advisory Committee (TAC). This structure was collaboratively designed and the members elected. The PMC are the primary Tripal Core developers, and they guide development that supports the full community. This is balanced by the TAC, whose membership is largely scientists and principal investigators that are invested in the future of Tripal and its member databases. The responsibilities of these two committees (Figure 3) and their communication and voting structures are outlined in the Tripal Community documentation [16]. The TAC, as implied by the name, acts in an advisory capacity only by providing recommendations for Tripal to the PMC, which can independently make decisions. This structure was designed to ensure Tripal development will be fairly steered to the benefit of all, not any single member database.
Figure 3 .
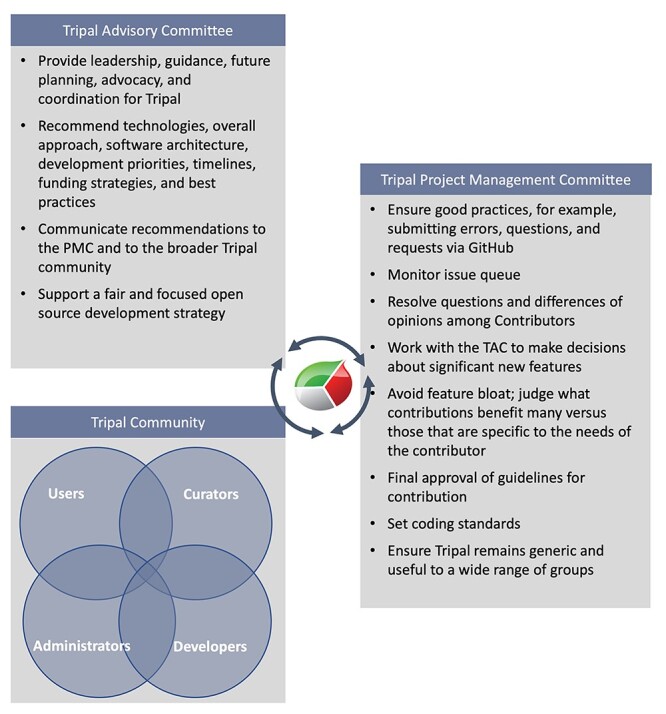
Tripal Governance Committee Responsibilities and their interaction with the Tripal Community. The structure of Tripal Governance is shown in detail with interaction occurring between the Tripal community, TAC and Tripal PMC. Specifically, the TAC surveys the needs of the Tripal Community to develop recommendations for the Tripal PMC. The PMC provides feedback on recommendations to the TAC as both work together to guide the future direction of Tripal. Additionally, the Tripal PMC interacts directly with the Tripal Community in respect to bug reports, feature requests and other code-focused concerns. The inclusivity of the Tripal Community is highlighted as it consists of all people who interact with Tripal either directly or indirectly.
Since many groups rely on Tripal for the longevity of their databases, Tripal has defined contribution guidelines for Tripal Core. The best practices are outlined in a detailed Developer’s Guide and include, for example: Drupal coding standards, GitHub pull request (PR) guidelines and unit testing requirements for new features. The code is hosted in GitHub and the contribution process is transparent and collaborative. All development, bugs, features and other contributions must start as a GitHub issue to provide the community a chance to comment and review. PRs (i.e. code additions or changes) must link to an issue and be tested by at least two developers. At least one of those developers must be a designated ‘trusted committer’, who checks that the PR meets the contribution guidelines, provides final approval and merges the modified code into Tripal Core.
Extension modules are not governed by the Tripal contribution guidelines, although the community encourages use of the guidelines. To encourage developers to create and maintain extension modules that benefit others, the Tripal community developed a module rating system in 2019. Developers are encouraged to self-assign a gold, silver or bronze badge on their modules, based on a list of best practices related to reusability and maintainability (Figure 4).
Figure 4 .
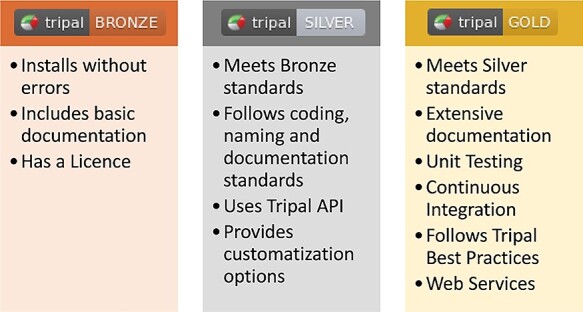
An overview of requirements for Tripal Extension Module Badges. These badges provide guidance to extension module developments for best practices on developing sharable extension modules as well as providing quality information to Tripal administrators looking to use these extension modules. The Tripal PMC awards badges to modules that meet the requirements and adds the badge to the official extension module listing on the tripal.info website.
Tripal use cases
Current databases leveraging Tripal mainly focus on data from plants and animals, including model organisms and agriculturally important organisms, and are hosted by scientists across the world. The diverse member communities illustrate the various ways Tripal supports different biological data portal needs. Each use case is provided in brief here with an expanded version in Supplementary File 1, see Supplementary Data available online at http://bib.oxfordjournal.org/.
Updating legacy code and databases
To contend with rapid technology changes, biological data portals eventually need to completely upgrade and refactor their code. TreeGenes, previously known as Dendrome when first funded in the early 1990s, recently converted their custom code base to Tripal, providing opportunity to expand to new data types, integrate ontologies and increase biocuration activities. Furthermore, Tripal provided an opportunity to develop a sophisticated web-based application housed within TreeGenes, CartograTree (https://treegenesdb.org/cartogratree). This resource enables the visualization and analysis of integrated genotype, phenotype and environmental data for georeferenced plants.
Adding breeding data to support growers
KnowPuse was established in 2010 for plant breeding, genetic and diversity data for legumes and pioneered support for germplasm, breeding and diversity data in Tripal. Unlike many biological data sites, KnowPulse leverages the power of the Drupal CMS to allow direct update of the resource by contributors. Long-term trusted curators add and edit the database directly, ensuring all users have the most up to date information. KnowPulse is a tangible example of both the ability to support diverse data types and extend Tripal to meet specific community needs.
Building a full-fledged database for a small research and breeding community
PeanutBase, launched in 2013, provides access to the genomic, diversity and marker-trait data for the peanut research community and industry. By leveraging existing Tripal modules, building new modules and collaborating with other databases, it has emerged as a full-fledged data portal, on par with sites serving far larger research communities. The approach taken by PeanutBase highlights how small development teams can meet the needs of stakeholders in a timely manner while making significant contributions to the community.
Supporting many communities for genome access and manual annotation
The i5k Workspace@NAL [23] supports community curation of genome annotations for over 70 non-model arthropod species. Tripal provided the initial support for housing the emerging genomes, and through a collaboration with another database, the Tripal HQ module was developed to enable community members to submit new data and for the data to undergo review by a database curator prior to acceptance. i5k has recently extended Tripal to link to and exchange data with Apollo, a popular genome annotation tool.
Monitoring quickly emerging genetic variants
The United States Swine Pathogen Database (US-SPD: https://swinepathogendb.org/) is the first centralized data repository for novel and endemic swine pathogens in the USA. Tripal provides a framework to monitor the patterns of genetic change of viruses in swine, enabling researchers to identify possible emerging threats and help control endemic viruses. The database is managed with only one developer yet offers a variety of data types as well as specialized tools, including the ability to classify porcine reproductive and respiratory syndrome virus 2 strains (PRRSV-2) by restriction fragment length polymorphism patterns.
Sharing development of common needs for many crops
The Main Lab hosts five major biological data portals: the Genome Database for Rosaceae (GDR), the Citrus Genome Database (CGD), the Pulse Crop Database (PCD), the Genome Database for Vaccinium (GDV) and CottonGen. Reliance on Tripal reduces developer time per site and enables modules developed for one to be quickly shared with the others. Recent development has standardized the storage of genetic map and trait association data in Chado for custom Tripal modules.
Community-driven Tripal functionality
The Core and extension module system is a powerful architecture for sharing code as well as developing custom functionality. Many of the most successful Tripal modules have relied on collaborative development across groups, including major updates of the Core module. The following module case studies illustrate collaborative development, innovative functionality and the flexibility inherent in this system.
Tripal BLAST: successful collaboration builds a module for everyone
The Tripal BLAST extension module (https://github.com/tripal/tripal_blast/) provides an intuitive UI to the NCBI Blast+ tools (Camacho et al., 2009). It features simple installation and integrates with any Drupal theme to ensure it stylistically matches each individual Tripal site. As one of the most installed Tripal extension modules, its ability to be installed on any Tripal site has been well tested and refined.
In addition to its popularity, another main success of Tripal BLAST is the community collaboration behind it. The Drupal 7, Tripal 2/3 version of this module was co-developed between the KnowPulse and Legume Federation development teams with sustained collaboration over a couple of years. There has also been substantial community input through both issue reports and PRs with 11 different organizations providing input and five organizations contributing directly to the codebase. These collaborations have been encouraged through clearly documented code, as well as through friendly, welcoming GitHub issue communication. Successful collaborative development of this module is also likely due to BLAST analysis being a very common and clearly defined process.
Tripal JBrowse: full integration of an external web application
JBrowse is an open source Javascript HTML5 genome browser, developed as a GMOD project, that is fast and scalable (Buels et al., 2016). The Tripal JBrowse extension module (https://github.com/tripal/tripal_jbrowse) facilitates embedding JBrowse within Tripal sites by providing listings of available JBrowse instances and, optionally, creating pages containing embedded JBrowse instances. This ensures that JBrowse is highlighted within the Tripal site, easily accessible, and facilitates a consistent user experience through shared branding and menus.
Tripal JBrowse also provides an administrative interface allowing multiple JBrowse instances to be managed through the Tripal administrative interface. This functionality couples the two web applications and provides a consistent interface to update both Tripal and JBrowse. Furthermore, registering JBrowse instances through Tripal JBrowse allows important metadata such as species and genome assembly to be captured.
Genotype data exploration with Gigwa
With the increasing number of genome-wide genotype datasets for large populations, community databases need to not only house this data, but present intuitive filtering and analysis options. The Gigwa application (Genotype Investigator for Genome-Wide Analyses) is an online tool to manage and explore one or more sets of genotypic data [36] and supports both the BrAPI (Breeding Application Programming Interface) [37] and GA4GH API (Global Alliance for Genomics and Health) [38] for programmatic data access. The Tripal Gigwa extension module integrates the features of a Gigwa server into a Tripal site. The administration interface enables data curators or administrators to load data and offers users an interface to select variants by genomic location, gene and variant effect (Figure 5).
Figure 5 .
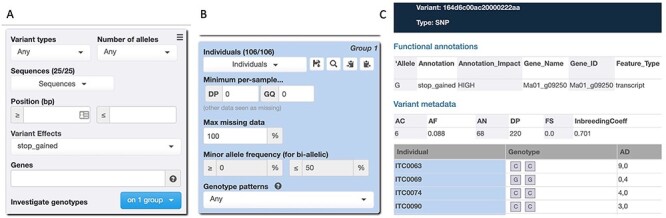
Gigwa interface on Musa Germplasm Information System offers users an interface to select variants by genomic location, gene and variant effect (A) and to filter individuals from a population based on various criteria (B). Results for each variant display the functional annotations due to the variant as well as the genotype calls and other metadata for individual accessions (C).
Cross-site searching with ElasticSearch interlinks community databases
Community data portals need to offer a fast, comprehensive keyword search for users; however, including content such as nucleotide sequences is not appropriate for traditional website search engines and leads to both slower performance and nonsensical results. (Sequences should be queried through a specialized algorithm such as the BLAST module described above.) The Tripal ElasticSearch engine solves this problem by natively integrating with Tripal, allowing a site administrator to use an administrative interface to determine what content to index and what to ignore [39].
With Tripal version 3, the search results were updated to provide a ‘faceted’ view that enables users to filter by the type of data. For example, after searching for ‘ash’, a user could then filter the results to only organism pages or to only gene feature pages. The Tripal Elasticsearch module also supports cross-site searching between Tripal 3 sites, enabling users to discover content from more than one community data portal. This is currently available on the Hardwood Genomics Project, TreeGenes, and CGD. This is faster than web services for large-scale searching and provides an exciting new method for Tripal database users to discover relevant content.
Drupal Display Suite customizes the look and feel of pages without programming
Drupal Display Suite is an extension to Drupal allowing a site manager to take full control over content page display using a drag and drop interface. With this tool, biological data pages can be easily rearranged and styled through the administrative UI without the need to delve into the Drupal templating system. Specifically, concrete pieces of content known as Fields (e.g. gene accession, genomic position, annotations) on a gene page can be moved independently and grouped into HTML div tags, tables, Tripal panes and fieldsets.
Tripal extension modules using the Tripal Fields API to provide content automatically integrate with the Drupal Display Suite without any additional programming. This allows content made available by Tripal extension modules to blend seamlessly into an existing biological content page. For example, germplasm pages on KnowPulse contain content provided by Tripal Core as well as three separate extension modules while still providing a consistent user experience (Figure 6A). These pages were created by administrators arranging content from diverse sources into Tripal panes to provide a coherent display (Figure 6B). Any Tripal site can leverage this integration to quickly and easily design biological data pages customized for their research communities.
Figure 6 .
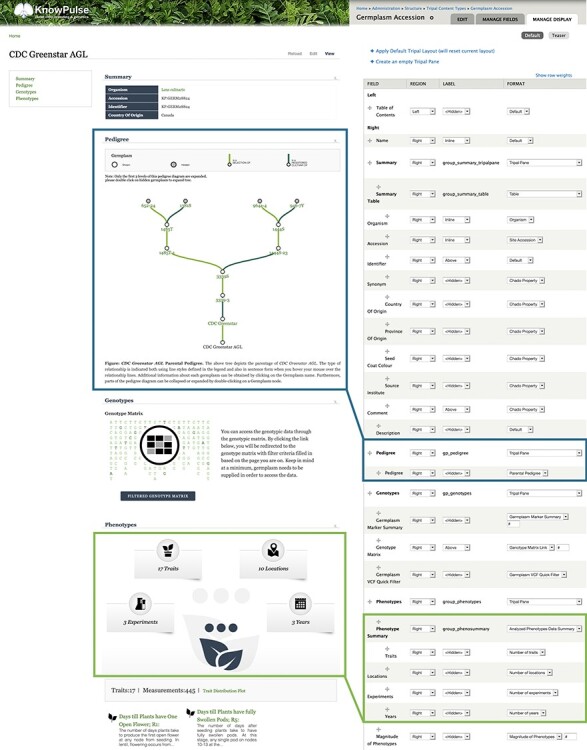
Tripal germplasm accession page designed using Display Suite integration. The KnowPulse CDC Greenstar AGL germplasm accession page (left) was designed using the Drupal Display Suite drag and drop administrative interface (right). Diverse data types provided by multiple Tripal extension modules were brought together into a cohesive display through data type focused categories. Each discrete piece of content is represented as a row in the administrative interface and can be rearranged and categorized easily by the administrator. The highlighted boxes indicate the configuration on the right with the generated display on the left. The configuration can be a single row as shown with the pedigree or multiple rows combined with a custom formatter as shown with the phenotypic summary depending on the level of configuration supported by the data type.
Integrating phylogenetics data into Tripal with MSAViewer and Phylotree.js
Comparative genomics is a powerful tool for researchers to explore the taxonomic and genomic history of related species. Users need to be able to explore multiple sequence alignments that form the basis for phylogenetic analysis and the resulting trees. Tripal now integrates with existing tools for these tasks: MSAViewer is a full-featured open source Javascript multiple sequence alignment viewer available on sites such as NCBI [40] and Phylotree.js is a full-featured Javascript web application used to display phylogenetic trees [41] (Figure 7). The Tripal MSAViewer and Tripal Phylotree.js extension modules quickly provide UIs to view and analyze alignment and tree data without programming expertise. By leveraging Drupal, the interfaces to these tools can be displayed alongside other content, such as gene, organism and gene family/orthogroup pages, to provide an intuitive user experience.
Figure 7 .
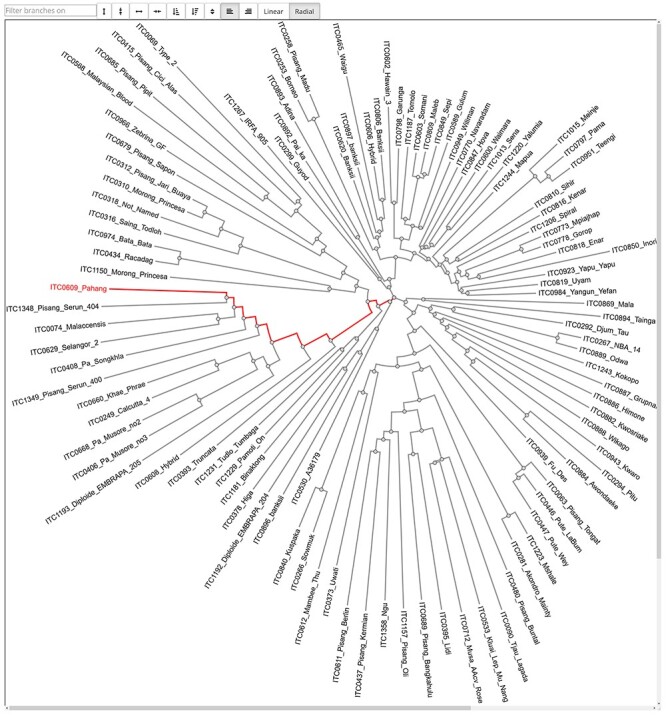
The Tripal Phylotree.js interface is capable of displaying large circular phylogenetic or taxonomic trees. Users can click to highlight paths and hover to see the distance measure between two leaves.
Breeding Information Management System
Breeding programs are producing increasingly large and complex amounts of data. Efficient management systems are needed to keep track of crosses, germplasm, pedigree, performance, geographical and image-based data as well as genotyping data. The Breeding Information Management System (BIMS, https://www.nrsp10.org/bims) was developed to provide breeders with autonomous control and management of their public or private breeding data in a secure setting. BIMS also allows breeders to integrate public data available in the community database with their private breeding data.
BIMS promotes the use and development of standard trait descriptors and metadata. The current functionality includes creating a breeding program, managing user permissions, managing trait/accession/cross/location data, bulk data import/export and individual or bulk data. Users can compare trait values across multiple datasets and visualize results as graphs with the distribution of the trait values for each dataset and a table with statistical values such as mean, maximum, minimum and standard deviation. The integration of breeding data with publicly available resources, as well as the integration of each breeder’s own genotypic and phenotypic data, maximizes the marker-assisted breeding utility for breeders and allied scientists. An upcoming version of BIMS will include BrAPI compliance and management of image files.
Sharing standardized breeding information with BrAPI
The Breeding API (BrAPI) project aims to enable interoperability among plant breeding databases, genetic databases and end user applications [37]. It provides a standardized RESTful web service API specification for cross-point communication. The BrAPI Tripal module implements BrAPI specifications to make Tripal sites BrAPI-compliant and has been implemented in MGIS [27]. It provides an administrative interface to configure each call to match the data storage backend specific to each Tripal site as well as manage user permissions Additional Javascript features allow a Tripal site to query other BrAPI end points and fetch breeding information. As such, the BrAPI Tripal module is a comprehensive solution providing both a secure BrAPI endpoint and a client implementation.
Galaxy module: integration of scientific workflows
Biological data portals offer access to very large biological datasets such as RNA/DNA read data or variant calls, but researchers are still faced with the challenges associated with accessing High Performance Computing resources and using the Linux command line. The popular open source analysis workflow system, Galaxy, solves many of these challenges [42] by offering an intuitive graphical UI through a web browser for users to select and pipeline software tools. A Tripal Galaxy module is available to link a Tripal site to one or more Galaxy instances [43]. This enables Tripal sites to offer users sophisticated analysis of large datasets housed by the database and/or with their own datasets. Tripal Galaxy can automatically render a Galaxy workflow as an intuitive web form on a Tripal site. The user can select datasets from the site or upload their own, alter parameters for each tool in the workflow, then submit the job, all within the ‘look and feel’ of the Tripal site. The jobs run on the Galaxy server and the results are returned to the user within the Tripal interface.
Tripal Galaxy also offers direct access to the Galaxy API via the blend4PHP library [44]. This can be used by any extension module to call for an analysis job via Galaxy while presenting a customized interface to the user. An example of a module leveraging this approach is Tripal CartograTree [45]. CartograTree enables users to use a map-based interface to select tree accessions, their associated genotype and/or phenotype data and layers of environmental data. All selected data can be integrated and analyzed by custom landscape genomics and association genomics analysis pipelines, which is accomplished programmatically by calling the Galaxy API.
Upgrading tripal core to drupal 9 through community development
Drupal is frequently updated with new features and improved security, and Tripal responds to leverage these advances. Community-driven development of Tripal 4, which will allow Tripal to run on Drupal 9, is currently underway and includes collaborators from a growing list of organizations. A large collection of resources and training materials has already been created.
Code is maintained in a GitHub repository, which features a detailed task list separated into easily approachable issues. Weekly group meetings help organize the work, develop collaborations and strengthen the community. Goals during these meetings include code review, discussions on software design and learning about tools to aid in development. State-of-the-art web development tools, such as symfony (https://symfony.com/), Composer (https://getcomposer.org/) and Twig (https://twig.symfony.com/), which are already integrated with Drupal 9, are being incorporated into Tripal to help the community leverage these technologies. Developers that contribute to Tripal Core development are not just helping the community; they gain familiarity with the new version that will help with upgrading their existing Tripal extension modules and creating new ones for Tripal 4.
Impact
The Tripal platform is used by at least 31 publicly available biological data portals and one in development (Supplementary Figure 1, see Supplementary Data available online at http://bib.oxfordjournal.org/, and Table 1). As the software is open source, there are likely other databases leveraging Tripal that are unknown to the broader Tripal community, as evidenced by the Drupal software module system reporting 130 publicly available installations. To quantify the impact of Tripal as well as the experiences and future development priorities of the Tripal community, two community surveys were developed in July of 2020. The first queried each database on details relating to data types stored, Tripal modules utilized, financial support, users served and personnel. The second was an anonymous survey for those utilizing Tripal in their work. These questions explored community opinions, including the current utility of Tripal, main challenges for developing and curating data, future technology priorities and more. The surveys and their results represent 21 databases and 26 individuals (Supplementary Files 2 and 3, see Supplementary Data available online at http://bib.oxfordjournal.org/). Aggregated across the databases responding to the survey, over 168 000 unique users interacted with Tripal sites in the past year. These databases employee 21.7 personnel as Tripal developers and 19.25 personnel as biocurators (fractional positions may reflect full-time positions split across different projects or students with part-time appointments).
The Tripal survey results highlight the international scope of funding resources, as well as the uneven split across specific funding sources, with government agencies and programs providing the majority of support compared with funding from industry and agricultural commodity boards. Of the 12 databases reporting financial support of development directly related to or building on Tripal, a total investment of over US $123 000 000 was reported, with the largest funding from the United States Department of Agriculture (USDA), the United States National Science Foundation (NSF) and Genome Canada. Other funding was obtained from Cotton Incorporated, Government of Saskatchewan, Saskatchewan Pulse Growers, United States National Pork Board, University of Saskatchewan and Western Grains Research Foundation (Supplementary File 2, see Supplementary Data available online at http://bib.oxfordjournal.org/). Another 10 databases responded to the survey by providing a list of funding agencies without financial specifics. Alphabetically, additional support was attributed to Belgian Directorate-general Development Cooperation (DGD), European Forest Institute, French CGIAR, French National Research Agency, Northern Pulse Growers, Stowers Institute for Medical Research, USDA, NSF, US National Institutes of Health, US Dry Pea and Lentil Council, and Washington Tree Fruit Research Commission.
Focus areas supporting the FAIR and TRUST principles
The FAIR (Findability, Accessibility, Interoperability and Reusability) reporting standards emphasizes that data should not only be stored, but also usable, and accessible, by external researchers [46]. These guidelines encourage individuals to identify tools and appropriate infrastructure to support the viability of their digital products [47]. These standards are further expanded by the recently published TRUST (Transparency, Responsibility, User focus, Sustainability and Technology) principles that describe the nature of the data repositories required to uphold FAIR [12]. These guidelines focus on the importance of repositories to offer a transparent method of data sharing that includes appropriate use and distribution, as well as long-term plans for preservation.
The Tripal infrastructure provides a strong foundation for the adoption of both FAIR and TRUST by supporting and encouraging open-source software, standards-based data storage and community-driven development. Tripal version 3 expanded this support with a focus on web services, backend platform flexibility and reliance on ontological frameworks.
The communities that implement Tripal databases have also responded with FAIR-minded extension modules that are intended to collect and curate data for improved distribution and integration. For example, Tripal Galaxy [43] is connecting data to analysis tools, thus paving the way for reproducible scientific workflows and more data reuse. Finally, Tripal’s recent adoption of governance committees supports many of the principles outlined by TRUST. Tripal is an example of user-driven and transparent development where community contributions are evaluated and adopted frequently. Today, Tripal is both a platform for independent databases as well as an integrated solution for cross-database communication and analysis.
The second anonymous Tripal survey asked administrators and developers to prioritize architectural considerations (Figure 8) and reflect on current challenges (Figure 9). The survey and the organization of the Tripal committees represents the Transparency aspect of TRUST. We relate the resulting objectives and challenges, as well as development objectives for Tripal v4, back to the remaining TRUST guidelines.
Figure 8 .
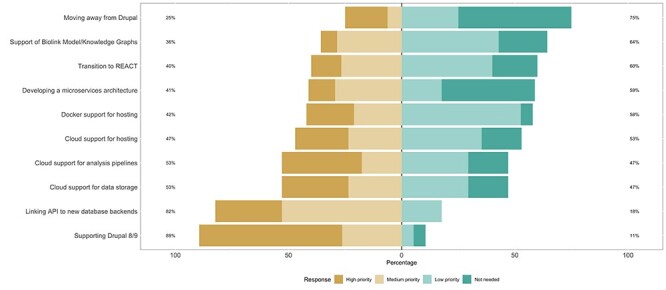
Anonymous survey results identifying architectural priorities for the community.
Figure 9 .
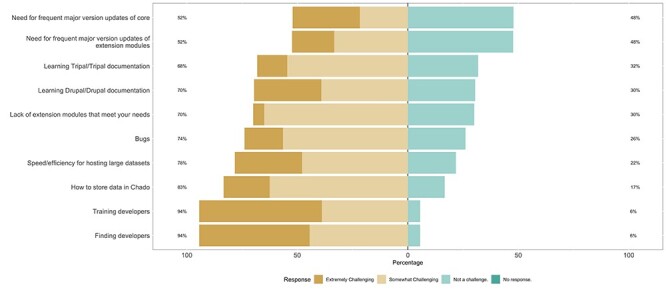
Anonymous survey results identifying common challenges to Tripal development.
(T)technology: support for multiple database schemas and engines
One of the ways Tripal strives to help biological data portals reach technological TRUST-worthiness is through flexible data storage support. Tripal core provides default integration with the GMOD Chado database schema that supports many biological data types [13]. Such integration allows Tripal to support many data types out of the box and the implementation of a common schema allows standards-based development of Tripal data loaders.
The highly normalized flexibility of Chado makes determining data storage best practices challenging for complex, interdependent data types (Figure 9). For example, quantitative trait loci (QTL) data associates phenotypic traits with genetic maps, which are both complex data types in their own right. The diverse Tripal community aids in development of robust, inclusive standards; however, finding common ground among diverse needs is still challenging. The Tripal community encourages groups to develop data importers for proposed data standards to facilitate evaluation and adoption by other biological data portals. There are currently a number of QTL-focused extension modules, each implementing different standards for community consideration. This approach ensures that community needs can be met quickly, while ideal data standards are being developed and evaluated.
In addition, some data are not well suited for relational models. For example, metabolic pathways are more intuitively stored in graph-based databases. These data highlight the need for support for additional data backends that provide flexibility beyond Chado’s highly normalized and relational structure. Tripal 3 supports additional data backends to allow storage flexibility; however, it remains dependent on Chado for storage of controlled vocabularies for its ontology-driven design. Since the Tripal community has indicated that this is a priority (Figure 8), Tripal 4 will retain the GMOD Chado default integration but also be completely database agnostic to allow maximum flexibility. This ensures that biological data portals using Tripal are able to use appropriate data storage mediums based on their data and associated standards.
(S)sustainability: backwards compatibility versus changing technologies
Quickly evolving web technologies create a dilemma: it is critical to support existing Tripal sites since new biological web portal funding for upgrades is not guaranteed, but it is often necessary to break backwards compatibility to leverage new technologies. Tripal has always guaranteed backwards compatibility of all Tripal APIs, even with the substantial changes between Drupal 7 and 8 that required a major rewrite of Tripal Core and many extension modules. Insulating the Tripal community from the difficulties associated with major Drupal upgrades was successful, with most developers reporting that the upgrade was not a major challenge (Figure 9); however, this was an immense task for the Tripal development team and not sustainable for future versions.
Fortunately, the Drupal software platform is maturing and reaching a new level of stability. Releases from Drupal 8 onward will follow a rolling deprecation scheme with a guarantee of backwards compatibility for any new functionality for at least two major versions. This allows progress in adopting new technologies but is implemented as smaller changes over a greater time period to ease developer workload. Tripal will now mirror the Drupal rolling deprecation philosophy to allow gradual adoption of new technologies.
Tripal will be fully upgraded to Drupal 9 (in Tripal 4, Figure 1) to provide site administrators and biocurators with a familiar administrative interface. Once this new version becomes available, the community will begin exploring new technologies prioritized by the community, such as microservices, additional data storage backends and tighter integration with other programming languages (Figure 8).
(R)responsibility: softening the Drupal learning curve
Both Drupal and Tripal application programmer interfaces (APIs) ensure that developers follow appropriate web development standards. While it is critical to implement these APIs within custom Tripal extensions, experienced developers are needed to do so. However, 94% of anonymous survey respondents identify hiring experienced developers and new developer training as somewhat or extremely challenging (Figure 9). Since the Tripal community also indicated their approval of Drupal as the primary platform for Tripal (Figure 8), steps are being taken with Tripal 4 to mitigate the Drupal learning curve and help developers become proficient with Tripal.
Specifically, documentation to teach Drupal from a Tripal perspective is underway. This will ensure that there is a single, complete source for learning both Drupal, Tripal and the connection between them, which should reduce the learning curve. It also eases the input required by principal investigators who will be able to point new trainees at a single resource that addresses the most common questions and links to relevant resources.
Additionally, Drupal 8 has adopted many popular open source web development projects including Composer, Symphony and Twig. This ensures that competent PHP developers will be able to leverage their existing knowledge within the Drupal ecosystem. Being able to hire from the wider pool of web developers with PHP experience versus only Drupal developers will greatly benefit the Tripal community. The adoption of these open source technologies makes learning Drupal more appealing to aspiring web developers. The Tripal community continues to welcome new developers with monthly conference calls and a welcoming GitHub issue queue housing questions and training alongside the typical bug reports.
(U)user focus: data biocuration
As biocurators face the continuing and growing challenge of preparing high-quality public data for archiving, integrating and sharing [48], the Tripal project will continue to support and streamline the curation process. Tripal is based on the content management framework Drupal to provide a foundation for manual curation of individual data objects without the need for programming. This ensures that curators are able to access user-friendly forms containing descriptions, help text and data specific form fields on any data page. These forms allow subject-area experts to correct data, while they are viewing it which greatly improves the likelihood of community curation.
Tripal has a number of other biocuration-friendly aspects. It provides loaders for common data formats, such as OBO (Open Biomedical Ontologies) and GFF (Generic Feature Format) files. Support for custom data is provided by a number of extension modules (https://tripal.readthedocs.io/en/latest/extensions/data_input.html) that can be used by biocurators to bulk import interrelated data into a Tripal site. This streamlined process goes beyond just importing the file, as it also focuses on collecting important metadata (i.e. species, cultivar), including analysis details such as analysis program and version.
Data need to be curated in growing in volume and complexity. This process can be streamlined by encouraging community contribution, where data generators perform the initial steps of submitting the data and providing relevant metadata. To ensure high quality and accuracy, submissions still need to be reviewed by professional curators. This process is facilitated by the Tripal HeadQuarters extension module (https://github.com/statonlab/tripal_hq), which allows registered users to submit their own data and metadata. Designated data curators have an administrative interface to view and accept or request changes to this triaged data. Data curation can also be more efficient with semi-automated (or machine-driven) curation. Examples of this are emerging in Tripal extension modules, including the Tripal Plant PopGen Submit Pipeline [5]. In this module, metadata standards such as the Minimum Information About a Plant Phenotyping Experiment and the associated ontological frameworks are assigned through the use of natural language processing-based approaches for the many of the sequence and phenotype objects.
These tools and approaches enable Tripal sites to provide high-quality data to their respective communities as efficiently as possible. However, the work and rewards of curation remain an under-appreciated commodity among funding agencies [49]. Biological data portals need long-term, well-trained biocurators.
(S)sustainability: funding for development of core and member data portals
Currently, Tripal is largely supported by member databases through a combination of federal grant dollars and industry/commodity funds. These funds support the database resources themselves, which in turn dedicate some developer time to Tripal as a support infrastructure. This model has proved challenging for funding the extensive effort needed for major refactoring and Core update efforts; current funding sources generally only support new infrastructure and innovation. As long-term software sustainability requires dedicated effort in maintenance and security updates, we foresee funding will remain a challenge for the Tripal community.
This model has additional major drawbacks for personnel. ‘Soft’ (i.e. temporary grant) funded that positions are by their very nature temporary, leading to developers coming and going in relatively short time frames (2–3 years). While this cycle of grant funding supports a training pipeline for students and temporary staff, it precludes true expertise from being developed and maintained in a system as complex as Tripal. Drupal is a substantially complex system with developers often requiring a long training period prior to productive code contribution. This is compounded by the need for the developer to understand biological entities. Tripal would benefit from full-time, dedicated developer positions that would not only maintain Core code but provide training and support to new developers.
In addition to maintaining Core, Tripal must support new data types and research methods to meet current research needs. The quantity and complexity of data will increase, as will the number of research communities needing online resources. Tripal can help through collaborative development and shared modules. However, these ambitious development efforts will need reliable funding, whether dedicated to a single resource or shared by dozens of resources.
The Tripal community has engaged in extensive discussions to develop recommendations for ongoing support. We recommend that granting agencies that fund member databases through short-term grants and/or long-term contracts/personnel consider funding long-term support personnel specifically for community database infrastructure. This would enable a more efficient, cost-effective, sustainable, shared and FAIR/TRUST-focused infrastructure to benefit all biological data portals and software systems, including Tripal. This support could also facilitate the migration of the many databases using different code bases to Tripal and other shared platforms. As Tripal is an international resource, facilitating shared support across borders is a challenge, but it should also be considered an opportunity. FAIR data that traverse borders and a shared infrastructure benefit the global research ecosystem.
Future directions
The Core architectural priority has always been to both ensure that Tripal is compatible with the most recent stable Drupal major version and that it leverages the new functionality provided by Drupal. With Tripal 4, the main goal is to become compatible with current Drupal release [9]. With the new release cycle, Tripal 4 can evolve into a system supporting the new functionality provided by Drupal 9 without requiring large rewrites of extension modules or extensive site upgrades. This current approach has been informed and approved by the community, which agrees that Drupal should continue to be the platform of choice (Figure 8).
Integration of additional storage backends (e.g. MongoDB, variant call format) has been a high priority since the initial development of Tripal 3 and as informed by the community (Figure 8), is one of the driving design principles behind Tripal 4. Core Tripal developers are also focused on easing the upgrade of both Tripal sites and Tripal extension modules during development of a new major version. Survey results also indicated a need for Core support of phenotypic breeding data, genome synteny and genetic data (Supplementary File 1, see Supplementary Data available online at http://bib.oxfordjournal.org/). With Tripal 3, Core data types were added, and with Tripal 4, additional support in the form of Core fields and data storage standards is planned. The PMC and TSC will continue to consult with the community through monthly user meetings and our Github issue queue to ensure emerging needs are addressed.
Many Tripal databases participate in the AgBioData consortium, a working group of representatives from biological databases and other software resources that meet regularly and facilitate cross-resource working groups to ensure standards and best practices for biological knowledge [17]. Outside of the agricultural systems, the Alliance for Genome Resources (AGR) project funded by the NIH National Human Genome Research Institute is an ongoing initiative facilitating a common infrastructure and union of data across the Gene Ontology consortium and five model organism databases. This project will highlight benefits and challenges that we can learn from and may also produce common data infrastructure that could be applied more broadly. The AgBioData consortium and AGR are in active discussions to facilitate more collaboration.
Conclusions
This review covers the current scope and impact of Tripal, as well as plans to sustain and expand its usefulness in supporting many biological data portals. Tripal provides tools for data storage, dissemination and discovery of genetic, genomic and breeding data. Research communities can leverage Tripal to build their own data web portals focusing on their specific needs while benefiting from the collaborative nature of Tripal and its community-derived extensions. Both Drupal and Tripal provide extensive APIs for portal customization and tool integration as highlighted in the Tripal community-driven functionality section. Furthermore, Tripal provides extensive documentation and training materials paired with a welcoming developer community and transparent governance structure.
Tripal currently underpins between 27 and 130 community databases, provides service to 168 000 unique users, employs at least 21.7 personnel and brings in over US$123 000 000 to member databases. The Tripal framework underpins a variety of successful community focused data web portals.
Tripal encourages its member databases to follow the guiding TRUST principles to develop trustworthy resources for their communities by leading through example. Development of Tripal focuses on (T)ransparency of governance, (R)esponsibility to adopt community standards, (U)ser focus on understanding and serving community needs, (S)ustainability of both Tripal and member databases, and regular (T)echnology upgrades. Tripal also provides tools for member databases to ensure data they house which is FAIR as discussed briefly here and more extensively in [12].
Tripal is currently funded entirely through its member databases. Given the importance of integrated big data and the growing database community, Tripal infrastructure would benefit from sustainable, reliable funding for Core efforts and coordination of this funding across international borders.
Key Points
Tripal is an open source, community developed software platform to build online, open access databases for biological knowledge, spanning genetic, genomic, trait and/or breeding data.
At its 11 year anniversary, Tripal serves as the base for over 30 public databases and has developed a unique, shared, inclusive governance structure to manage code development, communication and user support.
Through use cases and community surveys, we show the collaborative development, innovative functionality and the flexibility inherent in Tripal as well as its ongoing support for FAIR (Findable, Accessible, Interoperable and Reusable) data.
Supplementary Material
Margaret Staton earned her Ph.D. in plant and environmental science from Clemson University. Currently she is an Associate Professor of Bioinformatics in the Entomology and Plant Pathology Department at the University of Tennessee, Knoxville. Her interests include plant computational genomics, transcriptomics and epigenomics, as well as developing cyberinfrastructure platforms for genomics research data and supporting citizen science.
Ethalinda Cannon is a Bioinformatic Engineer with the USDA ARS, where she works on the maize community website, MaizeGDB. Her interests include collaborative software development, software engineering for genomic database projects, and management of genomic data and metadata.
Lacey-Anne Sanderson has been the Lead Developer of KnowPulse, a breeder-focused pulse database built at the University of Saskatchewan, for the past 12 years. She leads an interdisciplinary bioinformatics development team that produces high quality Tripal modules for use on KnowPulse and by the greater Tripal community. Additionally, she contributes to the core Tripal open-source platform as a member of both the Tripal Project Management Committee, Tripal Advisory Committee and as a core developer. She has contributed to publishing standards for agricultural biological databases to promote FAIR principles and sustainable databases through the AgBioData consortium.
Jill Wegrzyn is an Associate Professor in the Ecology and Evolutionary Biology Department at the University of Connecticut. She focuses on the computational analysis of genomic and transcriptomic sequences from non-model plant species by using machine learning and computational statistics approaches for gene finding, gene expression, transcriptome assembly, and conserved element identification. She also develops web-based applications that integrate genotype, phenotype, and environmental data to facilitate the forest geneticist or ecologist's ability to analyze, share, and visualize their data.
Tavis Anderson earned his Ph.D. in ecology and evolution from Rutgers University. Currently, he is a Research Biologist at the National Animal Disease Center, USDA-ARS. His research interests include the identification of genetic predictors of influenza A virus host range and virulence, and the use of sequence data to understand the genetic and antigenic variability of viruses infecting swine to generate applied solutions to preventing virus transmission.
Sean Buehler is a Research Assistant at the University of Tennessee, United States. He is working on the Tripal Project as a web developer. He is a member of Tripal's Project Management Committee (PMC). He is currently in a cooperative agreement with the USDA through their i5k project, which utilizes Tripal. He also collaborates with the University of Connecticut, United States, on the TreeGenes database, also a Tripal site. He received a BSs in Computer Science from Central Connecticut State University, United States.
Irene Cobo-Simón is a Postdoctoral Research Associate in the Department of Ecology and Evolutionary Biology at the University of Connecticut, USA. Her research interests include conservation genomics, transcriptomics, landscape genomics and association mapping. She received her Ph.D. in Biology from the Complutense University of Madrid, Spain.
Kay S. Faaberg is a Research Microbiologist of the USDA, Agricultural Research Service, National Animal Disease Center. Her research interests include molecular virology of RNA nidoviruses and evolution of porcine reproductive and respiratory syndrome virus.
Emily Grau is an independent Genomics Data Curator and Web Developer working with forest tree community database, TreeGenes. Her interests include collaborative Tripal software development and developing automated and manual curation pipelines for genomic data and metadata.
Valentin Guignon is a Bioinformatics Specialist working at the Montpellier office (France) of the Alliance of Bioversity International and CIAT (CGIAR) since 2010. He received his M.Sc. degree in Bioinformatics from the University of Montreal (Canada) in 2006. He worked in the French National Center for Scientific Research (CNRS, LIRMM) and then in the CIRAD before joining his current position. He designs and develops algorithms, computation pipelines, APIs, databases and information systems mainly focused on banana genetic resources.
Jessica Gunoskey was a Lab Manager at the University of Connecticut's Plant Computational Genomics laboratory. She received her B.Sc. from Washington College in 2021, specializing in Cell, Molecular, and Infectious Disease Biology. Her research interests include bacterial genomics and pathogenesis.
Blake Inderski is a Computational Biologist at the National Animal Disease Center, USDA-ARS, responsible for the development and maintenance of the United States Swine Pathogen Database. Interests include highly variable RNA viruses infecting swine and bioinformatics tools that describe this diversity.
Sook Jung is a Research Associate Professor at Washington State University. Her research focuses on the development of community databases using database platforms and tools that are adaptable and expandable. Her work also includes building data curation templates with specified metadata for various data types required to build crop databases. Her other area of research is studying the mode of evolution of Rosaceae species using sequence data.
Kelly Lager is Research Leader of the Virus Prion Research Unit, USDA, Agricultural Research Service. His research interests include the pathogenesis and control of swine viral diseases.
Dorrie Main is a Professor at Washington State University. Her highly collaborative, multi-disciplinary research program focuses development of database portals for crops, development of online computational tools including sequence analysis pipelines and generic database platforms, and discovery of genomic regions and markers controlling important traits. All of these efforts seek to provide genomic, genetic and breeding resources to enable basic, translational and applied research.
Monica Poelchau is a Geneticist at the USDA-Agricultural Research Service's National Agricultural Library. As a lead of the i5k Workspace@NAL database for insect genomics, her research interests include bioinformatics, text mining, and data management.
Risharde Ramnath is a Software Developer working on TreeGenes, CartograPlant and Tripal. He received his BSc and MSc from the University of the West Indies, Trinidad and Tobago. His research interests include real-time communications, web/mobile development and clustered computing.
Peter Richter is a Software Developer for the Plant Computational Genomics lab in the Department of Ecology and Evolutionary Biology at the University of Connecticut, USA. He is also a recent graduate from Boston University with a Master of Science in Artificial Intelligence. His interests include bioinformatics, software development, deep learning, and natural language processing.
Joe West worked on Tripal as an undergraduate in the Computer Science Department at the University of Tennessee, Knoxville. He graduated in 2021.
Stephen P. Ficklin is an Associate Professor in the Dept. of Hortidculture at Washington State University. His research program focuses on the development of computational methods towards development of multiomic models that are predictive of complex traits of agricultural importance by utilizing machine learning, systems-level models and cyberinfrastructure.
Contributor Information
Margaret Staton, University of Tennessee, Knoxville, TN USA.
Ethalinda Cannon, USDA-ARS, Corn Insects and Crop Genetics Research Unit, Ames, IA USA.
Lacey-Anne Sanderson, University of Saskatchewan, Saskatoon, SK Canada.
Jill Wegrzyn, University of Connecticut, Storrs, CT USA.
Tavis Anderson, USDA-ARS, National Animal Disease Center, Ames, IA, USA.
Sean Buehler, University of Tennessee, Knoxville, TN USA.
Irene Cobo-Simón, University of Connecticut, Storrs, CT USA.
Kay Faaberg, USDA-ARS, National Animal Disease Center, Ames, IA, USA.
Emily Grau, University of Connecticut, Storrs, CT USA.
Valentin Guignon, Bioversity International, Montpellier, France.
Jessica Gunoskey, University of Connecticut, Storrs, CT USA.
Blake Inderski, USDA-ARS, National Animal Disease Center, Ames, IA, USA.
Sook Jung, Washington State University, Pullman, WA USA.
Kelly Lager, USDA-ARS, National Animal Disease Center, Ames, IA, USA.
Dorrie Main, Washington State University, Pullman, WA USA.
Monica Poelchau, USDA-ARS, National Agricultural Library, Beltsville, MD, USA.
Risharde Ramnath, University of Connecticut, Storrs, CT USA.
Peter Richter, University of Connecticut, Storrs, CT USA.
Joe West, University of Tennessee, Knoxville, TN USA.
Stephen Ficklin, Washington State University, Pullman, WA USA.
Data availability statement
Tripal is open source software provided under the GNU General Public License v2.0 and can be found at https://github.com/tripal/tripal.
Funding
US Department of Agriculture, Agricultural Research Service, projects #5030-21000-068-00D, #8260-88888-003-00D, #5030-32000-118-00-D, #0500-00093-001-00-D; USDA National Institute of Food and Agriculture (NIFA) project NRSP10; USDA NIFA Mcintire-Stennis project WNP00009; USDA NIFA FACT #2019-67021-29920; USDA Animal and Plant Health Inspection Agency, projects #5030-32000-108-21I and #5030-32000-108-37I; Oak Ridge Institute for Science and Education, contract #DE-AC05-06OR23100; National Pork Board, project #16-222; US National Science Foundation (NSF) awards #1443040 and #1444573; USDA Hatch project 1014919; University of Saskatchewan through AGILE and EVOLVES projects; Genome Canada projects #8302 and #LSP19-16302; Government of Saskatchewan Agriculture Development Fund projects #20150331 and #20200026; Saskatchewan Pulse Growers project #BRE1516; Western Grains Research Foundation projects WGRF VarD 1538 and WGRF GC1903. For the US-Swine Pathogen Database, this work was supported by the US Department of Agriculture (USDA) Agricultural Research Service (ARS project number 5030-32000-118-00-D); USDA ARS project numbers 5030-32000-108-21I and 5030-32000-108-37I funded by Animal and Plant Health Inspection Agency; USDA Agricultural Research Service Research Participation Program of the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy (DOE) and USDA Agricultural Research Service (contract number DE-AC05-06OR23100 to B.I.); SCINet project of the USDA Agricultural Research Service (ARS project number 0500-00093-001-00-D); National Pork Board (NPB project number 16-222). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA, DOE or ORISE. USDA is an equal opportunity provider and employer.
Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and Employer.
The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or US Government determination or policy.
References
- 1. Oliver SG, Lock A, Harris MA, et al. Model organism databases: essential resources that need the support of both funders and users. BMC Biol 2016;14:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leonelli S, Ankeny RA. Re-thinking organisms: the impact of databases on model organism biology. Stud Hist Philos Sci Part C Stud Hist Philos Biol Biomed Sci 2012;43:29–36. [DOI] [PubMed] [Google Scholar]
- 3. Jiao Y, Peluso P, Shi J, et al. Improved maize reference genome with single-molecule technologies. Nature 2017;546.7659:524–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu J, Jung S, Cheng C-H, et al. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res 2014;42:D1229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falk T, Herndon N, Grau E, et al. Growing and cultivating the forest genomics database. TreeGenes Database 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thurmond J, Goodman JL, Strelets VB, et al. FlyBase 2.0: the next generation. Nucleic Acids Res 2019;47:D759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris TW, Arnaboldi V, Cain S, et al. WormBase: a modern Model Organism Information Resource. Nucleic Acids Res 2020;48.D1:D762–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huala E, Dickerman AW, Garcia-Hernandez M, et al. The Arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 2001;29:102–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Connor BD, Day A, Cain S, et al. GMODWeb: a web framework for the generic model organism database. Genome Biol 2008;9:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ficklin SP, Sanderson L-A, Cheng C-H, et al. Tripal: a construction toolkit for online genome databases. Database 2011;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanderson L-A, Ficklin SP, Cheng C-H, et al. Tripal v1.1: a standards-based toolkit for construction of online genetic and genomic databases. Database 2013;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spoor S, Cheng C-H, Sanderson L-A, et al. Tripal v3: an ontology-based toolkit for construction of FAIR biological community databases. Database 2019;2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics 2007;23:i337–46. [DOI] [PubMed] [Google Scholar]
- 14. Sites Using Tripal | Tripal .
- 15. Usage statistics for Tripal | Drupal.org .
- 16. Welcome to Tripal’s documentation ! Tripal 7.x-3.x documentation.
- 17. Harper L, Campbell J, Cannon EKS, et al. AgBioData consortium recommendations for sustainable genomics and genetics databases for agriculture. Database 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drupal - Open Source CMS . Drupal.org, 2018.
- 19. Droc G, Larivière D, Guignon V, et al. The Banana Genome Hub. Database 2013;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng Y, Wu S, Bai Y, et al. Cucurbit Genomics Database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res 2019;47:D1128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ficklin SP, Feltus FA. A systems-genetics approach and data mining tool to assist in the discovery of genes underlying complex traits in Oryza sativa. PLoS One 2013;8:e68551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jung S, Lee T, Cheng C-H, et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res 2019;47:D1137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poelchau M, Childers C, Moore G, et al. The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res 2015;43:D714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yue J, Liu J, Tang W, et al. Kiwifruit Genome Database (KGD): a comprehensive resource for kiwifruit genomics. Hortic Res 2020;7:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanderson L-A, Caron CT, Tan R, et al. KnowPulse: A web-resource focused on diversity data for pulse crop improvement. Front Plant Sci 2019;10:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dash S, Campbell JD, Cannon EKS, et al. Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Res 2016;44:D1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruas M, Guignon V, Sempere G, et al. MGIS: managing banana (Musa spp.) genetic resources information and high-throughput genotyping data. Database 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gong Y, Kang NK, Kim YU, et al. The NanDeSyn database for Nannochloropsis systems and synthetic biology. Plant J 2020;104:1736–45. [DOI] [PubMed] [Google Scholar]
- 29. Dash S, Cannon EKS, Kalberer SR, et al. PeanutBase and other bioinformatic resources for peanut. Peanuts. AOCS Press, 2016;241–52. [Google Scholar]
- 30. Accorsi A, Williams MM, Ross EJ, et al. Hands-on classroom activities for exploring regeneration and stem cell biology with planarians. Am Biol Teach 2017;79:208–23. [Google Scholar]
- 31. Nowotarski SH, Davies EL, Robb SMC, et al. The planarian anatomy ontology: a resource to connect data within and across experimental platforms. bioRxiv 2020. [DOI] [PMC free article] [PubMed]
- 32. Andrews RJ, Baber L, Moss WN. RNAStructuromeDB: a genome-wide database for RNA structural inference. Sci Rep 2017;7:17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zimmermann B, Robb SMC, Genikhovich G, et al. Sea anemone genomes reveal ancestral metazoan chromosomal macrosynteny. bioRxiv 2020.
- 34. Collins K, Zhao K, Jiao C, et al. SpinachBase: a central portal for spinach genomics. Database 2019;2019:baz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gui S, Yang L, Li J, et al. ZEAMAP, a comprehensive database adapted to the maize multi-omics era. iScience 2020;23:101241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sempéré G, Pétel A, Rouard M, et al. Gigwa v2—Extended and improved genotype investigator. GigaScience 2019;8:giz051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Selby P, Abbeloos R, Backlund JE, et al. BrAPI—an application programming interface for plant breeding applications. Bioinformatics 2019;35:4147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. The Global Alliance for Genomics and Health . A federated ecosystem for sharing genomic. clinical data Science 2016;352:1278–80. [DOI] [PubMed] [Google Scholar]
- 39. Chen M, Henry N, Almsaeed A, et al. New extension software modules to enhance searching and display of transcriptome data in Tripal databases. Database 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yachdav G, Wilzbach S, Rauscher B, et al. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics 2016;btw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shank SD, Weaver S, Kosakovsky Pond SL. phylotree.js - a JavaScript library for application development and interactive data visualization in phylogenetics. BMC Bioinformatics 2018;19:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 2005;15:1451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spoor S, Wytko C, Soto B, et al. Tripal and Galaxy: supporting reproducible scientific workflows for community biological databases. Database 2020;2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wytko C, Soto B, Ficklin SP. blend4php: a PHP API for galaxy. Database 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Herndon N, Richter P, Falk T, et al. Galaxy enables integrated analysis of phenotypic, genotypic, and environmental data for geo-referenced trees in CartograTree. F1000 Research 2018;7. [Google Scholar]
- 46. Wilkinson MD, Dumontier M, IjJ A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reiser L, Harper L, Freeling M, et al. FAIR: a call to make published data more findable, accessible, interoperable, and reusable. Mol Plant 2018;11:1105–8. [DOI] [PubMed] [Google Scholar]
- 48. International Society for Biocuration . Biocuration: distilling data into knowledge. PLoS Biol 2018;16:e2002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reiser L, Berardini TZ, Li D, et al. Sustainable funding for biocuration: the arabidopsis information resource (TAIR) as a case study of a subscription-based funding model. Database 2016;2016:baw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tripal is open source software provided under the GNU General Public License v2.0 and can be found at https://github.com/tripal/tripal.


